Abstract
Transcriptional regulation by estrogen receptor α (ERα) involves protein–protein interactions among the receptor, its associated coactivators and the RNA polymerase II transcriptional machinery. We have used an in vitro chromatin assembly and transcription system to examine the biochemistry of interactions among ERα, the SRC proteins and p300/CBP. Using polypeptides designed to block specific receptor– cofactor or cofactor–cofactor interactions, we show that interactions among ERα, its coactivators and the RNA pol II machinery are all required for ERα- mediated transcription. Furthermore, we show that ERα–SRC–p300/CBP interactions are necessary and sufficient for the targeted acetylation of nucleosomal histones on estrogen-responsive promoters in the absence of transcription. The protein–protein interactions required for histone acetylation constitute a subset of the interactions required for transcriptional activation. Finally, we show that the major role of SRC–p300/CBP interactions is to enhance ERα- mediated transcription initiation, and they have little or no role in stimulating subsequent rounds of transcription. Together, our results indicate a specific role for the SRC and p300/CBP coactivators, as well as targeted histone acetylation, in ERα-mediated transcription.
Keywords: chromatin/coactivator/estrogen receptor/histone acetylation/transcription
Introduction
Estrogenic hormones, such as 17β-estradiol (E2), play important roles in many physiological processes, including reproduction and development (Couse and Korach, 1999; Warner et al., 1999). The molecular actions of estrogens are mediated by two distinct nuclear estrogen receptor (ER) isoforms, ERα and ERβ (Couse and Korach, 1999; Warner et al., 1999). The ERs belong to a large conserved superfamily of nuclear hormone receptors that function as ligand-regulated, DNA-binding transcriptional activators (Mangelsdorf et al., 1995). In vivo, ERs bind to estrogen response elements (EREs) in the promoters or regulatory regions of estrogen-responsive genes assembled into chromatin in the nuclear environment of the cell. The assembly of genes into chromatin has important functional consequences for gene regulation by ERs and other transcriptional activators since chromatin acts as a general repressor of transcription by RNA polymerase II (RNA pol II) (Wolffe and Kurumizaka, 1998). A variety of cofactors have evolved to assist the ERs and other transcriptional activators in overcoming chromatin-mediated transcriptional repression to activate gene expression. These cofactors include chromatin remodeling complexes and coactivators (Kingston and Narlikar, 1999; Kornberg and Lorch, 1999; McKenna et al., 1999; Robyr et al., 2000).
Chromatin remodeling complexes use the energy stored in ATP to mobilize or structurally alter nucleosomes, allowing greater access of the transcriptional machinery to the DNA template (Kingston and Narlikar, 1999; Kornberg and Lorch, 1999). Coactivators serve at least three roles in nuclear receptor-mediated transcription: (i) they function as bridging factors to recruit other cofactors to chromatin-bound receptors; (ii) they acetylate nucleosomal histones and protein factors at the promoters of hormone target genes; and (iii) they recruit RNA pol II and other components of the basal transcriptional machinery to hormone-regulated promoters (McKenna et al., 1999; Leo and Chen, 2000; Robyr et al., 2000). Histone acetylation, which is thought to loosen chromatin structure and facilitate remodeling, is generally correlated with increased transcriptional activity (Mizzen and Allis, 1998; Kingston and Narlikar, 1999; Kornberg and Lorch, 1999). Current results indicate that the physical and functional interactions among ligand-activated, chromatin-bound ERs, coactivators [e.g. the steroid receptor coactivator (SRC) family of proteins and p300/CBP], chromatin remodeling complexes and RNA pol II underlie the transcriptional regulation of estrogen target genes (McKenna et al., 1999; Leo and Chen, 2000; Robyr et al., 2000) (Figure 1A). However, the requirement for the various factors and their precise contribution to the transcription process remain unclear.
