Abstract
Macroautophagy is a bulk degradation process induced by starvation in eukaryotic cells. In yeast, 15 Apg proteins coordinate the formation of autophagosomes. Several key reactions performed by these proteins have been described, but a comprehensive understanding of the overall network is still lacking. Based on Apg protein localization, we have identified a novel structure that functions in autophagosome formation. This pre-autophagosomal structure, containing at least five Apg proteins, i.e. Apg1p, Apg2p, Apg5p, Aut7p/Apg8p and Apg16p, is localized in the vicinity of the vacuole. Analysis of apg mutants revealed that the formation of both a phosphatidylethanolamine-conjugated Aut7p and an Apg12p– Apg5p conjugate is essential for the localization of Aut7p to the pre-autophagosomal structure. Vps30p/Apg6p and Apg14p, components of an autophagy- specific phosphatidylinositol 3-kinase complex, Apg9p and Apg16p are all required for the localization of Apg5p and Aut7p to the structure. The Apg1p protein kinase complex functions in the late stage of autophagosome formation. Here, we present the classification of Apg proteins into three groups that reflect each step of autophagosome formation.
Keywords: APG/autophagosome/autophagy/GFP/membrane dynamics
Introduction
During macroautophagy in yeast, a portion of the cytoplasm is non-selectively sequestered into double-membrane structures (autophagosomes) and delivered to the vacuole for degradation. Electron microscopic analysis revealed that autophagosomes fuse with the vacuolar membrane to release the inner membrane structure into the lumen (Baba et al., 1994). Autophagosomes are generated when a cup-shaped membrane sac (isolation membrane) extends to enwrap and seal cytoplasmic materials. This elongation process, accompanied by the assembly of membranous structures (Kirisako et al., 1999), is currently not well characterized. In mammalian cells, autophagosomes are generated in a similar manner. In rat hepatocytes, osmiophilic membranes designated as phagophores were observed under an electron microscope (Seglen, 1987; Fengsrud et al., 1995, 2000). In mouse embryonic stem cells, similar isolation membranes were identified by electron microscopy as precursors of autophagosomes (Mizushima et al., 2001). To understand the molecular mechanism controlling autophagy, we isolated 15 mutants defective in autophagy (apg) in yeast (Tsukada and Ohsumi, 1993) with disorders of bulk protein degradation, sporulation and the maintenance of viability under starvation conditions (Klionsky and Ohsumi, 1999). A vacuolar hydrolase aminopeptidase I (API) was constitutively transported to the vacuole via small double-membrane vesicles (Scott et al., 1997). This cytosol-to-vacuole targeting (Cvt) pathway runs under growing conditions. Extensive overlaps have been found in genes involved in the APG, AUT (autophagy) and CVT pathways (Harding et al., 1996). All apg mutants except apg17 are known to be defective in the Cvt pathway.
Recent characterization of the Apg proteins has identified two ubiquitin-like systems utilizing approximately half of the Apg proteins. Apg12p, a ubiquitin-like protein, is covalently attached to Apg5p to form an Apg12p– Apg5p conjugate controlled by the serial action of Apg7p and Apg10p (Mizushima et al., 1998). Apg7p, a member of the E1-like activating enzymes, forms a thioester linkage with Apg12p (Kim et al., 1999; Tanida et al., 1999; Yuan et al., 1999). Apg12p subsequently forms a thioester intermediate with Apg10p, an E2 conjugating enzyme (Shintani et al., 1999). Apg12p is then linked to Lys149 of Apg5p through an isopeptide bond with the C-terminal glycine. Apg16p then interacts through its C-terminal coiled-coil region to form homo-oligomers following direct interaction with the Apg12p–Apg5p conjugate (Mizushima et al., 1999). In mammalian cells, a homologue of the Apg12p–Apg5p conjugate transiently associates with the membranes of precursor autophagosomes (Mizushima et al., 2001).
Aut7p/Apg8p, a second ubiquitin-like protein involved in autophagy, is conjugated to phosphatidylethanolamine (PE; Aut7p–PE) by the serial action of three Apg proteins, Aut2p/Apg4p, Apg7p and Aut1p/Apg3p (Ichimura et al., 2000; Kirisako et al., 2000). The C-terminal Arg117 of Aut7p is removed through the action of Aut2p, a cysteine protease (Kirisako et al., 2000), to expose Gly116. Follow ing activation by Apg7p (E1), the processed Aut7p is transferred to Aut1p (E2). Aut7p is then covalently linked to PE. Aut2p further cleaves Aut7p–PE, releasing Aut7p, an essential step in the normal progression of autophagy (Kirisako et al., 2000). Aut7p is the first protein to localize to autophagosomes and intermediate structures (Kirisako et al., 1999; Huang et al., 2000); this characteristic of Aut7p allows us to use Aut7p to monitor autophagosome formation. LC3, the mammalian homologue of Aut7p, is also localized on the membranes of autophagosomal and precursor membranes (Kabeya et al., 2000; Mizushima et al., 2001).
Additional Apg proteins, not known to participate in ubiquitin-like systems, are also required for autophagy. Vps30p/Apg6p, in addition to a role in autophagy, functions in vacuolar protein sorting (Kametaka et al., 1998). Vps30p forms a specific phosphatidylinositol 3-kinase (PI3-kinase) complex required for autophagy, consisting of Vps30p, Apg14p, Vps34p and Vps15p. These data suggest that the dynamic membrane events mediated by the PI3-kinase complex are necessary for autophagy (Kihara et al., 2001a). A class of PI3-kinase and beclin, a human homologue of Vps30p, are also essential for autophagy in human cells (Liang et al., 1999; Petiot et al., 2000). Beclin physically interacts with the PI3-kinase complex on the trans-Golgi network (Kihara et al., 2001b).
Tor kinase negatively regulates the induction of autophagy; this kinase activity is inhibited by rapamycin (Noda and Ohsumi, 1998). As the inactivation of Tor activity induces autophagy, treatment with rapamycin mimics the starvation response. The inactivation of Tor activity causes a rapid dephospholylation of Apg13p (Abeliovich et al., 2000; Kamada et al., 2000). Apg13p and Apg17p associate with Apg1p protein kinase (Matsuura et al., 1997) to form the Apg1p protein kinase complex, an essential component of autophagy. Binding of dephosphorylated Apg13p to this complex enhances the kinase activity of Apg1p (Kamada et al., 2000).
The characteristics of each Apg protein are gradually being uncovered; their functional interrelationship, however, is poorly understood. This is due to a lack of markers allowing the identification of intermediate steps occurring before or during autophagosome formation. To obtain further insights into the functional relationship between the Apg proteins, we observed the behaviour of Apg proteins fused to fluorescent proteins utilizing a sensitive imaging system. We examined the dynamics of the Apg proteins involved in the two ubiquitin-like systems, Apg5p and Aut7p. Based on their modification and localization in apg mutants, we have defined a pre-autophagosomal structure containing Apg1p, Apg2p, Apg5p, Aut7p and Apg16p. This structure, involved in the production of autophagosomes, provides a clue for investigating the steps of autophagosome formation.
Results
A punctate structure on which Apg1p, Apg5p, Aut7p and Apg16p are colocalized
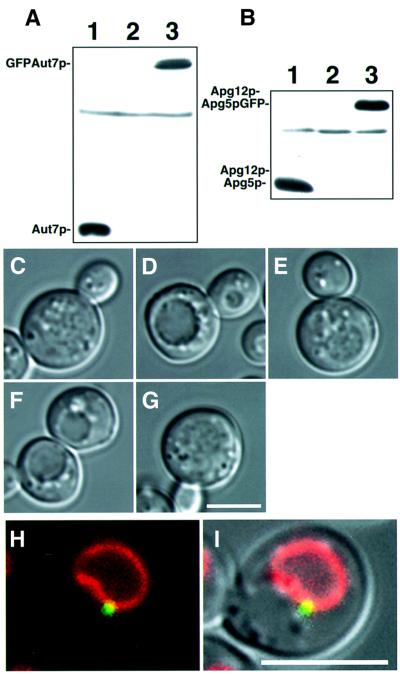
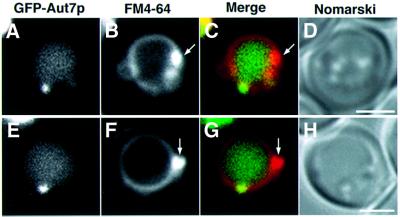
Apg proteins are involved in several essential reactions in autophagy, such as ubiquitin-like systems or phosphorylation reactions. The interrelationship between these reactions in autophagosome formation, however, remains unknown. It is critical to know the intracellular localization. Several Apg proteins, e.g. Apg5p (George et al., 2000), Aut7p/Apg8p (Kim et al., 2001) and Apg9p (Noda et al., 2000), are localized to perivacuolar punctate structures. To analyse the localization of Apg proteins to these punctate structures, we visualized green fluorescent protein (GFP)-fused Apg proteins using a sensitive microscope system. GFP fusions were expressed from centromeric plasmids under the control of natural promoters. In Δaut7 cells expressing physiological levels of a GFP–Aut7p fusion protein (Figure 1A), GFP–Aut7p complemented the defect of autophagy (Figure 1C–E). These molecules were localized to punctate structures, proximal to the vacuole (Figure 1H and I). Apg5p–GFP retained the ability to conjugate with Apg12p (Figure 1B); when introduced into deficient cells, Apg5p–GFP restored proper autophagic function (Figure 1C, F and G). The additional chimeric proteins used in this study were also functional in the autophagic process (data not shown).
