Abstract
The nuclear pore complex (NPC) is a large proteinaceous structure through which bidirectional transport of macromolecules across the nuclear envelope (NE) takes place. Nup153 is a peripheral NPC component that has been implicated in protein and RNP transport and in the interaction of NPCs with the nuclear lamina. Here, Nup153 is localized by immunogold electron microscopy to a position on the nuclear ring of the NPC. Nuclear reconstitution is used to investigate the role of Nup153 in nucleo- cytoplasmic transport and NPC architecture. NPCs assembled in the absence of Nup153 lacked several nuclear basket components, were unevenly distributed in the NE and, unlike wild-type NPCs, were mobile within the NE. Importin α/β-mediated protein import into the nucleus was strongly reduced in the absence of Nup153, while transportin-mediated import was unaffected. This was due to a reduction in import complex translocation rather than to defective receptor recycling. Our results therefore reveal functions for Nup153 in NPC assembly, in anchoring NPCs within the NE and in mediating specific nuclear import events.
Keywords: NPC/nuclear basket/nuclear import/nuclear pore complex Nup153
Introduction
Eukaryotic cells are characterized by their nuclear compartment, which provides a specialized environment for DNA replication, gene transcription and RNA processing. The nuclear envelope (NE) separates the nucleoplasm from the cytosol. Selective bidirectional traffic between the nucleus and the cytoplasm occurs through nuclear pore complexes, proteineacous structures of 125 MDa in vertebrates and 60 MDa in yeast (Reichelt et al., 1990; Yang et al., 1998). The NPCs are associated with intranuclear protein filaments (Franke and Scheer, 1970; Franke, 1970a,b; Maul, 1977; Cordes et al., 1993; Galy et al., 2000) and also contact the nuclear lamina, a meshwork of intermediate filaments underlying the NE (Akey, 1989; Goldberg and Allen, 1996). The lamina is thought to provide support for the NE and to mediate the attachment of the NE to interphase chromatin (for review see Gruenbaum et al., 2000).
Vertebrate NPCs exhibit 8-fold rotational symmetry. Their structural features include the central spoke-ring assembly and associated peripheral structures. On both NPC faces the peripheral structures are anchored to the central spoke-ring complex via a coaxial ring. On the cytoplasmic side there are eight short filaments (∼50 nm; Franke and Scheer, 1970; Richardson et al., 1988) and on the nuclear face eight longer filaments of 3–6 nm in diameter, which are joined at the distal ring, located ∼100 nm from the mid-plane of the NE, to form the NPC basket (Ris, 1989, 1991; Jarnik and Aebi, 1991; Goldberg and Allen, 1992; Cordes et al., 1993). Extensive analysis of yeast NPCs has identified 30 different stable nucleoporin constituents (Rout et al., 2000). It is presently unclear whether the larger size of vertebrate NPCs is accounted for by a more complex composition or by other factors. Several subcomplexes of both yeast and vertebrate NPCs have been defined and shown to have non-identical functional roles (for review see Doye and Hurt, 1997; Ohno et al., 1998; Ryan and Wente, 2000).
The effects of genetic depletion of a large number of yeast nucleoporins have been examined. Removal of individual nucleoporins has in this way been shown to specifically affect nuclear import or nuclear export pathways, the organization of the NE, the distribution of NPCs within the NE and aspects of intranuclear organization (Fabre and Hurt, 1997; Galy et al., 2000; Wente, 2000). In vertebrates, a very limited number of nucleoporins has also been analysed by genetic methods (van Deursen et al., 1996; Smitherman et al., 2000; von Kobbe et al., 2000; Wu et al., 2001). An alternative method, biochemical depletion of nucleoporins from Xenopus egg extract in which nuclear assembly on added chromatin templates takes place, has also been successfully applied to the production of nuclei whose NPCs lack specific components (Finlay and Forbes, 1990; Finlay et al., 1991; Powers et al., 1995; Grandi et al., 1997). The vertebrate studies have provided evidence for nucleoporin function in specific nucleocytoplasmic transport events (Finlay et al., 1991; van Deursen et al., 1996). Although some nucleoporins are well enough conserved to allow functional comparison between yeast and vertebrate homologues, many others are either poorly or not at all conserved across this evolutionary time span (Ohno et al., 1998). In addition, some gross organizational features of NPCs do not appear to be conserved. For example, while yeast NPCs have been reported to be mobile within the NE (Belgareh and Doye, 1997; Bucci and Wente, 1997), vertebrate NPCs are stably anchored in the membrane (Daigle et al., 2001).
The vertebrate nucleoporin Nup153 has no identifiable yeast homologue, but has been implicated in various aspects of NPC function. Nup153 has been localized to the nuclear side of the NPC (Sukegawa and Blobel, 1993; Cordes et al., 1993), and at higher resolution to the distal ring of the basket structure (Pante et al., 1994). It has been suggested that Nup153 might link to either the nuclear lamina, the Tpr-containing intranuclear filaments or both (Cordes et al., 1993; Bastos et al., 1996; Ullman et al., 1999). Support for the former hypothesis came initially from Nup153 overexpression studies where, among other phenotypes, the formation of both lamina-containing structures and membrane arrays within the nucleus were seen (Bastos et al., 1996). More recently, evidence for a direct interaction between Nup153 and lamin LIII, the major lamin form present in Xenopus oocytes and eggs, has been obtained (Smythe et al., 2000).
The primary structure of Nup153 can be roughly divided into three regions; a unique N-terminal region, a central domain consisting of four to five zinc fingers (depending upon species) and a C-terminal region containing ∼30 irregularly spaced FXFG repeats. Different classes of FG-containing repeats are found in many nucleoporins and often represent binding sites for nuclear import and export receptors (see Doye and Hurt, 1997; Ohno et al., 1998; Wente, 2000), and the C-terminal region of Nup153 has indeed been shown to interact with several such receptors (Moroianu et al., 1997; Shah and Forbes, 1998; Shah et al., 1998; Nakielny et al., 1999). Injection of anti-Nup153 antibodies into the nucleus of Xenopus oocytes blocked the nuclear export of most RNA substrates, but not tRNA (Ullman et al., 1999). Overexpression of the C-terminal domain of the protein in cultured somatic cells affected mRNA export (Bastos et al., 1996), while the addition of this region to nuclear import reactions inhibited importin α/β-mediated, but not transportin-mediated, import in vitro (Shah and Forbes, 1998). These studies were suggestive of a role for the C-terminal region in transport events, but could also be explained by a titration of importin β or other transport receptors in the reaction via their binding to the nucleoporin fragment. In agreement with this interpretation, Shah et al. (1998) reported an unusually stable complex between Nup153 and importin β in Xenopus egg extracts.
The non-repetitive N-terminal region of Nup153 binds to the import receptor transportin (Shah and Forbes, 1998; Nakielny et al., 1999), which is responsible for the import of proteins like hnRNPA1 that carry a nuclear localization signal (NLS) of the M9 domain class (Pollard et al., 1996). The function of this interaction is likely to be in the import of Nup153 into the nucleus, a requirement for its incorporation into the NPC (Bastos et al., 1996; Enarson et al., 1998; Shah and Forbes, 1998; Nakielny et al., 1999).
Nup153 interacts with homopolymeric RNAs in vitro (Ullman et al., 1999), and with DNA through its zinc finger domain (Sukegawa and Blobel 1993). The zinc finger region also interacts specifically with GDP-bound Ran (Nakielny et al., 1999). Ran is the GTPase that regulates transport receptor–transport cargo interaction and by so doing imposes directionality on nucleocytoplasmic transport (Mattaj and Englmeier, 1998; Görlich and Kutay, 1999). The functional significance of these Nup153 interactions is not yet clear.
We wished to investigate the proposed roles of Nup153 by the assembly of nuclei whose NPCs lacked Nup153. We reasoned that the reported peripheral location of Nup153 on the nuclear basket would permit this to be achieved without greatly disrupting the assembly or internal structure of the NPC. Our data, however, reveal an important role for Nup153 in the stable incorporation of at least two other nucleoporins into the NPC. NPCs lacking Nup153 exhibit a lack of anchoring within the NE as well as specific defects in nuclear import.
Results
Nup 153 is located at the base of the nuclear basket
Nup153 localization within the NPC has been examined previously (see Introduction). While all three previous studies agreed that Nup153 was on the nuclear face of the NPC, the reported localizations differed in detail. We wished to determine the precise location of Nup153 within the NPC by immunogold labelling of Xenopus oocyte NEs and electron microscopy. Polyclonal antibodies were generated against a purified recombinant N-terminal fragment of Xenopus Nup153 (xNup153) (see Materials and methods). After affinity purification the antibodies recognized a single band on western blots, which was superimposable on Nup153 visualized by the well characterized monoclonal antibody (mAb)414 (data not shown). mAb414 has been shown to react with a series of FXFG repeat-containing nucleoporins, including Nup358/ RanBP2, Nup214/CAN, Nup153 and p62 (Davis and Blobel, 1987).
