Abstract
decapentaplegic (dpp) encodes a Drosophila transforming growth factor-β homologue that functions as a morphogen in the developing embryo and in adult appendage formation. In the wing imaginal disc, a Dpp gradient governs patterning along the anteroposterior axis by inducing regional expression of diverse genes in a concentration-dependent manner. Recent studies show that responses to graded Dpp activity also require an input from a complementary and opposing gradient of Brinker (Brk), a transcriptional repressor protein encoded by a Dpp target gene. Here we show that Brk harbours a functional and transferable repression domain, through which it recruits the corepressors Groucho and CtBP. By analysing transcriptional outcomes arising from the genetic removal of these corepressors, and by ectopically expressing Brk variants in the embryo, we demonstrate that these corepressors are alternatively used by Brk for repressing some Dpp-responsive genes, whereas for repressing other distinct target genes they are not required. Our results show that Brk utilizes multiple means to repress its endogenous target genes, allowing repression of a multitude of complex Dpp target promoters.
Keywords: Brinker/CtBP/Dpp signalling/Groucho/transcriptional repression
Introduction
In multicellular organisms, the patterning of tissues depends on the intracellular integration and fine-tuning of multiple external signals, which evoke an assortment of cellular responses. Extrinsic cues are transduced by cytoplasmic effectors and, ultimately, information is relayed to the nucleus, where transcription factors are stimulated to either activate or repress target gene transcription. The transforming growth factor-β (TGF-β) superfamily members, which dictate a wide range of cellular outcomes such as proliferation, alteration of cell shape, apoptosis and cell fate specification, exemplify such signalling molecules (Raftery and Sutherland, 1999). One key Drosophila TGF-β homologue, encoded by the decapentaplegic (dpp) gene, has been shown to function as a long-range morphogen, specifying varied cell fates in a concentration-dependent manner (Lecuit et al., 1996; Nellen et al., 1996). In both embryonic and post-embryonic development, Dpp governs multiple patterning events, including the specification of cells along the dorsoventral (D/V) axis in the embryo and the patterning of the adult appendages (Podos and Ferguson, 1999). Thus, in the developing wing imaginal disc, dpp is expressed in a central narrow stripe of cells along the anteroposterior (A/P) compartment boundary, from where Dpp spreads towards the periphery, mediating both cell proliferation and A/P patterning (Burke and Basler, 1996; Lecuit et al., 1996; Nellen et al., 1996; Entchev et al., 2000; Teleman and Cohen, 2000).
A seemingly simple model provides a coherent framework for explaining how the external Dpp signal is transmitted by cytoplasmic components and how it brings about nuclear transcriptional outcomes: Dpp binds to a heteromeric type II/type I transmembrane serine/threonine kinase receptor complex, encoded by thickveins and punt, triggering the phosphorylation of Mad, the Drosophila receptor-specific Smad. Subsequently, phosphorylated Mad (pMad) associates with Medea, and the pMad– Medea complex enters the nucleus to activate Dpp-responsive genes (Raftery and Sutherland, 1999).
The recent cloning and characterization of brinker (brk), a resident Dpp target gene encoding a repressor protein that antagonizes Dpp-mediated activation (Campbell and Tomlinson, 1999; Jazwinska et al., 1999a,b; Minami et al., 1999), has enhanced our understanding of how Dpp is able to trigger specific target gene expression programs. Where Dpp signalling is active, the transcriptional regulator Schnurri (Shn) switches off brk expression, thus relieving a subset of Dpp-responsive genes from Brk repression (Marty et al., 2000). Consequently, brk is expressed in the ventrolateral regions of the embryo, abutting the dorsal dpp expression domain, whereas in the wing imaginal disc, brk is expressed at high levels only in the periphery of the disc, with its transcription diminishing towards the centre. Thus, two opposing and complementary gradients, i.e. activation mediated by Smads and repression by Brk, ensure that discrete thresholds for Dpp activity are attained. In this paper we explore the molecular basis underlying Brk repression.
Transcription factors negate gene expression in diverse manners (Mannervik et al., 1999). Some do so ‘passively’, by competing with, and occluding activators from binding to coincident cis-acting DNA elements. For other repressors, DNA binding is not sufficient. Rather, these repressors are fully reliant on tethered corepressors and act more instructively, by local ‘quenching’ of proximally-bound activators, interference at a distance with the basal transcription machinery or altering chromatin structure and organization (Johnson, 1995; Cai et al., 1996; Gray and Levine, 1996). To date, two prototypic Drosophila corepressors have been characterized that seem to be markedly distinct from each other, i.e. where tested, one assists negative transcriptional regulators acting at short-range while the other supports long-range repressors (Zhang and Levine, 1999). Thus, the C-terminal binding protein (CtBP; Nibu et al., 1998a,b; Poortinga et al., 1998) is a corepressor that acts in conjunction with repressors obstructing the function of activators bound up to 150 base pairs away (e.g. Gray et al., 1994; Arnosti et al., 1996). In addition, the corepressor Groucho (Gro; Fisher and Caudy, 1998; Parkhurst, 1998; Chen and Courey, 2000) is required by repressors capable of hindering promoter function at long-range, shutting off transcription even over distances of up to several thousand base pairs (Paroush et al., 1994; Cai et al., 1996; Barolo and Levine, 1997; Dubnicoff et al., 1997).
In this study we show that Brk contains a functional repression domain that accommodates Gro and CtBP recruitment motifs, and that Brk interacts physically with these cofactors. Although other Drosophila repressors are known to possess more than one repressor domain (e.g. Arnosti et al., 1996; Keller et al., 2000; Kobayashi et al., 2001), the biological relevance of this feature has not yet been genetically addressed. Here we investigate the functionality of Brk’s association with two corepressors and demonstrate that the mechanism of repression by Brk is dependent on promoter context. We show that Brk requires either or both Gro and CtBP for switching off some target genes, whereas for the silencing of others, it requires neither of these cofactors, presumably relying on its reported ability to outcompete activators from binding DNA. We surmise that the combinatorial use by Brk of these two corepressors provides a versatility that allows it to silence a variety of composite promoters in response to graded morphogenetic activity of Dpp.
Results
Dpp target genes are specifically repressed by overexpression of gro
Gro is ubiquitously expressed in the adult wing (Tata and Hartley, 1993) and mutations in gro have been identified in genetic screens for modifiers of various wing and eye phenotypes (e.g. Heitzler et al., 1996; Chanut et al., 2000), implicating Gro in advanced developmental stages. Indeed, Gro has been ascribed at least one specific role in the establishment of wing configuration, as a corepressor for the Enhancer of split basic-helix–loop–helix proteins acting downstream of Notch signalling in D/V wing patterning (Heitzler et al., 1996). To assess whether Gro also contributes in hitherto unrecognized ways to wing A/P axis formation, we analysed the expression of wing-patterning genes in marked clones of cells that either ectopically overexpress, or are mutant for, gro (Xu and Rubin, 1993; Pignoni and Zipursky, 1997; see Materials and methods). Overexpression of gro should enhance the silencing of genes normally repressed by Gro-dependent transcriptional regulators while, reciprocally, the loss of gro should result in derepression, and therefore in the ectopic induction of these genes.