Fig. 1. Functional GFP–Aut7p and Apg5p–GFP are expressed from natural promoters at physiological levels. (A) The expression level of GFP–Aut7p under growing conditions. Cell lysates were prepared as described in Materials and methods. (1) Wild-type (KA311A), (2) Δaut7 (YYK218) and (3) Δaut7 (YYK218) expressing GFP–Aut7p. (B) The Apg12p–Apg5p conjugate was produced normally in Δapg5 cells expressing Apg5p–GFP. Cell lysates were prepared as described in Materials and methods. (1) Wild-type (KA311A), (2) Δapg5 (GYS59) and (3) Δapg5 (GYS59) expressing Apg5p–GFP. (C–G) The accumulation of autophagic bodies was examined under a light microscope. Cells were incubated for 6 h in 0.17% yeast nitrogen base w/o amino acid and ammonium sulfate containing 1 mM PMSF. Nomarski images of (C) wild-type (KA311A), (D) Δaut7 (YYK218), (E) Δaut7 (YYK218) expressing GFP–Aut7p, (F) Δapg5 (GYS59) and (G) Δapg5 (GYS59) expressing Apg5p–GFP. (H–I) GFP–Aut7p visualized in Δaut7 cells (KVY5) under growing conditions. A punctate structure containing GFP–Aut7p is detected close to the vacuole, identified by FM4-64 labelling. (H) Fluorescence of GFP–Aut7p (green) and a FM4-64-labelled vacuole (red). (I) The Nomarski image is overlaid with GFP–Aut7p and FM4-64 fluorescence. The punctate structures, close to the vacuole, were detected in 43 of 206 cells (21%). Bar: 5 µm.
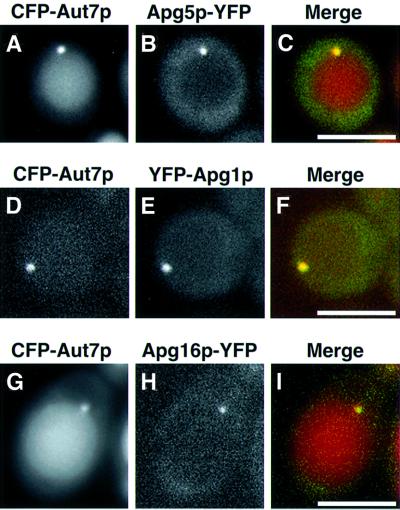
Next, we fused cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) to several different Apg proteins and examined their colocalization. Fluorescent Aut7p served as a marker of the punctate structures. Apg1p, Apg5p and Apg16p colocalized with Aut7p on a single punctate structure located close to the vacuole (Figure 2). After the addition of rapamycin, CFP–Aut7p was delivered to vacuoles in a time-dependent manner (Figure 2A, D and G). YFP–Apg1p was delivered to vacuoles after lengthy treatment with rapamycin (data not shown), whereas Apg5p–YFP and Apg16p–YFP were never transported to vacuoles (Figure 2B and H). Apg2p colocalizes with Aut7p on a perivacuolar punctate structure (Shintani et al., 2001). Consequently, the punctate structure is shown to contain Apg1p, Apg2p, Apg5p, Aut7p and Apg16p. Apg12p is likely to be colocalized with Apg5p and Apg16p as a component of the AApg12p– Apg5p–Apg16p complex. As Apg13p and Apg17p interact physically with Apg1p (Kamada et al., 2000), it is probable that these molecules also reside on the punctate structure. This is the first report demonstrating the colocalization of multiple Apg proteins.
Fig. 2. Colocalization of Apg1p, Apg5p, Apg16p and Aut7p on a punctate structure close to the vacuole. (A–C) Δapg5 cells (YNM119) expressing Apg5p–YFP and CFP–Aut7p were treated with rapamycin for 3 h: (A) Apg5p–YFP, (B) CFP–Aut7p, (C) merged image of Apg5p–YFP (green) and CFP–Aut7p (red). (D–F) Δapg1 cells (NNY20) expressing CFP–Aut7p and YFP–Apg1p were treated with rapamycin for 30 min: (D) CFP–Aut7p, (E) YFP–Apg1p, (F) merged image of CFP–Aut7p (red) and YFP–Apg1p (green). (G–I) Δapg16 cells (KVY117) expressing CFP–Aut7p and YFP–Apg16p were treated with rapamycin for 6 h: (G) CFP–Aut7p, (H) YFP–Apg16p, (I) merged image of CFP–Aut7p (red) and YFP–Apg16p (green). Bar: 5 µm.
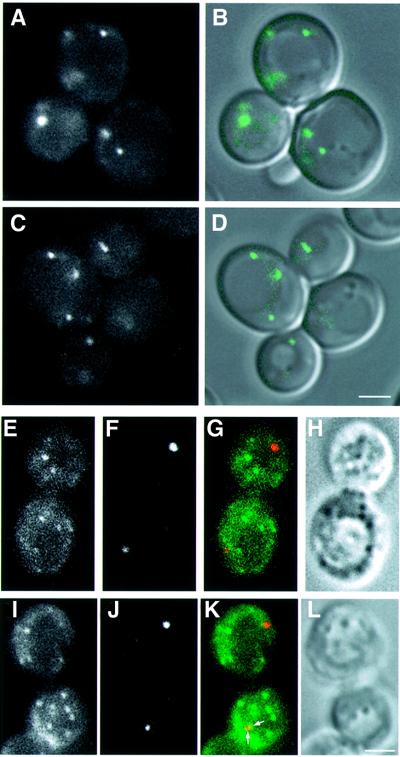
Apg9p is a putative membrane protein and is not cofractionated with typical endomembrane marker proteins, autophagosomes or Cvt vesicles (Noda et al., 2000). Apg9p–GFP exhibited an uneven, cytosolic distribution in addition to its localization on several punctate structures (Noda et al., 2000; Figure 3A and B). This pattern was not altered in other apg mutants (Figure 3C and D; data not shown). To examine the use of this protein as a punctate structure marker, we compared the localization of Apg9p–GFP and Aut7p by immunofluorescence microscopy. The punctate structures labelled with Aut7p seldom colocalized with those labelled by Apg9p–GFP (Figure 3E–L), although the two structures were occasionally in close proximity (arrows in Figure 3K). This pattern of Apg9p indicates that it is not a suitable marker for the punctate structure.
Fig. 3. Localization of Aut7p and Apg9p–GFP. (A and B) Apg9p–GFP was visualized in Δapg9 cells (CTD1) under growing conditions: (A) Apg9p–GFP, (B) the Nomarski image is overlaid with the fluorescence of Apg9p–GFP. (C and D) Apg9p–GFP in Δapg9Δapg14 cells (GYS29) was identified under growing conditions: (C) Apg9p–GFP, (D) the Nomarski image is overlaid with the fluorescence of Apg9p–GFP. (E–L) Δapg9 cells (CTD1) expressing Apg9p–GFP were treated with rapamycin for 2 h. Immunofluorescence microscopy was performed as described in Materials and methods. (E and I) Apg9p–GFP, (F and J) Aut7p, (G and K) merged images of Apg9p–GFP (green) and Aut7p (red) and (H and L) Nomarski images. Apg9p and Aut7p were occasionally found in close proximity (arrows in K). Bar: 2 µm.
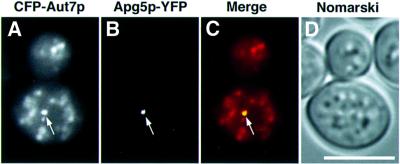
Electron microscopic analysis established that autophagosomes accumulate in the Δypt7 mutant (Kirisako et al., 1999). We therefore examined the localization of Apg5p–YFP and CFP–Aut7p in Δypt7 cells. Following rapamycin treatment of these cells, autophagosomes labelled with CFP–Aut7p appeared as dim dots in the cytosol (Figure 4A), whereas Apg5p–YFP was detected as a punctate pattern (arrow in Figure 4B). Every punctate structure labelled with Apg5p–YFP was associated with CFP–Aut7p (arrow in Figure 4C). CFP–Aut7p staining of the structure was of a greater intensity than the other CFP–Aut7p dots (arrow in Figure 4A). These observations suggest that this punctate structure containing several colocalized Apg proteins is not an autophagosome.
