Abstract
IS911 transposition involves a free circular transposon intermediate where the terminal inverted repeat sequences are connected. Transposase synthesis is usually driven by a weak promoter, pIRL, in the left end (IRL). Circle junction formation creates a strong promoter, pjunc, with a –35 sequence located in the right end and the –10 sequence in the left. pjunc assembly would permit an increase in synthesis of transposase from the transposon circle, which would be expected to stimulate integration. Insertion results in pjunc disassembly and a return to the low pIRL- driven transposase levels. We demonstrate that pjunc plays an important role in regulating IS911 transposition. Inactivation of pjunc strongly decreased IS911 transposition when transposase was produced in its natural configuration. This novel feedback mechanism permits transient and controlled activation of integration only in the presence of the correct (circular) intermediate. We have also investigated other members of the IS3 and other IS families. Several, but not all, IS3 family members possess pjunc equivalents, underlining that the regulatory mechanisms adopted to fine-tune transposition may be different.
Keywords: IS911/promoter/regulation/transposase/transposition
Introduction
Extensive analysis of a number of transposable elements has revealed that they have adopted a variety of different strategies to control their displacement. Even elements that use related transposases (Tpase) carrying a conserved triad of amino acids (the DDE motif; Fayet et al., 1990; Rowland and Dyke, 1990; Kulkosky et al., 1992; Rezsohazy et al., 1993) as part of their active site, and which therefore presumably share identical chemistry, show a wide diversity in the details of their transposition cycle (Mizuuchi, 1992; Haren et al., 1999). DDE Tpases catalyse the cleavage of a single DNA strand to generate a 3′-OH at the transposon end. This then acts as a nucleophile in the attack of the target DNA backbone. The major defining difference between the elements encoding such Tpases is the way in which they deal with the second strand (see Turlan and Chandler, 2000).
In one major strategy adopted by IS911 and other members of the IS3 family [IS911 (Ton-Hoang et al., 1997), IS3 (Sekine et al., 1994), IS2 (Lewis and Grindley, 1997) and IS150 (Welz, 1993)], a single-strand cleavage occurs initially at one end of the insertion sequence (IS). The free 3′-OH generated is then directed to attack the opposite end on the same DNA strand to generate a figure-of-eight form in which both ends are joined by a single-strand bridge (Polard and Chandler, 1995a). This is converted into a transposon circle (Polard and Chandler, 1995a; G.Duval-Valentin, unpublished results), which then undergoes the final step of integration. The transposon circle carries abutted left and right ends (IRL and IRR). Since such junctions have been observed for several ISs, including IS21 (Reimmann et al., 1989; Berger and Haas, 2001), IS30 (Olasz et al., 1993), IS256 (Lyon et al., 1987; M.Prudhomme, C.Turlan, J.-P. Claverys and M.Chandler, submitted) and ISL3 (Kallastu et al., 1998), members of these families may also transpose using a similar mechanism. In all cases IRL and IRR are separated by a short spacer whose length is a characteristic of the particular IS. These structures are several orders of magnitude more active as substrates in transposition than are plasmid donor molecules in which the two ends are distant (i.e. in their normal configuration). Transposition of these elements is thus also likely to pass through a circularization step.
In the case of IS3 family members, the transposition reactions are catalysed by two proteins, OrfAB and OrfA. The Tpase, OrfAB, is produced as a fusion protein by translational frameshifting between two consecutive and partially overlapping reading frames, orfA and orfB. OrfA, is the product of the upstream frame, orfA. OrfAB alone is capable of generating the bridged figure-of-eight form while both proteins are required for the reactions at the IRL–IRR junction leading to efficient insertion of the transposon circle (Ton-Hoang et al., 1997).
In addition to their inherently high recombination activity, the IRR–IRL junctions of several of these IS elements have been shown to constitute relatively strong promoters (pjunc) positioned to direct expression of the Tpase. The –35 element is located in IRR while the –10 hexamer is located in IRL (Ton-Hoang et al., 1997). This is illustrated for IS911 in Figure 1. The assembly of pjunc has lead to the notion of a novel regulatory circuit where the indigenous promoter, pIRL, maintains low levels of Tpase synthesis sufficient to generate low levels of figure-of-eight junctions. Processing these forms into transposon circles assembles the pjunc promoter leading to extensive expression of IS-encoded proteins, efficient Tpase-catalysed single-strand cleavage at each end and insertion into a target site. Insertion then separates the two promoter elements and again renders Tpase expression dependent on pIRL. This novel feedback model is illustrated in Figure 1.
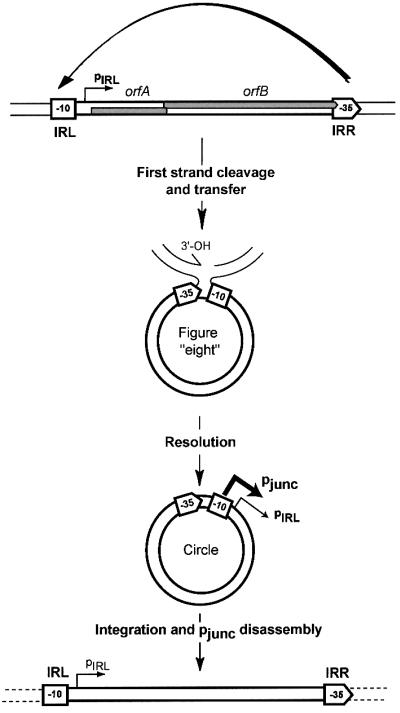
Fig. 1. IS911 circle formation. IS911 is shown in the top part of the figure together with its two overlapping reading frames, orfA and orfB (shaded boxes). The left and right terminal inverted repeats, IRL and IRR, are shown as a square and pointed box, respectively. The component –10 and –35 promoter elements of pjunc within these ends are also shown together with the weak pIRL promoter and the direction of transcription. In a first step, single-strand cleavage and transfer from one IR to the other is catalysed by OrfAB produced from the weak pIRL promoter. This is indicated at the top of the figure by the curved arrow. In this case, the right end (IRR) is shown attacking the left end (IRL). The resulting figure-of-eight form is drawn below and shows the free 3′-OH group generated on the flanking donor DNA sequence. In a second step, second-strand circularization occurs by an as yet undetermined mechanism involving host functions, but independent of transposon proteins (Turlan and Chandler, 2000). The resulting IRR–IRL junction carries suitably placed –35 and –10 hexamers, separated by the canonical 17 bp spacer and form a strong pjunc (bold arrow) promoter able to promote high levels of production of IS911 proteins. Integration of the circle results in disassembly of the promoter restoring low levels of expression from pIRL.
In the work described here we have addressed the role of pjunc in IS911 transposition. We have demonstrated that an IS911 derivative carrying a native pjunc junction promoter capable of driving Tpase expression in cis shows significantly higher transposition activity than a derivative in which pjunc has been inactivated. This difference is not observed if Tpase is provided in trans independently of pjunc. Since pjunc activity could be modulated by binding of IS911 transposition proteins, we have also analysed the effect of IS911-encoded proteins on expression from pjunc. We find that neither OrfA nor OrfAB is capable of repressing expression from pjunc. In contrast, an artificial OrfAB derivative, OrfAB[1–149], truncated at its C-terminal end and previously demonstrated to strongly bind isolated IRR and IRL sequences (Haren et al., 1998, 2000; Normand et al., 2001) is able to repress pjunc. Interestingly, while our results demonstrate that junction sequences of the IS3 family members IS911, IS2 and IS600 include a strong promoter, not all members of this family appear to share this property. Thus even though these elements form part of a highly homogenous family, the result underlines the diversity in strategies adopted by individual elements to fine-tune their transposition activities.