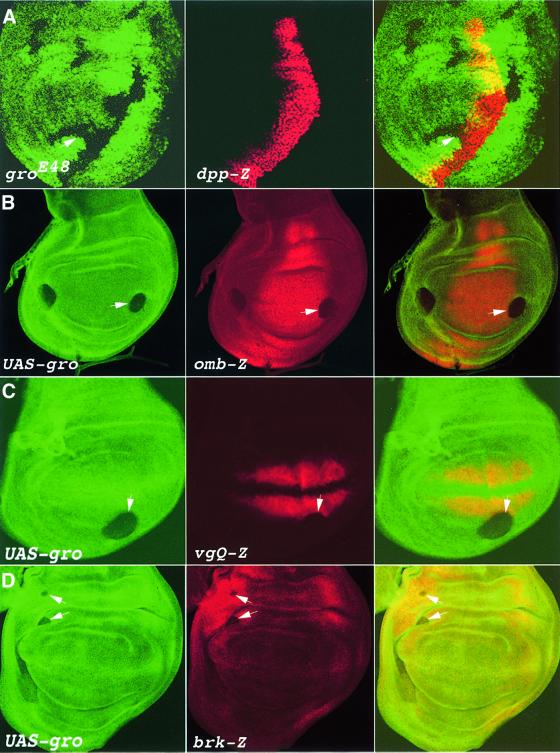
In the wing imaginal disc, cells in the posterior compartment are programmed by the engrailed selector gene product to secrete Hedgehog (Hh), which induces dpp in a stripe of anterior cells along the A/P boundary. Dpp then acts as a long-range morphogen that governs patterning across the entire imaginal disc field (Podos and Ferguson, 1999). To determine whether Gro participates in the implementation of Hh signalling, we stained clones overexpressing gro, or clones that are homozygous for the strong groE48 allele, for dpp-lacZ expression. In all clones, even those overlapping with the Hh activity domain, there are no noticeable alterations in the dpp expression pattern (Figure 1A; data not shown), indicating that Gro is not required downstream of Hh for dpp transcriptional regulation. In striking contrast, however, three distinct targets of the Dpp pathway, expressed either in the wing pouch (optomotor-blind; omb and vestigial; vg) or in the periphery of the wing disc (brk), are repressed in clones overexpressing gro (Figure 1B–D). Expression of omb-lacZ (Figure 1B), as well as that of a lacZ reporter driven by vg’s Dpp-responsive enhancer (vgQ-lacZ; Figure 1C), is completely abrogated in these clones, whereas expression of brk-lacZ is only reduced (Figure 1D; see below). All three Dpp targets are repressed in a cell autonomous manner, i.e. only in the clones but never in adjacent cells. These results, together with an extensive gro loss-of-function clonal analysis detailed below, implicate Gro specifically as a downstream effector of Dpp signalling.
Fig. 1. Overexpression of gro does not affect dpp expression, but brings about repression of Dpp target genes. (A–D) Third instar larval imaginal wing discs, stained for the πMyc or CD2 markers (left) and for β-galactosidase (centre); merge, right. In these, and subsequent figures, anterior is to the left and dorsal up. (A) dpp-lacZ expression (centre, red) is unaffected by groE48 mutant clones, marked by loss of the πMyc marker (left, green), or by gro overexpression (data not shown). In contrast, gro overexpression, in clones marked by loss of CD2 (left, green), leads to the complete repression of omb-lacZ (B) and vgQ-lacZ (C), and to a reduction in brk-lacZ (D) expression levels, in a cell-autonomous manner.
Brk interacts physically with two corepressors, Gro and CtBP
Recent genetic and molecular studies have shown that brk encodes a repressor acting downstream of the Dpp pathway, which helps define the low end of the Dpp gradient (Campbell and Tomlinson, 1999; Jazwinska et al., 1999a; Minami et al., 1999). In particular, the Dpp targets omb and vgQ are both derepressed in brk– mutant clones and in brk– wing imaginal discs, suggesting that they are normally subjected to Brk repression (Campbell and Tomlinson, 1999; Jazwinska et al., 1999a; Minami et al., 1999). More directly, Brk binds to specific sequences within defined omb and vgQ enhancer elements, bringing about their silencing by outcompeting the Mad–Medea complex, or some other activator, from binding to overlapping DNA sites (Sivasankaran et al., 2000; Kirkpatrick et al., 2001).
That putative Brk target genes are repressed in clones of cells with increased gro dosage strongly suggests that Brk is a Gro-dependent repressor. Accordingly, Brk’s proposed repression domain (RD) (Campbell and Tomlinson, 1999) harbours a potential Gro recruitment motif (FKPY), similar to the Gro-binding domains defined in the repressors Hairy (WRPW), Runt (WRPY) and Huckebein (FRPW), and identical to that in Even-skipped (Eve) (Paroush et al., 1994; Aronson et al., 1997; Goldstein et al., 1999; Kobayashi et al., 2001). It has been noted previously by others that Brk also contains a CtBP-binding domain (PMDLSLG; Jazwinska et al., 1999a). Below we show that Brk is in fact able to interact physically with both Gro and CtBP, and address the functional relevance of these associations to Brk’s in vivo repressor capacity.
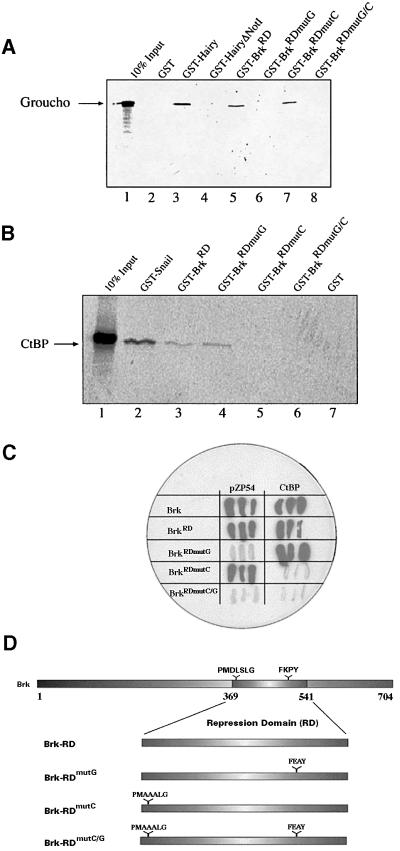
To demonstrate Brk’s ability to associate with the two corepressors in vitro, we fused the protein’s putative RD (amino acids 369–541) to glutathione S-transferase (GST), and incubated it with radioactively labelled Gro or CtBP (Figure 2; see Materials and methods). In GST pull-down assays, Brk’s RD (BrkRD), but not GST alone, readily retains [35S]methionine-labelled Gro (Figure 2A). To test further the specificity of this interaction, three mutant derivatives of the BrkRD, fused to the GST moiety, were generated in which the Gro recruitment domain (BrkRDmutG; FKPY to FEAY; Goldstein et al., 1999), the core of the CtBP-binding motif (BrkRDmutC; DLS to AAA; Zhang and Levine, 1999) or both (BrkRDmutC/G) were altered (Figure 2D). As shown in Figure 2A, Brk’s binding to Gro is impaired by the modifications in the FKPY motif. Significantly, however, Gro associates with the GST– BrkRDmutC construct as strongly as it does with the native GST–BrkRD fusion. GST–BrkRD also binds labelled CtBP in vitro (Figure 2B) and, although the binding of Brk to CtBP is weak in this assay, the specificity of the interaction is clearly evident: the association between the two proteins is abolished by mutations in the CtBP recruitment domain but is unaffected by alterations in the Gro recruitment motif (Figure 2B).