Fig. 4. Localization of Aut7p and Apg5p in Δypt7 cells. Δypt7Δapg5 cells (YAK3) expressing CFP–Aut7p and Apg5p–YFP were treated with rapamycin for 5 h. (A) Autophagosomes stained with CFP–Aut7p; (B) Apg5p–YFP; (C) merged image of CFP–Aut7p (red) and Apg5p–YFP (green); (D) Nomarski image: CFP–Aut7p and Apg5p–YFP colocalized on a punctate structure (arrows). Bar: 5 µm.
To investigate the involvement of the endocytic pathway in punctate structure formation, we examined the class E compartment, an exaggerated prevacuolar compartment in class E vacuolar protein sorting (vps) mutants (Raymond et al., 1992). The class E compartment is labelled with FM4-64, a lipophilic stylyl dye, as an intensely stained membrane-enclosed structure (Vida and Emr, 1995) and morphologically similar to the punctate structure labelled with Aut7p. We observed the localization of GFP–Aut7p in a class E vps mutant, Δvps4 (Raymond et al., 1992). The GFP–Aut7p punctate structure did not colocalize with the class E compartment in this mutant (Figure 5). These data suggest that the punctate structure is not organized through the endocytic pathway. Therefore, this punctate structure is a novel structure localized proximal to the vacuole.
Fig. 5. Localization of Aut7p and the class E compartment. The class E compartment of Δvps4 cells (MBY3) expressing GFP–Aut7p was labelled with FM4-64 as described in Materials and methods, following treatment with rapamycin for 2 h. (A and E) GFP–Aut7p. (B and F) The class E compartments stained with FM4-64 (arrows). (C and G) Merged images of GFP–Aut7p (green) and FM4-64 (red). (D and H) Nomarski images. Bar: 2 µm.
Genetic analysis of APG genes required for the organization of the punctate structure
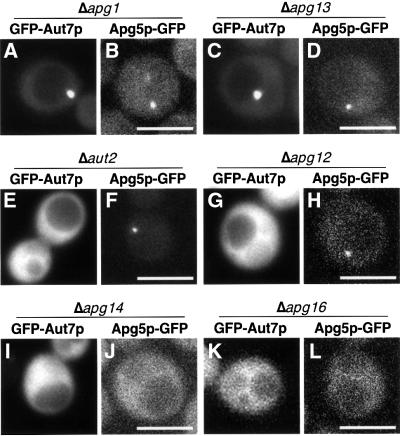
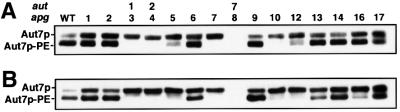
We have found a novel punctate structure in which at least five Apg proteins are concentrated. We examined the organization within this punctate structure by studying the localization of Apg5p–GFP and GFP–Aut7p in apg mutants (Figure 6; summarized in Table I). We also assessed the quantities of PE-conjugated Aut7p (Ichimura et al., 2000; Kirisako et al., 2000) in each apg mutant by SDS–PAGE in the presence of 6 M urea (Figure 7). Based on these observations, we grouped the apg mutants into three categories. These classes corresponded well to defined functional units, including the Apg1p protein kinase complex, the two ubiquitin-like systems and the PI3-kinase complex necessary for autophagy. This classification provides several new insights into the interaction between these classes, detailed in the following sections.
Fig. 6. Localization of GFP–Aut7p and Apg5p–GFP in the apg mutants grown in SD + CA medium. (A) Δapg1 cells (NNY20). GFP–Aut7p was detected on a punctate structure close to the vacuole in 30 of 161 cells (18%). Δapg13 (C; TFD13W2), Δapg17 (YYK111), apg2 (MT2-4-4) cells exhibited an identical phenotype. (B) Δapg1 cells (YYK36) expressing Apg5p–GFP. Punctate structures were detected in 11 of 91 cells (12%). Δapg13 (D; TFD13W2), Δapg17 (YYK111) and apg2 (MT2-4-4) cells demonstrated an identical phenotype. (E) This image of a Δaut2 cell (GYS6) expressing GFP–Aut7p is representative of the Aut7 system. The punctate structures were detected in one of 78 cells (1%). Δapg7 (GYS9) and Δaut1 (GYS5) cells exhibited the same phenotype. (F) Δaut2 cells (GYS6) expressing Apg5p–GFP. A single punctate structure was detected in 11 of 52 cells (21%). Δaut1 (GYS5) and Δaut7 (YYK218) cells demonstrated a similar phenotype. (G) This Δapg12 cell (GYS13) expressing GFP–Aut7p is representative of mutants of the Apg12 system. Punctate structures were detected in one of 84 cells (1%). Δapg10 (TFD10-L1) and Δapg5 (SKD5-1D) cells possessed an identical phenotype. (H) A Δapg5Δapg12 cell (YNM117) expressing Apg5p–GFP. A single punctate structure was detected in 16 of 104 cells (15%). Δapg7 (GYS9), Δapg10 (TFD10-L1) and Δapg5 (SKD5-1D) cells all exhibited an identical phenotype. (I) A Δapg14 cell (SKD14–1C) expressing GFP–Aut7p. None of the 51 cells displayed a punctate structure. Δvps30 cells (SKD6-1D) and Δapg9 cells (CTD1) possessed an identical phenotype. (J) A Δapg14 cell (AKY12) expressing Apg5p–GFP under growing conditions. Punctate structures were observed in one of 248 cells (<1%). Δvps30 cells (AKY74) and Δapg9 cells (CTD1) exhibited an identical phenotype. (K) A Δapg16 cell (KVY117) expressing GFP–Aut7p. No punctate structures were observed in any of the 77 cells. (L) A Δapg5Δapg16 cell (YNM126) expressing Apg5p–GFP under growing conditions. No punctate structures were observed in the 184 cells examined. Bar: 5 µm.
Table I. Localization of GFP–Aut7p and Apg5p–GFP on the punctate structure in apg mutants.
| Genotype | GFP–Aut7p | Apg5p–GFP |
|---|---|---|
| Δapg1 | + | + |
| apg2 | + | + |
| Δaut1/Δapg3 | – | + |
| Δaut2/Δapg4 | – | + |
| Δapg5 | – | + (WT) |
| Δvps30/Δapg6 | – | – |
| Δapg7 | – | + |
| Δaut7/Δapg8 | + (WT) | + |
| Δapg9 | – | – |
| Δapg10 | – | + |
| Δapg12 | – | + |
| Δapg13 | + | + |
| Δapg14 | – | – |
| Δapg16 | – | – |
| Δapg17 | + | + |
| Δapg1Δapg7 | – | not examined |
| Δapg1Δvps30 | – | not examined |
| Δapg1Δapg9 | – | not examined |
| Δaut2Δapg10 | ||
| expressing GFP–Aut7FGp | – | not examined |
| Δaut2Δapg12 | ||
| expressing GFP–Aut7FGp | – | not examined |
| Δvps38 | + | not examined |
+: GFP–Aut7p or Apg5p–GFP is localized to the punctate structure.
–: GFP–Aut7p or Apg5p–GFP shows a diffuse cytoplasmic staining pattern.
WT: wild type.
Fig. 7. Levels of Aut7p–PE present in each apg mutant. Lysates were prepared by glass bead disruption prior to SDS–PAGE as described in Materials and methods. (A) Cells in vegetative growth. (B) Cells following a 4.5 h starvation in SD (–N) medium. We examined the phenotype of wild type (SEY6210), Δapg1 (GYS102), apg2 (MT2-4-4), Δaut1 (KVY113), Δaut2 (KVY13), Δapg5 (KVY142), Δvps30 (KVY135), Δapg7 (KVY118), Δaut7 (KVY5), Δapg9 (KVY114), Δapg10 (KVY136), Δapg12 (KVY115), ΔΔapg13 (KVY116), Δapg14 (GYS115), Δapg16 (KVY117) and Δapg17 (YYK111) cells.
Analyses of class A apg mutants provide evidence that the punctate structure is involved in autophagosome formation
The class A mutants possess both Apg5p–GFP and GFP–Aut7p localized to the punctate structure as in wild-type cells (Table I). The class A genes consist of APG1, APG2, APG13 and APG17. Apg13p and Apg17p are regulatory factors of the Apg1p protein kinase. As the class A mutants demonstrate normal punctate structures, the Apg1p protein kinase complex is unlikely to be involved in the formation of the punctate structure. The class A mutants all possessed normal levels of Aut7p–PE (Figure 7).