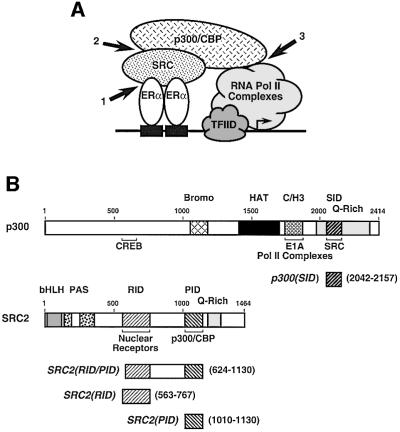
Fig. 1. A subset of protein–protein interactions leading to ligand-regulated transcription by ERα. (A) A schematic representation of some of the interactions involved in ERα-mediated transcription, including interactions between (1) ERα and SRC proteins, (2) SRC proteins and p300/CBP and (3) p300/CBP and the RNA pol II machinery. (B) Schematic representations of p300, SRC2 and the protein fragments used in these studies. Top panel: schematic diagram of human p300. Specific regions of p300 are indicated: CREB-binding region, bromodomain (Bromo), histone acetyltransferase (HAT) domain, C/H3 domain/E1A-binding region, glutamine-rich region (Q-rich) and SRC interaction domain (SID). The residues included in the p300(SID) polypeptide are indicated. Bottom panel: schematic diagram of mouse SRC2. Specific regions of SRC2 are indicated: basic helix–loop–helix region (bHLH), Per-Arnt-Sim domain (PAS), nuclear receptor interaction domain (RID), p300/CBP interaction domain (PID) and glutamine-rich region (Q-rich). The residues included in the SRC2(RID/PID), SRC2(RID) and SRC2(PID) polypeptides are indicated.
The SRC family of proteins contains three structurally and functionally related members unified under the nomenclature of SRC1, SRC2 and SRC3 (Li and Chen, 1998; referred to collectively herein as SRC) that function primarily, but not exclusively, as coactivators for nuclear receptors (McKenna et al., 1999; Leo and Chen, 2000). The SRC proteins bind directly to liganded nuclear receptors via α-helical motifs related to the sequence Leu-X-X-Leu-Leu (referred to as LXXLL motifs or NR boxes) (Heery et al., 1997; Torchia et al., 1997). The NR boxes, which are located in the receptor interaction domain (RID) of the SRC proteins (Figure 1B, bottom), interact with a hydrophobic groove on the surface of the receptor ligand-binding domains (Darimont et al., 1998; Nolte et al., 1998; Shiau et al., 1998). The SRC proteins contribute to transcriptional activation via distinct activation domains, one of which functions as a p300 and CBP interaction domain (PID) (see for example Li and Chen, 1998; Voegel et al., 1998; Sheppard et al., 2001) (Figure 1B, bottom). Furthermore, some SRC family members may possess a weak intrinsic histone acetyltransferase (HAT) activity (compared with p300, CBP and PCAF) (Chen et al., 1997; Spencer et al., 1997), although such an activity has not been detected universally for the SRC family (Voegel et al., 1998; Sheppard et al., 2001).
p300 and CBP are large, highly related, multifunctional coactivators that share many structural and functional attributes and are referred to collectively as p300/CBP (Goodman and Smolik, 2000; Vo and Goodman, 2001). p300/CBP functions as a coactivator for many DNA-binding transcriptional activator proteins, including nuclear receptors and other signal-regulated activators (Goodman and Smolik, 2000; Vo and Goodman, 2001). The conserved motifs and functional domains in p300/CBP include a bromodomain, three cysteine–histidine (C/H)-rich regions (C/H1, C/H2, C/H3), a glutamine (Q)-rich region, an intrinsic acetyltransferase (AT) activity and an SRC interaction domain (SID) (Figure 1B, top). The bromodomain is a histone-interacting module found in many chromatin- and transcription-related factors (Winston and Allis, 1999) and is required for the direct interaction of p300/CBP with chromatin (Manning et al., 2001). The C/H3 region is the site of interaction of a number of different transcription-related factors, including RNA pol II complexes (Nakajima et al., 1997a,b), the adenovirus E1A oncoprotein (Yang et al., 1996; Felzien et al., 1999; O’Connor et al., 1999), TFIIB (Felzien et al., 1999; O’Connor et al., 1999) and PCAF (Yang et al., 1996). The Q-rich region, which has features similar to the Q-rich transcriptional activation domains found in some transcriptional activator proteins, contains the SID (Kamei et al., 1996). The intrinsic AT activity is capable of acetylating free or nucleosomal histones, as well as SRC family members and some transcriptional activator proteins (Bannister and Kouzarides, 1996; Ogryzko et al., 1996; Chen et al., 1997; Vo and Goodman, 2001). All of these domains of p300/CBP are required, to varying degrees, for ERα-mediated transcription with chromatin templates (Kraus et al., 1999).
