Abstract
Ribonucleoproteins (RNPs) consisting of derivatives of a ribozyme and an RNA-binding protein were designed and constructed based upon high-resolution structures of the corresponding prototype molecules, the Tetrahymena group I self-splicing intron RNA and two proteins (bacteriophage λN and HIV Rev proteins) containing RNA-binding motifs. The splicing reaction proceeds efficiently only when the designed RNA associates with the designed protein either in vivo or in vitro. In vivo mutagenic protein selection was effective for improving the capability of the protein. Kinetic analyses indicate that the protein promotes RNA folding to establish an active conformation. The fact that the conversion of a ribozyme to an RNP can be accomplished by simple molecular design supports the RNA world hypothesis and suggests that a natural active RNP might have evolved readily from a ribozyme.
Keywords: group I intron/in vivo selection/ribonucleoprotein/ribozyme/RNA–protein interaction
Introduction
The splicing reaction of group I intron RNA has been shown to depend on its own catalytic activity (Cech, 1990). The catalysis is dependent on the absolutely conserved core region in the highly sophisticated structure of the intron RNA (Jaeger et al., 1996). The tertiary structure of the core is stabilized by its own internal RNA–RNA interactions and long-range interactions with peripheral domains (Jaeger et al., 1996). Several group I introns have been shown to require specific binding proteins as splicing factors in vivo. Factors such as Neurospora CYT-18, yeast CBP2 or Aspergillus AnCOB group I intron maturase are known to promote active conformation of the intron RNAs, as in the case of the RNA–RNA interactions (Weeks, 1997; Ho and Waring, 1999; Lambowitz et al., 1999).
According to the ‘RNA world’ hypothesis (Gilbert, 1986), the ‘world’ initially consisted of only RNA molecules and evolved into the ‘RNA–protein world’ through gradual replacement of RNA structural elements with those of proteins (Lambowitz and Perlman, 1990). Mohr et al. (1994) provided evidence supporting this hypothesis by demonstrating that Neurospora CYT-18 can replace a peripheral domain, P5abc, in the Tetrahymena ribozyme (Inoue, 1994). Another indication of RNA– protein replacements is the suggestion that during the evolution of the bovine mitochondrial ribosome, half of the rRNA was replaced with proteins, because the mitochondrial ribosomes have approximately the same molecular weight as Escherichia coli ribosomes but the ratio of RNA to protein of mitochondrial and E.coli ribosomes is 1:2 and 2:1, respectively (Matthews et al., 1982).
That RNA–RNA or RNA–protein interactions can enhance the activity of ribozymes indicates that these two types of interaction can be functionally equivalent and likely interchangeable. Based on this line of thought, we designed a ribonucleoprotein (RNP) in which an RNA–RNA long-range interaction in the Tetrahymena intron ribozyme is replaced with new RNA–protein interactions.
Results
RNP design
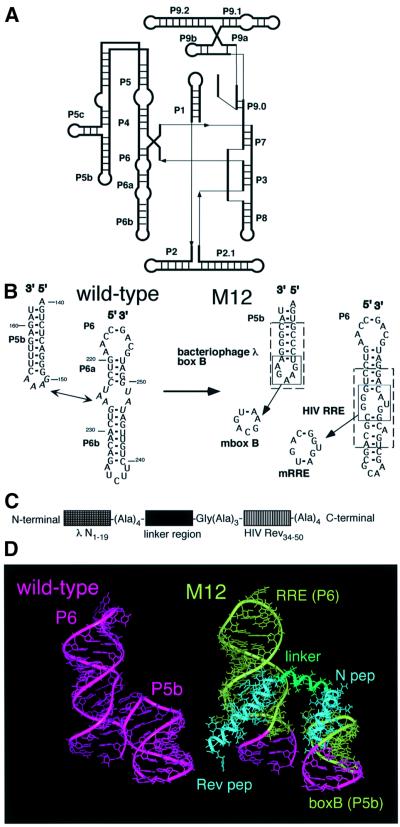
Composition of a designed RNP was planned so that an RNA component would act as a ribozyme upon formation of a complex with an RNA-binding protein. A molecular modeling program was employed for the design by taking advantage of the high-resolution structures of a ribozyme and two RNA–peptide complexes. For the design of the RNA component in the RNP, the Tetrahymena self-splicing intron ribozyme whose subdomain P4–P6 has been characterized by X-ray crystallography was employed as the starting RNA (Figure 1A) (Cate et al., 1996a). The design of the RNA-binding protein was based upon two arginine-rich motifs derived from bacteriophage λN and HIV Rev proteins because the structures of the respective peptide–RNA complexes have been elucidated by NMR spectroscopy (Battiste et al., 1996; Legault et al., 1998).
Fig. 1. Design of the RNP. (A) Schematic representation of the secondary structure of the Tetrahymena ribozyme. (B) Secondary structures of P5b and P6 domains of the Tetrahymena ribozyme and its derivative (M12) used in this study. Nucleotides involved in the RNA–RNA interactions are italicized. Bacteriophage λ boxB and HIV RRE in M12 RNA are boxed by dotted lines. The mutants (mboxB and mRRE) lacking one of the binding sites are also indicated. (C) Schematic representation of the protein employed in this study. (D) Three-dimensional model of the RNP consisting of M12 RNA and the designed protein. The linker region of the protein is represented by a tentatively inserted 12mer α-helix (green).
The principle for designing the RNP was to fix two structural elements in the RNA component, which contain newly introduced peptide-binding motifs, with an RNA-binding protein. For the RNA component, an inactive derivative of the Tetrahymena ribozyme (denoted M12) was devised by replacing the terminal loop in the P5b element and an internal loop in the P6 element of the wild type with two binding sites, boxB and RRE from bacteriophage λ and HIV, respectively (Figure 1B) (Murphy and Cech, 1994). In the protein component, two terminal regions consisting of two RNA-binding motifs, λN1–19 and HIV Rev34–50, which respectively bind box B and RRE with high affinity without assistance from a large protein framework, were joined with a short linker (Figure 1C) (Tan et al., 1993; Tan and Frankel, 1995).