To examine whether the punctate structure is physiologically functional, we generated an apg1ts allele by polymerase chain reaction (PCR) mutagenesis. Auto phagic activities were measured by monitoring the alkaline phosphatase (ALP) activity in cells expressing a cytosolic proform of the ALP (Pho8Δ60p), which is transported to vacuoles and becomes mature (Noda et al., 1995). We estimated the temperature sensitivity of this allele from the maturation of API (Figure 8A) and the ALP assay (Figure 8B) using Δapg1 cells carrying the apg1ts plasmid (apg1ts cells). At 23°C, the apg1ts cells contained normal levels of mature API (Figure 8A) and possessed significant autophagic activity (Figure 8B). At 37°C, however, both the maturation of API and autophagic activity were completely blocked (Figure 8A and B). Autophagy in the apg1ts cells at 30°C occurred at the same levels as at 23°C. The activity of ALP increased linearly with time for 6 h following incubation in nitrogen-free medium at 23°C (Figure 8C). On transferring the cells to 37°C, autophagy was immediately blocked (Figure 8C). When the cells were transferred from 37°C to 23°C, the autophagic activity was recovered without a lag period (Figure 8C). Apg1tsp was detectable by immunoblot even after an overnight incubation at 37°C (Figure 8A), demonstrating that apg1ts is a reversible temperature-sensitive allele defective in autophagy.
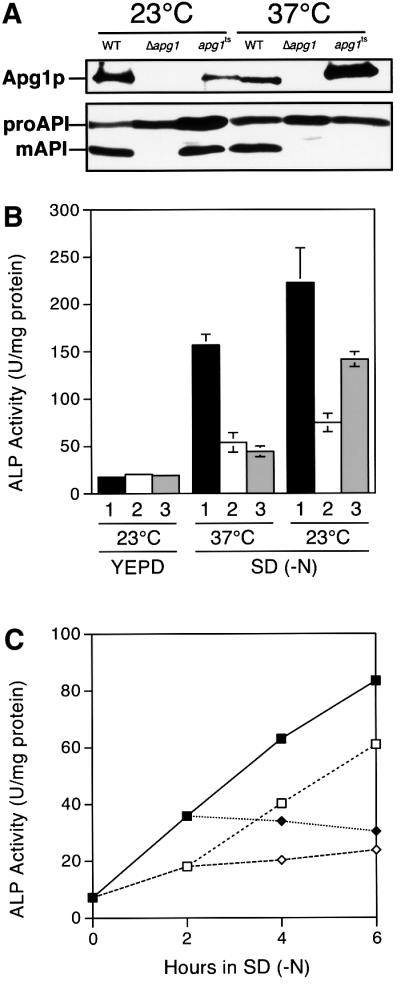
Fig. 8. apg1ts mutant possessing functional temperature sensitivity. (A) Although temperature-sensitive Apg1p was detectable, the maturation of API was completely blocked at 37°C in apg1ts cells. The API proform (proAPI) is processed to mature API (mAPI) in an Apg1p activity-dependent manner. Wild-type cells (TN125), Δapg1 cells (YYK126) and Δapg1 cells (YYK126) carrying the apg1ts plasmid were grown in YEPD medium at 23°C or 37°C overnight. Apg1p and API were detected as described in Materials and methods. (B) Δapg1 cells (YYK126) carrying (1) the APG1 plasmid, (2) the vector or (3) the apg1ts plasmid were grown in YEPD medium at 23°C. They were then transferred into SD (–N) medium at 23°C or 37°C and incubated for 6 h before ALP activity was measured. (C) Δapg1 cells (YYK126) carrying the apg1ts plasmid were grown in YEPD medium at 23°C. The cells were then transferred into SD (–N) medium and incubated at 23°C or 37°C. Closed squares, cells starved continuously at 23°C. Open squares, cells incubated for 2 h at 23°C and then transferred to 37°C. Open lozenges, cells starved continuously at 37°C. Closed lozenges, cells incubated for 2 h at 37°C and then transferred to 23°C.
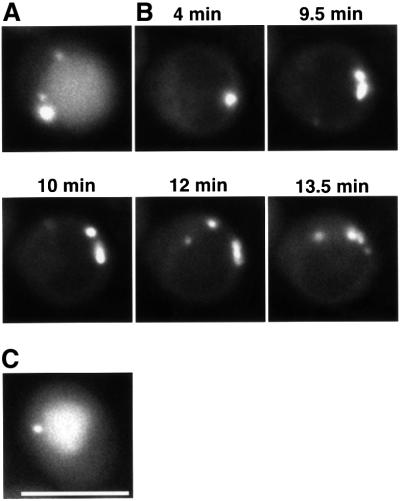
On shifting the temperature from 37°C to 30°C, we followed the dynamics of GFP–Aut7p in apg1ts cells. At 30°C, GFP–Aut7p was visualized in the punctate structure and the vacuole after rapamycin treatment, as seen in wild-type cells (Figure 9A). At 37°C, the staining intensity of the punctate structure increased, accompanied by a lack of vacuolar staining after rapamycin treatment (Figure 9B, 4 min). Next, we examined the progression of GFP–Aut7p staining following temperature decrease in real time using time-lapse microscopy. Within 10 min, several dots labelled with GFP–Aut7p separated from the punctate structure (Figure 9B). During 30 min of incubation at 30°C, fluorescence intensity of vacuoles became brighter as these dots disappeared, suggesting that they are autophagosomes. We concluded that the punctate structure actually participates in the formation of autophagosomes.
Fig. 9. Time-lapse microscopy of apg1ts cells expressing GFP–Aut7p. Δapg1 cells (NNY20) carrying the apg1ts and GFP–Aut7p plasmids were used. (A) Cells treated with rapamycin for 3 h at 30°C. (B) Time-lapse images following temperature decrease. Cells were treated with rapamycin for 4 h at 37°C before decreasing the temperature to 30°C. (C) Cell incubated for 2 h at 30°C after temperature decrease. Bar: 5 µm.
We next treated apg1ts cells with cycloheximide for 20 min, following a 4 h incubation at 37°C in the presence of rapamycin. On decreasing the temperature, similar dots appeared, leading to fluorescent staining of the vacuole (data not shown). This experiment demonstrates that the emergence of autophagosomes from the punctate structure depends on the activity of Apg1p; de novo protein synthesis, however, is not necessary. As Apg1p is localized to the punctate structure (Figure 2D–F), the Apg1p protein kinase complex functions in autophagosome formation at a later step than the organization of the structure. We therefore designated this punctate structure the ‘pre-autophagosomal structure’.
Class B apg mutants have a defect in the localization of Aut7p on the pre-autophagosomal structure
In class B mutants Δaut1, Δaut2, Δapg5, Δapg7, Δapg10 and Δapg12, Apg5p–GFP was detected on the pre-auto phagosomal structure; GFP–Aut7p was not. Interestingly, these are the genes involved in the two ubiquitin-like conjugation systems: the Aut7 system (Δaut2, Δapg7 and Δaut1) and the Apg12p–Apg5p conjugation system (Δapg12, Δapg7, Δapg10 and Δapg5). In the Δapg1Δapg7 double mutant, GFP–Aut7p did not localize to the pre-autophagosomal structure (Table I). This result demonstrates that APG7 (class B) is epistatic to APG1 (class A) for the localization of Aut7p, suggesting that the class B APG genes govern the localization of Aut7p to the pre-autophagosomal structure prior to class A gene function. As previously reported, mutants of the Aut7 system lacked detectable Aut7p–PE (Figure 7; Ichimura et al., 2000). These studies demonstrated that the localization of GFP–Aut7p to the pre-autophagosomal structure is tightly coupled to Aut7p lipidation. The association of Aut7p with the pre-autophagosomal structure occurs with the PE-conjugated form. In Δaut1 or Δaut2 mutant, the Apg12p– Apg5p conjugate is formed normally (Mizushima et al., 1998); Apg5p–GFP was localized on the pre-autophagosomal structure, as seen in wild-type cells (Table I). Therefore, the lipidation of Aut7p is essential for the localization of Aut7p, but not required for localization of (Apg12p–)Apg5p to the pre-autophagosomal structure.
Interestingly, Apg5p–GFP was detected on the pre-autophagosomal structure in mutants of the Apg12 system (Δapg12 and Δapg10; Table I), suggesting that the conjugation of Apg12p is not necessary for Apg5p localization. In Apg12 system mutants (Δapg12, Δapg10 and Δapg5), we could not detect GFP–Aut7p on the pre-autophagosomal structure (Table I). In addition, these mutants demonstrated severely reduced levels of Aut7p–PE (Figure 7). A defect in the membrane association of Aut7p in Apg12p conjugation mutants (Kim et al., 2001) may be explained by decreases in the total level of Aut7p–PE in the absence of the conjugate.