Results
Inactivation of IS911 pjunc by directed mutagenesis
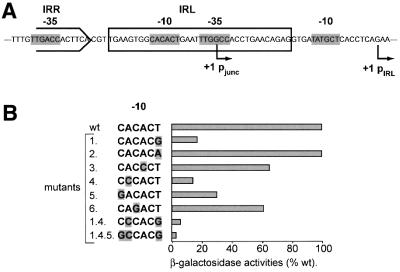
To determine the role of pjunc in regulating IS911 transposition, the putative –10 hexamer (CACACT) located in IRL (Figures 1 and 2A) was subjected to directed mutagenesis in order to reduce or eliminate pjunc activity. We chose to introduce transversions at the most conserved bases in the canonical sequence TATAAT. Five single point mutants were generated (Figure 2B) at positions that are not conserved in IRR. The mutant IRL ends were substituted for the wild-type end in an IS911 derivative carrying a lacZ reporter gene (pCN100; Normand et al., 2001). Promoter activity was measured after transposon circle formation in the presence of OrfAB (Materials and methods) using a standard β-galactosidase assay. Three of the mutations exhibited only a slight reduction in activity compared with the wild-type promoter (mutants 2, 3 and 6; see Figure 2), while three others showed significantly lower activities (17%, mutant 1, CACACG; 14%, mutant 4, CCCACT; and 30%, mutant 5, GACACT) compared with the wild-type pjunc. In an attempt to further reduce activity, all three mutations were introduced into a single –10 hexamer. The resulting triple mutant (IRL1.4.5) exhibited a residual activity of ∼3% compared with the wild-type pjunc promoter. This residual expression is presumably due to the presence of the weak pIRL promoter in IRL in these plasmid constructs (Figure 2A), which has previously been shown to have an activity of ∼2–3% that of pjunc (Ton-Hoang et al., 1997).
Fig. 2. The pIRL and pjunc promoters. (A) Representation of pIRL and pjunc promoters in the IS911 circle junction. Arrows show the position of transcription initiation sites. Potential –35 and –10 consensus sequences are boxed in grey. (B) Inactivation of pjunc by directed mutagenesis. This was performed on the –10 hexamer of the promoter and the effect was assessed directly on the lacZ reporter gene located downstream. Circles were formed in vivo and the effect of single mutations were tested by β-galactosidase activities. Mutant bases are shown with a grey background. Triple mutant 1.4.5 was selected for use in all following experiments. Note that pIRL represents a back ground of ∼2–3% compared with the active junction in the presence of the fully active pjunc, and probably represent the residual activity of mutant 1.4.5.
Effect of the mutations on the inherent recombination activity of IRL
Since the mutations introduced into pjunc lie within IRL, it is possible that they directly affect the inherent transposition activity in addition to their effect on promoter activity. Although no obvious difference in the global amounts of transposon circles produced by the different mutants could be detected (data not shown), it was possible that they exerted more subtle effects. To examine this, the possible direct effect on transposition activity was determined in detail for several steps in the transposition cycle. These included the efficiency of production of transposition intermediates (figure-of-eight and transposon circles), circle junction integration and the overall transposition activity when transposition enzymes were supplied in trans to the mutated ends.
Kinetics of figure-of-eight formation in vitro. To test their possible effect on the presumed first step of the transposition cycle, figure-of-eight formation of an artificial transposon carrying the mutant IRL1.4.5 and a correctly orientated wild-type IRR sequence was measured in vitro. In this system, which is dependent on OrfAB alone, the recombination reaction does not lead to transposon circles but is arrested after figure-of-eight formation. Plasmids pAPT56.2 (carrying wild-type IRR and IRL sequences) and pDV27 (carrying a wild-type IRR and IRL1.4.5) (Figure 3A) were used in a standard in vitro reaction (Materials and methods) in the presence of Mg2+. Samples were removed at intervals, digested with EcoRV, to convert the figure-of-eight into a χ form, and analysed by electrophoresis through a 0.8% agarose gel. This allows separation of the figure-of-eight from the parent plasmid (Materials and methods; Polard and Chandler, 1995b). Figure 3B shows a SyBr green-stained gel. Quantification of the bands by fluorimagery showed that both mutant and wild-type ends display indistinguishable activities in figure-of-eight formation (Figure 3C).
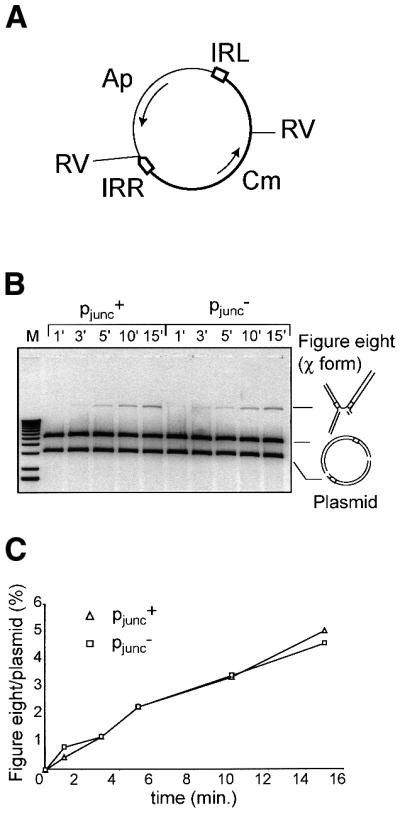
Fig. 3. Kinetics of figure-of-eight formation in vitro. (A) Plasmid substrates. Two plasmids, pAPT56.2 and pDV27 containing, respectively, IRLwt (pjunc+) and IRL1.4.5 (pjunc–) were used. The general structure of these plasmids is shown. The position of EcoRV restriction sites used in subsequent analysis is indicated (RV) as are the positions and direction of transcription of the chloramphenicol (Cm) and ampicillin (Ap) resistance genes. The transposable element is drawn in bold. The left and right inverted repeats (IRL, IRR) are indicated. (B) Results of incubation of plasmid DNA with OrfAB in vitro as a function of time. Intermediates were separated on a 0.8% agarose gel after EcoRV digestion. Incubation periods in minutes are indicated on the top of the gel. The χ form derived from figure-of-eight molecules and the two fragments derived from the parental plasmid are indicated. (C) Quantification of figure-of-eight levels by fluorimaging after staining with Sybr green I.
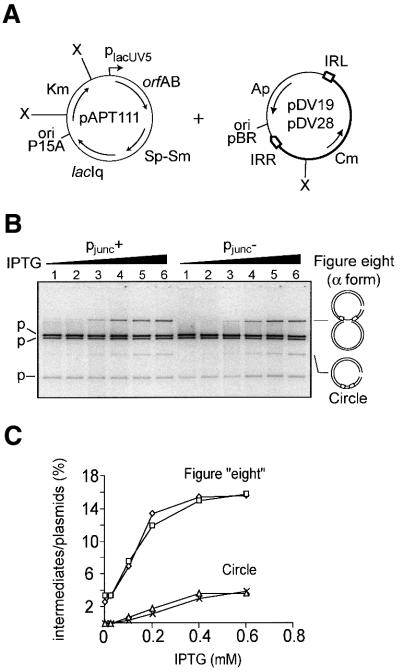
Figure-of-eight and transposon circle formation in vivo. Recombination activity of IRL1.4.5 was also examined in vivo by determining the level of transposition intermediates (figure-of-eight and transposon circle molecules) produced under standard growth conditions. In these experiments two IS911 derivatives carried by pDV19 and pDV28 were used (Figure 4A). These plasmids carry the terminal 52 bp of the wild-type (pDV19) or mutated left end (pDV28) together with a wild-type right end flanking an orfA–lacZ translational fusion. OrfAB was provided in trans under control of the placUV5 promoter from a second compatible plasmid, pAPT111. The plasmid content of cultures grown in increasing levels of IPTG (to induce OrfAB synthesis; Materials and methods) was analysed by agarose gel electrophoresis following digestion with XhoI to convert the figure-of-eight into an α form and to linearize the transposon circle (Figure 4A). No significant difference in levels of transposition intermediates could be detected between IS911 derivatives carrying the wild-type (pDV19) or triple mutant (pDV28) ends (Figure 4B and C).