The P4–P6 domain of the Tetrahymena ribozyme containing an RNA–RNA interaction between P5b and P6 provides a scaffold for the correct assembly of the rest of the ribozyme (Downs and Cech, 1996; Doherty and Doudna, 1997; Sclavi et al., 1998; Treiber et al., 1998). It is known that disruption of the interaction does not cause a global conformational change of the ribozyme, although it reduces the ribozymatic activity (Laggerbauer et al., 1994; Cate et al., 1996b). Because the role of the interaction is to facilitate the folding of the ribozyme, we hoped that the protein would promote correct folding of the RNA and/or finely tune the active conformation upon binding to the RNA as designed.
In the molecular model for the RNP, the structures of the λN1–19–boxB and HIV Rev34–50–RRE complexes were superimposed on those of P5b and P6 of the Tetrahymena ribozyme, respectively (Figure 1D). The two peptide elements were connected with a linker whose size was determined based on the distance between the C-terminus of λN1–19 and the N-terminus of HIV Rev34–50, approximately equal to that of a 12-residue α-helix. At the ends of the linker, three to four alanine residues were added next to λN1–19 and HIV Rev34–50 because alanine extensions at peptide termini are known to enhance their binding affinity by increasing the α-helix content (Tan et al., 1993; Tan and Frankel, 1995). (For technical reasons, a glycine residue was added next to three consecutive alanine residues to generate a restriction enzyme recognition site.) In the middle of the linker, a specific sequence is presumably needed to allow the two RNA-binding domains to interact appropriately with the RNA. Due to the absence of an applicable example, we selected two distinctively different sequences for the segment: Ala–Ala–Ala–Ala, which presumably forms part of a stable helix; and Gly–Gly–Gly–Gly, which is structurally flexible. The proteins containing the alanines or glycines are denoted as pep A or pep G, respectively.
Self-splicing reaction with the designed proteins
The splicing reactions of the precursor RNA containing the designed intron ribozyme were tested in the presence of the designed protein, varying its concentration in the presence of 2 mM MgCl2 and 80 mM KCl at 37°C (Figures 2 and 7). Under the optimal protein concentration, the final extent of the splicing reaction with pep A or pep G was ∼8- or 4-fold higher, respectively, than that of M12 alone, indicating that the efficiency of the reaction is under the direction of the binding protein (Figure 2).
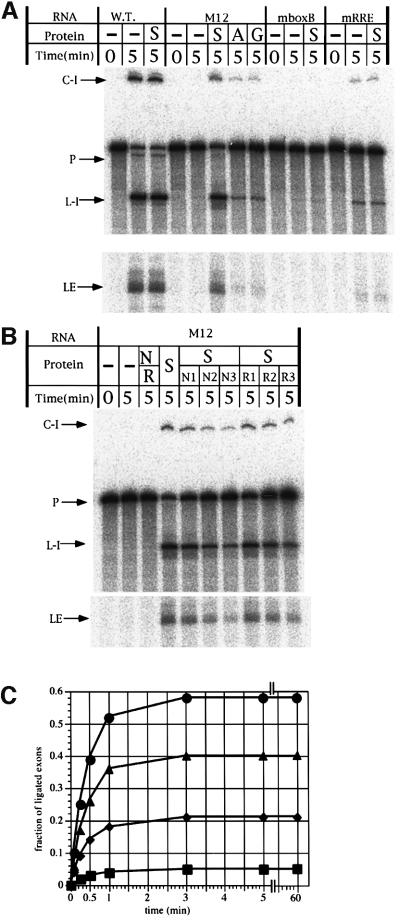
Fig. 2. The in vitro splicing reaction of the designed RNP. (A) Schematic representation of the proteins employed in this study. Mutations in RNA binding sites of pep AmN and pep AmRev are also indicated using the one-letter code. (B) An autoradiogram is shown for the splicing reactions with 10 nM 32P-labeled precursor RNA in the presence or absence of 3 µM protein. G, pep G; A, pep A; N, pep AmN; R, pep AmRev; D, pep Δlinker; C-I, circular form of spliced intron; L-I, linear form of spliced intron; LE, ligated exons.
Fig. 7. The splicing reaction of the designed RNP was tested in the presence of various concentrations of protein. The final extent of the splicing reaction of M12 with pep A, pep G or pep S is shown with closed triangles, closed circles and open circles, respectively, and that of the wild type or M12 is shown with dotted lines.
To see whether the protein utilizes both RNA-binding sites, the splicing reaction of M12 was tested in the presence of pep A mutants in which one of the binding motifs is missing (denoted pep AmN and pep AmRev; Figure 2A). In addition, splicing was also attempted with a protein lacking eight amino acids in the linker region (denoted pep Δlinker; Figure 2A). Enhancement of the reaction was observed with none of these proteins, indicating that both RNA-binding sites and the linker region are required for efficient splicing (Figure 2). The splicing reaction of the wild-type ribozyme was also attempted with pep A and pep G. Neither had an effect on the kobs value or the final extent of the reaction, indicating that the protein is ineffective without the binding sites in the RNA (data not shown). These results demonstrate the feasibility of the RNP design.
To investigate the role of the linker region, the splicing reaction was attempted in the presence of pep AmN and pep AmRev; they are not physically connected but are separately able to bind to the two binding sites in the RNA. No enhancement of the splicing reaction was observed in the presence of 1 µM each of the two proteins (Figure 5B), indicating the necessity for the linker. (Note, enhancement was not observed by increasing or decreasing the concentration of the proteins to 3 or 0.1 µM, respectively; data not shown.)