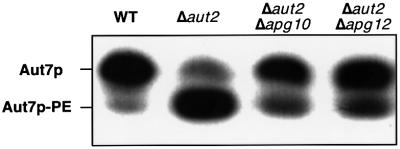
To examine the effects of the disruption of the Apg12p–Apg5p conjugate on Aut7p localization, we estimated the levels of Aut7p–PE in Δaut2 cells expressing the carboxy-terminal processed form of Aut7p (Aut7FGp; Ichimura et al., 2000; Kirisako et al., 2000). Aut2p/Apg4p, a cysteine protease, is essential for Aut7p processing and Aut7p–PE deconjugation. Aut7p–PE accumulates in Δaut2 cells expressing Aut7FGp because of the inability of these cells to deconjugate Aut7p–PE (Kirisako et al., 2000). The levels of Aut7p–PE in Δaut2Δapg10 and Δaut2Δapg12 cells were reduced from the levels in Δaut2 cells, but Aut7p–PE was actually generated in Δaut2Δapg10 and Δaut2Δapg12 cells (Figure 10). This result indicates the Apg12p–Apg5p conjugate is not essential for Aut7p–PE formation. We then constructed a GFP–Aut7FG plasmid, which we expressed in Δaut2Δapg10 and Δaut2Δapg12 cells. Although significant amounts of Aut7p–PE were produced in these mutants (Figure 10), GFP–Aut7FGp did not localize to the pre-autophagosomal structure (Table I). This result indicates that the Apg12p–Apg5p conjugate is absolutely required for the proper localization of Aut7p. The role of the conjugate is not restricted to the localization of Aut7p, however, as Δapg5 cells possess more severe defects in API maturation than the Δaut7 cells (Abeliovich et al., 1999, 2000).
Fig. 10. Amounts of Aut7p–PE in Δaut2 cells expressing Aut7FGp during vegetative growth. Lysates were prepared by glass bead disruption and subjected to SDS–PAGE as described in Materials and methods. The strains used in this experiment are as follows: wild type (SEY6210), Δaut2 (KVY13), Δaut2Δapg10 (KVY150) and Δaut2Δapg12 (KVY148).
Class C apg mutants do not deliver either Apg5p or Aut7p to the pre-autophagosomal structure
In class C mutants, Δvps30, Δapg9, Δapg14 and Δapg16, neither Apg5p–GFP nor GFP–Aut7p localized to the pre-autophagosomal structure. VPS30 and APG14 are the genes involved in the autophagy-specific PI3-kinase complex. Double disruptant Δapg1Δvps30 or Δapg1 Δapg9, expressing GFP–Aut7p demonstrated a diffuse cytoplasmic staining pattern (Table I). This result indicates that VPS30 and APG9 (class C) are epistatic to APG1 (class A) in the organization of the pre-autophagosomal structure. Therefore, the class C APG genes play roles in the organization of Apg5p and Aut7p on the pre-autophagosomal structure in a class A gene-independent manner. We further analysed the levels of Aut7p–PE in these mutants. Δapg16 cells contained significant levels of Aut7p–PE during vegetative growth (Figure 7A). The ratio of Aut7p–PE to unmodified Aut7p, however, decreased after nitrogen starvation (Figure 7B). The levels of Aut7p–PE in growing cells and starved cells were similar (data not shown), indicating that the synthesis of Aut7p during starvation resulted in a decrease in the relative levels of Aut7p–PE in Δapg16 cells. In contrast, Δvps30, Δapg9, Δapg14 cells possessed normal levels of Aut7p– PE, regardless of the nutrient conditions (Figure 7).
Apg16p is a coiled-coil protein linking the Apg12p– Apg5p conjugates (Mizushima et al., 1999). Although the Apg12p–Apg5p conjugate was formed, we did not observe a dot structure of Apg5p–GFP in Δapg16 cells (Table I). This observation suggests that the interaction of Apg5p and Apg16p is necessary for the proper localization of Apg5p to the pre-autophagosomal structure. Although Aut7p–PE was formed in the Δapg16 mutant during vegetative growth (Figure 7A), we could not observe the localization of Aut7p to the pre-autophagosomal structure (Table I). This phenotype is similar to that of Δaut2 Δapg10 or Δaut2Δapg12 cells expressing GFP–Aut7FGp (Table I), which contain significant levels of Aut7p–PE (Figure 10). This result indicates that Apg12p–Apg5p– Apg16p complex formation is needed for the localization of Aut7p. As the Δapg16 mutant failed to maintain Aut7p–PE levels during starvation, the Apg12p–Apg5p– Apg16p complex must be necessary for the maintenance of Aut7p–PE levels.
Two types of PI3-kinase complex contain Vps34p, Vps15p and Vps30p/Apg6p as common components (Kihara et al., 2001a). In these complexes, Apg14p and Vps38p are the components specifying autophagy or the carboxypeptidase Y (CPY), a vacuolar resident enzyme, sorting pathway, respectively. In Δapg14 cells, Aut7p and Apg5p were located diffusely throughout the cytoplasm, whereas in Δvps38 cells, the pre-autophagosomal structure was detectable (Table I). In Δvps30 and Δapg14 strains, Aut7p–PE and the Apg12p–Apg5p conjugate were produced normally (Figure 7; Mizushima et al., 1998). These results indicate that the autophagy-specific PI3-kinase complex is essential to recruit Aut7p–PE and the Apg12p–Apg5p conjugate to the pre-autophagosomal structure. The Δapg9 mutant also demonstrates class C phenotype. A physical interaction of Apg9p with other Apg proteins, however, has not been identified. Our results suggest that Apg9p has some relationship with the autophagy-specific PI3-kinase complex.
Discussion
We and other groups have reported the intracellular localization of several Apg proteins forming a punctate structure close to the vacuole. The physiological significance of this punctate structure, however, has not been examined in detail. Here, we systematically investigated the localization of GFP-tagged Apg proteins when expressed at physiological levels. These analyses identified a novel ‘pre-autophagosomal structure’, in which Apg1p, Apg2p, Apg5p, Aut7p and Apg16p are assembled. Apg5p and Aut7p are recruited to the structure by the action of Apg16p and the autophagy-specific PI3-kinase complex. The Apg1p protein kinase complex is also localized to the structure. This kinase functions in concert with the Apg12p–Apg5p–Apg16p complex and Aut7p to play a crucial role in generating autophagosomes. Our characterization of this structure found that Apg proteins exist in three functional groups, the Apg1p protein kinase complex, the ubiquitin-like systems (the Apg12 system and the Aut7 system) and the autophagy-specific PI3-kinase complex. Epistatic analyses of the apg mutant phenotypes clearly uncovered the interrelationship on the pre-autophagosomal structure between these three groups. Although the functions of the APG2 and APG9 genes remain unknown, we obtained results suggesting that these genes encode proteins acting close to the Apg1p protein kinase complex and the autophagy-specific PI3-kinase complex, respectively.
The pre-autophagosomal structure
Following rapamycin treatment, the fluorescence of GFP–Aut7p and other GFP-fused Apg proteins visualizing the pre-autophagosomal structure rapidly increases in intensity (data not shown). The amount of Aut7p increases during autophagy, whereas there is no significant increase in Apg1p, Apg2p, Apg5p or Apg16p. These results suggest the further recruitment of Apg proteins to the pre-autophagosomal structure by unknown mechanisms. This change in the composition of the pre-autophagosomal structure may initiate autophagosome formation.
Repeated cell fractionation analyses of Apg proteins have not uncovered a biochemical identification of the pre-autophagosomal fraction. It is likely that low levels of each Apg protein are recruited onto the structure; in addition, the structure may be unstable in cell lysates, leading to difficulties in its biochemical characterization. Aut7p is concentrated on premature autophagosomes, isolation membranes and Aut7p-enriched regions near the vacuole, as determined by immunoelectron microscopy (Kirisako et al., 1999). This Aut7p-enriched region is one candidate for the pre-autophagosomal structure. Our intensive examination of Aut7p-concentrated structures, however, has not succeeded in identifying a definite membrane structure corresponding to the pre-autophagosomal structure. The structure may not be observable under an electron microscope as a defined membrane compartment. At present, our analyses, based on fluorescence microscopy, are the most promising investigations dissecting the process of autophagosome formation.
We demonstrated that the pre-autophagosomal structure is a functional structure rather than a dead-end structure. Temperature shift experiments using the apg1ts strain indicated that several dots separated from the pre-autophagosomal structure at a permissive temperature (Figure 9B) to subsequently stain the vacuole (Figure 9C). This result indicates that the pre-autophagosomal structure is an organizing centre for autophagosomes. Each dot corresponds to the pre-autophagosomal structure or an autophagosome; the weakly stained dots seen in later stages (12 min and 13.5 min in Figure 9B) may be autophagosomes.
The organization of the pre-autophagosomal structure is not affected in the class A mutants, Δapg1, apg2, Δapg13 or Δapg17. Furthermore, we demonstrated that Apg1p is a component of the pre-autophagosomal structure (Figure 2D–F). Apg13p and Apg17p are also likely components of the structure, as Apg1p interacts directly with Apg13p and Apg17p (Kamada et al., 2000). In response to nutrient depletion, enhanced activity of the Apg1p kinase complex controls switching from Cvt vesicle formation to autophagosome formation. Aut7p is normally induced in Δapg1 cells by rapamycin or starvation (Kirisako et al., 1999). Therefore, the Apg1p kinase complex plays a role in both the induction of autophagy and the initiation of autophagosome formation. The delivery of GFP–Apg1p to the vacuolar lumen during rapamycin treatment (data not shown) also supports the involvement of Apg1p in the latter stage of autophagosome formation.