Fig. 4. Figure-of-eight and circle formation in vivo as a function of increasing concentrations of IPTG. (A) Relevant features of pAPT111, which was used to produce OrfAB under control of the placUV5 promoter in trans and of the synthetic omegon-carrying plasmids pDV19 (pjunc+ with IRLwt) and pDV28 (pjunc– with IRL1.4.5) are shown. Also indicated are: the position of the XhoI sites used in the analysis (X); the positions and direction of transcription of the transposase, orfAB, the spectinomycin-streptomycin (Sp-Sm), kanamycin (Km), chloramphenicol (Cm) and amicillin (Ap) resistance genes and the lac repressor gene (lacIq) used to control placUV5 (which drives OrfAB expression); and the two compatible origins of replication (ori P15A and pBR322). The transposable element carried by pDV19 and pDV28 is drawn in bold. (B) DNA samples were prepared by the cleared lysate procedure and digested by XhoI to separate all intermediates and plasmids. Samples were separated by electrophoresis on a 0.8% TAE-agarose gel. Concentrations of IPTG added in lanes 1–6 were: 0, 0.025, 0.1, 0.2, 0.4 and 0.6 mM, respectively. (C) Detection and quantitation of figure-of-eight molecules and circles were performed by fluorimaging after Sybr green I staining.
Circle junction integration activity in vivo. Although the IRL1.4.5 mutant had no measurable effect on the production of figure-of-eight intermediates and transposon circles, it was possible that it affects the final step in the transposition cycle: integration of the circle.
In a first set of experiments, the effect of the mutant end on insertion activity of the junction was determined. Transposon donor plasmids were constructed by cloning the IRR–IRL junction into a suitable plasmid backbone. Purified transposon circles carrying the wild-type or mutant IRR–IRL junctions derived from pCN100 (Normand et al., 2001) and pDV11 respectively, were cloned into pAPT120 as described previously (Ton-Hoang et al., 1997; see Materials and methods). IS911 proteins were provided in trans under the control of a placUV5 promoter either from reading frames with a wild-type configuration in which OrfAB is produced by frameshifting (pAPT112, expressing the wild-type complement of IS911 proteins; Polard and Chandler, 1995b), or from a fused-frame derivative that produces OrfAB alone without frameshifting (pAPT111; Polard and Chandler, 1995b). The activity of the junctions was measured in a standard mating-out assay using pOX38Km as the conjugative target plasmid (Galas and Chandler, 1982; Chandler and Galas, 1983). The results are presented in Figure 5.
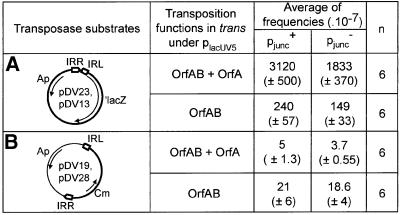
Fig. 5. Analysis of the effects of mutation on the integration step and overall transposition activity. (A) Integration frequencies using a preformed IRL–IRR junction. The general structure of the donor plasmids pDV13 (pjunc+) and pDV23 (pjunc–) is shown in the left hand column. (B) Overall transposition using a substrate with distant ends. The general structure of the donor plasmids pDV19 (pjunc+) and pDV28 (pjunc–) is shown in the left hand column. Symbols are those used in the legends to Figures 3 and 4. IS911 proteins were supplied in two different configurations: a wild-type configuration of the open reading frames in which the transposase was produced by translational frameshifting (OrfAB + OrfA) or with OrfAB alone. The values obtained represent the average of six distinct experiments (n = 6). Standard errors are shown in parentheses.
As found previously (Ton-Hoang et al., 1997), integration using preformed IRR–IRL junctions occurred at a relatively low frequency in the presence of OrfAB alone, but was stimulated by a factor of 12–13 when a wild-type complement of proteins was used (Figure 5A). This was observed both for junctions composed of the wild-type or mutant ends and reflects a requirement for OrfA observed also in an in vitro integration reaction (Ton-Hoang et al., 1998). In both cases, the mutant junction exhibited an integration efficiency of ∼0.6 that of the wild-type junction. Thus the combination of the three mutations, which reduces promoter activity to at least 30-fold compared with the wild-type junction, has a <2-fold effect on integration activity.
Overall transposition activity in vivo. In the experiments described above, the two transposon ends were retained close together in a preformed junction structure. This configuration of ends is highly active but represents only the final step in the transposition cycle. In the preceding steps, IRL and IRR must be assembled into a paired end synaptic complex (catalysed by OrfAB alone; Haren et al., 1998, 2000; Normand et al., 2001), strand cleavage and transfer must take place and the resulting figure-of-eight molecules must be resolved into transposon circles carrying the junction. To determine whether the triple mutation in IRL affects these initial steps in the transposition cycle, an artificial IS911 derivative, including IRL1.4.5 at one end and a wild-type IRR at the other, was constructed in a plasmid donor and its behaviour in the mating-out assay was measured. The derivative used was composed of a chloramphenicol resistance gene (CmR) introduced between the wild-type IRL and IRR ends (pDV19) or between the triple mutant IRL1.4.5 and a wild-type IRR (pDV28). Again, IS911 proteins were supplied in trans. The results are presented in Figure 5B. No significant effect of the IRL mutations on transposition frequency was observed with either OrfAB alone or with the wild-type configuration of proteins.
Moreover, these results confirm previous findings (Ton-Hoang et al., 1997) which showed that overall transposition frequencies are significantly lower than circle junction-mediated integration (∼10-fold with OrfAB and 5–6 × 102-fold with a wild-type complement of proteins). This presumably reflects the fact that synapsis between IRR and IRL, figure-of-eight formation or resolution into transposon circles are limiting steps in IS911 transposition relative to integration using the circle junction.
Effect of supplying IS911 proteins in cis and the role of pjunc
If pjunc is indeed involved in regulating IS911 transposition activity, it would be expected to exhibit its effects only when present in cis, i.e. where it would drive OrfA and OrfAB synthesis from the element whose transposition is being monitored. In order to determine whether this is the case, elements were constructed in which the natural configuration of orfA and orfAB, or a fused-frame configuration, was placed under control of pIRL. In these derivatives a CmR gene was also included between the ends as a selectable marker. It was cloned downstream of orfB in such a way as not to disrupt the orfB frame, which normally continues into IRR (Polard et al., 1994). Activity was measured using the mating-out assay.
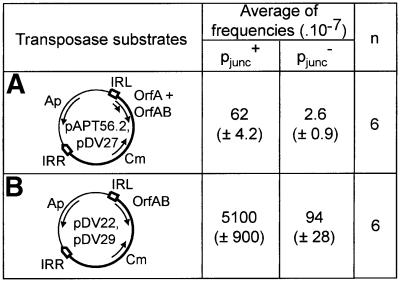
The results obtained with a wild-type complement of proteins supplied in cis from the weak pIRL promoter are presented in Figure 6A. Under these conditions, a mutant IRL1.4.5 end (pDV27) produced a marked effect on overall activity compared with the wild-type end (pAPT56.2). It resulted in an ∼20-fold reduction in transposition frequency. This is in contrast to the results obtained when a similar configuration of transposition proteins were supplied in trans (Figure 5B). Thus, pjunc clearly provides a regulatory function in its natural context when driving expression of transposition proteins in cis.