Fig. 5. The splicing reaction with the selected protein in vitro. (A) An autoradiogram is shown for the splicing reactions with 10 nM 32P-labeled precursor RNA in the presence or absence of 100 nM protein. WT, wild-type Tetrahymena ribozyme; S, pep S; A, pep A; G, pep G; C-I, circular form of spliced intron; L-I, linear form of spliced intron; LE, ligated exons. (B) An autoradiogram is shown for the splicing reactions with 10 nM 32P-labeled precursor RNA in the presence or absence of various concentrations of proteins. Lane S, 100 nM pep S; lane N/R, 1 µM pep AmN plus 1 µM pep AmRev; lanes S/N1, S/N2, S/N3, 100 nM pep S plus 100, 200 or 400 nM pep AmN, respectively; lanes S/R1, S/R2, S/R3, 100 nM pep S plus 100, 200 or 400 nM pep AmRev, respectively. (C) Time-courses of the splicing reactions with 10 nM 32P-labeled M12 precursor RNA in the presence or absence (squares) of 3 µM pep S (circles), pep A (triangles) or pep G (diamonds).
In vivo selection for improvement of the designed protein
The results show that at least two different sequences can be used in the middle segment of the linker region. To optimize the protein function, we performed in vivo selection employing a pool of modified proteins in which the sequences of the four amino acids at the site were randomized. Plasmids were constructed to express both RNA and protein under the control of a lac promoter. The DNA template for the RNA was inserted into a β-galactosidase α-fragment gene in the pTZ18U vector in order to direct production of mature mRNAs following splicing (Williamson et al., 1989). The protein library was constructed in plasmids derived from an expression vector, pSTV28. The growth rate of E.coli cells harboring these two plasmids incubated in M9 media containing lactose as a sole carbon source should reflect the level of β-galactosidase expression that in turn depends on the efficacy of splicing. Thus, the cells expressing the activator proteins should grow faster than those expressing abortive proteins.
After one round of selection from 1 × 106 transformants, fast growing cells were isolated. Among 20 randomly picked selectants, 19 were found to contain Gly–Val–Gly–Arg (denoted pep S) with the remaining clone encoding Cys–Ser–Val–Gly.
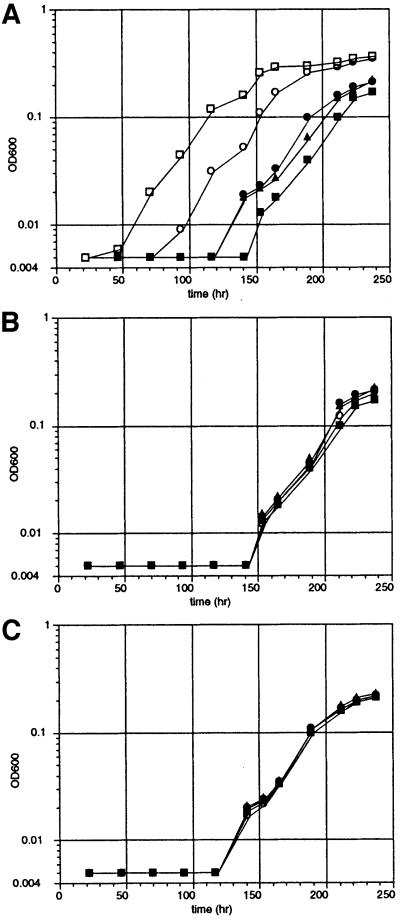
The growth rates were compared for cells transformed with plasmids encoding pep S, pep A, pep G or a control peptide, N pep, consisting of λN1–19 with four alanines at its C-terminus (Figure 3A). The rate with pep A or pep G was faster than that of those harboring the N pep plasmid (whose rate was equal to that of cells without the plasmid). The rate of the cells harboring the selected pep S plasmid was faster than that of the rest, albeit slower than that of those with the wild-type ribozyme. Cells harboring ΔP5abc ribozyme, a derivative lacking P5abc that is inactive under low Mg2+ concentrations, and pep S plasmid were unable to grow under the same conditions (Joyce et al., 1989). These results indicate that growth depends on the accumulation of the ligated exons produced by the splicing reaction.
Fig. 3. Time-courses for the growth of E.coli cells containing the derivative of the Tetrahymena intron ribozyme and the activator protein. Cells were incubated in M9 medium containing lactose at 37°C. (A) Time-courses for cells containing M12 plasmid transformed with N pep (closed squares), pep A (closed triangles), pep G (closed circles) and pep S (open circles) plasmids and cells containing wild-type plasmid transformed with N pep plasmid (open squares). (B) Time-courses for cells containing the mboxB plasmid transformed with the respective proteins described for (A). (C) Time-courses for cells containing the mRRE plasmid transformed with the respective proteins as described for (A).
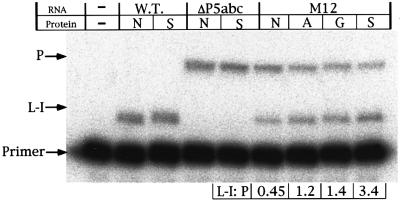
Primer extension assays were carried out to compare the ratio of the precursor and the spliced linear intron (L IVS) RNA in cells (Figure 4) (Zhang et al., 1995). An oligo nucleotide complementary to the 5′ end of the intron was used to determine the amount of the precursor and L IVS RNAs in total cellular RNAs. In this experiment, the reverse transcription was designed to stop at the first G upstream of the 5′ splice site for the precursor RNA in the presence of ddCTP. Therefore, the termination of the transcription for the spliced RNA should be at the 5′ end of the L IVS, resulting in a product four nucleotides shorter than that from the precursor. For M12, the ratio of the L IVS to the precursor was 2.7-, 3.0- or 7.5-fold higher than that with N pep when pep A, pep G or pep S was used, respectively. For the wild-type and ΔP5abc RNA, the ratio remained constant in the presence or absence of pep S. These results indicate that the designed proteins, especially pep S, significantly enhance the splicing reaction in vivo.