The anti-xNup153 antibody was used to localize the protein on isolated Xenopus oocyte NEs by immunogold labelling and electron microscopy. We reasoned that the best resolution in the plane of the NE, and parallel to it, could be obtained by field emission in-lens scanning electron microscopy (FEISEM; Allen et al., 1997), and the best resolution perpendicular to the NE by transmission electron microscopy (TEM). To this end, spreads of NEs manually dissected from Xenopus oocytes were labelled with anti-Nup153 antibody and 10 nm colloidal gold-conjugated secondary antibody. After processing for FEISEM (see Materials and methods), individual NPCs were imaged at high resolution, and the mean distance of the colloidal gold from the centre of the NPC was determined to be 47.0 ± 8.8 nm (n = 100) (Figure 1A). A control with only secondary Ab showed no staining (data not shown). For TEM, samples were fixed after antibody labelling, and embedded in epoxy resin. Thin sections of 70 nm were imaged and mid-sections through the NPC were recorded. The distance of the gold particles from the mid-plane of the NE was determined to be 44.6 ± 13.9 nm (n = 84) (Figure 1B). Again, no significant labelling was observed in controls (data not shown). From the TEM micrographs, pictures of central sections were selected and the median radial position of the gold particles from the centre of the pore was determined to be 49.15 ± 14 nm. Statistical analysis (Mann–Whitney U test) showed that there was no significant difference from the localization obtained by FEISEM (P = 0.4149). Therefore the two data sets can be incorporated into a single model, and suggest a localization of Nup153 adjacent to the NE at the base of the nuclear pore basket or on the nucleoplasmic coaxial ring (Figure 1D).
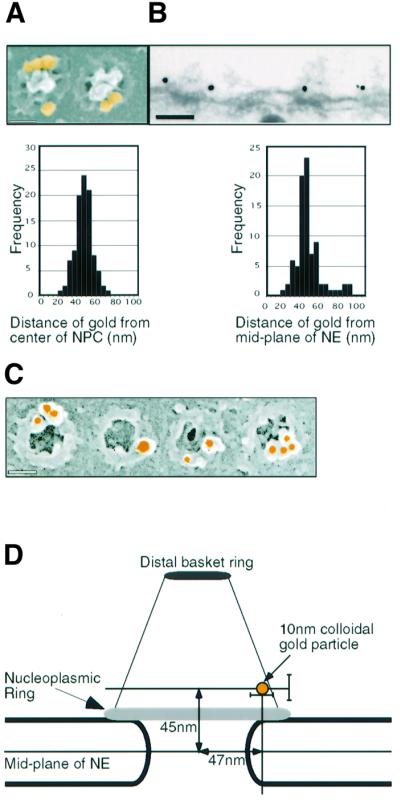
Fig. 1. Localization of Nup153 within the NPC. Fixed Xenopus oocyte nuclear envelopes were incubated with affinity-purified anti-Nup153 primary antibody and 10 nm colloidal gold labelled secondary antibodies and analysed using electron microscopy. (A) Upper panel: representative FEISEM image of NPCs decorated with gold particles (false coloured yellow). Gold particles were identified using the backscatter electron image. Lower panel: distribution of distances to the centre of the NPC (in nm) of 100 NPC-associated gold particles. Bar, 100 nm. (B) Upper panel: representative TEM image of a stretch of nuclear envelope (NE) decorated with anti-Nup153 primary antibodies and gold labelled secondary antibodies. Bar, 100 nm. Lower panel: distribution of distances to the mid-plane of the NE (in nm) of 85 NE-associated gold particles. (C) Representative images of immunogold localization of Nup153 on NPCs after stripping of the nucleoplasmic basket. Bar, 100 nm. (D) Schematic representation of the localization of Nup153-specific gold labelling on a diagram of the NPC basket. Note that two antibody molecules (not shown in the diagram) are interposed between Nup153 and the gold particle. Bars indicate standard deviations of measurement.
Recently the dimension of a single FXFG sequence has been reported to be as much as 2 nm (Bayliss et al., 2000). Given the fact that Nup153 contains ∼30 of these repeat sequences and that our antibody recognized the N-terminal region of Nup153, it was possible that Nup153 extended from the nuclear ring to the terminal distal ring of the NPC basket. To exclude this possibility, Xenopus NEs were treated with EDTA and with high salt to remove the nuclear baskets. After this treatment, immunogold labelling using the specific anti-Nup153 antibody was performed and FEISEM images examined. No nuclear baskets or residual nuclear filaments were visible. In spite of this loss of material the nuclear coaxial ring was labelled even more strongly than in the absence of treatment (2- to 3-fold increase in gold particles per NPC; Figure 1C and data not shown), indicating that the accessibility of the Nup153 epitope was increased. This very strongly suggests that Nup153 is part of the nucleo plasmic coaxial ring of the NPC.
Depletion of nup153 from reconstituted nuclei
In order to investigate the function of Nup153 we made use of nuclear assembly in extracts derived from activated Xenopus eggs. In this system a cytosolic fraction, a membrane fraction and an energy regenerating system combine to form a functional nucleus around exogenously added chromatin in vitro (Forbes et al., 1983; Lohka and Masui, 1983; Newport, 1987). In all our assays demembranated sperm were used as the source of chromatin.
As described previously by Forbes and coworkers (Finlay and Forbes, 1990; Finlay et al., 1991; Powers et al., 1995; Grandi et al., 1997), nucleoporins can be immunodepleted from the soluble fraction and nuclei reconstituted whose NPCs lack the depleted nucleoporin(s). Figure 2A shows a western blot with mAb414 of all the component fractions of the assembly assay. Neither the membrane fraction nor the demembranated sperm contained detectable amounts of xNup153 (Figure 2A, lanes 4 and 5). The soluble extracts could be efficiently immunodepleted of xNup153 (Figure 2A, compare lanes 6 and 8), without significantly affecting the levels of the other FXFG repeat-containing Nups, Nup358/RanBP2, Nup214/CAN or p62.
Fig. 2. Generation of Nup153-deficient nuclei. (A) Western blot probed with mAb414. Left panel: 1 µl of recombinant hNup153 (lane 1) compared with 2 and 1 µl of undepleted Xenopus egg cytosol fraction (lanes 2 and 3). Right panel: 2 µl of Xenopus egg membrane fraction, 2 µl of sperm chromatin, 1 µl of either mock or Nup153-depleted nuclear assembly reaction with or without addition of recombinant human Nup153, as indicated above the lanes. Molecular weight markers (kDa) are shown on the left. Positions of mAb414-reactive nucleoporins are shown on the right. (B) Immunofluorescence staining of nuclei formed in the mock-depleted or the Nup153-depleted (ΔNup153) extracts using monospecific anti-Nup153 antibodies or monoclonal S49 against lamin LIII (Lourim and Krohne, 1993, 1998). As secondary antibodies, Alexa-546 conjugates were used (red). Decondensed sperm chromatin is stained with DAPI (blue). Bar, 10 µm.
To test for the specificity of the effects of depletion in functional assays, we wished to add back recombinant Nup153 produced in Escherichia coli (Figure 2A). For this, human (h) Nup153, expressed as a GST fusion protein, was used, since the N-terminus of Xenopus Nup153 has not been sequenced to date and since we were unable to express the truncated xNup153 fragment in any expression system tested. The amount of recombinant hNup153 added back and its stability during the assembly reaction is shown in Figure 2 A (lanes 1, 7 and 9). The lack of detectable xNup153 in a nucleus assembled in Nup153-depleted extract was demonstrated by immunofluorescence (Figure 2B, top panels).
Depletion of Nup153 leads to clustering of NPCs
During the course of the characterization of Nup153-depleted nuclei we observed that the remaining nucleoporins were not evenly distributed within the NE. To characterize this, we collected stacks of confocal sections of the nuclei labelled with mAb414 or with anti-Nup214/CAN antibodies and reconstructed the images in three dimensions. Projections of these reconstructions demonstrated that the nucleoporins were not uniformly distributed within the NE of depleted nuclei. An example is shown in Figure 3A. This phenomenon is reminiscent of, but apparently less severe than, the clustering phenomenon observed in yeast strains carrying mutations in one of a number of specific nucleoporin genes (Fabre and Hurt, 1997).