Fig. 6. Role of pjunc with IS911 proteins supplied in cis. Transposition frequencies were analysed by mating-out. (A) Analysis performed with the wild-type configuration (OrfA + OrfAB) or (B) OrfAB alone. The values obtained represent the average of six distinct experiments (n = 6). Symbols are those used in the legends to Figures 3 and 4. Standard errors are shown in parentheses.
To extend this analysis, the effect of pjunc-driven OrfAB alone rather than the wild-type protein complement was also measured. Previous attempts to introduce a mutation which fuses orfA and orfB into IS911 have been unsuccessful (Polard et al., 1992). Constitutive synthesis of OrfAB (i.e. without frameshifting) from its native promoter within the IS resulted in a high level of deletions and rearrangements in the vector plasmid. In order to facilitate construction of this type of IS911 derivative, we exploited the observation that OrfAB is inactive at 42°C (Haren et al., 1997). Bacterial hosts were therefore maintained at 42°C during the appropriate stages of plasmid construction and at all times preceding the mating-out assays. The results of mating-out assays are presented in Figure 6B. They clearly show an effect of pjunc on transposition. Comparison of transposition frequencies obtained with the wild-type IRL (pDV22) and IRL1.4.5 (pDV29) showed a 54-fold decrease in the case of the mutant. This con firms that the transient pjunc promoter plays an important stimulatory role in the transposition cycle, which operates only in the presence of the correct (circular) intermediate.
It should be underlined that the cis effect of pjunc is to amplify Tpase production. This is distinct from the cis preference observed for other ISs (see Mahillon and Chandler, 1998, for a review) as well as for IS911 (Polard et al., 1992), which implies that Tpase acts preferentially on the element from which it is synthesized.
Effects of IS911 proteins on pjunc activity
Since IS911 transposase must bind to the IRR–IRL junction, it is possible that the transient activity of pjunc could be further modulated by these proteins. To determine whether OrfA or OrfAB might modify pjunc activity, their effect on pjunc-driven expression of a lac reporter gene was determined. The reporter plasmid was similar to pDV23 described above, but the wild-type IRR–IRL junction was replaced with a junction in which both terminal 5′-CA-3′ dinucleotides were mutated to 5′-AG-3′. This modification inactivates the ends as donors in transposition and was introduced to prevent OrfAB-catalysed cleavage of the junction and therefore loss of pjunc (Ton-Hoang et al., 1997). These mutations, localized in the pjunc spacer, have no effect on the activity of the promoter as previously shown (Ton-Hoang et al., 1997) and confirmed here by β-galactosidase dosage (Figure 7). IS911 proteins were supplied in trans under the control of placUV5.
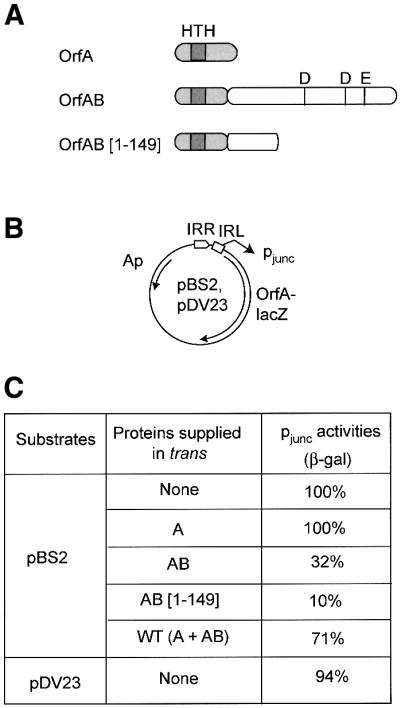
Fig. 7. Effect of IS911-encoded proteins on pjunc activity measured by β-galactosidase assays. (A) The proteins. Four protein configurations were tested: OrfA, OrfAB, a truncated derivative of OrfAB, OrfAB[1–149], deleted for the C-terminal catalytic domain and OrfA + OrfAB (wild-type configuration). The proteins are shown schematically illustrating the relative position of the catalytic DDE motif and of a helix–turn–helix motif potentially involved in DNA binding. The proteins were supplied in trans under control of placUV5. from plasmids pAPT156 (OrfA), pAPT111 (OrfAB), pLH114 (OrfAB[1–149]), and pAPT112 (OrfA + OrfAB). pAPT110 was used as a control without IS911 proteins. (B) The test plasmids. pBS2 carries mutations in the terminal dinucleotides at both ends in the IRR–IRL junction. pDV23 carries a wild-type IRR–IRL junction. Symbols are as described in Figures 3, 4 and 5. (C) β-galactosidase activities. The right hand column indicates β-galactosidase activities as a percentage relative to pjunc activity in the absence of any proteins in trans (100%). These results are the average of three independant experiments with a standard error of ∼15%. pDV23 (active junction) was tested in the absence of proteins supplied in trans to ensure that the terminal dinucleotide mutations (5′-CA to 5′-AG) introduced to inactivate integration activity of the junction did not affect pjunc activity.100% = 17 500 Miller units.
Three IS911-derived proteins were tested in these studies: the full-length transposase, OrfAB (pAPT111); a derivative deleted for the C-terminal catalytic domain, OrfAB(1–149) (pLH114); and OrfA (pAPT156). Previous in vitro studies have shown that OrfAB binds poorly to IRR and IRL but that OrfAB(1–149) binds strongly and generates a paired-end complex. OrfA showed no significant binding (Haren et al., 1998, 2000).
The results of these experiments are presented in Figure 7. OrfA alone had no detectable effect on pjunc efficiency. On the other hand, a clear inhibition of ∼90% of β-galactosidase levels was observed in the presence of OrfAB[1–149]. This is consistent with the observation that OrfA fails to bind either IS911 end in vitro, whereas OrfAB[1–149] binds strongly (Haren et al., 2000). An inhibition of 70% was observed in the presence of OrfAB. Although we cannot rule out the possibility that part of this may be due to trace levels of a truncated OrfAB derivative protein, OrfAB*, which is often detected in these preparations (Ton-Hoang et al., 1997; Haren et al., 1998), this result is compatible with the low but measurable binding activity of OrfAB. The wild-type configuration of IS911 proteins (OrfA + OrfAB; pAPT112) showed a small but reproducible inhibitory effect (30%). It should be noted that the level of OrfAB in this configuration is significantly lower than that of OrfA. These results raise the possibility that transposase exerts a feedback control on pjunc activity in addition to its catalytic activity.
Pjunc formation with other ISs?
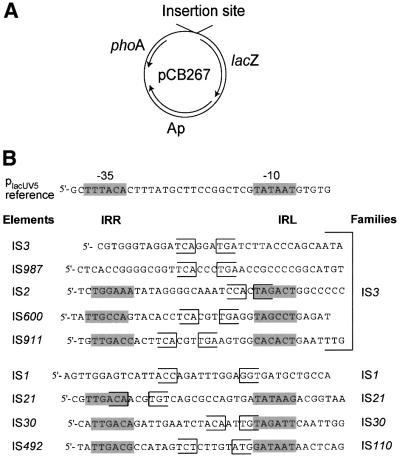
The presence of a junction promoter has been described in IS elements belonging to other families including IS21 (Reimmann et al., 1989) and IS30 (Dalrymple, 1987). To determine how general this phenomenon is, we surveyed the sequence of the ends of known ISs for the capacity to form such promoters. Although putative –35 and –10 elements may be present in IRR and IRL, respectively, two limitations of this analysis are: whether the IS naturally generates a structure with two abutted ends and, if so, whether the spacer length is compatible with an active promoter. Several of these are presented in Figure 8B. Some have been demonstrated to generate IRR–IRL junctions (IS1, IS2, IS3, IS21, IS30, IS911, IS492), whereas others have not (IS600, IS987).