Here, we have examined the biochemistry of a subset of ERα–coactivator interactions leading to estrogen-regulated gene transcription, specifically those involving the receptor, the SRC proteins and p300/CBP (Figure 1A). We have focused on these coactivators because (i) they are recruited to the receptor either directly or indirectly in a ligand-dependent manner (McKenna et al., 1999; Leo and Chen, 2000); (ii) they localize to estrogen-regulated promoters in vivo (Chen et al., 1999; Shang et al., 2000); (iii) there is good biochemical evidence that they directly stimulate ERα-mediated transcription (Kraus and Kadonaga, 1998; Kraus et al., 1999); and (iv) their functional domains, especially those of p300/CBP, are well characterized (Goodman and Smolik, 2000; Leo and Chen, 2000; Vo and Goodman, 2001). However, the details of the functional interactions between SRC and p300/CBP at estrogen-regulated promoters and their contribution to various steps in the transcription process are unclear. We have examined the functional consequences of SRC-mediated recruitment of p300/CBP to liganded, chromatin-bound ERα using an in vitro chromatin assembly and transcription system that accurately recapitulates the known ligand-dependent transcriptional activity of ERα (Kraus and Kadonaga, 1998). Our results indicate that the recruitment of p300/CBP by SRC, as well as the resulting targeted acetylation of nucleosomal histones, plays a specific role in the process of ERα- mediated transcription, namely to stimulate the formation of a stable transcription pre-initiation complex and subsequent transcription initiation.
Results
SRC2(PID), but not p300(SID), has autonomous transcriptional activation activity
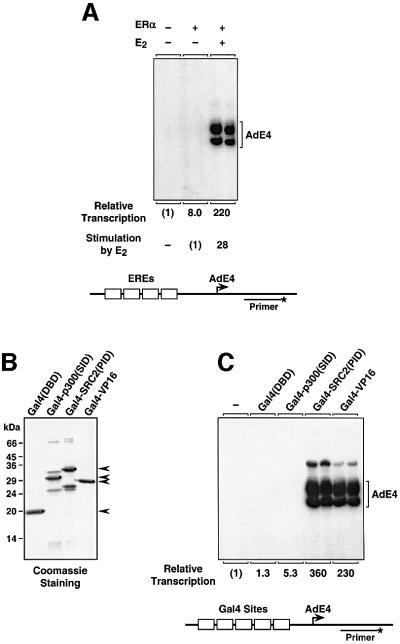
We used an in vitro chromatin assembly and transcription system (Kraus and Kadonaga, 1998, 1999) to examine the biochemistry of ERα-mediated transcription. This assay recapitulates the known ligand-dependent transcriptional activity of ERα on a test template containing multiple EREs upstream of the adenovirus E4 (AdE4) promoter (Figure 2A). Numerous studies have demonstrated a role for coactivators, including SRC and p300/CBP, in the transcriptional activity of ERα and other nuclear receptors (McKenna et al., 1999; Leo and Chen, 2000; Robyr et al., 2000). Based on the model shown in Figure 1A and previous mutagenesis of p300 (Kraus et al., 1999), we hypothesized that the protein surfaces mediating interactions between SRC and p300/CBP [i.e. the SRC(PID) and the p300/CBP(SID); see Figure 1B] might play critical roles in their activities. Previous transient transfection analyses have shown that the SRC(PID) has autonomous transcriptional activity (see e.g. Li and Chen, 1998; Voegel et al., 1998; Sheppard et al., 2001). We sought to determine whether the p300/CBP(SID) also has autonomous transcriptional activity, and whether both SRC(PID) and p300/CBP(SID) might function as autonomous activation domains in the context of chromatin. We reasoned that such an experiment might reveal distinct roles for SRC and p300/CBP in the transcription process, as well as suggest an order for the assembly of an SRC–p300/CBP coactivator complex.