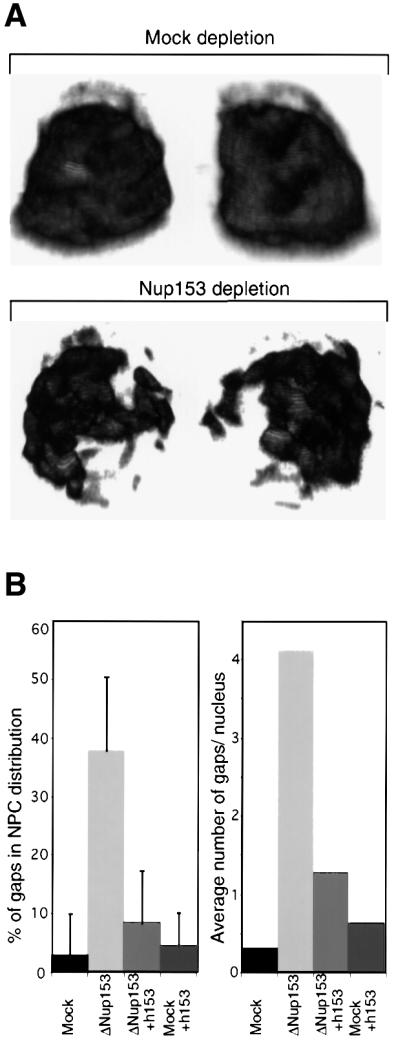

Fig. 3. Clustering of Nup153-deficient NPCs. Nuclei were assembled in vitro from mock-depleted or Nup153-depleted Xenopus egg cytosol and NPC distribution analysed using confocal immunofluorescence. (A) 3D reconstruction of confocal sections through a wild-type (Mock depletion) and Nup153-deficient (Nup153 depletion) nucleus stained with anti-Nup214/CAN primary and Alexa-488 conjugated secondary antibodies. Left hand images are rotated 180° as compared with right hand images. (B) Quantitation of NPC clustering. Nuclei were assembled from mock or Nup153-depleted (ΔNup153) Xenopus egg extracts in the presence (+h153) or absence of recombinant hNup153, and immunostained with mAb414 and Alexa-488 conjugated secondary antibody. Mid-plane confocal images were recorded and NEs were analysed for discontinuities. Both the percentage of NE length without nucleoporin staining (left panel) and the number of nucleoporin gaps in the nucleoporin staining per nucleus (right panel) are represented. Bars represent standard deviations. (C) Representative immunofluorescence images of mid-plane confocal sections of synthetic nuclei analysed in (B). Nucleoporin staining is in red, chromatin staining (DAPI) in blue.
In optical cross-sections, the nucleoporin staining along the rim of the Nup153-deficient nuclei appeared as a broken line (Figure 3C, bottom left panel). In order to analyse this quantitatively, several optical mid-sections of nuclei from each reaction were collected and two parameters were measured: the percentage of each NE that did not show a detectable mAb414 signal, and the number of apparent gaps in NPC distribution along the nuclear rim. Representative pictures of nuclei obtained in Nup153-depleted or mock-depleted reconstitution reactions, with or without the addition of recombinant Nup153, are shown in Figure 3C.
As shown in Figure 3B, depletion of Nup153 resulted in almost 40% of the NE being free of detectable NPCs. The addition of recombinant hNup153 to the nuclear reconstitution reactions reversed this effect to 9% NPC-free NE, demonstrating that the clustering was specifically caused by depletion of Nup153 (Figure 3B and C, bottom right). In a mock-depleted nuclear reconstitution reaction only a small fraction of nuclei contained gaps in their nuclear rim staining (average = 0.3, n = 21). However, an average of 4.1 gaps per nuclear rim were present in Nup153-depleted nuclei (n = 18) (Figure 3B, right). Again, this effect was almost completely reversed by adding recombinant hNup153 to the reaction (average = 0.6, n = 18) (Figure 3B). The addition of recombinant hNup153 to mock-depleted reconstitution reactions had no significant effect (Figure 3B and C).
Depletion of Nup153 results in loss of several Nups from the nuclear basket
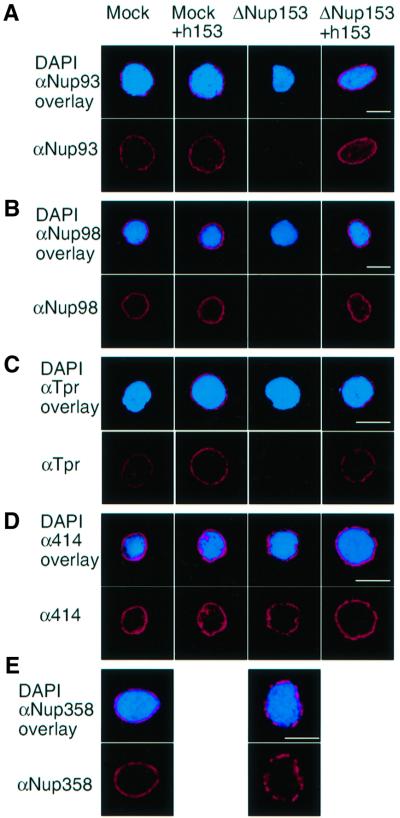
In order to gain more insight into the non-uniform NPC distribution in Nup153-deficient NPCs, we wished to test the effect of depletion on the incorporation of other components of the NPC basket as well as associated proteins. To this end, monospecific antibodies were used to assay the distribution of Nup98, Nup93 and Tpr by confocal microscopy. Images were recorded using the same settings for all reactions within one experiment. Nup93 and Nup98 have been localized to the nuclear basket structure of the NPC (Radu et al., 1995; Grandi et al., 1997). Tpr has been detected at or adjacent to the distal ring of the NPC basket and forms filaments emanating from the NPC and reaching into the nucleus (Cordes et al., 1993, 1997).
Nup93, Nup98 and Tpr were all almost undetectable by immunofluorescence of Nup153-depleted nuclei. None of these proteins was co-depleted with Nup153 however (data not shown), and their loss from the nucleus was completely reversed by the addition of recombinant Nup153 to the assembly reactions (Figure 4A–C). The removal and restoration of Nup93, Nup98 and Tpr by depletion and readdition of Nup153 indicates that Nup153 is required for either the formation or the structural integrity of the nuclear face of the NPC, and suggests that a large part of the nuclear basket may be missing in Nup153-depleted NPCs. In a study of the effects of lamins on Nup153 assembly into the NPC, Smythe et al. (2000) reported that Nup93 was detectable in nuclei in which Nup153 incorporation was not detected. We do not understand this apparent discrepancy. The loss of Nup93 after depletion of Nup153 was reproducible in our hands, and we therefore assume that differences in the experimental conditions, the obvious major one being that Nup153 was still present in the assembly reactions of Smythe and coworkers, account for the result obtained.
Fig. 4. Nup153-deficient NPCs lack several other components. Nuclei were assembled in vitro from mock-depleted (Mock) or Nup153-depleted (ΔNup153) Xenopus egg cytosol in the presence (+h153) or absence of recombinant hNup153, as indicated. Assembled nuclei were fixed and immunostained with antibodies recognizing Nup93 (A), Nup98 (B), Tpr (C), with mAb414 (D) or Nup358/RanBP2 (E). Secondary antibodies were conjugated to Alexa-546 (red). Decondensed sperm chromatin is stained with DAPI (blue). Bar, 10 µm.
To test whether nucleoporins localizing to the central or cytoplasmic regions of the NPC were also absent from Nup153-depleted NPCs, we performed immunofluorescence with mAb414. Immunofluorescence with this antibody reflects the distribution of the centrally located p62 and to a lesser extent the two cytoplasmic face nucleoporins Nup358/RanBP2 and Nup214/CAN, as well as Nup153 itself (Figure 2A; Davis and Blobel, 1987). The nuclear rim signal obtained was only slightly reduced in the Nup153-depleted nuclei, indicating that the bulk of the recognized proteins remains associated with the NE (Figure 4D, see also 3C). Nup358/RanBP2 and Nup214/CAN were also shown to be present at normal levels in Nup153-depleted NPCs by staining with monospecific antibodies (Figure 4E and data not shown). Other structural features of the synthetic nuclei, for example the nuclear lamina (Figure 2B), remained unchanged in Nup153-depleted extracts.