Fig. 8. Analysis of junctions of other IS elements. (A) The promoter–probe vector pCB267. Junctions were constructed using synthetic oligonucleotides which were cloned into this plasmid upstream of a promoterless β-galactosidase gene (lacZ). The plasmid also carries a promoterless phoA gene which was not used in these studies. (B) Junctions of different ISs used. Boundaries of IRR and IRL are shown as brackets and putative –35 or –10 boxes are boxed in grey when close to the canonical hexamers. Five families were represented (IS3, IS1, IS21, IS30, IS110) and nine different junctions including IS911 were cloned in front of the lacZ gene. The promoter used as reference in these experiments was placUV5.
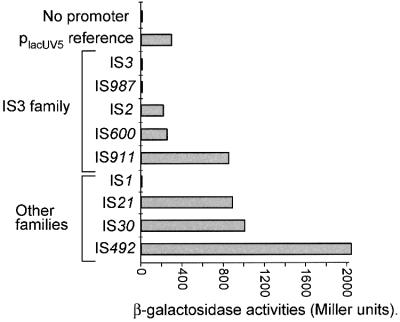
These junctions were synthesized and cloned into a standard lac reporter gene system (Figure 8A). The β–galactosidase activities specified by the resulting plasmids were measured and are presented in Figure 9. The results show that the IS3 family members, IS2 and IS600, exhibit a promoter activity comparable to placUV5, IS911 has an activity 3-fold greater than placUV5, while two other members of the family, IS3 and IS987, have no detectable promoter activity. Thus junction promoter formation does not appear to have been chosen as a control strategy for all members of this family.
Fig. 9. Promoter activities of nine different IS junctions. Results of measurements of β-galactosidase activities of the nine constructions shown in Figure 8 are given in Miller units and represent the average of three separate experiments with a standard error of ∼15%.
On the other hand, members of other families also show strong junction promoter activities. These include IS21 (IS21 family; 3-fold placUV5), IS30 (IS30 family; 3-fold placUV5) and IS492 (IS110 family; >6.5-fold placUV5). IS1 (IS1 family), on the other hand, does not appear to exhibit junction promoter activity.
Discussion
The experiments described here were designed to determine whether the pjunc promoter, assembled by circularization of IS911, plays a role in regulating transposition activity as proposed in the model presented in Figure 1. Although the pjunc –10 and –35 hexamers do not entirely fit the consensus Escherichia coli promoter elements, the promoter does include an extended –10 TG motif (Burr et al., 2000). This presumably contributes to the relatively high activity of this promoter (2.8-fold greater than that of placUV5; Figure 9). In order to significantly reduce pjunc activity, it was found to be necessary to introduce three mutations into the probable –10 hexamer located in IRL. The residual activity was found to be 3% that of wild-type pjunc, a value similar to that maintained by the resident pIRL (Ton-Hoang et al., 1998), also present in these constructions. To minimize any direct effects on transposition activity of the mutated end, these mutations were chosen to modify base pairs that are not conserved between the two IRs. No detectable difference in recombination activity of the triple mutant, IRL1.4.5 and its wild-type counterpart was observed either in vitro, in figure-of-eight formation (Figure 3), or in vivo in both figure-of-eight and transposon circle formation (Figure 4).
Using a more sensitive genetic assay to measure integration activity of the preformed IRR–IRL junction (Figure 5), the mutated IRL1.4.5 exhibited at most a 1.5-fold lower activity compared with the wild-type IRL. This small effect may indeed be due to slight differences in transposition activity or in the environment of the IR, but is minimal compared with the reduction of at least 97% in promoter activity.
Pjunc regulation of transposition
To determine whether pjunc plays a role in regulating overall transposition activity, the mutant IRL was placed at its natural position where it would be capable of driving transposase synthesis and its activity was compared with that obtained with a wild-type end. Two types of derivative were constructed. In one, pjunc would drive a wild-type reading frame configuration following transposon circularization, while in the other, the promoter would drive a fused-frame derivative producing OrfAB alone. The derivative carrying the fused-frame mutant had previously been shown to be particularly unstable (Polard et al., 1994). This problem was overcome here by exploiting the temperature-sensitive nature of OrfAB activity (Haren et al., 1997). The corresponding plasmid was constructed and maintained by imposing a high growth temperature (42°C) at all times.
The results of mating experiments using these donor plasmids (Figure 6) capable of forming a functional pjunc indicated that the activity of the element expressing the OrfAB fusion protein was significantly higher than that expressing the wild-type complement of proteins (pjunc+: 5 × 10–4 compared with 6 × 10–6). In both cases, the presence of the mutated pjunc, pjunc– reduced activity by a factor of 22 for the wild-type complement of proteins and 54-fold for the fused-frame derivative. This difference in overall transposition activity compares well with the ratio of 45 obtained by measurement of pjunc and pIRL activities in a standard β-galactosidase assay (Ton-Hoang et al., 1997). Suppression of pjunc activity therefore decreases the transposition frequency by a factor corresponding to the basal level of pIRL activity compared with pjunc. These results demonstrate an important role of pjunc in the regulation of IS911 transposition in its natural configuration.
Regulation of expression in cis and preferential cis activity
Pjunc-mediated expression in cis as observed here is distinct from the previously described preferential cis activity of various transposases (see Mahillon and Chandler, 1998). Although preferential cis activity has been concluded to occur in some circumstances for IS911, the experiments described here do not address this question directly. However, even in the absence of pjunc, OrfAB alone retained a 5-fold higher activity in cis (94 × 10–7; Figure 6B) under the control of the weak pIRL compared with its activity in trans under control of the strong placUV5 (18.6 × 10–7; Figure 5B). In the case of the wild-type protein complement, it is possible that the absence of a measurable effect results from a combination of inherently low activity together with the difference in promoter strength. A true comparison would necessitate the use of promoters with identical strengths in both situations.
Differential activities of OrfAB alone and the wild-type protein complement
The results presented in Figures 5 and 6 also reiterate previous observations that OrfAB alone does not have the same effect at different steps of the transposition process as does the wild-type complement of proteins. This has been described in situations where these proteins were supplied in trans (Ton-Hoang et al., 1997). Thus the frequency of integration of a preformed junction is 12- to 13-fold lower with OrfAB alone (Figure 5A) whereas, while globally lower, OrfAB alone sustains a 4-fold higher level of transposition in a situation requiring prior formation of the junction (Figure 5B). The stimulatory effect of OrfAB in this situation is more pronounced when it is provided in cis either with a functional pjunc (82-fold) or in the absence of pjunc (36-fold; Figure 6A and B).
Regulation of pjunc activity by transposition proteins
Although regulation of pjunc activity presumably occurs primarily by promoter assembly and disassembly, a more subtle modulation could occur by repression involving binding of IS911 proteins to the abutted IRs. The effect on pjunc expression of different IS911 proteins supplied in trans (Figure 7) clearly indicated that a truncated derivative of the OrfAB transposase (OrfAB[1–149]) represses pjunc activity. This is consistent with the DNA-binding properties of the protein (Haren et al., 2000; Normand et al., 2001). Full-length transposase, which appears to bind less well, exhibits a measurably lower but significant effect. Although part of this repression could be due to the presence of truncated derivatives of OrfAB often found following partial purification of the protein (Ton-Hoang et al., 1997; Haren et al., 1998), this observation raises the possibility that pjunc activity could be controlled by feedback repression in addition to assembly and disassembly.