Fig. 4. Primer extension analysis of the precursor and intron. Ribozymes and proteins were overexpressed for 10 min prior to the isolation of the total RNA. P, cDNA from the RNA containing an intact 5′ splice site; L-I, cDNA from linear intron RNA; Primer, unreacted primer; WT, wild-type Tetrahymena ribozyme; ΔP5abc, ΔP5abc ribozyme; N, N pep; S, pep S; A, pep A; G, pep G.
The splicing reaction was attempted in vitro by employing the designed proteins (Figure 5A). Pep S (100 nM) was capable of promoting the splicing of the M12 ribozyme (10 nM) in the presence of 2 mM MgCl2 and 80 mM KCl at 37°C. The splicing activity was ∼50% that of the wild type, and 8-fold higher than that of the inactive derivative of the ribozyme (Figure 5A). Pep A and pep G (100 nM) had barely any effect under the same conditions. Pep S was unable to act on variant RNA in which one peptide-binding motif is missing, or on the wild-type ribozyme (Figure 5A).
Characterization of the RNA–protein interactions
To see whether pep S utilizes both protein-binding sites in vivo, we attempted to grow cells harboring the template for pep S in the presence of plasmids expressing a precursor RNA with a modified intron in which one of the binding motifs is missing (denoted mboxB and mRRE; Figure 1A) (Figure 3B and C) (Tan and Frankel, 1995; Jain and Belasco, 1996). No acceleration of the growth rate was observed in these cells, indicating that both sites are required for ribozyme activation.
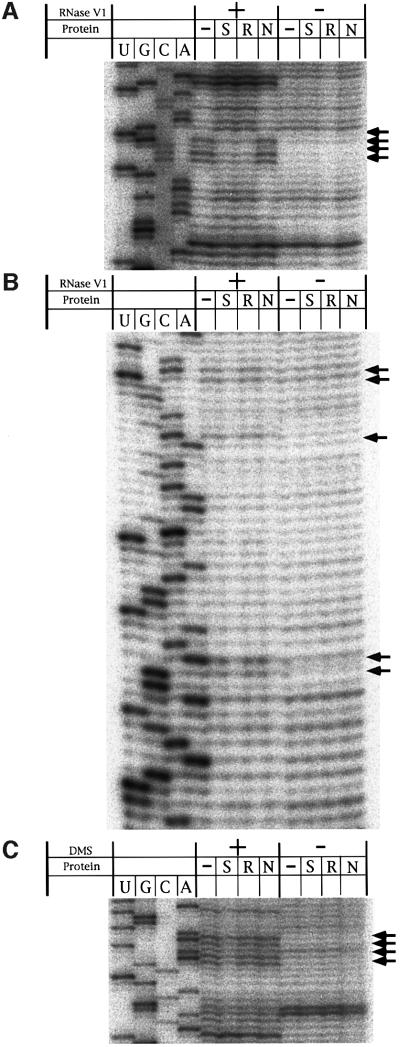
RNA footprinting was attempted for the putative RNP by employing RNase V1 in vitro (Figure 6) (Kjems et al., 1991; Chattopadhyay et al., 1995). M12 RNA was partially digested in the presence or absence of pep S, pep AmN or pep AmRev. The 5′ strand of the box B sequence was protected from RNase digestion when pep S or pep AmRev was present (Figure 6A). Protection was also observed with five nucleotides of RRE in the presence of pep S or pep AmN. We found that the activity of M12 in the presence of pep S was retarded by the presence of either pep AmN or pep AmRev (Figure 5B). These results indicate that the two RNA-binding sites of the designed protein interact with their target sites to form a stable RNP complex as we had expected.
Fig. 6. RNase and chemical footprints of M12 in the presence or absence of the designed proteins. Protection studies were carried out with RNase V1 or DMS. Accessibility was monitored by primer extension using RNase- or DMS-treated samples as templates. Autoradiograms are shown for RNase footprints of box B (A) and RRE (B) region and chemical footprints of the A-rich bulge (C). Arrows indicate sites that show protection from RNase or DMS.
Kinetic analysis
The splicing reaction of the precursor RNA containing the designed intron ribozyme was attempted in the presence of various concentrations of the protein, 2 mM MgCl2 and 80 mM KCl at 37°C. The final extent of the reaction was 58, 40 or 21% for pep S, pep A or pep G, respectively, under conditions where M12 is saturated with the protein (denoted as FE value; Figures 5C and 7) (the value was 82 or 5% for the wild-type ribozyme or M12 alone, respectively).
Hill’s coefficient and Kd (the overall dissociation constant) were determined by Hill’s plot. The fraction of the saturation of the specific protein binding can be written as the final extent value under each protein concentration normalized by FE value. Hill’s coefficient was 1.0, 1.1 or 1.0 for pep S, pep A or pep G, respectively, indicating that one molecule of the RNA associates with one molecule of the protein although the data do not exclude the possibility that M12 has other independent binding sites for the protein. The Kd values were 63, 602 or 141 nM for pep S, pep A or pep G, respectively. The drastic decrease in the Kd value for pep S implies that the selected protein can effectively bind to the RNA due to the optimized relative positions of the two RNA-binding sites.
The disruption of the long-range interaction between the loop of P5b and P6 does not cause a global conformational change of the ribozyme. However, it reduces the ribozymatic activity, indicating an effect on RNA folding (Laggerbauer et al., 1994; Cate et al., 1996b). To examine the effect(s) of Mg2+ on M12, we induced its folding in the presence of 5 mM Mg2+ and checked its activity at 2 and 5 mM Mg2+ (Zaug et al., 1993). The final extent of the reaction with M12 increased to ∼50% in both reaction conditions (data not shown), indicating that the disruption affects the RNA folding but does not influence the activity of the folded RNA.
The presence or absence of the protein had little effect on kcat and KmGTP values. This implies that the active conformation of M12 is essentially identical in the presence or absence of protein factors and that the proteins aid the folding of the ribozyme to decrease the fraction of the RNA trapped in misfolded form(s) (Table I) (Weeks, 1997). The Kd and FE values for pep S are better than those of pep A or pep G, indicating that pep S is capable of preventing more ribozymes from misfolding.