Nup153-deficient NPCs become mobile within the NE
The results presented above demonstrate that the composition of both the nuclear basket of the NPC and that of the associated Tpr filaments is significantly altered by the lack of Nup153, and that these NPCs exhibit an abnormal distribution within the NE. Taken together, the results suggest that Nup153-deficient NPCs might lack the connection that anchors NPCs within the NE (Daigle et al., 2001). We therefore tested the effect of Nup153 on the mobility of NPCs by fluorescence recovery after photobleaching (FRAP). NPCs were labelled with Oregon Orange–wheatgerm agglutinin (WGA*). WGA has been shown to bind stably to several nucleoporins through residues modified by O-linked N-acetylglucos amine (Finlay et al., 1987).
Assembled nuclei were collected on coverslips and free WGA* was removed. A stripe of the nuclear rim fluorescence signal was bleached (Figure 5A). In nuclei assembled in mock-depleted extracts no redistribution of the WGA* fluorescence back into the bleached region was seen within 40 min (Figure 5A, top panels; see also Daigle et al., 2001). This indicated that the NPCs were immobile in the NE during the time course of the experiment and that WGA did not detectably dissociate from, and rebind to, NPCs during this period. In contrast, when nuclei assembled in Nup153-depleted extracts were monitored for the recovery of fluorescence in the bleached area, they recovered a large fraction of their initial intensity (Figure 5A, lower panels). The results of three independent experiments were quantified (Figure 5B). NPC fluorescence returned to ∼50% of its initial level after 30 min in the bleached area, demonstrating that NPCs lacking Nup153 were mobile within the NE.
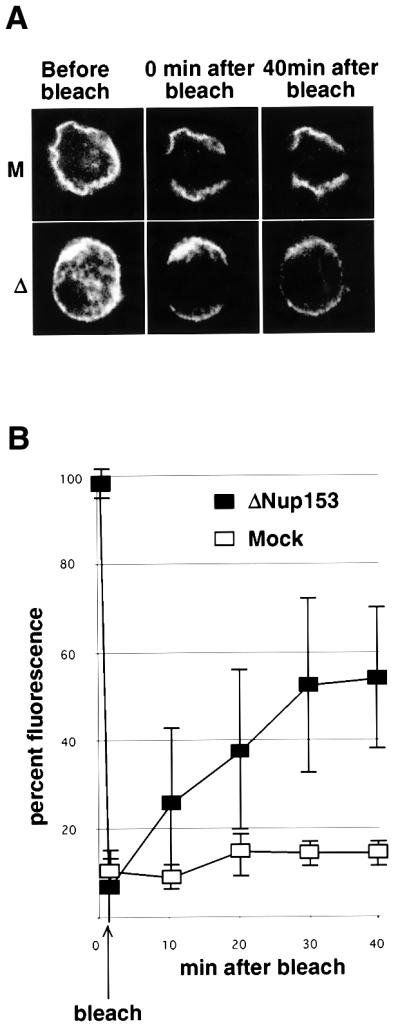
Fig. 5. Nup153-deficient NPCs display increased mobility within the NE. (A) Representative fluorescence recovery after photobleaching (FRAP) series of nuclei assembled in vitro from mock-depleted (M, upper panels) or Nup153-depleted (Δ, lower panels) Xenopus egg extracts, stained with Oregon Orange labelled wheatgerm agglutinin. (B) Quantitation of FRAP in three independent experiments using mock-depleted (open squares) or Nup153-depleted (solid squares) assembled nuclei.
Nup153 is required for basic NLS-mediated nuclear protein import but not for M9-mediated import
We wished to determine the effect of Nup153 depletion on NPC function during protein import into the nucleus. Two import pathways were compared, using model substrates. The first was a peptide containing the SV40 large T antigen NLS sequence cross-linked in vitro to bovine serum albumin (BSA) (Palacios et al., 1996). This binds to the importin α/β heterodimer, which mediates its translocation through the pore (Görlich and Kutay, 1999). Since BSA alone is already larger than the exclusion limit of the NPC, no diffusion into the nucleus occurs, as can be seen in reactions incubated at 4°C (Palacios et al., 1996).
The second substrate was a fusion protein of the nucleoplasmin core and the M9 domain of hnRNP A1 (Englmeier et al., 1999). Since the nucleoplasmin core domain forms pentamers this substrate is also too big to enter the nucleus by diffusion. The M9 domain is recognized and imported into the nucleus by transportin (Pollard et al., 1996). In both cases, nuclear import could be monitored by measuring the fluorescence of the nuclei by confocal microscopy. For each experiment, several fields containing multiple nuclei were recorded and the fluorescence quantified.
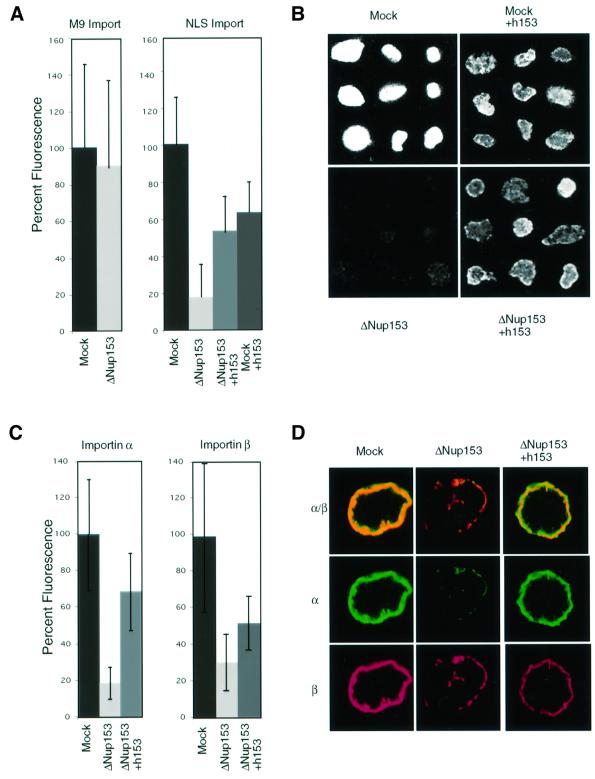
Depletion of Nup153 had no effect on the import of the M9-containing substrate (Figure 6A, left). In contrast, in nuclei devoid of Nup153 the import of BSA–NLS was reduced to 15–20% of that in a control reaction (Figure 6A, right, and B). This effect was not due to a co-depletion of importin β or Ran with Nup153, as determined by western blots of the depleted extract (data not shown). In the depleted nuclei both the intranuclear signal and the rim staining at the NE, indicative of substrate docking at the NPC, were reduced (Figure 6B). Addition of recombinant hNup153 to a control reaction diminished the level of nuclear import of BSA–NLS (Figure 6A and B), presumably due to the presence of C-terminal fragments of Nup153 in the hNup153 preparation that bind to importin β and inhibit its interaction with the NPC (Shah and Forbes, 1998). In spite of this problem, the effect of Nup153 depletion on BSA–NLS import could be restored to the level of nuclei formed in a mock reaction containing recombinant hNup153 by the addition of hNup153 to the reconstitution reaction (Figure 6A and B). Thus, removal of Nup153 had a drastic and specific effect on importin α/β-mediated import. Since M9-dependent protein import into the Nup153-depleted nuclei was not decreased, it appears highly unlikely that the observed effect on NLS-mediated import can be accounted for by a reduction in the number of NPCs in the depleted nuclei.
Fig. 6. Nup153-deficient NPCs display a selective protein import defect. Nuclei were assembled in vitro from mock-depleted (Mock) or Nup153-depleted (ΔNup153) Xenopus egg cytosol in the presence (+h153) or absence of recombinant hNup153 as indicated. (A) Quantitation of import of fluorescently labelled M9 (left panel) or classical NLS (right panel) import substrates in randomly selected nuclei from different experiments. Import is represented as the percentage of the nuclear fluorescence in mock-depleted nuclei. Bars show standard deviations. (B) Representative images of fluorescently labelled BSA–NLS import into nuclei as analysed in (A). Note the lack of nuclear rim staining in the bottom left panel. (C) Quantitation of nuclear importin α and importin β by immunofluorescence and direct fluorescence measurement. (D) Representative images of nuclei stained for importin α (α) or importin β (β), as quantified in (C).