Fig. 2. Brk interacts physically with Gro and CtBP. (A and B) In vitro pull-down assays. 35S-labelled Gro (A) or dCtBP (B) were incubated with GST–BrkRD derivatives immobilized on glutathione–agarose beads and, following washing, retained 35S-labelled protein was subjected to SDS–PAGE (not shown) and autoradiography. (A) Gro binds specifically to GST–BrkRD (lane 5) and to GST–BrkRDmutC (lane 7), but not to GST–Brk variants in which Brk’s FKPY motif is mutated (lanes 6 and 8). GST–Hairy (lane 3) serves as a positive control, whereas Hairy lacking its C-terminal Gro-binding domain (HairyΔNotI, lane 4) and GST alone (lane 2) are negative controls. (B) CtBP binds specifically to GST–BrkRD (lane 3) and BrkRDmutG (lane 4), although to a lesser extent than to GST–Snail (lane 2), an established CtBP partner (Nibu et al., 1998a). Mutating Brk’s CtBP recruitment core motif abolishes CtBP binding (lanes 5 and 6). GST alone, lane 7. (A and B) 10% of input-labelled protein was run in lane 1. Arrows indicate positions of full-length Gro (A) and CtBP (B). (C) Yeast two-hybrid assay. Full-length Brk, or just its RD, interacts strongly in yeast with full-length Gro (not shown), with pZP54 (Gro251–719) and with CtBP (white colonies indicate lack of interactions). Mutating either the Gro or CtBP recruitment motif in Brk’s RD results in loss of interactions between Brk and the corresponding corepressor. (D) A schematic representation of Brk’s RD (residues 369–541) and derived constructs. The Brk corepressor recruitment motifs, and respective mutant versions, are indicated.
We confirmed Brk’s ability to interact with Gro and CtBP, particularly the specificity of the associations, using the yeast two-hybrid system (Figure 2C; see Materials and methods). The full-length Brk, or only its RD portion, interacts strongly with Gro and pZP54 (Gro251–719; Paroush et al., 1994), as with CtBP (Figure 2C; data not shown). Here too, mutations in one recruitment motif selectively obliterate the binding of Brk to only the single respective corepressor but not to the other.
Brk contains a functional repression domain that depends on corepressors
Brk has been reported to negate transcription by competing with activators, such as Mad/Medea, for overlapping DNA target sites, thereby preventing them access to target promoters (Sivasankaran et al., 2000; Kirkpatrick et al., 2001; Rushlow et al., 2001). Its direct interactions with Gro and CtBP, however, suggest that Brk acts in a more instructive manner. While in the former ‘passive’ mechanism Brk is expected to rely solely on its competitive DNA-binding activity, the latter ‘active’ mechanism predicts that it accommodates an innate RD that depends on the recruitment of corepressors.
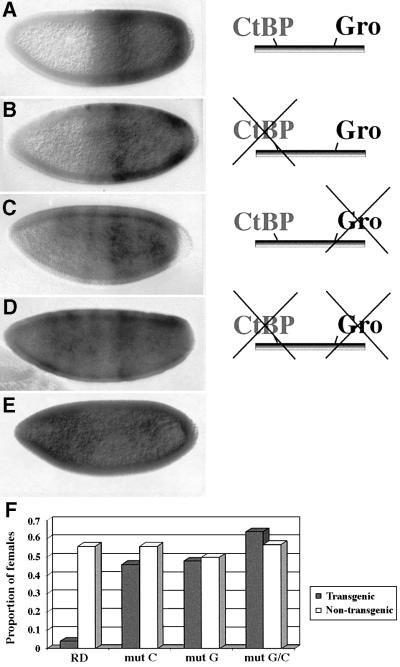
To establish whether Brk contains a functional RD that can silence gene expression, separable from its DNA-binding domain, we employed an in vivo assay that relies on repression of the sex-determining Sex-lethal (Sxl) gene by ectopic expression of the pair-rule gene hairy (Parkhurst et al., 1990; Jiménez et al., 1997). Sxl is normally expressed only in female embryos whereas, in males, it is repressed by Deadpan (Dpn), an autosomally encoded Hairy-related repressor protein. When Hairy is expressed prematurely, under the hunchback (hb) promoter, it mimics Dpn’s repressor function and eradicates Sxl transcription in the anterior of syncytial blastoderm female embryos. Because Sxl is essential for dosage compensation in females, this repression subsequently leads to female-specific lethality (Parkhurst et al., 1990). A form of Hairy, lacking its own RD, is inert in this assay. However, fusion of heterologous RDs to the truncated Hairy protein restores its ability to repress Sxl (Jiménez et al., 1997; Goldstein et al., 1999). Indeed, the equivalent expression of a hb-Hairy-BrkRD transgene results in an effective repression of Sxl in the anterior halves of female embryos (Figure 3A) and female-specific lethality ensues (Figure 3F; see Materials and methods). Thus, the region in Brk spanning the Gro- and CtBP-binding domains promotes potent repression in embryos.
Fig. 3. Brk requires both Gro and CtBP for full repression of Sxl. (A) Expression of Hairy-BrkRD blocks Sxl expression in the anterior halves of female embryos and, consequently, strong female-specific lethality ensues (F). The ability of the BrkRD to repress Sxl in female embryos is compromised when the CtBP-binding motif is mutated (B) and is completely abolished by mutations in the Gro recruitment domain (C and D). Female-specific lethality is, correspondingly, affected (F). (E) Wild-type expression of Sxl. (A–E) Early Sxl expression was monitored using the Sxl-Pe:lacZ reporter strain (Estes et al., 1995). Equivalent results were obtained by staining embryos with a monoclonal antibody specific to the active form of Sxl. (F) The proportional number of transgenic (black) and non-transgenic (white) females of representative lines indicates the magnitude of female-specific lethality (see Materials and methods).
The ability to selectively disrupt Brk binding to each individual corepressor allowed us to start exploring the dependence of its repressor potential on Gro and/or CtBP in vivo. As both Gro- and CtBP-mediated repression can be detected in the Sxl-repression assay (Jiménez et al., 1997), we fused truncated Hairy to the three derivatives of the Brk RD, mutated in the Gro, CtBP or both recruitment motifs (see Figure 2D) and placed them under hb promo ter regulation. In female embryos expressing Hairy-BrkRDmutC, Sxl is substantially repressed, although not as effectively as by Hairy-BrkRD (Figure 3B). Furthermore, this repression still leads to statistically significant female-specific lethality (Figure 3F; p = 0.001; see Materials and methods). Thus, blocking CtBP binding does not completely abolish activity of the Brk RD. In comparison, mutating the Gro recruitment domain causes only residual Sxl repression (Figure 3C) and no apparent female-specific lethality (Figure 3F). Finally, Sxl expression is seen throughout female embryos expressing hb-Hairy-BrkRDmutC/G (Figure 3D), and no female-specific lethality is observed (Figure 3F). Thus, Brk relies mainly on Gro for repressing Sxl. Nevertheless, since mutating the CtBP recruitment motif in Brk’s RD attenuates Sxl repression (Figure 3B and F), we conclude that, for full potency as a negative transcriptional regulator, Brk requires both corepressors.
omb and spalt are repressed by Brk independently of Gro and CtBP
The above data indicate that the interactions between Brk and the corepressors Gro and CtBP are indispensable for maximal repression of Sxl in vivo. We next sought to establish whether Brk requires both cofactors for repression of its endogenous target genes. Below (Figures 4–7) we show that, for repression of distinct target genes, Brk requires Gro and/or CtBP differentially, presumably as a function of specific promoter topology and architecture (see Discussion).
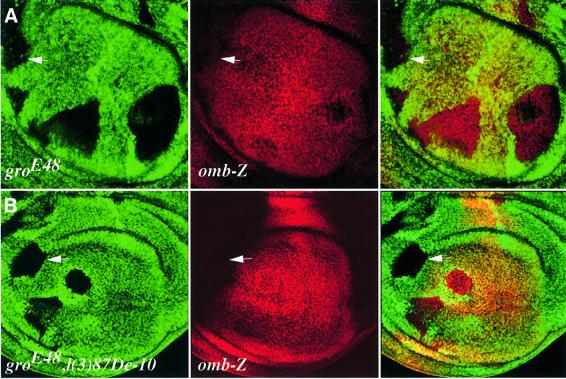
Fig. 4. Brk represses omb independently of Gro and CtBP. Wing imaginal discs, bearing gro– single (A) or CtBP–, gro– double mutant (B) clones, discernible by loss of the πMyc marker and by the appearance of a nearby twin-spot (left, green), do not show elevation of omb-lacZ expression (centre, red; arrows); merge, right.