Table I. Kinetic data of splicing reactiona.
| RNA | Protein | kcat (min–1) | KmGTP (µM) | FEb (%) | Kd (nM) |
|---|---|---|---|---|---|
| WT | none | 2.4 | 4.3 | 82 | |
| M12 | none | 2.2 | 8.9 | 5 | |
| M12 | pep A | 2.2 | 8.9 | 40 | 602 |
| M12 | pep G | 2.3 | 8.7 | 21 | 141 |
| M12 | pep S | 2.3 | 8.3 | 58 | 63 |
aGTP concentrations used in these experiments were 0.5, 1, 2, 5, 10 and 30 µM. Reaction times were 0.083, 0.25, 0.5, 1, 3, 5 and 60 min.
bThe final extent of the reaction under conditions where the RNA was saturated with one of the proteins.
To assess the structural integrity of the P4–P6 domain of M12, chemical probing experiments were carried out by using dimethyl sulfate (DMS) to detect unpaired A and C residues in M12 RNA in the presence of pep S, pep AmN, pep AmRev or no protein. Protection of the A-rich bulge, which was observed for the canonical form of the P4–P6 domain that directs the formation of active ribozyme, was also observed when pep S was present, indicating that pep S can assist correct folding of the P4–P6 domain (Figure 6C) (Flor et al., 1989).
The designed proteins that assist the correct folding of M12 are likely to be dispensable after the RNA has folded because the kcat and KmGTP values of M12 remain the same in the presence or absence of the proteins. We found that the activity of M12 that was enhanced by pep S did not decrease after rigorous digestion with proteinase K (data not shown). Unless the pep S associated with M12 is extremely resistant to proteinase, the results indicate that the correctly folded ribozyme is still active by itself without the protein.
We attempted to see whether the proteins assist the folding of the ribozyme by resolving misfolded RNAs like RNA chaperones (Coetzee et al., 1994). None of the proteins was able to enhance the splicing activity of renatured M12 RNA (Table I) (data not shown), indicating that the proteins are unable to resolve misfolded RNA.
Discussion
A self-splicing RNA–protein complex was designed based upon the crystal structure of the Tetrahymena self-splicing intron and the NMR structures of two known RNA-binding peptides by employing a molecular modeling program. The splicing reaction by the RNA was facilitated by the binding of the protein, in vivo and in vitro. The activity of the complex was further enhanced by a modified protein obtained from in vivo protein selection. Kinetic analysis indicates that the protein aids the correct folding of the ribozyme (Weeks, 1997).
Molecular designs based on the structures of natural products have been reported for both protein and RNA. For protein, size reduction or alteration of properties of enzymes have been successfully accomplished (Liu et al., 1997; MacBeath et al., 1998; DeGrado et al., 1999). For RNA, allosteric ribozymes have been constructed by introducing small molecule-binding domains to a hammerhead ribozyme (Tang and Breaker, 1997; Soukup and Breaker, 1999). The activity of the resulting ribozymes is regulated by conformational changes induced by ligand interaction with the binding domains. For the combination of RNA and protein, a hairpin ribozyme was engineered to associate with a protein; however, the protein had no influence on the activity of the ribozyme in this case (Burke, 1996). Our work demonstrates that the catalytic activity of the designed RNA is under the regulation of the designed protein.
Two group I intron splicing factors, CYT-18 and CBP2, are known to bind to specific regions of the group I introns and stabilize their active structures during the reaction. It has been suggested that these proteins also contribute to the folding step of the ribozymes: CYT-18 binds to the P4–P6 region and facilitates formation of the P7–P3–P8 domain of the intron (Caprara et al., 1996; Saldanha et al., 1996), and CBP2 binds to a partially folded tertiary structure of the RNA and captures the transiently associated P5–P4–P6 and P7–P3–P8 domains (Weeks and Cech, 1996).
The differences between our proteins and the natural group I intron splicing factors (CBP2 and CYT-18) are as follows. The designed proteins significantly aid the appropriate RNA folding that determines the final extent of the reaction as shown in Figure 7, but very weakly affect the kcat of the ribozyme (Table I). In contrast, CBP2 accelerates a particular folding step of the ribozyme but does not significantly affect the final extent of the reaction under the conditions employed (Weeks and Cech, 1995, 1996); however, it increases the kcat value up to several hundred times when compared with the ribozyme alone in the presence of low concentrations of magnesium ions (Weeks and Cech, 1995). The nature of CYT-18 has been thought to resemble that of CBP2, although comparable kinetic analyses have not been performed. It is known that the protein facilitates formation of the P7–P3–P8 domain by binding to the P4–P6 region (Caprara et al., 1996; Saldanha et al., 1996).
Our proteins are not involved in the reaction step(s) although they can facilitate the correct folding of the ribozyme. Thus, we think that they can be regarded as a primitive form of the splicing factors or a novel form of RNA chaperones that promote the correct folding of the RNA via non-specific RNA–protein interactions and are dispensable for the reaction (Coetzee et al., 1994).
According to the ‘RNA world’ hypothesis for early molecular evolution, proteins would have gradually taken over the structural elements in ribozymes. As it has previously been shown that several non-specific RNA-binding proteins can enhance RNA function, the emergence of non-specific RNA-binding proteins may have been an early step in the transition from the RNA world to the RNA–protein world as suggested by Herschlag (1995). It has been proposed that certain self-splicing intron RNAs that happened to bind to proteins in cells became the splicing RNPs and subsequently the RNA components lost their self-autonomous activity due to accumulated mutations (Lambowitz and Perlman, 1990). Although the peptide-binding motifs were introduced to the RNA prior to the binding of the protein in our system, the success of the simple design of an RNP shows that an active RNP might have evolved easily from a ribozyme.
We have shown that the molecular design of a functional RNP is simple and effective by employing known structures of an RNA and peptides in a molecular modeling program. This strategy of molecular building will have broad applications for constructing various forms of functional RNPs in the fields of science and engineering.