At least three explanations for the specific inhibitory effect of Nup153 depletion on importin β-mediated transport can be proposed. The receptor–cargo translocation step might be affected, the transport complexes might not be disassembled from the NPC, thereby blocking subsequent import steps, or the recycling of the receptor components after their dissociation in the nucleus might be inhibited. In the first case we would predict that the steady-state level of the receptor in the nucleus would decrease, in the second that there would be a strong accumulation of the receptors at the nuclear rim, and in the third that the steady-state nuclear level of the receptors would increase. To distinguish the import step affected, the localization of importin α and β in nuclei lacking Nup153 was determined. Importin α was localized by indirect immunfluorescence and importin β by the use of fluorescently labelled protein in the nuclear assembly reaction. Representative experiments are shown in Figure 6D and a quantitation of the nuclear fluorescence is provided in Figure 6C. The nuclear levels of importin α were reduced to 19 ± 9% in Nup153-depleted nuclei and those of importin β to 30 ± 15% (Figure 6C and D). These defects could be partly restored by the addition of recombinant hNup153 (Figure 6C and D). Note that the restoration of the importin β localization on addition of Nup153 was less efficient than the restoration of importin α or of BSA–NLS import (Figure 6A). The observed specific import inhibition of ‘classical’ NLS-containing transport complexes in the absence of Nup153 is therefore very unlikely to be due to a defect in importin α or β recycling or to their trapping at the NPC, but rather to an effect on import complex translocation through the NPC.
Discussion
We have localized Nup153 within the NPC and used a nuclear reconstitution assay based on affinity-depleted Xenopus egg extract (Finlay and Forbes, 1990), to analyse its function in a biochemically amenable system. Nup153 is localized at the base of the nuclear basket of the NPC on the coaxial ring, adjacent to the NE. Consistent with this localization, depletion of Nup153 leads to a lack of stable association of other components of the nuclear basket and of NPC-associated filamentous structures. Lack of Nup153 also results in mobilization of NPCs within the NE and, perhaps as a consequence, a non-uniform distribution of NPCs within the NE. NPCs lacking Nup153 exhibit a defect in importin α/β-mediated nuclear protein import but not in transportin-mediated import, demonstrating that these different receptors require different specific interactions at the NPC in order to function optimally.
Localization of Nup153 in Xenopus oocyte NPCs
The localization of Nup153 within the NPC was investigated by two independent EM methods. We obtained images from immunogold labelled Xenopus oocytes with a high resolution in the plane of the NE by utilizing FEISEM, and in the perpendicular direction using TEM. In both cases the localization was determined relative to easily recognizable structures of the NPC. Moreover, the preparation conditions used were essentially the same until the final stages of the procedures, justifying the integration of the data from both methods into a single model (Figure 1C). This is also reflected by the fact that there is no significant difference in the localization of the gold particles in the plane parallel to the NE as determined by the two methods. Nup153 is localized to the NE end of the NPC basket on the nucleoplasmic ring structure. This is in part consistent with previous electron microscopy data: Sukegawa and Blobel (1993) used a polyclonal antibody raised against an N-terminal and a C-terminal fragment of rat Nup153 to localize it to the nuclear side of the NPC. Using a monoclonal antibody, a second study located Nup153 to the nuclear side of the NE, at a mean distance of 50–55 nm from the centre of the NPC (Cordes et al., 1993). A third study assigned most Nup153 to the terminal ring of the nuclear basket of Xenopus oocyte NPCs (Pante et al., 1994). However, the antibody used for localization in this case was raised against a C-terminal 10 amino acid peptide of the human Nup153 protein. Sequencing of Xenopus Nup153 (Shah et al., 1998) revealed that only two of the 10 amino acids were conserved in both proteins, providing a likely explanation for the discrepancy. Since the 10 amino acids included an FXFG sequence, recognition of other Xenopus nucleoporins by the antibody seems possible.
The role of Nup153 in anchoring the NPC
We analysed the composition of the NPCs in Nup153-depleted nuclei by immunofluorescence and found that at least two additional components of the nuclear basket, Nup98 and Nup93 (Radu et al., 1995; Grandi et al., 1997), were not correctly localized. Tpr, which is part of filamentous structures attached to the base of the NPC and reaching into the nucleoplasm (Cordes et al., 1993, 1997), was also not detectable in Nup153-depleted nuclei. Importantly, none of these proteins was co-depleted with Nup153 (data not shown) and their absence could be reversed by the addition of bacterially expressed Nup153. No defects in nucleoporins that reside further from Nup153 within the NPC were observed. Nup214/CAN, Nup358/RanBP2 and the nucleoporins recognized by the general FXFG nucleoporin antibody mAb414 were not measurably affected by Nup153 depletion.
In nuclei that lack Nup153, Nup98, Nup93 and Tpr, NPCs were rendered mobile in the plane of the NE. This is in contrast to the normal vertebrate situation, where NPCs are anchored within the NE (Daigle et al., 2001; Figure 5A). Anchoring could be mediated by any of the missing proteins, i.e. Nup93, Nup98, Tpr or Nup153 itself, forming an interaction with a stable nuclear structure, like chromatin, the Tpr-containing intranuclear filaments or the lamina. Recently, an interaction between Nup98 and Tpr has been described (Fontoura et al., 2001). In addition, in the yeast Saccharomyces cerevisiae, the homologue of Nup93 (Nic96) has been reported to interact with one of two yeast Tpr homologues (Mlp2p) (Strambio-de-Castillia et al., 1999; Kosova et al., 2000). The loss of these interactions could rationalize the absence of these nucleoporins after depletion of Nup153, assuming that at least one of them needs Nup153 for its incorporation.
Both interaction with chromatin and with the intranuclear filaments is thought to be mediated by Tpr (Cordes et al., 1993, 1997; Zimowska et al., 1997; Galy et al., 2000). Since Tpr is absent from the nucleus when Nup153 is depleted from NPCs, a lack of Tpr filaments could contribute to the observed mobility of NPCs within the NE. Concerning a possible role of Tpr in anchoring the NPC in the NE it should however be borne in mind that yeast NPCs, in which the interaction of Mlp2 and Nic96 is intact, are mobile within the NE (Belgareh and Doye, 1997; Bucci and Wente, 1997).
Several previous observations suggest that the nuclear lamina could mediate the anchoring and spacing of NPCs in higher eukaryotes. The insertional Dm0 mutant of the Drosophila lamin gene causes abnormal NPC distribution in the NE (Lenz-Bohme et al., 1997). Similarly, prevention of expression of the only identifiable lamin gene in Caenorhabditis elegans by double-stranded RNA interference (RNAi) leads to a clustering of NPCs (Liu et al., 2000). It has recently been reported that Nup153 interacts directly with at least one lamin protein (Smythe et al., 2000), further supporting the hypothesis that NPCs might be anchored, at least in part, by an interaction between Nup153 and the lamina. S.cerevisiae in contrast, which has mobile NPCs, contains no discernible genes homologous to lamins and also no obvious Nup153 homologue.
Taken together, this allows us to formulate several models for Nup153 function in anchoring NPCs. In one scenario, Nup153 interacts directly with the lamina and holds the NPCs in place. Disruption of this interaction will lead to increased mobility of the NPCs, which as a result non-specifically stick together. Alternatively, Nup153 interacts directly or indirectly with Tpr, which in turn forms filaments protruding into the nucleus and interacts with the chromatin, thereby anchoring the NPCs. Although the latter possibility is not supported by evolutionary arguments, we cannot at present rule out the possibility that Tpr interaction causes, or contributes to, NPC anchoring in vertebrates. Thirdly, Nup153 is required for the assembly of components of the nuclear basket, and the basket structure makes the contacts required to immobilize the NPCs. These different models are not mutually exclusive.
The role of Nup153 in nuclear protein import
Nup153 has been shown to interact with a variety of factors required for different protein import pathways: importin β, importin α, RanGDP and transportin (Moroianu et al., 1997; Shah and Forbes, 1998; Shah et al., 1998; Nakielny et al., 1999). The interaction with importin β in Xenopus egg extracts appears particularly stable, being easily detected by co-immunoprecipitation (Shah et al., 1998). Due to this finding and the localization of Nup153 on the nuclear face of the NPC, it has been proposed that Nup153 might constitute the last step in importin β-mediated nuclear protein import and/or the first step of recycling of importin β back to the cytoplasm (Shah et al., 1998). It is therefore interesting that we find that the depletion of Nup153 results in a drastic reduction of importin α/β-mediated import, although it must be noted that more than just Nup153 is absent from the defective NPCs. Genetic or biochemical depletion of the two nucleoporins that we found to be missing from Nup153-depleted NPCs has been performed and does not support a specific role for either Nup98 or Nup93 in importin α/β-mediated import. Genetic deletion of Nup98 leads to a reduced import activity of both classical and M9-containing import substrates. In this case, however, the cytoplasmic nucleoporins Nup214/CAN and Nup358/RanBP2 are also found to be depleted from the NPC (Wu et al., 2001). The data presented here make it probable that the reported defect in import of M9-containing cargoes is an indirect effect of the deletion of Nup98. In addition, antibody depletion of Nup98 from a nuclear reconstitution system has shown that Nup98 is not required for nuclear import of cargoes containing a ‘classical’ NLS (Powers et al., 1995).