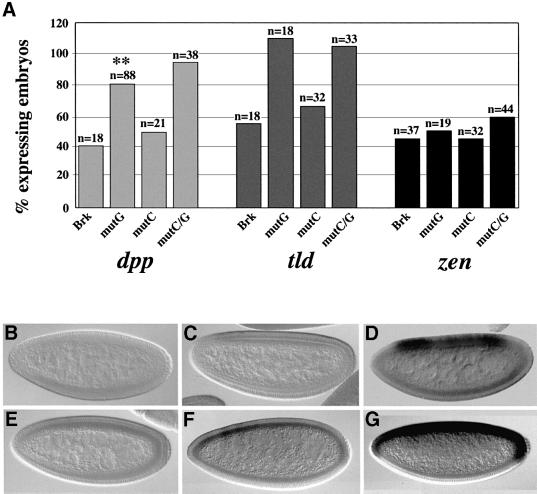
Fig. 7. Brk requires corepressors for its repressor function in the embryo. (A) Transgenic embryos overexpressing full-length Brk (from UAS-brk transgenes, driven by maternal Gal4VP16), either in its native form or with its corepressor-binding domains mutated, were stained for the expression of zen (right), tld (centre) and dpp (left) during mid- to late-cellularization. The Y-axis designates the percentage of embryos expressing a given target gene, calculated relative to the number of expressing wild-type embryos, referred to as 100%. n = number of embryos, at the appropriate stage, that were scored. (B–G) Representative embryos. (B) UAS-BrkmutC/G, stained for zen; (C) UAS-BrkmutC, stained for tld; (D) UAS-BrkmutG, stained for tld; (E) UAS-BrkmutC, stained for dpp; (F) UAS-BrkmutG, stained for dpp; (G) UAS-BrkmutC/G, stained for dpp. Repression of zen is independent of corepressors (A and B) whereas that of tld is strictly Gro dependent (A, C and D). As for the silencing of dpp, Brk relies mainly on Gro (A and E–G). Nevertheless, since the level of dpp expression is significantly lower in BrkmutG-expressing embryos (A, marked by ‘**’; F) when compared with wild-type embryos or with embryos expressing the BrkmutC/G transgene (G), CtBP must also be contributing to maximal Brk repressor ability.
Brk competes with an activator for binding to an omb wing enhancer (Sivasankaran et al., 2000), suggesting that, for this promoter, Brk should act independently of corepressors. Consistent with this, we find that omb-lacZ is not ectopically expressed in cells homozygous for groE48 (hereafter referred to as gro– clones) (Figure 4A), nor is it affected by CtBP loss-of-function clones, generated using the l(3)87De-10 allele (CtBP–; data not shown), or by CtBP–, gro– double mutant clones (Figure 4B). Thus, single and double mutant clones for gro and CtBP do not phenocopy the omb derepression seen in brk– clones (Campbell and Tomlinson, 1999; Jazwinska et al., 1999a; Minami et al., 1999), implying that Brk can repress omb even in the absence of these corepressors. Repression of the Dpp target gene spalt (sal; Nellen et al., 1996) is also independent of Gro and CtBP (data not shown). Nonetheless, in gro overexpression clones, omb is repressed (Figure 1B), suggesting that, even for the omb promoter, Gro reinforces Brk repressor function (see Discussion).
Brk requires Gro, but not CtBP, for repressing vgQ
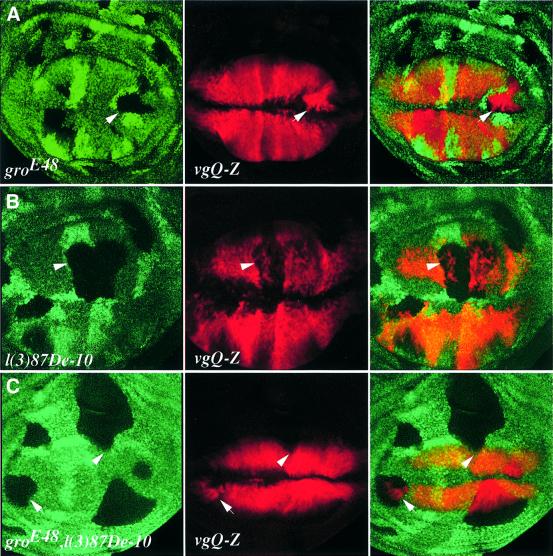
To establish whether Brk represses vgQ via Gro, CtBP or both, we monitored vgQ-lacZ expression in gro– and CtBP– single, and CtBP–, gro– double mutant clones. In this instance, we find a mandatory requirement for Gro, but not for CtBP; in gro– clones, vgQ is upregulated (Figure 5A). Importantly, as is the case for brk– clones (data not shown; Campbell and Tomlinson, 1999), the cell-autonomous upregulation of vgQ is seen only in gro– clones close to the periphery of the disc, suggesting that the observed effects are Brk dependent. In contrast, in CtBP– mutant clones vgQ expression is downregulated, in the Brk territory but also outside it, at the centre of the disc (Figure 5B), indicating that these effects are Brk independent and that CtBP is positively required for vg expression (see Discussion). CtBP–, gro– double mutant clones show a composite effect: ectopic expression and upregulation of vgQ in clones in the brk expression domain, and a phenotype resembling that of CtBP– clones at the middle of the disc, where brk is not expressed (Figure 5C). Thus, Brk repression of vgQ is Gro- but not CtBP-dependent.
Fig. 5. Brk requires Gro, but not CtBP, for repressing vgQ. (A) Clones of cells mutant for gro and marked by the loss of πMyc (left, green) show ectopic vgQ expression (centre, red). In contrast, CtBP– clones show a reduction in levels of vgQ expression (B). Note that gro– clones show a phenotype only when in the periphery of the disc, as seen in brk mutant clones (Campbell and Tomlinson, 1999), while the downregulation of vgQ in CtBP– clones is observed regardless of their position, indicating that these effects are Brk independent. (C) A composite phenotype in CtBP–, gro– double mutant clones resembles that of gro– in peripheral clones (arrow) and that of CtBP– in central clones (arrowhead).
Negative autoregulation of brk requires either Gro or CtBP
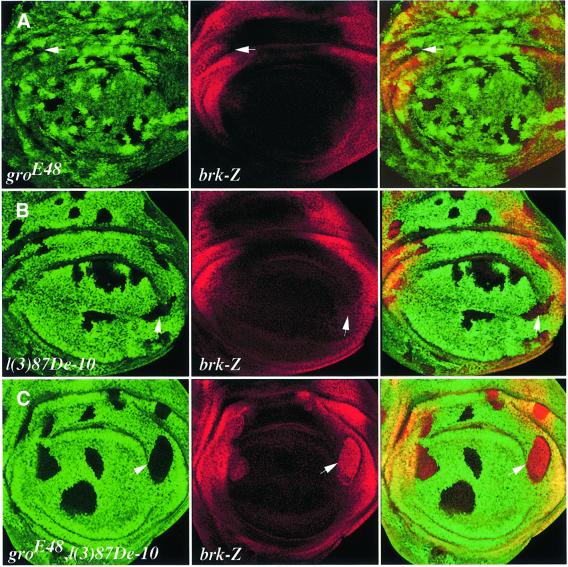
omb and vgQ expression is completely shut off in clones of cells overexpressing gro, whereas that of brk is only reduced (Figure 1), suggesting that Brk might be repressing its own transcription via a negative autoregulatory loop (B.Müller and K.Basler, unpublished results). To establish whether, in negating its own expression, Brk is assisted by Gro and/or CtBP, we stained gro– and CtBP– single (Figure 6A and B), or CtBP–, gro– double (Figure 6C) mutant clones for brk-lacZ expression. Figure 6A and B shows that brk is never ectopically expressed in any of the single mutant clones, whereas ectopic brk expression is clearly observable in double mutant clones (Figure 6C). Thus, in the absence of one corepressor, repression is adequately mediated by the other, suggesting that negative autoregulation by Brk is robust, relying on either Gro or CtBP.