The presence of pjunc in other elements
IS911 is not unique in carrying a junction promoter. These have been described for several elements belonging to different IS families including IS21 (Reimmann et al., 1989), IS30 (Dalrymple, 1987) and IS492 (Perkins-Balding et al., 1999). In a limited survey of IS3 family members, three, including IS911, exhibited a strong pjunc and two did not (Figures 8 and 9). It remains possible that the absence of promoter activity in certain cases is due to the choice of an incorrect spacer length between IRL and IRR or that the sequences of the ends themselves were incorrect. For two members of the IS3 family, IS987 and IS600, the formation of an IRR–IRL junction has yet to be formally demonstrated. Indeed, more extensive studies on IS3 in which the length of the spacer was varied also failed to reveal a pjunc activity (Sekine et al., 1999). Of those elements from families other than IS3 whose transposition activity includes the formation of an IRR–IRL junction, IS492 exhibits exceptionally high activity (see also Perkins-Balding et al., 1999), whereas pjunc activity of IS30 and IS21 is similar to that of IS911 (Figures 8 and 9) and IS1 (with an 8 bp spacer as found in IS1 transposon circles; Turlan and Chandler, 1995) shows no activity. Again, more extensive results using 6, 7, 8 and 9 bp spacers also failed to reveal an IS1 pjunc (Shiga et al., 1999). Although it has not been examined here, it is interesting to note that IS256 also generates transposon circles and tandem IS dimers (M.Prudhomme, C.Turlan, J.-P.Claverys and M.Chandler, submitted). Inspection of such junctions (which carry a 5 or 6 bp spacer) reveals the presence of a nearly canonical –35 element together with a canonical –10. These elements are separated in a majority of cases by the optimal 17 bp spacing and, like the IS911 pjunc, many include the conserved extended –10 TG motif (Burr et al., 2000).
Taken together, these results clearly demonstrate that IS911 has adopted a novel strategy for modulating its transposition activity via the assembly of a strong promoter intimately linked to the formation of a circular transposition intermediate and its disassembly upon integration of the circle. This assures that high levels of transposase expression occur only in the presence of the correct integration substrate, transposon circles. Once integrated, such circles lie ‘dormant’ and are activated only rarely to generate circles. Thus, the system is set up to ensure integration once these rare circles are formed and to limit transposition following their integration. Moreover, the strategy of activation of transposition by creation of a transitory promoter able to boost transposase expression appears to be widespread, since several other IS elements tested also share this property. It is not universal, however, even within a single IS family. These observations therefore underline the fact that although different elements may share an overall transposition mechanism, the strategies that have been chosen to fine-tune transposition activities are not necessarily identical. This further emphasizes the diversity of transposable elements.
Materials and methods
Bacterial strains and media
JS219 (MC1061, recA1, lacIq) has been described previously (Cam et al., 1988) and was used as a donor strain in mating-out experiments. The recipient strain for mating-out experiments was MC240 [XA103, F–, ara, del(lac pro), gyrA (nalR), metB, argEam, rpoB, thi, supF]. NM522 mutS [F′, lacIq Δ(lacZ)M15 proA+B+/supE thi Δ(lac-proAB) Δ(hsdMS-mcrB)5 (rK– mK– mcrBC–) (mutS::Tn10)] was used for directed mutagenesis as described in Deng and Nickoloff (1992). DH5α [F′, endA1, hsdR17 (rK–mK+) supE44, thi1, recA1, gyrA (NalR), relA1, Δ(lacI2YA-argF) U169, deoR (φ80dlacΔ(lacZ) M15] was used for β-galactosidase assays. Cultures were grown in Luria broth supplemented, where necessary, with ampicillin (Ap, 100 µg/ml), oxacillin or methicillin (Ox or Met, 0.8 g/ml), chloramphenicol (Cm, 30 µg/ml), kanamycin (Km, 25 µg/ml), nalidixic acid (Nal, 20 µg/ml), rifampicin (Rif, 50 µg/ml), streptomycin (Sm, 20 µg/ml), spectinomycin (Sp, 30 µg/ml) and tetracycline (Tc, 15 µg/ml).
Mutagenesis
Directed mutagenesis was based on the unique site elimination procedure developed by Deng and Nickoloff (1992) using the U.S.E. Mutagenesis Kit (Pharmacia Biotech). This approach utilizes two primers. One is used to introduce the desired mutation, the second eliminates a unique non-essential restriction site in the plasmid permitting selection of the mutant plasmid. A set of oligonucleotides (described in Figure 2B) was used to obtain simple, double or triple mutations in the –10 box of pjunc located in IRL. The substrate plasmid pCN99 was constructed by replacing the BamHI–PstI fragment from pBR322 by the BamHI–PstI from pAPT166 (Polard and Chandler, 1995b) bearing a wild-type IRL.
Plasmids
Transposase substrates. Plasmids carrying separated pairs of mutant IRL and wild-type IRR ends were obtained by directed mutagenesis of pCN99 as described above. The BamHI–PstI bearing a wild-type or mutant IRL1.4.5 was cloned into pCN100 as described in Normand et al. (2001) to generate pVK13 and pDV11, respectively.
Plasmids carrying an IRR–IRL junction, pDV23 [wild-type IRL (IRLwt)] and pDV13 (IRL1.4.5) were assembled in two steps. First, transposon circles were generated in vivo from pVK13 and pDV11 using pAPT111 as a source of transposase (Polard and Chandler, 1995b). After agarose gel separation and isolation, the circular transposon DNAs were linearized by XbaI and treated with DNA polymerase I Klenow fragment. This generates a linear molecule carrying the junctions upstream a OrfA–lacZ gene fusion. This fragment was then cloned into the gel-purified 3325 bp BamHI–HindIII fragment obtained from pAPT120 (Ton-Hoang et al., 1997), which had been treated with DNA polymerase I Klenow fragment and dephosphorylated with Shrimp phosphatase.
Plasmids carrying IS911 derivatives with the wild-type configuration of transposon genes in cis were constructed as follows. pDV27 (IRL1.4.5) was obtained by partial digestion of pAPT56.2 (Polard et al., 1994) with BalI, purification of the 4640 bp fragment containing the transposon orfs and a downstream CmR gene, and ligation with the purified 3586 bp fragment obtained after BalI restriction of pDV11. Clones in which the insert was in the required orientation were distinguished by PstI–HindIII digestion.
Plasmids carrying the fused-frame transposase OrfAB gene, pDV22 and pDV29, were obtained by substitution of the 987 bp SnaBI–PmeI fragment of pAPT111 (containing the orfAB fusion; Polard and Chandler, 1995b) into pAPT56.2 (IRLwt) or pDV27 (IRL1.4.5). All cultures were grown at 42°C to inactivate the transposase during successive transformations.
Derivatives of pAPT56.2 and pDV27 devoid of transposon genes but retaining the CmR gene were constructed by deletion between PmeI and SnaBI sites and recircularization of the resulting 7241 bp fragment. This generated pDV19 and pDV28 containing, respectively, IRLwt and IRL1.4.5 ends.
pBST2 carrying a mutation on the terminal 5′-CA-3′ of IRL and IRR in the junction to inactivate integration has been described previously (Ton-Hoang et al., 1997).
Plasmids supplying transposition functions in trans. pAPT110, pAPT111 and pAPT112, which express no IS911 proteins, OrfAB alone or OrfA + OrfAB (wild-type configuration), respectively, under control of the placUV5 promoter have been described previously (Polard and Chandler, 1995b). In addition, experiments designed to test the inhibition effect of IS911 proteins on the activity of pjunc, also employed pLH114 (OrfAB[1–149]; Haren et al., 1998) and pAPT156 (OrfA; Ton-Hoang et al., 1998). More detailed descriptions of the construction of these plasmids are available on request.
Alkaline and cleared lysates
Alkaline lysates were prepared as described previously (Birnboim and Doly, 1979). Cleared lysates were prepared as described previously (Clewell and Helinski, 1969) with the following modifications: the cleared supernatants were collected and incubated with proteinase K (0.5 mg/ml) and SDS (0.5%) for 1 h at 50°C. DNA was concentrated by isopropanol precipitation and resuspended in 100 µl of TE buffer and purified by using a Qiaquick kit as described by the manufacturer (Qiagen).