When a complex containing Nup93 was biochemically depleted the resulting nuclei also showed no defect for importin α/β-mediated protein import (Grandi et al., 1997). These data make it unlikely that either Nup93 or Nup98 is responsible for the import defect observed in Nup153-depleted nuclei, and suggest that the defect is either directly due to the lack of Nup153 or to the loss of other, as yet unidentified, NPC components in the absence of Nup153.
Searching for a mutant form of Nup153 that would restore the NPC targeting of other nucleoporins, but retain the defect in importin β-mediated protein import, would be a way of further elucidating the mechanism of Nup153 action in nuclear protein import, but is made dificult by problems in expressing recombinant forms of Nup153, whether wild-type or mutant, for use in reconstitution studies. The fact that transportin-mediated import into the Nup153-depleted nuclei was not defective supports the hypothesis (Nakielny et al., 1999) that the interaction between Nup153 and transportin has the function of delivering Nup153 into the nucleus rather than being required for import of transportin’s substrates.
Given the stable, RanGTP-reversible, interaction between importin β and Nup153 mentioned above, and the fact that an importin β mutant that cannot bind RanGTP interacts tightly with a site or sites on the NPC and, when bound, acts as a dominant inhibitor of NPC translocation (Kutay et al., 1997), it has been proposed that Nup153 might represent the site at which importin β-containing import complexes are dissociated by RanGTP (Shah et al., 1998; Görlich and Kutay, 1999). If this were the case, the depletion of Nup153 would be expected to result in the inefficient recycling of importin β and importin α from the nucleus to the cytoplasm and thereby the reduction of NLS-mediated import activity. Alternatively, a strong binding site for importin β on Nup153 could be required on the nuclear side of the NPC to allow for efficient movement of importin β complexes away from the weaker binding sites that line the translocation channel to a site at which they could be dissociated by RanGTP (Melchior and Gerace, 1998; Rout et al., 2000). Since we do not see a build up of either BSA–NLS import complexes or of importin β or importin α in the nucleus or the nuclear rim in the absence of Nup153, our data suggest that the defect in NLS protein import is at the stage of NPC translocation rather than in recycling or trapping of import complexes on the NPC.
It is possible that an import defect in Nup153-depleted nuclei is the cause of the lack of detectable nuclear Nup93, Nup98 and Tpr, if these proteins are imported into the nucleus via a receptor whose function requires Nup153. A model can even be imagined in which the order of import into the nucleus and assembly into the nuclear face of the NPC of nucleoporins could be determined by a cascade of interactions, in which the import and assembly of one nucleoporin is a prerequisite for the function of the import receptor required to bring the next nucleoporin to the inner side of the NPC. Further analysis of this interesting possibility is merited.
In summary, we show that Nup153 is crucial for anchoring the NPCs in the NE by assembling the nuclear basket structure and allowing, directly or indirectly, for interaction of the NPC with the nuclear lamina and/or intranuclear filaments. Moreover, Nup153 is crucial for the efficient functioning of at least one nuclear protein import pathway. This latter observation supports further the hypothesis originally proposed on the basis of a mutation in the yeast nucleoporin Nsp1 (Nehrbass et al., 1993), that different import receptors require different, specific interactions with nucleoporins for their efficient translocation through the NPC.
Materials and methods
Nup153-specific antibodies
To generate monospecific antibodies against Xenopus Nup153, an N-terminal fragment spanning amino acids 1–149 of the published sequence (Shah et al., 1998) was expressed as a His-tagged fusion protein from pQE30 (Qiagen) in E.coli strain BL21 [pRep4], and purified under denaturing conditions. Antibodies were raised in rabbits. The antibodies were affinity purified using the antigen cross-linked to Affigel-10 (Bio-Rad). To generate a resin for the depletion from Xenopus egg extracts, saturating amounts of antibody were bound to protein G–Sepharose (Pharmacia) and cross-linked with 10 mM dimethyl pimelimidate (Sigma).
Immunogold labelling and FEISEM
For the immunolocalization of Nup153, Xenopus oocyte nuclei were isolated in buffer (83 mM KCl, 17 mM NaCl, 10 mM HEPES pH 7.5) and prefixed in 2% formaldehyde in V buffer (1 mM MgCl2, 150 mM sucrose, 80 mM PIPES pH 6.8) for 10 min. Samples were incubated in 100 mM glycine in phosphate-buffered saline (PBS) for 10 min and blocked in 1% fish scale gelatine in PBS for 1 h. To strip nuclear baskets, NEs were first incubated in 10 mM HEPES pH 7.4, 1 mM EDTA for 30 min and subsequently in the same buffer containing 500 mM KCl. NEs were labelled with affinity-purified anti-Nup153 antibody for 1 h, washed six times with PBS and incubated with 10 nm colloidal gold-conjugated secondary antibody (Amersham) overnight at 4°C. Controls were performed using only secondary antibodies, or an irrelevant primary antibody. For TEM analysis, nuclear envelopes were fixed in 2% paraformaldehyde, 0.5% glutaraldehyde in 0.2% tannic acid for 1 h, and stained in 1% OsO4, 0.2 M Na cacodylate pH 7.4 and 0.1% uranyl acetate for 10 min each. After dehydration and embedding in epoxy resin, 70 nm sections were cut, collected and visualized in a JEOL 1220 TEM.
For FEISEM analysis, NEs were isolated on to silicon chips (PLANO), postfixed, stained and dehydrated as above and subjected to critical point drying. The samples were then sputter-coated with 4 nm chromium and visualized using a Topcon DS130F FEISEM at 30 kV accelerating voltage. Colloidal gold particles were imaged by using a solid state backscattered electron detector (KE developments). All measurements were made using AnalySIS (SIS) software.
Xenopus egg extract preparation and depletion
Fractionated egg extracts were prepared as described (Hetzer et al., 2000). The 10 000 g crude extract was fractionated by ultracentrifugation for 30 min at 4°C into a yellowish phase, containing clear cytosol with a few membrane bands, a membrane phase underneath and a pellet fraction containing glycogen. The yellowish phase was diluted with 0.3 vol of S250 buffer (50 mM KCl, 2.5 mM MgCl2, 20 mM HEPES pH 7.5) and fractionated into a cytosolic fraction and a membrane pellet by ultracentrifugation at 200 000 g for 30 min at 4°C. The cytosolic fraction was incubated twice with an equal volume of mock or anti-Nup153 resin, which was pre-blocked with 5% BSA for 1 h. Glycogen was added to the supernatant to 3% v/v and 50 µl aliquots were frozen and stored in liquid nitrogen. The membranes were diluted 20-fold in S250 and centrifuged through a cushion of S250 with 0.5 M sucrose, divided into 10 µl aliquots, frozen and stored in liquid nitrogen.
Sperm chromatin preparation
Sperm was recovered from Xenopus testis in SuNaSp buffer (250 mM sucrose, 75 mM NaCl, 0.5 mM spermidine tetrachloride, 0.15 mM spermine tetrachloride) and chromatin was prepared to a final concentration of 1 × 103 sperm/µl as described (Gurdon, 1976).
Expression and purification of recombinant Nup153
GST was fused to the N-terminus of full-length human Nup153 in pGEX2T and expressed in BL21 codonplus (Stratagene). Cultures were grown from a single colony to an OD600 of 0.7 and induced with 1 mM IPTG for 4 h at room temperature. GST–Nup153 was purified on G–Sepharose (Pharmacia) following the manufacturer’s instructions. The fractions obtained were dialysed against S250, snap frozen and stored at –80°C.
Nuclear assembly, import assay and indirect immunofluorescence
Assembly, import and immunofluorescence reactions were performed as described (Hetzer et al., 2000). In brief, for a 20 µl reaction, 13.5 µl cytosol, 1.5 µl energy mix (10 mM ATP, 100 mM creatine phosphate, 2 mg/ml creatine kinase), 20 mg/ml glycogen (USB, Amersham) and either 4 µl S250 buffer or recombinant Nup153 were mixed. One microlitre of sperm chromatin followed by 2 µl of membrane fraction were added and the reaction incubated for 2 h at 20°C. For nuclear import reactions a fluorescently labelled BSA–NLS conjugate (Palacios et al., 1996) or NPC–M9–M10 (Englmeier et al., 1999) was added and further incubated for 40 min. The samples were subsequently fixed with 4% formaldehyde and centrifuged through a cushion of 30% w/v sucrose on to an l-polylysine-coated coverslip and either monitored directly by confocal microscopy or further processed for immunofluorescence as described (Arts et al., 1997). Importin β was prepared as described (Palacios et al., 1997), labelled with Alexa-546 (Molecular Probes) according to the manufacturer’s specifications and tested for functionality in interaction assays.