After completion of our work, an example of the design of a specific RNA-binding dimer was reported by Campisi et al. (2001). In this study, a dimeric RNA–peptide complex was designed and constructed based on the NMR structure of the BIV Tat–TAR complex by employing a computer modeling method.
Materials and methods
Design and construction of the templates for mutant Tetrahymena ribozymes and designed proteins
Molecular modeling of the RNA was performed by using Insight II on a Silicon Graphics workstation. A molecular model of M12 was constructed from the coordinates of the crystal structure of the P4–P6 domain of the Tetrahymena ribozyme (Protein Data Bank ID 1GID; Cate et al., 1996a), the NMR structures of the bacteriophage λN peptide–boxB RNA complex (1QFQ; Legault et al., 1998) and HIV Rev peptide–RRE RNA complex (1ETF; Battiste et al., 1996). Plasmids encoding the derivatives of the Tetrahymena ribozyme were prepared from pTZIVSU (Williamson et al., 1989) using PCR (Imai et al., 1991) and verified by sequencing. Templates for pep A, pep G and their derivatives were constructed on a pSTV28 (Takara) derivative pSTVpep, in which the gene coding for lacZα′ was replaced by a gene encoding the protein.
Preparation of the precursor RNAs
Template DNAs for the precursor RNAs were generated by performing 25 cycles of PCR using KOD DNA polymerase (TOYOBO). All RNAs employed in this study were prepared by in vitro transcription with T7 RNA polymerase (Milligan et al., 1987). Following purification in 5% polyacrylamide denaturing gels (Milligan et al., 1987), the isolated RNAs were passed over a Sephadex G-50 spin column. For preparation of 32P-labeled RNAs, the transcription was performed in the presence of [α-32P]ATP.
Preparation of the activator proteins
The proteins were synthesized from expression plasmid pTYB1 derivatives in E.coli strain ER2566 followed by purification with the IMPACT™ T7 System (Chong et al., 1997) (New England Biolabs). Protein concentrations were determined by the Bradford assay (Bio-Rad).
In vitro splicing assays and kinetic analysis
32P-labeled precursor RNAs (10 nM) were dissolved in distilled water followed by incubation at 80°C for 5 min. After cooling and incubation at 37°C for 1 min, 10× concentrated reaction buffer and protein in dilution buffer (20 mM Tris–HCl pH 7.5, 40 mM KCl, 50% glycerol) were simultaneously added at 37°C. (Note, in the experiments in which the proteins were added to the renatured RNA, the protein was added at 5, 10 or 15 min after the addition of 10× concentrated reaction buffer.) After pre-incubation at 37°C for 5 min, reactions were started with 200 µM GTP at 37°C in the presence of 40 mM Tris–HCl pH 7.5, 2 mM MgCl2, 80 mM KCl and 2.5% glycerol. [Note, pre-incubation time (10, 15 or 20 min) did not influence kobs and the final extent of the splicing reaction.] Aliquots were removed at specified times. The reactions were terminated by addition of an equal volume of stop solution (150 mM EDTA, 70% formamide, 0.25% xylenecyanol) followed by electrophoresis on 5% polyacrylamide denaturing gels. RNAs were quantitated with a Bio Imaging Analyzer (BA2500; Fuji Film). kcat and KmGTP were determined from time-courses of splicing reactions at different GTP concentrations. The data were fitted to a single exponential, (fraction of ligated exons)t = (fraction of ligated exons)t=∞ – (fraction of ligated exons)t=∞ × exp(–kobst), to obtain the rate constant; kobs⋅kcat and KmGTP were determined from Hanes–Woolfe plots of kobs as a function of GTP concentration.
The in vivo mutagenic protein selection
The protein library was constructed on pSTVpep with degenerate oligonucleotides, 5′-GTGGAAAGCCGCTGCAGCG(NNK)4GGCGCC GCAGCGGCGCAGA-3′, where N is an A:C:T:G mixture (1:1:1:1 ratio) and K is a G:T mixture (1:1) (Harada et al., 1997). A primer (5′-TCTGCGCCGCTGCGGCGCC-3′) was annealed to the oligonucleotide and double-stranded DNAs were synthesized using KOD DNA polymerase (TOYOBO). The resulting DNAs were ligated into the PstI and KasI sites of pSTVpep. Escherichia coli JM109 containing plasmids for the RNAs were transformed with the ligation mixture. The cells were incubated in 200 ml of M9 medium (Silhavy et al., 1984) containing 400 mg/l lactose, 50 mg/l ampicillin and 50 mg/l chloramphenicol with shaking at 37°C for 4 days.
Growth rate assays
M9 medium (10 ml) containing lactose was inoculated with ∼1 × 106 cells from overnight cultures and the resulting mixtures were incubated at 37°C. Aliquots were removed at specific times and their OD600 measured (Roth, 1970). All assays were repeated at least three times and gave highly reproducible results.
RNA isolation and primer extensions
Escherichia coli JM109 containing the plasmids for the RNA and proteins were grown at 37°C until their OD600 reached 0.2 in LB media containing 50 mg/l ampicillin and 50 mg/l chloramphenicol. Isopropyl-β-d-thiogalactopyranoside was added to a final concentration of 0.3 mM and the resulting cultures were grown for an additional 10 min. Isolation of cellular RNA and primer extension analysis were performed with the following modifications (Zhang et al., 1995). Reaction mixtures were incubated for 15 min at 50°C with ReverTra Ace (TOYOBO). Products were separated on 15% polyacrylamide denaturing gels and quantitated with a Bio Imaging Analyzer (BA2500; Fuji Film).