Organization of the pre-autophagosomal structure
Aut7p–PE is an essential component of the pre-autophagosomal structure (Table I and Figure 7). We demonstrated that the absence of the Apg12p–Apg5p conjugate results in a severe decrease in Aut7p–PE levels (Figure 7). GFP–Aut7FGp is not detected on the pre-autophagosomal structure in Δaut2Δapg10, Δaut2Δapg12 cells (Table I) despite significant levels of Aut7p–PE (Figure 10). These data suggest that the Apg12p–Apg5p conjugate must play a role in the localization of Aut7p–PE. On treatment with rapamycin, however, Δaut7 cells demonstrate some degree of API maturation, whereas Δapg5 cells possess no mature API (Abeliovich et al., 1999, 2000; Kirisako et al., 2000). This suggests that the conjugate is essential for both autophagosome formation and the maintenance of Aut7p–PE localized to the pre-autophagosomal structure.
Apg16p forms a complex with the Apg12p–Apg5p conjugate (Mizushima et al., 1999). GFP–Aut7p exhibits a diffuse staining pattern in Δapg16 cells, regardless of the normal levels of Aut7p–PE present during vegetative growth (Figure 7A). Δapg16 mutant failed to increase the level of Aut7p–PE during starvation (Figure 7B). Apg16p may participate in the maintenance of the Aut7p–PE levels during starvation. Apg5p–GFP also possesses a diffuse pattern throughout the cytosol in Δapg16 cells (Figure 6L). The localization of Apg16p to the structure (Figure 2G–I) suggests that Apg16p plays a role in targeting the AApg12p–Apg5p conjugate to the pre-autophagosomal structure. In Δapg16 cells, Aut7p–PE may not be localized on the pre-autophagosomal structure due to a lack of the AApg12p–Apg5p-Apg16p complex.
We could not identify GFP–Aut7p punctate structures in growing Δapg9 cells (Table I); the punctate structures are reported in Δapg9 cells under nitrogen-starved conditions (Kim et al., 2001). To investigate the localization of GFP–Aut7p during autophagy, we examined GFP–Aut7p in rapamycin-treated Δapg9 cells. We observed a similar punctate staining pattern similar to that described by Kim et al. (2001; data not shown). These punctate structures, however, were less distinct and of a lower intensity than those seen in Δapg1 cells (data not shown). This result suggests that Apg9p plays an important role in the recruitment of Aut7p to the pre-autophagosomal structure.
The autophagy-specific PI3-kinase complex controls the organization of the pre-autophagosomal structure but the PI3-kinase complex specific to the CPY pathway does not. In autophagy-specific PI3-kinase complex mutants, both Aut7p–PE and the Apg12p–Apg5p conjugate are produced normally, indicating that these conjugation reactions occur in the absence of the complex. One possible function of the autophagy-specific PI3-kinase complex is to select cargo during vesicle budding (Kihara et al., 2001a). The complex may load Aut7p–PE and/or the Apg12p–Apg5p conjugate into vesicles to facilitate their transport to the pre-autophagosomal structure. A second possibility is that the complex plays a role in vesicular sorting, allowing those containing Aut7p–PE and/or the Apg12p–Apg5p conjugate to be delivered to the pre-autophagosomal structure. There is also a third possibility, that the PI3-kinase complex provides the pre-autophagosomal structure with phosphatidylinositol 3-phosphate (PI3P); PI3P then mediates the localization of the Apg12p–Apg5p conjugate and Aut7p–PE to the structure. Analysis of the autophagy-specific PI3-kinase during the organization of the pre-autophagosomal structure will further the understanding of general membrane dynamics mediated by PI3-kinases.
Here, we define a pre-autophagosomal structure, which is directly involved in the formation of autophagosomes. We also propose specific interrelationships between the many APG genes. This report provides insights into the precise functions of Apg proteins.
Materials and methods
Yeast strains and media
Yeast strains used in this study are listed in Table II. We used standard methods for yeast manipulation (Kaiser et al., 1994). Cells were grown in either YEPD (1% yeast extract, 2% peptone and 2% glucose) or SD + CA medium (0.17% yeast nitrogen base without amino acid and ammonium sulfate, 0.5% ammonium sulfate, 0.5% casamino acid and 2% glucose) supplemented with 0.002% adenine sulfate, 0.002% tryptophan and 0.002% uracil if necessary. Autophagy was induced in growth media containing either 0.5 or 1 µg/ml rapamycin (Sigma). To induce nitrogen starvation, the cells were incubated in SD (–N) medium (0.17% yeast nitrogen base without amino acid or ammonium sulfate, with 2% glucose). Yeast extract, yeast nitrogen base without amino acid and ammonium sulfate, and casamino acid were obtained from Difco. Peptone, glucose, ammonium sulfate, adenine sulfate, tryptophan and uracil were purchased from Wako Pure Chemical Industries, Ltd.
Table II. Strains used in this study.
| Strain | Genotype | Source |
|---|---|---|
| SEY6210 | MATα ura3 leu2 his3 trp1 lys2 suc2 | Darsow et al. (1997) |
| YW5-1B | MATa ura3 leu2 trp1 | Tsukada and Ohsumi (1993) |
| KA311A | MATa ura3 leu2 his3 trp1 | K.Irie (Osaka University, Japan) |
| KA311B | MATα Δura3 leu2 his3 trp1 | K.Irie (Osaka University, Japan) |
| BJ2168 | MATa ura3 leu2 trp1 pep4-3 prb1-1122 prc1-407 | Yeast Genetic Stock Center |
| TN125 | MATa ade2 ura3 leu2 his3 trp1 lys2 pho8::pho8Δ60 | Noda and Ohsumi (1998) |
| GYS102 | SEY6210 Δapg1::LEU2 | this study |
| MT2-4-4 | MATα Δleu2 ura3 apg2 | Tsukada and Ohsumi (1993) |
| KVY113 | SEY6210 Δaut1::LEU2 | this study |
| KVY13 | SEY6210 Δaut2::LEU2 | Kirisako et al. (2000) |
| KVY142 | SEY6210 Δapg5::LEU2 | this study |
| KVY135 | SEY6210 Δvps30::LEU2 | Kirisako et al. (2000) |
| KVY118 | SEY6210 Δapg7::HIS3 | Kirisako et al. (2000) |
| KVY5 | SEY6210 Δaut7::HIS3 | Kirisako et al. (1999) |
| KVY114 | SEY6210 Δapg9::TRP1 | this study |
| KVY136 | SEY6210 Δapg10::HIS3 | this study |
| KVY115 | SEY6210 Δapg12::HIS3 | this study |
| KVY116 | SEY6210 Δapg13::TRP1 | this study |
| GYS115 | SEY6210 Δapg14::LEU2 | this study |
| KVY117 | SEY6210 Δapg16::LEU2 | this study |
| NNY20 | YW5-1B Δapg1::LEU2 | Matsuura et al. (1997) |
| GYS5 | YW5-1B Δaut1::TRP1 | Ichimura et al. (2000) |
| GYS6 | YW5-1B Δaut2::LEU2 | Ichimura et al. (2000) |
| SKD5-1D | YW5-1B Δapg5::LEU2 | Kametaka et al. (1996) |
| SKD6-1D | YW5-1B Δvps30::LEU2 | Kametaka et al. (1998) |
| GYS9 | YW5-1B Δapg7::LEU2 | Ichimura et al. (2000) |
| CTD1 | YW5-1B Δapg9::TRP1 | Noda et al. (2000) |
| TFD10-L1 | YW5-1B Δapg10::LEU2 | Shintani et al. (1999) |
| GYS13 | YW5-1B Δapg12::TRP1 | this study |
| TFD13W2 | YW5-1B Δapg13::TRP1 | Funakoshi et al. (1997) |
| SKD14–1C | YW5-1B Δapg14::LEU2 | Kametaka et al. (1998) |
| YYK36 | KA311A Δapg1::LEU2 | Kamada et al. (2000) |
| GYS59 | KA311A Δapg5::HIS3 | this study |
| YNM119 | KA311B Δapg5::HIS3 | Mizushima et al. (1998) |
| AKY74 | KA311A Δvps30::LEU2 | this study |
| YYK218 | KA311A Δaut7::TRP1 | this study |
| AKY12 | KA311A Δapg14::LEU2 | this study |
| YYK111 | KA311A Δapg17::HIS3 | Kamada et al. (2000) |
| YYK190 | BJ2168 Δapg1::LEU2 | this study |
| YYK126 | TN125 Δapg1::LEU2 | Kamada et al. (2000) |
| YAK3 | SEY6210 Δypt7::HIS3 Δapg5::LEU2 | this study |
| MBY3 | SEY6210 Δvps4::TRP1 | Babst et al. (1997) |
| YNM117 | MATa ura3 leu2 trp1 Δapg5::LEU2 Δapg12::HIS3 | Mizushima et al. (1999) |
| YNM126 | KA311B Δapg5::HIS3 Δapg16::LEU2 | Mizushima et al. (1998) |
| GYS22 | YW5-1B Δapg1::LEU2 Δapg7::HIS3 | this study |
| GYS23 | YW5-1B Δapg1::URA3 Δvps30::LEU2 | this study |
| GYS24 | YW5-1B Δapg1::LEU2 Δapg9::TRP1 | this study |
| GYS29 | YW5-1B Δapg9::TRP1 Δapg14::LEU2 | this study |
| KVY150 | SEY6210 Δapg4::LEU2 Δapg10::HIS3 | this study |
| KVY148 | SEY6210 Δapg4::LEU2 Δapg12::HIS3 | this study |
Plasmids
In this study, centromeric plasmids were used to produce physiological levels of expression. The pRS316 GFP–AUT7 plasmid was created by substituting the 3× haemagglutinin sequence immediately after the start codon of the AUT7 open reading frame (ORF) in pTK108 (Kirisako et al., 1999) with a modified GFP sequence (S65T), following digestion by BamHI. The pRS316 GFP–AUT7FG plasmid was created by substituting the 3× myc sequence immediately after the start codon of the pRS316 AUT7FG (Kirisako et al., 2000). The pRS314 ECFP–AUT7 plasmid was created by cloning an enhanced CFP (ECFP) fragment, amplified from pECFP (Clontech), into pRS316 GFP–AUT7 digested with BamHI. From the resulting pRS316 ECFP–AUT7, we subcloned an excised SacI–XhoI fragment into pRS314. The pRS314 APG5–GFP and the pRS416 APG9– GFP plasmids were the generous gifts of Dr D.J.Klionsky (University of Michigan). Site-directed mutagenesis using QuikChange (Stratagene) was used to insert a BamHI site into pYAPG507, immediately before the stop codon of the APG5 ORF (Kametaka et al., 1998) to create the pRS316 APG5-EYFP plasmid. An enhanced YFP (EYFP) fragment was amplified from pEYFP (Clontech), digested with BamHI and cloned into the BamHI site of pYAPG507.