Images were recorded with a Zeiss LSM 510 or a Leica TCS confocal microscope. 3D reconstructions were performed using Huygens (Bitplane) software for deconvolution and Imaris (Bitplane) for projection. Quantitation of import reactions used an NIH image macro. The mean fluorescence of 10 or more images containing several nuclei each was determined. The identity of nuclei was checked by DAPI staining for chromatin.
Fluorescence recovery after photobleaching (FRAP)
Nuclei were assembled as described above and subsequently incubated for 10 min with 4 µl of 1 mg/ml Oregon Orange–WGA (Molecular Probes). The samples were diluted in 300 µl S250 and spun through a 30% w/v sucrose cushion on to l-polylysine-coated coverslips and mounted in chambers containing acetate buffer (100 mM KAc, 3 mM MgAc, 5 mM EGTA, 20 mM HEPES–KOH pH 7.4, 150 mM sucrose, 1 mM DTT) for in situ imaging. Photobleaching was performed using the LSM 510 microscope and the accompanying software. Quantitative analysis was performed, after background subtraction, with NIH image software. Average intensities were corrected for fluorescence loss during the bleach.
Acknowledgments
Acknowledgements
We would like to thank Volker Cordes, Jan van Deursen, Ed Hurt and Georg Krohne for providing reagents, Jens Rietdorf for help with confocal microscopy and 3D reconstructions, Sandra Rutherford for help with electron microscopy and Victoria Juarez and Pauli Peraesalmi for help with animal work. Special thanks to Jan Ellenberg for experimental advice, sharing unpublished results and critical comments, Martin Hetzer for experimental advice, and Peter Askjaer, Daniel Bilbao-Cortes, Ludwig Englmeier, Oliver J.Gruss, Martin Hetzer, Margy Koffa, Mutsuhito Ohno, Christoph Schatz and Alexandra Segref for critical comments on the manuscript. The work was funded by EMBL and a Human Science Frontier Programme Organization Research grant to I.W.M.
References
- Akey C.W. (1989) Interactions and structure of the nuclear pore complex revealed by cryo-electron microscopy. J. Cell Biol., 109, 955–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen T.D., Bennion,G.R., Rutherpord,S.A., Reipert,S., Ramalho,A., Kiseleva,E. and Goldberg,M.W. (1997) Macromolecular substructure in nuclear pore complexes by in-lens field-emission scanning electron microscopy. Scanning, 19, 403–410. [DOI] [PubMed] [Google Scholar]
- Arts G.J., Englmeier,L. and Mattaj,I.W. (1997) Energy- and temperature-dependent in vitro export of RNA from synthetic nuclei. Biol. Chem., 378, 641–649. [DOI] [PubMed] [Google Scholar]
- Bastos R., Lin,A., Enarson,M. and Burke,B. (1996) Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol., 134, 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayliss R., Littlewood,T. and Stewart,M. (2000) Structural basis for the interaction between FxFG nucleoporin repeats and importin-β in nuclear trafficking. Cell, 102, 99–108. [DOI] [PubMed] [Google Scholar]
- Belgareh N. and Doye,V. (1997) Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol., 136, 747–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucci M. and Wente,S.R. (1997) In vivo dynamics of nuclear pore complexes in yeast. J. Cell Biol., 136, 1185–1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes V.C., Reidenbach,S., Kohler,A., Stuurman,N., van Driel,R. and Franke,W.W. (1993) Intranuclear filaments containing a nuclear pore complex protein. J. Cell Biol., 123, 1333–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes V.C., Reidenbach,S., Rackwitz,H.R. and Franke,W.W. (1997) Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J. Cell Biol., 136, 515–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N., Beaudouin,J., Hartnell,L., Imreh,G., Hallberg,E., Lippincott-Schwartz,J. and Ellenberg,J. (2001) Nuclear pore complex turnover and network in live mammalian cells. J. Cell Biol., 154, 71–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis L.I. and Blobel,G. (1987) Nuclear pore complex contains a family of glycoproteins that includes p62: glycosylation through a previously unidentified cellular pathway. Proc. Natl Acad. Sci. USA, 84, 7552–7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doye V. and Hurt,E. (1997) From nucleoporins to nuclear pore complexes. Curr. Opin. Cell Biol., 9, 401–411. [DOI] [PubMed] [Google Scholar]
- Enarson P., Enarson,M., Bastos,R. and Burke,B. (1998) N-terminal sequences that direct nucleoporin nup153 to the inner surface of the nuclear envelope. Chromosoma, 107, 228–236. [DOI] [PubMed] [Google Scholar]
- Englmeier L., Olivo,J.C. and Mattaj,I.W. (1999) Receptor-mediated substrate translocation through the nuclear pore complex without nucleotide triphosphate hydrolysis. Curr. Biol., 9, 30–41. [DOI] [PubMed] [Google Scholar]
- Fabre E. and Hurt,E. (1997) Yeast genetics to dissect the nuclear pore complex and nucleocytoplasmic trafficking. Annu. Rev. Genet., 31, 277–313. [DOI] [PubMed] [Google Scholar]
- Finlay D.R. and Forbes,D.J. (1990) Reconstitution of biochemically altered nuclear pores: transport can be eliminated and restored. Cell, 60, 17–29. [DOI] [PubMed] [Google Scholar]
- Finlay D.R., Newmeyer,D.D., Price,T.M. and Forbes,D.J. (1987) Inhibition of in vitro nuclear transport by a lectin that binds to nuclear pores. J. Cell Biol., 104, 189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay D.R., Meier,E., Bradley,P., Horecka,J. and Forbes,D.J. (1991) A complex of nuclear pore proteins required for pore function. J. Cell Biol., 114, 169–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontoura B.M., Dales,S., Blobel,G. and Zhong,H. (2001) The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc. Natl Acad. Sci. USA, 98, 3208–3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forbes D.J., Kirschner,M.W. and Newport,J.W. (1983) Spontaneous formation of nucleus-like structures around bacteriophage DNA microinjected into Xenopus eggs. Cell, 34, 13–23. [DOI] [PubMed] [Google Scholar]
- Franke W.W. (1970a) Nuclear pore flow rate. A characteristic for nucleocytoplasmic exchange of macromolecules and particles. Naturwissenschaften, 57, 44–45. [DOI] [PubMed] [Google Scholar]
- Franke W.W. (1970b) On the universality of nuclear pore complex structure. Z. Zellforsch. Mikrosk. Anat., 105, 405–429. [DOI] [PubMed] [Google Scholar]
- Franke W.W. and Scheer,U. (1970) The ultrastructure of the nuclear envelope of amphibian oocytes: a reinvestigation. I. The mature oocyte. J. Ultrastruct. Res., 30, 288–316. [DOI] [PubMed] [Google Scholar]
- Galy V., Olivo-Marin,J.C., Scherthan,H., Doye,V., Rascalou,N. and Nehrbass,U. (2000) Nuclear pore complexes in the organization of silent telomeric chromatin. Nature, 403, 108–112. [DOI] [PubMed] [Google Scholar]
- Goldberg M.W. and Allen,T.D. (1992) High resolution scanning electron microscopy of the nuclear envelope: demonstration of a new, regular, fibrous lattice attached to the baskets of the nucleoplasmic face of the nuclear pores. J. Cell Biol., 119, 1429–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M.W. and Allen,T.D. (1996) The nuclear pore complex and lamina: three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J. Mol. Biol., 257, 848–865. [DOI] [PubMed] [Google Scholar]
- Görlich D. and Kutay,U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell. Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- Grandi P., Dang,T., Pane,N., Shevchenko,A., Mann,M., Forbes,D. and Hurt,E. (1997) Nup93, a vertebrate homologue of yeast Nic96p, forms a complex with a novel 205-kDa protein and is required for correct nuclear pore assembly. Mol. Biol. Cell, 8, 2017–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y., Wilson,K.L., Harel,A., Goldberg,M. and Cohen,M. (2000) Review: nuclear lamins—structural proteins with funda mental functions. J. Struct. Biol., 129, 313–323. [DOI] [PubMed] [Google Scholar]
- Gurdon J.B. (1976) Injected nuclei in frog oocytes: fate, enlargement and chromatin dispersal. J. Embryol. Exp. Morphol., 36, 523–540. [PubMed] [Google Scholar]
- Hetzer M., Bilbao-Cortes,D., Walther,T.C., Gruss,O.J. and Mattaj,I.W. (2000) GTP hydrolysis by Ran is required for nuclear envelope assembly. Mol. Cell, 5, 1013–1024. [DOI] [PubMed] [Google Scholar]