Fig. 6. Negative autoregulation of brk requires either Gro or CtBP. Single gro– (A) or CtBP– (B) clones do not affect brk expression whereas CtBP–, gro– double mutant clones show ectopic brk expression (C). The phenotype in (C) is seen only in the periphery of the disc, suggesting that it is Brk dependent and Schnurri independent.
Strikingly, the effects on brk expression are seen only in double mutant clones found at the periphery of the disc, but not at the centre where Shn is active (Figure 6C; Marty et al., 2000), supporting the notion that the effects are, indeed, Brk- but not Shn-dependent. Furthermore, the fact that double mutant clones at the middle of the disc do not ectopically express brk suggests that Shn-mediated repression of brk transcription must be taking place even in the absence of both corepressors.
Corepressors are required for Brk repression in the embryo
In embryogenesis, Brk plays a comparable role to that in the wing disc, i.e. it blocks low- and intermediate-level Dpp target gene expression (Jazwinska et al., 1999b; Ashe et al., 2000; Rushlow et al., 2001; Zhang et al., 2001). We next wanted to establish whether, in the developing embryo, Brk is also reliant on corepressors for the silencing of its downstream targets. A direct assessment of Brk’s dependence on Gro using germ-line clones devoid of maternal gro is hindered, however, by the prior requirement for Gro by Dorsal, which in embryonic D/V axis formation represses several Brk subordinate targets before brk is expressed and functional (Dubnicoff et al., 1997; Jazwinska et al., 1999b). For example, in gro– germ-line clones Dorsal fails to repress dpp and zerknullt (zen), and these expand ventrally (Dubnicoff et al., 1997), making it impossible to distinguish between the loss of Brk activity from that of Dorsal’s.
To be able to compare Brk’s dependence on Gro and CtBP in the embryo, we undertook an alternative approach, of overexpressing full-length Brk in its native form or with its corepressor-binding domains mutated (Figure 2D), using UAS-brk transgenes driven by maternal GAL4 (Brand and Perrimon, 1993). This experimental design is inapplicable for studying Brk’s targets in the wing, as ectopic expression of brk prevents proliferation and survival of imaginal disc cells (data not shown), but is nevertheless effective in the embryo (Jazwinska et al., 1999b). Rushlow et al. (2001) have shown that ectopic Brk represses zen and dpp in mid- to late-cellularizing embryos but not earlier, so we analysed endogenous Brk targets in transgenic embryos at comparable stages of development. As shown in Figure 7, ectopic expression of all three mutant forms of Brk in embryos brings about repression of zen to the same extent as does native Brk (cf. Brk and BrkmutC/G in Figure 7A, right; Figure 7B). This result suggests that Brk represses zen independently of corepressors, in keeping with published reports showing that Brk acts on the zen promoter by competing with pMad over DNA-binding sites (Rushlow et al., 2001). In contrast, Brk requires corepressors for negating transcription of both tolloid (tld) and dpp. Thus, abolishing Brk’s interactions with Gro (BrkmutG), but not with CtBP (BrkmutC), completely relieves tld repression (Figure 7A, centre; Figure 7C and D), indicating that Brk repression of tld is strictly Gro dependent, as is repression of pannier (pnr; Zhang et al., 2001). Similarly, dpp is repressed in embryos expressing BrkmutC, but is still transcribed in embryos expressing Brk with its Gro recruitment motif mutated (Figure 7A, left; Figure 7E–G). In the case of dpp, however, CtBP must also be contributing to Brk repression, since the level of dpp expression is significantly lower in BrkmutG-expressing embryos (marked by ‘**’ in Figure 7A), in comparison with wild-type embryos or embryos expressing BrkmutC/G (cf. Figure 7F and G). We thus conclude that, for repression of dpp, Brk rests mainly on Gro, yet for maximal repressor activity it also requires CtBP. While these experiments were in progress, Zhang et al. (2001) reported that mutating Brk’s Gro recruitment motif relieves dpp repression only partially, consistent with an additional input into dpp regulation. Our results suggest that this input might be in the form of Brk repression mediated by CtBP. In any case, our data indicate that Brk utilizes different means of repression for silencing its downstream targets in the embryo, as in the adult.
Discussion
Dpp activity converges on target genes as an assimilation of (i) transcriptional activation mediated by the pMad– Medea effector complex, and (ii) silencing induced by the sequence-specific repressor Brk (Podos and Ferguson, 1999). Thus, discrete thresholds induced by Dpp signalling output are generated in practice by a summation of two opposite and antagonistic gradients, with the holistic nature of the Dpp patterning process guaranteed by the fact that brk itself is a Dpp target. Here we have shown that Brk recruits two corepressors, namely Gro and CtBP, and that these cofactors are differentially required by Brk for repressing a subset of its targets while, for negating other promoters, they are entirely dispensable. Collectively, our results suggest that Brk utilizes multiple means to repress divergent Dpp-responsive genes in a promoter context-dependent manner.
Gro in adult patterning
The Gro corepressor participates in diverse embryonic developmental settings, such as neurogenesis, segmentation, sex determination and terminal patterning (Fisher and Caudy, 1998; Parkhurst, 1998; Chen and Courey, 2000). Previous work has also shown gro function to be critical for normal eye and wing patterning. In both these tissues, Gro is clearly involved in the Notch pathway, in silencing expression of proneural genes (e.g. Heitzler et al., 1996; Chanut et al., 2000). However, gro also contributes to organogenesis independently of the neurogenic pathway (de Celis and Ruiz Gomez, 1995; Heitzler et al., 1996). In this paper we implicate Gro as an effector of the Dpp pathway in patterning of the wing, specifically as a cofactor for the Brk repressor. Significantly, we also show CtBP to be required for full implementation of Brk repression.
Two additional points are implied indirectly from our data. First, the effects caused by overexpressing gro (Figure 1) suggest that although gro is uniformly expressed throughout the wing imaginal disc (Tata and Hartley, 1993), its levels must be limiting. Secondly, as in the embryo, Gro is required in the adult only for a subset of repressors. gro is not involved in dpp transcriptional regulation, since the dpp pattern itself is unaffected by the absence of gro or by its overexpression, and Dpp target genes are repressed in gro overexpression clones even outside and away from the central dpp expression domain (Figure 1; data not shown). Thus, for Cubitus interruptus acting downstream of Hh and upstream of dpp, as for Shn acting upstream of brk (Figure 6), Gro (and CtBP) is non-essential.
Functional interactions between Brk and two corepressors
The Brk RD includes two distinct corepressor recruitment motifs, with which it recruits both Gro (this study; Zhang et al., 2001) and CtBP (this study) to target promoters. It is still unclear how, once tethered to DNA, Gro and CtBP elicit repression of activated and basal transcription. Both complex with histone deacetylases (Sundqvist et al., 1998; Chen et al., 1999), indicating that they suppress gene expression at least partly by influencing chromatin organization via histone deacetylation. CtBP probably also harbours a unique enzymatic activity (Nibu et al., 1998b; Poortinga et al., 1998). Whatever their precise molecular mode of action, the Gro (long-range) and CtBP (short-range) corepressors must be mediating repressor functions via separate mechanisms (Zhang and Levine, 1999). By acting with both, Brk uses multiple molecular means of repression, which should invigorate its regulatory activity.