The pRS316 YFP–APG1 plasmid was created by inserting the BamHI cassette of EYFP immediately after the start codon of the APG1 ORF. Site-directed mutagenesis then inserted a BglII site using QuikChange The pRS314 APG16–YFP plasmid was created by inserting the NotI cassette of EYFP immediately before the stop codon of the APG16 ORF (Mizushima et al., 1999).
Immunoblot analysis
To detect either the GFP–Aut7p or Apg12p–Apg5p–GFP conjugate, lysates were prepared from cells grown in SD + CA medium to a density of ∼OD600 = 1 by NaOH/2-mercaptoethanol extraction with a slight modification (Horvath and Reizman, 1994). Briefly, anti-Aut7p anti serum (Kirisako et al., 2000) and anti-Apg12 antiserum (A.Kuma, N.Mizushima, N.Ishihara and Y.Ohsumi, in preparation), respectively, were used as the primary antibodies. To detect Apg1p and API, we utilized anti-Apg1p antiserum (Matsuura et al., 1997) and anti-aminopeptidase I antiserum (gift from Dr D.J.Klionsky) as the primary antibodies, respectively. Aut7p–PE was detected following SDS–PAGE in the presence of 6 M urea, as described (Kirisako et al., 2000). We collected cells that had been either grown in YEPD medium to a density of ∼OD600 = 1 or then starved in SD (–N) medium. Harvested cells were then washed with distilled water and disrupted with glass beads (Sigma) for 5 min in a buffer containing 50 mM Tris–HCl pH 7.5, 150 mM NaCl, 5 mM EDTA, 5 mM EGTA, 1 mM phenylmethanesulfonyl fluoride (PMSF; Sigma) and a protease inhibitor cocktail (Complete; Roche). Cell lysates were boiled in SDS sample buffer for 5 min. Equal protein amounts (20 µg protein) were subjected to electrophoresis, transferred to PVDF membrane (Millipore) and detected with a combination of anti-Aut7p antibody and peroxidase-conjugated goat anti-rabbit IgG antibody (Seikagaku Kogyo) by the ECL system (Amersham Pharmacia Biotech). Protein concentration was measured using a BCA reagent kit (Pierce).
Microscopy
Fluorescence microscopy was performed using a DeltaVision microscope (Applied Precision). FM4-64 (Molecular Probes) was used to identify the vacuolar membrane under a fluorescence microscope. Growing cells were incubated with 1 µg/ml FM4-64 for 15 min and then chased for 30 min at 30°C. The labelled cells were visualized by fluorescence microscopy using a Texas Red filter. To observe GFP-tagged proteins under a fluorescence microscope, we employed a FITC filter. A dual band filter set for CFP and YFP (86002; Chroma Technology Corp.), was used to detect the localization of CFP- and YFP-tagged proteins. Culture temperatures were controlled with a ΔTC3 culture dish system (Bioptechs) during time-lapse video microscopy.
Immunofluorescence microscopy was performed as previously described (Nishikawa et al., 1994). Cells harboring the APG9–GFP plasmids were grown in SD + CA medium to a density of ∼OD600 = 1.5. Cultures were then treated with 0.5 µg/ml rapamycin for 2 h and subsequently fixed in 5% formaldehyde. Cells were then converted into spheroplasts and permeabilized with PBS containing 0.5% Triton X-100 for 10 min at room temperature. Next, the cells were dropped onto multiwell slide glasses (Cel-line Associates), pre-coated with polylysine (Sigma). Affinity-purified anti-Aut7p antibody was used as the primary antibody, followed by incubation with the Cy5-conjugated secondary antibody (Amersham). For double labelling with GFP-tagged proteins, we used Cy5, a fluorescent dye (Ex: 649 nm and Em: 670 nm), to minimize the bleeding of the fluorescence into the GFP channel (Ex: 489 nm and Em: 511 nm). GFP and Cy5 were visualized with the FITC and the Cy5 filter sets, respectively.
ALP assay
Autophagic activities were estimated by measuring ALP activity in cells expressing a cytosolic proform of the phosphatase (Pho8Δ60p; Noda et al., 1995). α-naphthyl phosphate (Sigma) was used as a substrate. Pho8Δ60p is transported to the vacuole via autophagy and processed in the vacuolar lumen to become active.
Generation of the apg1ts allele
APG1 was mutagenized by previously described PCR procedures (Cadwell and Joyce, 1995). The mutagenized gene was co-transformed into YYK190 cells with a gapped pRS316 plasmid, containing portions homologous to both ends of the PCR product. Two hundred and four colonies were picked and patched onto YEPD plates, which were then incubated overnight at 30 and 37°C. Cells were then subjected to treatment with 0.4 µg/ml rapamycin at each temperature. We examined the generation of autophagic bodies under a phase-contrast microscope. Plasmids recovered from the 22 colonies that passed the initial screening were used to re-transform YYK190 cells to confirm the temperature-sensitive phenotype.
Eight strains that passed the initial screening were subjected to an ALP assay to quantify autophagic activity. YYK126 cells were transformed with each candidate plasmid. Cells were cultured in YEPD medium at 23°C; ALP activities were measured after a 6 h nitrogen starvation at 23 or 37°C. Five strains were finally confirmed as temperature-sensitive mutants. Two of the five strains exhibiting clear temperature sensitivity were used for further analysis. The plasmids recovered from these two strains possessed identical sequences. This allele was used as apg1ts. The temperature sensitivity resulted from the following amino acid substitutions: L88H, F112L and S158P.
Acknowledgments
Acknowledgements
We would like to thank Dr Daniel J.Klionsky for kindly providing plasmids. We would also like to thank Mr Takaji Nemoto, Drs Tokuko Haraguchi and Yasushi Hiraoka for their technical advice on fluorescence microscopy. We would like to thank Dr Scott D.Emr, Ms Akiko Kuma, Mr Yoshinobu Ichimura, Drs Akio Kihara and Takahiro Shintani for their donation of cell strains, as well as their helpful discussions. We would also like to thank NIBB Center for Analytical Instruments for their technical assistance. This work was supported in part by Grants-in-Aids for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan. K.S. was supported by the Research Fellowships of the Japan Society for the Promotion of Science for Young Scientists.