Mating-out assays
The transposition frequencies of the different IS911 synthetic elements was measured by standard mating-out assay (Galas and Chandler, 1982) using the conjugative plasmid target pOX38Km (Chandler and Galas, 1983). Strain JS219 containing pOX38Km was transformed with the different compatible derivatives as source of OrfAB and/or OrfA, synthetic elements carrying the chloramphenicol acetyltransferase gene (Cm) or plasmids carrying IRR–IRL junction with the β-lactamase gene (Ap) as phenotypic markers. Overnight cultures grown at 42°C to reduce OrfAB activity, which is temperature sensitive (Haren et al., 1997), from a single colony in LB supplemented with Ap, Km, Sp and Sm (and Cm when present), were diluted into LB without antibiotics at an OD540 of 0.05 at 37°C, with shaking. Growth was continued to an OD540 of 0.5 and followed by 1 h incubation without agitation to allow pili formation. Donor cells were then mixed at a 1:1 ratio with the recipient XA103 cells (NalR, RifR) also grown at 37°C. Suitable dilutions were plated on LB agar plates supplemented with Nal, Rif, Km (scoring for transfer) and Nal, Rif, Km, Cm (or Ap and Met instead of Cm for plasmids carrying junctions) for scoring transposition or integration, and incubated overnight at 37°C.
Transposase purification and in vitro activity assay
OrfAB was purification following expression from pAPT158 (Ton-Hoang et al., 1998) under the control of the placUV5 promoter and bacteriophage T7 φ 10 ribosome-binding site sequence from pAR30–39 (Studier and Moffatt, 1986).
Figure-of-eight formation as a function of time was performed at 30°C for 1–15 min in a final volume of 50 µl containing 2.5 µg of DNA substrates (pDV19 or pDV28) and 2 µg of transposase, in buffer B (20 mM HEPES pH 7.5, 5 mM DTT, 200 mM KCl, 10 mM MgCl2 and 10% glycerol). Aliquots (10 µl) were removed at intervals and the reaction was stopped with 20 mM EDTA on ice. The samples were then treated with 0.5 mg/ml (final concentration) of proteinase K 1 h at 50°C, and purified with Qiaquick purification minicolumns as described by the manufacturer (Qiagen).
Figure-of-eight and circle formation in vivo
Titration experiments as a function of isopropyl-β-D-thiogalactopyranoside (IPTG) concentration (used to induce OrfAB production) were performed to compare the level of figure-of-eight and transposon circle molecules with IRLwt or IRL1.4.5. Overnight cultures at 42°C (to inactive residual transposase) of MC1061 recA carrying the constructions to be tested were diluted in fresh LB medium at 42°C at an OD600 of 0.05. After ∼90 min of growth (OD600 = 0.3–0.4), cultures were transferred to 30°C and after 5 min incubation, to equilibrate the temperature, increasing concentrations of IPTG were added (0, 0.025, 0.05, 0.1, 0.2, 0.5 mM). Cultures were grown for a further 60 min at 30°C and stopped by centrifugation at 4°C. DNA was then extracted by the cleared lysate method, digested with XhoI restriction enzymes and loaded on a 0.8% agarose gel.
Cloning junctions of a selection of ISs
Both previously identified and potential IRR–IRL junctions from various IS elements were analysed by measurement for the presence and strength of a junction promoter. These include the IS3 family members, IS2 (L.A.Lewis and N.D.Grindley, personal communication), IS3 itself (Sekine et al., 1994), IS987 (Hermans et al., 1991), IS600 (Matsutani et al., 1987), IS911 (this work), and members of other IS families such as IS1 (Turlan and Chandler, 1995), IS21 (Reimmann et al., 1989; S.Schmid and D.Haas, personal communication), IS30 (Olasz et al., 1993) and IS492 (Perkins-Balding et al., 1999), whose transposition mechanisms have been postulated to proceed via formation of tandem or circles creating junctions of their ends. The junctions were synthesized with an extension of 5–11 bases on either side of the –35 and –10 consensus sequences so as to include the complete sequence of potential promoters. Double-stranded fragments were constructed by annealing complementary synthetic oligonucleotides from 30 to ∼55 nucleotides with protruding ends compatible with BamHI and XbaI restriction sites. About 50 pmol of single-stranded fragments were mixed with a ratio of 1/1.5 (in order to completely anneal one of them) in TEN buffer (10 mM Tris–HCl pH 8, 1 mM EDTA, 50 mM NaCl), boiled for 5 min, and annealed by slowly decreasing the temperature for 4–5 h. The double-stranded fragments were then 5′-phosphorylated with T4 kinase and cloned with T4 DNA ligase (Gibco-BRL) into plasmid pCB267 (Schneider and Beck, 1986), linearized by digestion with BamHI and XbaI. This promoter–probe vector carries a multiple cloning site in upstream of a lacZ marker without promoter. DNA of 10 clones were extracted by alkaline lysates and directly sequenced with Amersham kit. The placUV5 promoter was also cloned upstream of the lacZ gene and used as reference for β-galactosidase dosage experiments.
β-galactosidase assays
Cultures of DH5α cells transformed with the different constructions to be tested were grown overnight at 37°C in LB medium supplemented with ampicillin (100 µg/ml) and methicillin (1 mg/ml), and then diluted into fresh medium at an OD600 of 0.05. In experiments where IS911 proteins were supplied in trans, IPTG (1 mM final concentration) was added to induce production under control of the lacUV5 promoter. After ∼2 h of growth (OD600 = 0.3–0.4), β-galactosidase was measured according to the method of Miller (1972), except that cultures were performed in LB medium instead of VB supplemented with glucose and casamino acids, BSA (1 mg/ml) was included in the reaction, and lysis was accomplished by SDS-chloroform treatment. The lysates were centrifuged before measurement at OD420.
Acknowledgments
Acknowledgements
We thank members of the Mobile Genetic Elements group both past (M.Betermier, O.Fayet, L.Haren, P.Polard and M.-F.Prère) and present (R.Alazard, C.Loot, P.Rousseau, B.Ton-Hoang and C.Turlan) for suggestions during the course of this work, and for critically reading the manuscript. This work benefited from grants from the CNRS (UPR9007/UMR5100), the Association pour la Recherche contre le Cancer, the Region Midi-Pyrenées, the ‘Programme Physique et Chimie du vivant’ (CNRS) and the ‘Programme microbiologie’ (MENRST). This paper is dedicated to C.H.Dalton.