Several Drosophila repressors have been shown genetically and molecularly to possess more than one discrete RD (e.g. Aronson et al., 1997; Keller et al., 2000; Kobayashi et al., 2001). What might be the biological significance of multiple RDs, or in the case of Brk two autonomous corepressor binding motifs, within a single negative transcriptional regulator? These domains could be acting jointly on each individual target gene, required both in tandem for full repressor competence (Kobayashi et al., 2001). Alternatively, each distinct RD could alone be mediating the full repression of a specific, distinguishable subset of targets. We have addressed this issue genetically, and find that Brk manifests full repression of some of its endogenous targets even in the absence of one corepressor or the other. Thus, for maintaining its negative autoregulation, Brk relies on the recruitment of either Gro or CtBP (Figure 6) whereas, for repressing vgQ (as well as tld and pnr in the embryo), Brk obligatorily rests on Gro, whether or not CtBP is present (Figures 5 and 7; Zhang et al., 2001). Importantly, for repressing dpp (and Sxl) in the embryo, Brk’s two disparate corepressor recruitment domains are cooperatively required to promote maximal repression (Figures 3 and 7). Thus, Brk requires Gro and CtBP differentially for repressing its endogenous target genes.
As a regulatory cofactor, CtBP is bifunctional in nature, acting either as a coactivator or as a corepressor (Phippen et al., 2000). In mediating Brk transcriptional silencing, CtBP seems to function as a dedicated corepressor; it can effectively mediate Brk autoregulation even when Gro is lacking (Figure 6), and it augments repression of some promoters for which Brk repressor activity is largely Gro dependent, such as dpp and Sxl (Figures 3 and 7). Notably, like Brk, the Hairy repressor also binds both CtBP and Gro; however, in this context CtBP plays an antagonistic role, suppressing Hairy’s Gro-dependent repression (Zhang and Levine, 1999; Phippen et al., 2000). Given the Brk-independent reduction in vgQ expression observed in CtBP– clones (Figure 5B and C), we propose that, as with Hairy, in conjunction with some as yet unknown transcription factor CtBP participates as a coactivator in vg regulation.
Versatile employment of multiple repressor mechanisms by Brk in Dpp signalling
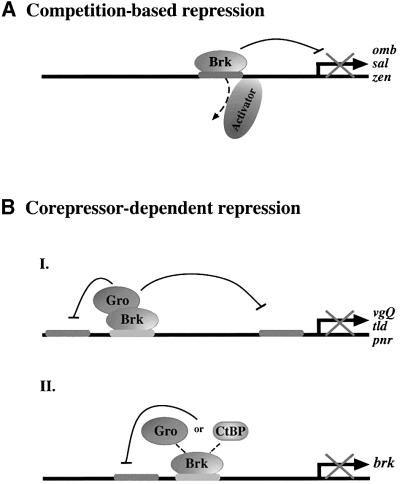
Gro and CtBP mediate gene silencing in qualitatively different ways (Zhang and Levine, 1999). Gro potentiates long-range repressors that function at a distance and that are able to block, in a dominant fashion, complex modular promoters consisting of multiple enhancer elements (Cai et al., 1996). In contrast, CtBP-dependent short-range repressors inhibit activators only locally, thereby permitting enhancer autonomy in a compound promoter (Gray et al., 1994; Nibu et al., 1998b). By virtue of its ability to recruit both Gro and CtBP, together with its capacity to outcompete pMad and other activators from binding DNA, Brk is competent to repress a multitude of complex Dpp target promoters, which receive positive inputs from manifold signalling pathways. We propose that, for promoters with low-affinity Mad-binding sites, the driving repressor force is direct competition between Brk and pMad for DNA binding, whereas for Dpp target promoters that contain high-affinity Mad-binding sites, corepressors are essential for mediating Brk repression. For this latter class of promoters, Brk relies on one or both of its cognate corepressors, depending on the particular promoter topology.
Brk utilizes a self-reliant mechanism, which need not depend on tethered corepressors, by competing with activators over coinciding DNA-binding sites (Figure 8; Sivasankaran et al., 2000; Kirkpatrick et al., 2001; Rushlow et al., 2001). We find that, in the absence of both Gro and CtBP, Brk represses not only omb and zen, but also sal (Figures 4 and 7; data not shown), suggesting that Brk-binding site(s) in the sal promoter overlap with those employed by activators. Transcription of both sal and vgQ requires activation by Mad (Kim et al., 1996; Nellen et al., 1996), yet, although both promoters are exposed to identical levels of pMad, the sal expression domain is spatially more restricted than that of vgQ, presumably because activation of sal requires higher levels of pMad than that of vgQ. Hence, ‘passive’ competition-based repression should efficiently block activation of sal but may not be sufficient for promoters like vgQ, which are activated even by low amounts of Mad. For silencing such promoters, alternative mechanisms such as recruitment of corepressors have evolved and are employed.
Fig. 8. Brk employs multiple repressor mechanisms in Dpp signalling. (A) Competition-based repression should suffice for promoters that bind Mad, or some other activator, at low affinity. Even for such promoters, recruitment of corepressors probably reinforces repression (see text). (B) For promoters with high activator-binding affinity, or when Brk-binding sites do not overlap with those of Mad, corepressors are employed to allow ‘active’ repression. Gro mediates dominant, long-range repression (BI), whereas either Gro or CtBP act at short-range (BII). Notably, Brk repression of dpp (and Sxl) is Gro dependent; however, for maximal repression CtBP is also required.
We find that Brk represses its distinct endogenous target genes by recruiting Gro and/or CtBP differentially (Figure 8). For the silencing of many target promoters, Gro alone is sufficient (vg, tld and pnr) but, for fully repressing others, Brk depends on both corepressors. Thus, in the case of dpp and Sxl, when CtBP is lacking, a decrease in Brk’s overall repressor capacity is apparent and, in the absence of Gro, repression is almost completely impaired. Importantly, for negating its own transcription, Brk can utilize either corepressor.
The majority of activator and repressor binding sites in most Dpp-responsive enhancers have yet to be precisely mapped. We nevertheless propose that lengthy and complex promoters, which respond to several signalling inputs, will be found to be strictly silenced in a Gro-dependent manner. Thus, in repressing the vgQ enhancer, a composite cis-acting regulatory sequence with multiple elements that integrate information relayed by the dpp, wingless and EGF receptor signalling pathways, Brk is fully reliant on Gro, but not on CtBP. For other more simple promoters, short-range repression should be adequate and will be mediated by either corepressor, as exemplified by the robust Brk autoregulation, for which either Gro or CtBP is sufficient; CtBP and Gro are presumably interchangeable in this context, compensating for each other’s absence.
Significantly, the overexpression of gro results in ectopic omb repression (Figure 1), suggesting that, even for promoters that are switched off in a ‘passive’, competitive manner, excess Gro can over-potentiate Brk-mediated negative transcriptional regulation. Thus, Gro and/or CtBP might reinforce Brk repression of those promoters on which it initially acts by competing with activators for binding to DNA (Sivasankaran et al., 2000; Kirkpatrick et al., 2001; Rushlow et al., 2001), via recruitment of histone deacetylases and alterations to chromatin structure, or by some other mechanism.
In summary, our data suggest that Brk uses multiple means to negate target gene expression, such as competition and the varied recruitment of long- and short-range corepressors. We propose that this versatility is, biologically, most significant given Brk’s role in Dpp signalling, as it facilitates its negative regulation of diverse, complex Dpp target promoters.