References
- Abeliovich H., Darsow,T. and Emr,S.D. (1999) Cytoplasm to vacuole trafficking of aminopeptidase I requires a t-SNARE-Sec1p complex composed of Tlg2p and Vps45p. EMBO J., 18, 6005–6016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeliovich H., Dunn,W.A., Jr, Kim,J. and Klionsky,D.J. (2000) Dissection of autophagosome biogenesis into distinct nucleation and expansion steps. J. Cell Biol., 151, 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M., Takeshige,K., Baba,N. and Ohsumi,Y. (1994) Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. J. Cell Biol., 124, 903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M., Sato,T.K., Banta,L.M. and Emr,S.D. (1997) Endosomal transport function in yeast requires a novel AAA-type ATPase, Vps4p. EMBO J., 16, 1820–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadwell R.C. and Joyce,G.F. (1995) Mutagenic PCR. In Dieffenbach,C.W. and Dveksler,G.S. (eds), PCR Primer. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 583–589.
- Darsow T., Rieder,S.E. and Emr,S.D. (1997) A multispecificity syntaxin homologue, Vam3p, essential for autophagic and biosynthetic protein transport to the vacuole. J. Cell Biol., 138, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fengsrud M., Roos,N., Berg,T., Liou,W., Slot,J.W. and Seglen,P.O. (1995) Ultrastructural and immunocytochemical characterization of autophagic vacuoles in isolated hepatocytes: effects of vinblastine and asparagine on vacuole distributions. Exp. Cell Res., 221, 504–519. [DOI] [PubMed] [Google Scholar]
- Fengsrud M., Erichsen,E.S., Berg T.O., Raiborg,C. and Seglen,P.O. (2000) Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. Eur. J. Cell Biol., 79, 871–882. [DOI] [PubMed] [Google Scholar]
- Funakoshi T., Matsuura,A., Noda,T. and Ohsumi,Y. (1997). Analyses of APG13 gene involved in autophagy in yeast, Saccharomyces cerevisiae. Gene, 192, 207–213. [DOI] [PubMed] [Google Scholar]
- George M.D., Baba,M., Scott,S.V., Mizushima,N., Garrison,B.S., Ohsumi,Y. and Klionsky,D.J. (2000) Apg5p functions in the sequestration step in the cytoplasm-to-vacuole targeting and macroautophagy pathways. Mol. Biol. Cell, 11, 969–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding T.M., Hefner-Gravink,A., Thumm,M. and Klionsky,D.J. (1996) Genetic and phenotypic overlap between autophagy and the cytoplasm to vacuole protein targeting pathway. J. Biol. Chem., 271, 17621–17624. [DOI] [PubMed] [Google Scholar]
- Horvath A. and Reizman,H. (1994) Rapid protein extraction from Saccharomyces cerevisiae. Yeast, 10, 1305–1310. [DOI] [PubMed] [Google Scholar]
- Huang W.P., Scott,S.V., Kim,J. and Klionsky,D.J. (2000) The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem., 275, 5845–5851. [DOI] [PubMed] [Google Scholar]
- Ichimura Y. et al. (2000) A ubiquitin-like system mediates protein lipidation. Nature, 408, 488–492. [DOI] [PubMed] [Google Scholar]
- Kabeya Y., Mizushima,N., Ueno,T., Yamamoto,A., Kirisako,T., Noda,T., Kominami,E., Ohsumi,Y. and Yoshimori,T. (2000) LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J., 19, 5720–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C., Michaelis,S. and Mitchell,A. (1994) Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Kamada Y., Funakoshi,T., Shintani,T., Nagano,K., Ohsumi,M. and Ohsumi,Y. (2000) Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol., 150, 1507–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kametaka S., Matsuura,A., Wada,Y. and Ohsumi,Y. (1996) Structural and functional analyses of APG5, a gene involved in autophagy in yeast. Gene, 178, 139–143. [DOI] [PubMed] [Google Scholar]
- Kametaka S., Okano,T., Ohsumi,M. and Ohsumi,Y. (1998) Apg14p and Apg6/Vps30p form a protein complex essential for autophagy in the yeast, Saccharomyces cerevisiae. J. Biol. Chem., 273, 22284–22291. [DOI] [PubMed] [Google Scholar]
- Kihara A., Noda,T., Ishihara,N. and Ohsumi,Y. (2001a) Two distinct Vps34 PtdIns 3-kinase complexes function in autophagy and CPY sorting in Saccharomyces cerevisiae.J. Cell Biol., 152, 519–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kihara A., Kabeya,Y., Ohsumi,Y. and Yoshimori,T. (2001b) Beclin-phosphatidylinositol 3-kinase complex functions at the trans-Golgi network. EMBO Rep., 2, 330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Dalton,V.M., Eggerton,K.P., Scott,S.V. and Klionsky,D.J. (1999) Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy and peroxisome degradation pathways. Mol. Biol. Cell, 10, 1337–1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Huang,W.P. and Klionsky,D.J. (2001) Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p and the autophagy conjugation complex. J. Cell Biol., 152, 51–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba,M., Ishihara,N., Miyazawa,K., Ohsumi,M., Yoshimori,T., Noda,T. and Ohsumi,Y. (1999) Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol., 147, 435–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T. et al. (2000) The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol., 151, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J. and Ohsumi,Y. (1999) Vacuolar import of proteins and organelles from the cytoplasm. Annu. Rev. Cell. Dev. Biol., 15, 1–32. [DOI] [PubMed] [Google Scholar]
- Liang X.H., Jackson,S., Seaman,M., Brown,K., Kempkes,B., Hibshoosh,H. and Levine,B. (1999) Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature, 402, 672–676. [DOI] [PubMed] [Google Scholar]
- Matsuura A., Tsukada,M., Wada,Y. and Ohsumi,Y. (1997) Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene, 192, 245–250. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda,T., Yoshimori,T., Tanaka,Y., Ishii,T., George,M.D., Klionsky,D.J., Ohsumi,M. and Ohsumi,Y. (1998) A protein conjugation system essential for autophagy. Nature, 395, 395–398. [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda,T. and Ohsumi,Y. (1999) Apg16p is required for the function of the Apg12p–Apg5p conjugate in the yeast autophagy pathway. EMBO J., 18, 3888–3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Yamamoto,A., Hatano,M., Kobayashi,Y., Kabeya,Y., Suzuki,K., Tokuhisa,T., Ohsumi,Y. and Yoshimori,T. (2001) Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. J. Cell Biol., 152, 657–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa S., Hirata,A. and Nakano,A. (1994) Inhibition of endoplasmic reticulum (ER)-to-Golgi transport induces relocalization of binding protein (BiP) within the ER to form the BiP bodies. Mol. Biol. Cell, 5, 1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noda T. and Ohsumi,Y. (1998) Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem., 273, 3963–3966. [DOI] [PubMed] [Google Scholar]
- Noda T., Matsuura,A., Wada,Y. and Ohsumi,Y. (1995) Novel system for monitoring autophagy in the yeast Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun., 210, 126–132. [DOI] [PubMed] [Google Scholar]
- Noda T., Kim,J., Huang,W.P., Baba,M., Tokunaga,C., Ohsumi,Y. and Klionsky,D.J. (2000) Apg9p/Cvt7p is an integral membrane protein required for transport vesicle formation in the Cvt and autophagy pathways. J. Cell Biol., 148, 465–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petiot A., Ogier-Denis,E., Blommaart,E.F., Meijer,A.J. and Codogno,P. (2000) Distinct classes of phosphatidylinositol 3′-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem., 275, 992–998. [DOI] [PubMed] [Google Scholar]
- Raymond C.K., Howald-Stevenson,I., Vater,C.A. and Stevens,T.H. (1992) Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol. Biol. Cell, 3, 1389–1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott S.V., Baba,M., Ohsumi,Y. and Klionsky,D.J. (1997) Amino peptidase I is targeted to the vacuole by a nonclassical vesicular mechanism. J. Cell Biol., 138, 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seglen P.O. (1987) Regulation of autophagic protein degradation in isolated liver cells. In Glaumann,H. and Ballard,F.J. (eds), Lysosomes: Their Role in Protein Breakdown. Academic Press, London, pp. 371–414.
- Shintani T., Mizushima,N., Ogawa,Y., Matsuura,A., Noda,T. and Ohsumi,Y. (1999) Apg10p, a novel protein-conjugating enzyme essential for autophagy in yeast. EMBO J., 18, 5234–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T., Suzuki,K., Kamada,Y., Noda,T. and Ohsumi,Y. (2001) Apg2p functions in autophagosome formation on the perivacuolar structure. J. Biol. Chem., 276, 30452–30460. [DOI] [PubMed] [Google Scholar]
- Tanida I., Mizushima,N., Kiyooka,M., Ohsumi,M., Ueno,T., Ohsumi,Y. and Kominami,E. (1999) Apg7p/Cvt2p: A novel protein-activating enzyme essential for autophagy. Mol. Biol. Cell, 10, 1367–1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada M. and Ohsumi,Y. (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett., 333, 169–174. [DOI] [PubMed] [Google Scholar]
- Vida T.A. and Emr,S.D. (1995) A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol., 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W., Strømhaug,P.E. and Dunn,W.A., Jr (1999) Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein. Mol. Biol. Cell, 10, 1353–1366. [DOI] [PMC free article] [PubMed] [Google Scholar]