References
- Berger B. and Haas,D. (2001) Transposase and cointegrase: specialized transposition proteins of the bacterial insertion sequence IS21 and related elements. Cell Mol. Life Sci., 58, 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim H.C. and Doly,J. (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res., 7, 1513–1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr T., Mitchell,J., Kolb,A., Minchin,S. and Busby,S. (2000) DNA sequence elements located immediately upstream of the –10 hexamer in Escherichia coli promoters: a systematic study. Nucleic Acids Res., 28, 1864–1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam K., Bejar,S., Gil,D. and Bouche,J.P. (1988) Identification and sequence of gene dicB: translation of the division inhibitor from an in-phase internal start. Nucleic Acids Res., 16, 6327–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler M. and Galas,D.J. (1983) Cointegrate formation mediated by Tn9. II. Activity of IS1 is modulated by external DNA sequences. J. Mol. Biol., 170, 61–91. [DOI] [PubMed] [Google Scholar]
- Clewell D.B. and Helinski,D.R. (1969) Supercoiled circular DNA–protein complex in Escherichia coli: purification and induced conversion to an open circular DNA form. Proc. Natl Acad. Sci. USA, 62, 1159–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalrymple B. (1987) Novel rearrangements of IS30 carrying plasmids leading to the reactivation of gene expression. Mol. Gen .Genet., 207, 413–420. [DOI] [PubMed] [Google Scholar]
- Deng W.P. and Nickoloff,J.A. (1992) Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal. Biochem., 200, 81–88. [DOI] [PubMed] [Google Scholar]
- Fayet O., Ramond,P., Polard,P., Prere,M.F. and Chandler,M. (1990) Functional similarities between retroviruses and the IS3 family of bacterial insertion sequences? Mol. Microbiol., 4, 1771–1777. [DOI] [PubMed] [Google Scholar]
- Galas D.J. and Chandler,M. (1982) Structure and stability of Tn9-mediated cointegrates. Evidence for two pathways of transposition. J. Mol. Biol., 154, 245–272. [DOI] [PubMed] [Google Scholar]
- Haren L., Betermier,M., Polard,P. and Chandler,M. (1997) IS911-mediated intramolecular transposition is naturally temperature sensitive. Mol. Microbiol., 25, 531–540. [DOI] [PubMed] [Google Scholar]
- Haren L., Normand,C., Polard,P., Alazard,R. and Chandler,M. (2000) IS911 transposition is regulated by protein–protein interactions via a leucine zipper motif. J. Mol. Biol., 296, 757–768. [DOI] [PubMed] [Google Scholar]
- Haren L., Polard,P., Ton-Hoang,B. and Chandler,M. (1998) Multiple oligomerisation domains in the IS911 transposase: a leucine zipper motif is essential for activity. J. Mol. Biol., 283, 29–41. [DOI] [PubMed] [Google Scholar]
- Haren L., Ton-Hoang,B. and Chandler,M. (1999) Integrating DNA: transposases and retroviral integrases. Annu. Rev. Microbiol., 53, 245–281. [DOI] [PubMed] [Google Scholar]
- Hermans P.W., van Soolingen,D., Bik,E.M., de Haas,P.E., Dale,J.W. and van Embden,J.D. (1991) Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect. Immun., 59, 2695–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallastu A., Horak,R. and Kivisaar,M. (1998) Identification and characterization of IS1411, a new insertion sequence which causes transcriptional activation of the phenol degradation genes in Pseudomonas putida.J. Bacteriol., 180, 5306–5312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J., Jones,K.S., Katz,R.A., Mack,J.P. and Skalka,A.M. (1992) Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol., 12, 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L.A. and Grindley,N.D. (1997) Two abundant intramolecular transposition products, resulting from reactions initiated at a single end, suggest that IS2 transposes by an unconventional pathway. Mol. Microbiol., 25, 517–529. [DOI] [PubMed] [Google Scholar]
- Lyon B.R., Gillespie,M.T. and Skurray,R.A. (1987) Detection and characterization of IS256, an insertion sequence in Staphylococcus aureus. J. Gen. Microbiol., 133, 3031–3038. [DOI] [PubMed] [Google Scholar]
- Mahillon J. and Chandler,M. (1998) Insertion sequences. J. Bacteriol., 180, 5306–5312.9765560 [Google Scholar]
- Matsutani S., Ohtsubo,H., Maeda,Y. and Ohtsubo,E. (1987) Isolation and characterization of IS elements repeated in the bacterial chromosome. J. Mol. Biol., 196, 445–455. [DOI] [PubMed] [Google Scholar]
- Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbour, NY.
- Mizuuchi K. (1992) Transpositional recombination: mechanistic insights from studies of mu and other elements. Annu. Rev. Biochem., 61, 1011–1051. [DOI] [PubMed] [Google Scholar]
- Normand C., Duval-Valentin,G., Haren,L. and Chandler,M. (2001) The terminal inverted repeats of IS911: requirements for synaptic complex assembly and activity. J. Mol. Biol., 308, 853–871. [DOI] [PubMed] [Google Scholar]
- Olasz F., Stalder,R. and Arber,W. (1993) Formation of the tandem repeat (IS30)2 and its role in IS30-mediated transpositional DNA rearrangements. Mol. Gen. Genet., 239, 177–187. [DOI] [PubMed] [Google Scholar]
- Perkins-Balding D., Duval-Valentin,G. and Glasgow,A.C. (1999) Excision of IS492 requires flanking target sequences and results in circle formation in Pseudoalteromonas atlantica. J. Bacteriol., 181, 4937–4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P. and Chandler,M. (1995a) Bacterial transposases and retroviral integrases. Mol. Microbiol., 15, 13–23. [DOI] [PubMed] [Google Scholar]
- Polard P. and Chandler,M. (1995b) An in vivo transposase-catalyzed single-stranded DNA circularization reaction. Genes Dev., 9, 2846–2858. [DOI] [PubMed] [Google Scholar]
- Polard P., Prere,M.F., Fayet,O. and Chandler,M. (1992) Transposase-induced excision and circularization of the bacterial insertion sequence IS911. EMBO J., 11, 5079–5090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polard P., Seroude,L., Fayet,O., Prere,M.F. and Chandler,M. (1994) One-ended insertion of IS911. J. Bacteriol., 176, 1192–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimmann C., Moore,R., Little,S., Savioz,A., Willetts,N.S. and Haas,D. (1989) Genetic structure, function and regulation of the transposable element IS21. Mol. Gen. Genet., 215, 416–424. [DOI] [PubMed] [Google Scholar]
- Rezsohazy R., Hallet,B., Delcour,J. and Mahillon,J. (1993) The IS4 family of insertion sequences: evidence for a conserved transposase motif. Mol. Microbiol., 9, 1283–1295. [DOI] [PubMed] [Google Scholar]
- Rowland S.J. and Dyke,K.G. (1990) Tn552, a novel transposable element from Staphylococcus aureus. Mol. Microbiol., 4, 961–975. [DOI] [PubMed] [Google Scholar]
- Schneider K. and Beck,C.F. (1986) Promoter-probe vectors for the analysis of divergently arranged promoters. Gene, 42, 37–48. [DOI] [PubMed] [Google Scholar]
- Sekine Y., Aihara,K. and Ohtsubo,E. (1999) Linearization and transposition of circular molecules of insertion sequence IS3. J. Mol. Biol., 294, 21–34. [DOI] [PubMed] [Google Scholar]
- Sekine Y., Eisaki,N. and Ohtsubo,E. (1994) Translational control in production of transposase and in transposition of insertion sequence IS3. J. Mol. Biol., 235, 1406–1420. [DOI] [PubMed] [Google Scholar]
- Shiga Y., Sekine,Y. and Ohtsubo,E. (1999) Transposition of IS1 circles. Genes Cells, 4, 551–561. [DOI] [PubMed] [Google Scholar]
- Studier F.W. and Moffatt,B.A. (1986) Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol., 189, 113–130. [DOI] [PubMed] [Google Scholar]
- Ton-Hoang B., Betermier,M., Polard,P. and Chandler,M. (1997) Assembly of a strong promoter following IS911 circularization and the role of circles in transposition. EMBO J., 16, 3357–3371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton-Hoang B., Polard,P. and Chandler,M. (1998) Efficient transposition of IS911 circles in vitro. EMBO J., 17, 1169–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlan C. and Chandler,M. (1995) IS1-mediated intramolecular rearrangements: formation of excised transposon circles and replicative deletions. EMBO J., 14, 5410–5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turlan C. and Chandler,M. (2000) Playing second fiddle: second-strand processing and liberation of transposable elements from donor DNA. Trends Microbiol., 8, 268–274. [DOI] [PubMed] [Google Scholar]
- Welz C. (1993) Functionelle analyse des Bakteriellen Insertionelements IS150. PhD thesis. Albert-Ludwigs-Universität Freiburg, Freiburg, Germany.