Materials and methods
Fly culture
Flies were cultured and crossed on yeast–cornmeal–molasses–malt extract–agar medium at 25°C.
Generating clones
Clones of cells lacking functional gro (groE48), CtBP [CtBPl(3)87De-10] or both were generated using Flp-mediated mitotic recombination (Xu and Rubin, 1993), and identified by the loss of πMyc and the concurrent appearance of a twin-spot. Marked clones of cells overexpressing gro were generated by crossing UAS-gro flies with actin>CD2>Gal4 transgenic flies bearing an appropriate marker (Pignoni and Zipursky, 1997). After 2 days of egg laying, flies were removed and 2–3 days later larvae were heat-shocked (30 min at 34°C), dissected, fixed and stained.
In situ hybridization and antibody staining of Drosophila embryos
In situ hybridizations were performed as previously described (Goldstein et al., 1999). Sxl expression was monitored in three independent transgenic lines for each hb-Hairy-Brk variant, using the Sxl-Pe:lacZ reporter strain (Estes et al., 1995), and by staining with a monoclonal antibody specific to the active form of Sxl as described previously (Jiménez et al., 1997). Embryos were mounted in methacrylate (JB-4; Polyscience) and examined under Nomarski optics.
Calculating female-specific lethality
To establish whether female-specific lethality is caused by the expression of a given transgene, the Mantel–Haenszel test for association was used to determine whether the proportion of transgene-containing females differed from that of non-transgenic females. The expected proportion of females was calculated from the non-transgenic sibling group and female-specific lethality in a transgenic fly group was computed by the following ratio: [(expected minus observed)/expected] female flies. P-values indicate that there is a significant association between the proportion of females and group (transgenic yes/no) in the hb-Hairy-BrkRD and hb-Hairy-BrkRDmutC types, but not in the other two groups.
Plasmids
Molecular manipulations were conducted according to standard protocols. Constructs containing full-length Brk, or modified versions, were prepared by inserting fragments generated by standard PCR amplification into pBluescript (Stratagene) and, following sequencing, into appropriate vectors and sites. pGEX-Hairy and -HairyΔNot, pET-Gro and LexA-Gro have been described previously (Paroush et al., 1994). hb-Hairy-BrkRD and derivatives were constructed as described in Jiménez et al. (1997). The mutations in corepressor recruitment motifs were generated using Stratagene’s QuikChange™ Site-Directed Mutagenesis Kit.
Yeast two-hybrid interaction assays
Yeast two-hybrid interaction assays were performed as described (Goldstein et al., 1999). lacZ reporter expression was analysed on appropriate indicator plates containing X-Gal.
In vitro GST pull-down assays
Pull-down assays were performed essentially as described (Goldstein et al., 1999), with the exception that for experiments using GST–CtBP, the pull-down buffer contained 20 mM HEPES pH 7.9, 0.2 M NaCl, 1 mM EDTA, 1 mM MgCl2, 1 mM dithiothreitol, 0.5% NP-40 and 10% bovine serum albumin.
Acknowledgments
Acknowledgements
This collaboration was born out of a visit by P.H. to Zürich, made possible by an EMBO short-term fellowship (#ASTF9422), for which we are most grateful. We should like to thank members of our laboratories for continued help, discussions and encouragement during this project. We thank Anne Shalom for generating transgenic flies, L.Rosen for biostatistical analysis, David Ish-Horowicz, Susan Parkhurst, Benny Shilo and Talila Volk for insightful discussions and constructive comments on the manuscript, and K.Arora, D.Bopp, O.Cook, P.Gergen, A.Michelson, S.Roth, B.Shilo, R.Stanley, D.St Johnston, S.Parkhurst and J.Thomas for probes, antibodies and fly stocks. We apologize for the incomplete referencing due to rigid space constraints. This work was supported by grants from the Israel Science Foundation (#116/00-1), the Israel Cancer Research Fund (RCDA), and the United States–Israel Binational Science Foundation (#96-108) to Z.P., and by grants from the Swiss National Science Foundation and the Kanton of Zürich to K.B. Z.P. is also supported by the Jan M. and Eugenia Krol Charitable Foundation, and is a Joseph H. and Belle R.Braun Lecturer in Medicine.
References
- Arnosti D.N., Gray,S., Barolo,S., Zhou,J. and Levine,M. (1996) The gap protein knirps mediates both quenching and direct repression in the Drosophila embryo. EMBO J., 15, 3659–3666. [PMC free article] [PubMed] [Google Scholar]
- Aronson B.D., Fisher,A.L., Blechman,K., Caudy,M. and Gergen,J.P. (1997) Groucho-dependent and -independent repression activities of Runt domain proteins. Mol. Cell. Biol., 17, 5581–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashe H., Mannervik,M. and Levine,M. (2000) Dpp signaling thresholds in the dorsal ectoderm of the Drosophila embryo. Development, 127, 3305–3312. [DOI] [PubMed] [Google Scholar]
- Barolo S. and Levine,M. (1997) hairy mediates dominant repression in the Drosophila embryo. EMBO J., 16, 2883–2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brand A.H. and Perrimon,N. (1993) Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development, 118, 401–415. [DOI] [PubMed] [Google Scholar]
- Burke R. and Basler,K. (1996) Dpp receptors are autonomously required for cell proliferation in the entire developing Drosophila wing. Development, 122, 2261–2269. [DOI] [PubMed] [Google Scholar]
- Cai H.N., Arnosti,D.N. and Levine,M. (1996) Long-range repression in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 93, 9309–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell G. and Tomlinson,A. (1999) Transducing the Dpp morphogen gradient in the wing of Drosophila: regulation of Dpp targets by brinker. Cell, 96, 553–562. [DOI] [PubMed] [Google Scholar]
- Chanut F., Luk,A. and Heberlein,U. (2000) A screen for dominant modifiers of ro(Dom), a mutation that disrupts morphogenetic furrow progression in Drosophila, identifies Groucho and Hairless as regulators of atonal expression. Genetics, 156, 1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G. and Courey,A.J. (2000) Groucho/TLE family proteins and transcriptional repression. Gene, 249, 1–16. [DOI] [PubMed] [Google Scholar]
- Chen G., Fernandez,J., Mische,S. and Courey,A.J. (1999) A functional interaction between the histone deacetylase Rpd3 and the corepressor Groucho in Drosophila development. Genes Dev., 13, 2218–2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis J.F. and Ruiz Gomez,M. (1995) groucho and hedgehog regulate engrailed expression in the anterior compartment of the Drosophila wing. Development, 121, 3467–3476. [DOI] [PubMed] [Google Scholar]
- Dubnicoff T., Valentine,S.A., Chen,G., Shi,T., Lengyel,J.A., Paroush,Z. and Courey,A.J. (1997) Conversion of Dorsal from an activator to a repressor by the global corepressor Groucho. Genes Dev., 11, 2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Entchev E., Schwabedissen,A. and Gonzalez-Gaitan,M. (2000) Gradient formation of the TGF-β homolog Dpp. Cell, 103, 981–991. [DOI] [PubMed] [Google Scholar]