RNase and chemical probing and primer extension analysis of the modified RNA
L-21-ScaI M12 RNAs (100 nM) were dissolved in distilled water followed by incubation at 80°C for 5 min. After cooling and incubation at 37°C for 1 min, 10× concentrated reaction buffer and protein (3 µM) were added. After pre-incubation for 5 min, M12 RNA or RNA–protein complexes were digested by addition of 1 µl of RNase V1 (0.5 U/ml; Amersham Pharmacia Biotech) or incubated with 1 µl of DMS diluted to 1:600, followed by incubation for 10 min at 37°C. Chemical probing reactions were stopped with 12.5 µl of 2 M β-mercaptoethanol. The RNAs were subsequently extracted with phenol and ethanol precipitated. RNase cleavage sites or modified nucleotides were detected by primer extension using two oligonucleotide primers, complementary to positions 199–222 and 262–288 of the wild-type ribozyme. One-third of the modified RNAs was extended by ReverTra Ace (TOYOBO). Products were separated on 6% polyacrylamide denaturing gels and quantitated with a Bio Imaging Analyzer (BA2500; Fuji Film).
Acknowledgments
Acknowledgements
We thank Dr Ruth T.Yu for critical reading of the manuscript, Dr Kazuei Igarashi for the gift of RNase V1 and members of the Inoue laboratory for their helpful advice. This work was supported by Grants-in-Aids for Scientific Research on Priority Areas from the Ministry of Education, Science, Sports and Culture, Japan.
References
- Battiste J.L., Mao,H., Rao,N.S., Tan,R., Muhandiram,D.R., Kay,L.E., Frankel,A.D. and Williamson,J.R. (1996) α helix–RNA major groove recognition in an HIV-1 rev peptide–RRE RNA complex. Science, 273, 1547–1551. [DOI] [PubMed] [Google Scholar]
- Burke J.M. (1996) Structural analysis and modifications of the hairpin ribozyme. In Eckstein,F. and Lilley,D.M.J. (eds), Catalytic RNA. Springer, Berlin, Germany, pp. 129–143.
- Campisi D.M., Calabro,V. and Frankel,A.D. (2001) Structure-based design of a dimeric RNA–peptide complex. EMBO J., 20, 178–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprara M.G., Mohr,G. and Lambowitz,A.M. (1996) A tyrosyl-tRNA synthetase protein induces tertiary folding of the group I intron catalytic core. J. Mol. Biol., 257, 512–531. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996a) Crystal structure of a group I ribozyme domain: principles of RNA packing. Science, 273, 1678–1685. [DOI] [PubMed] [Google Scholar]
- Cate J.H., Gooding,A.R., Podell,E., Zhou,K., Golden,B.L., Szewczak,A.A., Kundrot,C.E., Cech,T.R. and Doudna,J.A. (1996b) RNA tertiary structure mediation by adenosine platforms. Science, 273, 1696–1699. [DOI] [PubMed] [Google Scholar]
- Cech T.R. (1990) Self-splicing of group I introns. Annu. Rev. Biochem., 59, 543–568. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay S., Garcia-Mena,J., DeVito,J., Wolska,K. and Das,A. (1995) Bipartite function of a small RNA hairpin in transcription antitermination in bacteriophage λ. Proc. Natl Acad. Sci. USA, 92, 4061–4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S. et al. (1997) Single-column purification of free recombinant proteins using a self-cleavable affinity tag derived from a protein splicing element. Gene, 192, 271–281. [DOI] [PubMed] [Google Scholar]
- Coetzee T., Herschlag,D. and Belfort,M. (1994) Escherichia coli proteins, including ribosomal protein S12, facilitate in vitro splicing of phage T4 introns by acting as RNA chaperones. Genes Dev., 8, 1575–1588. [DOI] [PubMed] [Google Scholar]
- DeGrado W.F., Summa,C.M., Pavone,V., Nastri,F. and Lombardi,A. (1999) De novo design and structural characterization of proteins and metalloproteins. Annu. Rev. Biochem., 68, 779–819. [DOI] [PubMed] [Google Scholar]
- Doherty E.A. and Doudna,J.A. (1997) The P4–P6 domain directs higher order folding of the Tetrahymena ribozyme core. Biochemistry, 36, 3159–3169. [DOI] [PubMed] [Google Scholar]
- Downs W.D. and Cech,T.R. (1996) Kinetic pathway for folding of the Tetrahymena ribozyme revealed by three UV-inducible crosslinks. RNA, 2, 718–732. [PMC free article] [PubMed] [Google Scholar]
- Flor P.J., Flanegan,J.B. and Cech,T.R. (1989) A conserved base pair within helix P4 of the Tetrahymena ribozyme helps to form the tertiary structure required for self-splicing. EMBO J., 8, 3391–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert W. (1986) The RNA world. Nature, 319, 618. [Google Scholar]
- Harada K., Martin,S.S., Tan,R. and Frankel,A.D. (1997) Molding a peptide into an RNA site by in vivo peptide evolution. Proc. Natl Acad. Sci. USA, 94, 11887–11892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herschlag D. (1995) RNA chaperones and the RNA folding problem. J. Biol. Chem., 270, 20871–20874. [DOI] [PubMed] [Google Scholar]
- Ho Y. and Waring,R.B. (1999) The maturase encoded by a group I intron from Aspergillus nidulans stabilizes RNA tertiary structure and promotes rapid splicing. J. Mol. Biol., 292, 987–1001. [DOI] [PubMed] [Google Scholar]
- Imai Y., Matsushima,Y., Sugimura,T. and Terada,M. (1991) A simple and rapid method for generating a deletion by PCR. Nucleic Acids Res., 19, 2785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T. (1994) Time to change partners. Nature, 370, 99–100. [DOI] [PubMed] [Google Scholar]
- Jaeger L., Michel,F. and Westhof,E. (1996) The structure of group I ribozymes. In Eckstein,F. and Lilley,D.M.J. (eds), Catalytic RNA. Springer, Berlin, Germany, pp. 33–51.