- Jarnik M. and Aebi,U. (1991) Toward a more complete 3-D structure of the nuclear pore complex. J. Struct. Biol., 107, 291–308. [DOI] [PubMed] [Google Scholar]
- Kosova B., Pante,N., Rollenhagen,C., Podtelejnikov,A., Mann,M., Aebi,U. and Hurt,E. (2000) Mlp2p, a component of nuclear pore attached intranuclear filaments, associates with nic96p. J. Biol. Chem., 275, 343–350. [DOI] [PubMed] [Google Scholar]
- Kutay U., Izaurralde,E., Bischoff,F.R., Mattaj,I.W. and Görlich,D. (1997) Dominant-negative mutants of importin-β block multiple pathways of import and export through the nuclear pore complex. EMBO J., 16, 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz-Bohme B., Wismar,J., Fuchs,S., Reifegerste,R., Buchner,E., Betz,H. and Schmitt,B. (1997) Insertional mutation of the Drosophila nuclear lamin Dm0 gene results in defective nuclear envelopes, clustering of nuclear pore complexes and accumulation of annulate lamellae. J. Cell Biol., 137, 1001–1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Ben-Shahar,T.R., Riemer,D., Treinin,M., Spann,P., Weber,K., Fire,A. and Gruenbaum,Y. (2000) Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression and spatial organization of nuclear pore complexes. Mol. Biol. Cell, 11, 3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourim D. and Krohne,G. (1993) Membrane-associated lamins in Xenopus egg extracts: identification of two vesicle populations. J. Cell Biol., 123, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lourim D. and Krohne,G. (1998) Chromatin binding and polymerization of the endogenous Xenopus egg lamins: the opposing effects of glycogen and ATP. J. Cell Sci., 111, 3675–3686. [DOI] [PubMed] [Google Scholar]
- Lohka M.J. and Masui,Y. (1983) Formation in vitro of sperm pronuclei and mitotic chromosomes induced by amphibian ooplasmic components. Science, 220, 719–721. [DOI] [PubMed] [Google Scholar]
- Mattaj I.W. and Englmeier,L. (1998) Nucleocytoplasmic transport: the soluble phase. Annu. Rev. Biochem., 67, 265–306. [DOI] [PubMed] [Google Scholar]
- Maul G.G. (1977) The nuclear and the cytoplasmic pore complex: structure, dynamics, distribution and evolution. Int. Rev. Cytol. Suppl., 6, 75–186. [PubMed] [Google Scholar]
- Melchior F. and Gerace,L. (1998) Two-way trafficking with Ran. Trends Cell Biol., 8, 175–179. [DOI] [PubMed] [Google Scholar]
- Moroianu J., Blobel,G. and Radu,A. (1997) RanGTP-mediated nuclear export of karyopherin α involves its interaction with the nucleoporin Nup153. Proc. Natl Acad. Sci. USA, 94, 9699–9704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S., Shaikh,S., Burke,B. and Dreyfuss,G. (1999) Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J., 18, 1982–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U., Fabre,E., Dihlmann,S., Herth,W. and Hurt,E.C. (1993) Analysis of nucleo-cytoplasmic transport in a thermosensitive mutant of nuclear pore protein NSP1. Eur. J. Cell Biol., 62, 1–12. [PubMed] [Google Scholar]
- Newport J. (1987) Nuclear reconstitution in vitro: stages of assembly around protein-free DNA. Cell, 48, 205–217. [DOI] [PubMed] [Google Scholar]
- Ohno M., Fornerod,M. and Mattaj,I.W. (1998) Nucleocytoplasmic transport: the last 200 nanometers. Cell, 92, 327–336. [DOI] [PubMed] [Google Scholar]
- Palacios I., Weis,K., Klebe,C., Mattaj,I.W. and Dingwall,C. (1996) RAN/TC4 mutants identify a common requirement for snRNP and protein import into the nucleus. J. Cell Biol., 133, 485–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios I., Hetzer,M., Adam,S.A. and Mattaj,I.W. (1997) Nuclear import of U snRNPs requires importin β. EMBO J., 16, 6783–6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N., Bastos,R., McMorrow,I., Burke,B. and Aebi,U. (1994) Interactions and three-dimensional localization of a group of nuclear pore complex proteins. J. Cell Biol., 126, 603–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard V.W., Michael,W.M., Nakielny,S., Siomi,M.C., Wang,F. and Dreyfuss,G. (1996) A novel receptor-mediated nuclear protein import pathway. Cell, 86, 985–994. [DOI] [PubMed] [Google Scholar]
- Powers M.A., Macaulay,C., Masiarz,F.R. and Forbes,D.J. (1995) Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J. Cell Biol., 128, 721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A., Blobel,G. and Moore,M.S. (1995) Identification of a protein complex that is required for nuclear protein import and mediates docking of import substrate to distinct nucleoporins. Proc. Natl Acad. Sci. USA, 92, 1769–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichelt R., Holzenburg,A., Buhle,E.L.,Jr, Jarnik,M., Engel,A. and Aebi,U. (1990) Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J. Cell Biol., 110, 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson W.D., Mills,A.D., Dilworth,S.M., Laskey,R.A. and Dingwall,C. (1988) Nuclear protein migration involves two steps: rapid binding at the nuclear envelope followed by slower translocation through nuclear pores. Cell, 52, 655–664. [DOI] [PubMed] [Google Scholar]
- Ris H. (1989) Three-dimensional imaging of cell ultrastructure with high resolution low voltage SEM. Inst. Phys. Conf. Ser., 98, 657–662. [Google Scholar]
- Ris H. (1991) The three-dimensional structure of the nuclear pore complex as seen by high voltage electron microscopy and high resolution low voltage scanning electron microscopy. EMSA Bull., 21, 54–56. [Google Scholar]
- Rout M.P., Aitchison,J.D., Suprapto,A., Hjertaas,K., Zhao,Y. and Chait,B.T. (2000) The yeast nuclear pore complex: composition, architecture and transport mechanism. J. Cell Biol., 148, 635–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan K.J. and Wente,S.R. (2000) The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr. Opin. Cell Biol., 12, 361–371. [DOI] [PubMed] [Google Scholar]
- Shah S. and Forbes,D.J. (1998) Separate nuclear import pathways converge on the nucleoporin Nup153 and can be dissected with dominant-negative inhibitors. Curr. Biol., 8, 1376–1386. [DOI] [PubMed] [Google Scholar]
- Shah S., Tugendreich,S. and Forbes,D. (1998) Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J. Cell Biol., 141, 31–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smitherman M., Lee,K., Swanger,J., Kapur,R. and Clurman,B.E. (2000) Characterization and targeted disruption of murine Nup50, a p27(Kip1)-interacting component of the nuclear pore complex. Mol. Cell. Biol., 20, 5631–5642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smythe C., Jenkins,H.E. and Hutchison,C.J. (2000) Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J., 19, 3918–3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel,G. and Rout,M.P. (1999) Proteins connecting the nuclear pore complex with the nuclear interior. J. Cell Biol., 144, 839–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa J. and Blobel,G. (1993) A nuclear pore complex protein that contains zinc finger motifs, binds DNA and faces the nucleoplasm. Cell, 72, 29–38. [DOI] [PubMed] [Google Scholar]
- Ullman K.S., Shah,S., Powers,M.A. and Forbes,D.J. (1999) The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell, 10, 649–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Deursen J., Boer,J., Kasper,L. and Grosveld,G. (1996) G2 arrest and impaired nucleocytoplasmic transport in mouse embryos lacking the proto-oncogene CAN/Nup214. EMBO J., 15, 5574–5583. [PMC free article] [PubMed] [Google Scholar]
- von Kobbe C., van Deursen,J.M., Rodrigues,J.P., Sitterlin,D., Bachi,A., Wu,X., Wilm,M., Carmo-Fonseca,M. and Izaurralde,E. (2000) Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin nup98. Mol. Cell, 6, 1243–1252. [DOI] [PubMed] [Google Scholar]
- Wente S.R. (2000) Gatekeepers of the nucleus. Science, 288, 1374–1377. [DOI] [PubMed] [Google Scholar]
- Wu X., Kasper,L.H., Mantcheva,R.T., Mantchev,G.T., Springett,M.J. and van Deursen,J.M. (2001) Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc. Natl Acad. Sci. USA, 98, 3191–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Rout,M.P. and Akey,C.W. (1998) Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol. Cell, 1, 223–234. [DOI] [PubMed] [Google Scholar]
- Zimowska G., Aris,J.P. and Paddy,M.R. (1997) A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J. Cell Sci., 110, 927–944. [DOI] [PubMed] [Google Scholar]