- Estes P.A., Keyes,L.N. and Schedl,P. (1995) Multiple response elements in the Sex-lethal early promoter ensure its female-specific expression pattern. Mol. Cell. Biol., 15, 904–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher A.L. and Caudy,M. (1998) Groucho proteins: transcriptional corepressors for specific subsets of DNA-binding transcription factors in vertebrates and invertebrates. Genes Dev., 12, 1931–1940. [DOI] [PubMed] [Google Scholar]
- Goldstein R.E., Jiménez,G., Cook,O., Gur,D. and Paroush,Z. (1999) Huckebein repressor activity in Drosophila terminal patterning is mediated by Groucho. Development, 126, 3747–3755. [DOI] [PubMed] [Google Scholar]
- Gray S. and Levine,M. (1996) Transcriptional repression in develop ment. Curr. Opin. Cell Biol., 8, 358–364. [DOI] [PubMed] [Google Scholar]
- Gray S., Szymanski,P. and Levine,M. (1994) Short-range repression permits multiple enhancers to function autonomously within a complex promoter. Genes Dev., 8, 1829–1838. [DOI] [PubMed] [Google Scholar]
- Heitzler P., Bourouis,M., Ruel,L., Carteret,C. and Simpson,P. (1996) Genes of the Enhancer of split and achaete–scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development, 122, 161–171. [DOI] [PubMed] [Google Scholar]
- Jazwinska A., Kirov,N., Wieschaus,E., Roth,S. and Rushlow,C. (1999a) The Drosophila gene brinker reveals a novel mechanism of Dpp target gene regulation. Cell, 96, 563–573. [DOI] [PubMed] [Google Scholar]
- Jazwinska A., Rushlow,C. and Roth,S. (1999b) The role of brinker in mediating the graded response to Dpp in early Drosophila embryos. Development, 126, 3323–3334. [DOI] [PubMed] [Google Scholar]
- Jiménez G., Paroush,Z. and Ish-Horowicz,D. (1997) Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev., 11, 3072–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A.D. (1995) The price of repression. Cell, 81, 655–658. [DOI] [PubMed] [Google Scholar]
- Keller S. et al. (2000) dCtBP-dependent and -independent repression activities of the Drosophila Knirps protein. Mol. Cell. Biol., 20, 7247–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Sebring,A., Esch,J.J., Kraus,M.E., Vorwerk,K., Magee,J. and Carroll,S.B. (1996) Integration of positional signals and regulation of wing formation and identity by Drosophila vestigial gene. Nature, 382, 133–138. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick H., Johnson,K. and Laughon,A. (2001) Repression of Dpp targets by binding of Brinker to Mad sites. J. Biol. Chem., 276, 18216–18222. [DOI] [PubMed] [Google Scholar]
- Kobayashi M., Goldstein,E.R., Fujioka,M., Paroush,Z. and Jaynes,B.J. (2001) Groucho augments the repression of multiple Even-skipped target genes in establishing parasegment boundaries. Development, 128, 1805–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecuit T., Brook,W., Ng,M., Calleja,M., Sun,H. and Cohen,S. (1996) Two distinct mechanisms for long-range patterning by Dpp in the Drosophila wing. Nature, 381, 387–393. [DOI] [PubMed] [Google Scholar]
- Mannervik M., Nibu,Y., Zhang,H. and Levine,M. (1999) Transcriptional coregulators in development. Science, 284, 606–609. [DOI] [PubMed] [Google Scholar]
- Marty T., Muller,B., Basler,K. and Affolter,M. (2000) Schnurri mediates dpp-dependent repression of brinker transcription. Nature Cell Biol., 2, 745–749. [DOI] [PubMed] [Google Scholar]
- Minami M., Kinoshita,N., Kamoshida,Y., Tanimoto,H. and Tabata,T. (1999) brinker is a target of Dpp in Drosophila that negatively regulates Dpp-dependent genes. Nature, 398, 242–246. [DOI] [PubMed] [Google Scholar]
- Nellen D., Burke,R., Struhl,G. and Basler,K. (1996) Direct and long-range action of a DPP morphogen gradient. Cell, 85, 357–368. [DOI] [PubMed] [Google Scholar]
- Nibu Y., Zhang,H., Bajor,E., Barolo,S., Small,S. and Levine,M. (1998a) dCtBP mediates transcriptional repression by Knirps, Krüppel and Snail in the Drosophila embryo. EMBO J., 17, 7009–7020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibu Y., Zhang,H. and Levine,M. (1998b) Interaction of short-range repressors with Drosophila CtBP in the embryo. Science, 280, 101–104. [DOI] [PubMed] [Google Scholar]
- Parkhurst S.M. (1998) Groucho: making its Marx as a transcriptional co-repressor. Trends Genet., 14, 130–132. [DOI] [PubMed] [Google Scholar]
- Parkhurst S.M., Bopp,D. and Ish-Horowicz,D. (1990) X:A ratio, the primary sex-determining signal in Drosophila, is transduced by helix–loop–helix proteins. Cell, 63, 1179–1191. [DOI] [PubMed] [Google Scholar]
- Paroush Z., Finley,R.L.,Jr, Kidd,T., Wainwright,S.M., Ingham,P.W., Brent,R. and Ish-Horowicz,D. (1994) Groucho is required for Drosophila neurogenesis, segmentation and sex determination and interacts directly with hairy-related bHLH proteins. Cell, 79, 805–815. [DOI] [PubMed] [Google Scholar]
- Phippen T., Sweigart,A., Moniwa,M., Krumm,A., Davie,J. and Parkhurst,S. (2000) Drosophila C-terminal binding protein functions as a context-dependent transcriptional co-factor and interferes with both Mad and Groucho transcriptional repression. J. Biol. Chem., 275, 37628–37637. [DOI] [PubMed] [Google Scholar]
- Pignoni F. and Zipursky,S.L. (1997) Induction of Drosophila eye development by Decapentaplegic. Development, 124, 271–278. [DOI] [PubMed] [Google Scholar]
- Podos S. and Ferguson,E. (1999) Morphogen gradients: new insights from DPP. Trends Genet., 15, 396–402. [DOI] [PubMed] [Google Scholar]
- Poortinga G., Watanabe,M. and Parkhurst,S. (1998) Drosophila CtBP: a Hairy-interacting protein required for embryonic segmentation and Hairy-mediated transcriptional repression. EMBO J., 17, 2067–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raftery L. and Sutherland,D. (1999) TGF-β family signal transduction in Drosophila development: from Mad to Smads. Dev. Biol., 210, 251–268. [DOI] [PubMed] [Google Scholar]
- Rushlow C., Colosimo,P., Lin,M.M., Xu,M. and Kirov,N. (2001) Transcriptional regulation of the Drosophila gene zen by competing Smad and Brinker inputs. Genes Dev., 15, 340–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankaran R., Vigano,M., Muller,B., Affolter,M. and Basler,K. (2000) Direct transcriptional control of the Dpp target omb by the DNA binding protein Brinker. EMBO J., 19, 162–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundqvist A., Sollerbrant,K. and Svensson,C. (1998) The carboxy-terminal region of adenovirus E1A activates transcription through targeting of a C-terminal binding protein–histone deacetylase complex. FEBS Lett., 429, 183–188. [DOI] [PubMed] [Google Scholar]
- Tata F. and Hartley,D.A. (1993) The role of the enhancer of split complex during cell fate determination in Drosophila. Dev. Suppl., 139–148. [PubMed] [Google Scholar]
- Teleman A. and Cohen,S. (2000) Dpp gradient formation in the Drosophila wing imaginal disc. Cell, 103, 971–980. [DOI] [PubMed] [Google Scholar]
- Xu T. and Rubin,G.M. (1993) Analysis of genetic mosaics in developing and adult Drosophila tissues. Development, 117, 1223–1237. [DOI] [PubMed] [Google Scholar]
- Zhang H. and Levine,M. (1999) Groucho and dCtBP mediate separate pathways of transcriptional repression in the Drosophila embryo. Proc. Natl Acad. Sci. USA, 96, 535–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Levine,M. and Ashe,H. (2001) Brinker is a sequence-specific transcriptional repressor in the Drosophila embryo. Genes Dev., 15, 261–266. [DOI] [PMC free article] [PubMed] [Google Scholar]