- Jain C. and Belasco,J.G. (1996) A structural model for the HIV-1 Rev–RRE complex deduced from altered-specificity rev variants isolated by a rapid genetic strategy. Cell, 87, 115–125. [DOI] [PubMed] [Google Scholar]
- Joyce G.F., van der Horst,G. and Inoue,T. (1989) Catalytic activity is retained in the Tetrahymena group I intron despite removal of the large extension of element P5. Nucleic Acids Res., 17, 7879–7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems J., Brown,M., Chang,D.D. and Sharp,P.A. (1991) Structural analysis of the interaction between the human immunodeficiency virus Rev protein and the Rev response element. Proc. Natl Acad. Sci. USA, 88, 683–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B., Murphy,F.L. and Cech,T.R. (1994) Two major tertiary folding transitions of the Tetrahymena catalytic RNA. EMBO J., 13, 2669–2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A.M. and Perlman,P.S. (1990) Involvement of aminoacyl-tRNA synthetases and other proteins in group I and group II intron splicing. Trends Biochem. Sci., 15, 440–444. [DOI] [PubMed] [Google Scholar]
- Lambowitz A.M., Caprara,M.G., Zimmerly,S. and Perlman,P.S. (1999) Group I and group II ribozymes as RNPs: clues to the past and guides to the future. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 451–485.
- Legault P., Li,J., Mogridge,J., Kay,L.E. and Greenblatt,J. (1998) NMR structure of the bacteriophage λ N peptide/boxB RNA complex: recognition of a GNRA fold by an arginine-rich motif. Cell, 93, 289–299. [DOI] [PubMed] [Google Scholar]
- Liu D.R., Magliery,T.J., Pastrnak,M. and Schultz,P.G. (1997) Engineering a tRNA and aminoacyl-tRNA synthetase for the site-specific incorporation of unnatural amino acids into proteins in vivo. Proc. Natl Acad. Sci. USA, 94, 10092–10097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacBeath G., Kast,P. and Hilvert,D. (1998) Redesigning enzyme topology by directed evolution. Science, 279, 1958–1961. [DOI] [PubMed] [Google Scholar]
- Matthews D.E., Hessler,R.A., Denslow,N.D., Edwards,J.S. and O’Brien,T.W. (1982) Protein composition of the bovine mitochondrial ribosome. J. Biol. Chem., 257, 8788–8794. [PubMed] [Google Scholar]
- Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr G., Caprara,M.G., Guo,Q. and Lambowitz,A.M. (1994) A tyrosyl-tRNA synthetase can function similarly to an RNA structure in the Tetrahymena ribozyme. Nature, 370, 147–150. [DOI] [PubMed] [Google Scholar]
- Murphy F.L. and Cech,T.R. (1994) GAAA tetraloop and conserved bulge stabilize tertiary structure of a group I intron domain. J. Mol. Biol., 236, 49–63. [DOI] [PubMed] [Google Scholar]
- Roth J.R. (1970) Genetic techniques in studies of bacterial metabolism. Methods Enzymol., 17, 3–35. [Google Scholar]
- Saldanha R., Ellington,A. and Lambowitz,A.M. (1996) Analysis of the CYT-18 protein binding site at the junction of stacked helices in a group I intron RNA by quantitative binding assays and in vitro selection. J. Mol. Biol., 261, 23–42. [DOI] [PubMed] [Google Scholar]
- Sclavi B., Sullivan,M., Chance,M.R., Brenowitz,M. and Woodson,S.A. (1998) RNA folding at millisecond intervals by synchrotron hydroxyl radical footprinting. Science, 279, 1940–1943. [DOI] [PubMed] [Google Scholar]
- Silhavy T.J., Berman,M.L. and Enquist,L.W. (1984) Experiments with Gene Fusions. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Soukup G.A. and Breaker,R.R. (1999) Design of allosteric hammerhead ribozymes activated by ligand-induced structure stabilization. Struct. Fold. Des., 7, 783–791. [DOI] [PubMed] [Google Scholar]
- Tan R. and Frankel,A.D. (1995) Structural variety of arginine-rich RNA-binding peptides. Proc. Natl Acad. Sci. USA, 92, 5282–5286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan R., Chen,L., Buettner,J.A., Hudson,D. and Frankel,A.D. (1993) RNA recognition by an isolated α helix. Cell, 73, 1031–1040. [DOI] [PubMed] [Google Scholar]
- Tang J. and Breaker,R.R. (1997) Rational design of allosteric ribozymes. Chem. Biol., 4, 453–459. [DOI] [PubMed] [Google Scholar]
- Treiber D.K., Rook,M.S., Zarrinkar,P.P. and Williamson,J.R. (1998) Kinetic intermediates trapped by native interactions in RNA folding. Science, 279, 1943–1946. [DOI] [PubMed] [Google Scholar]
- Weeks K.M. (1997) Protein-facilitated RNA folding. Curr. Opin. Struct. Biol., 7, 336–342. [DOI] [PubMed] [Google Scholar]
- Weeks K.M. and Cech,T.R. (1995) Efficient protein-facilitated splicing of the yeast mitochondrial bI5 intron. Biochemistry, 34, 7728–7738. [DOI] [PubMed] [Google Scholar]
- Weeks K.M. and Cech,T.R. (1996) Assembly of a ribonucleoprotein catalyst by tertiary structure capture. Science, 271, 345–348. [DOI] [PubMed] [Google Scholar]
- Williamson C.L., Desai,N.M. and Burke,J.M. (1989) Compensatory mutations demonstrate that P8 and P6 are RNA secondary structure elements important for processing of a group I intron. Nucleic Acids Res., 17, 675–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaug A.J., McEvoy,M.M. and Cech,T.R. (1993) Self-splicing of the group I intron from Anabaena pre-tRNA: requirement for base-pairing of the exons in the anticodon stem. Biochemistry, 32, 7946–7953. [DOI] [PubMed] [Google Scholar]
- Zhang F., Ramsay,E.S. and Woodson,S.A. (1995) In vivo facilitation of Tetrahymena group I intron splicing in Escherichia coli pre-ribosomal RNA. RNA, 1, 284–292. [PMC free article] [PubMed] [Google Scholar]