Fig. 2. SRC2(PID), but not p300(SID), has potent autonomous transcriptional activity with chromatin templates. (A) pERE (see schematic, bottom) was assembled into chromatin in the presence of ERα and 17β-estradiol (E2) as noted and subjected to in vitro transcription analysis. The resulting RNA products were analyzed by primer extension. (B) SDS–PAGE of the purified recombinant Gal4(DBD) fusion proteins used in (C). (C) pGIE0 (see schematic, bottom) was assembled into chromatin in the presence of purified recombinant Gal4 DNA-binding domain (DBD) alone or Gal4(DBD) fusions as indicated and subjected to in vitro transcription analysis.
We used fusions of the Gal4 DNA-binding domain (DBD) with SRC2(PID) or p300(SID), which were expressed in Escherichia coli, purified (Figure 2B) and then tested for autonomous transcriptional activity in the context of chromatin using the in vitro chromatin assembly and transcription system described above (Figure 2C). Gal4(DBD) and Gal4-VP16 were used as negative and positive controls, respectively. In this assay, SRC2(PID), but not p300(SID), functioned as a very potent transcriptional activation domain, eliciting a transcriptional response stronger than the well-characterized VP16 activation domain. These results indicate that SRC2(PID) has autonomous transcriptional activity and is sufficient for activation in the context of chromatin. In addition, they support the model that recruitment of p300/CBP by SRC(PID) is an important mechanism underlying coactivation by the SRC proteins.
Polypeptide inhibitors of interactions among ERα, SRC, p300/CBP and the RNA pol II machinery block ERα-mediated transcription
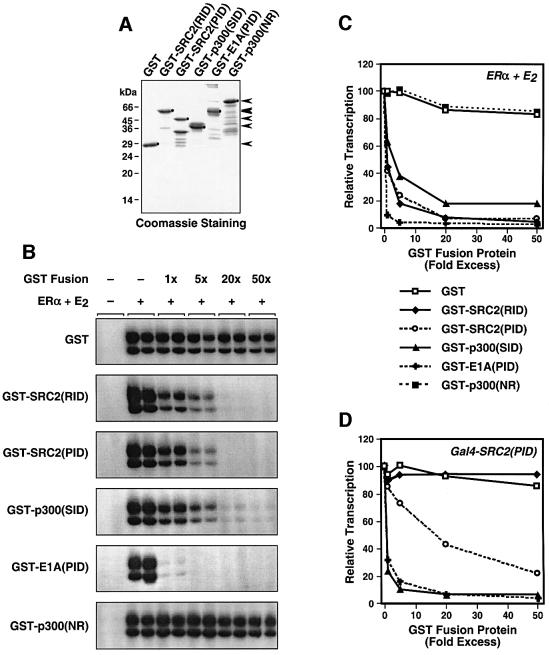
A critical question related to coactivators is whether they are required for nuclear receptor-mediated transcription or whether they simply serve an enhancing role. Thus, we developed a set of inhibitory polypeptides that could specifically block protein–protein interactions leading to ERα-mediated transcription with chromatin (see Figure 1A; ERα–SRC, SRC–p300/CBP and p300/CBP–RNA pol II machinery interactions). We used previously defined factor interaction domains, including SRC2(RID), SRC2(PID), p300(SID) and E1A(PID) (Figure 1B and not shown), which we reasoned might be able to function as dominant-negative inhibitors of the protein–protein interactions shown in Figure 1A. These factor interaction domains are well characterized and have been shown to interact with their cognate binding partners in a variety of assays (McKenna et al., 1999; Leo and Chen, 2000; Robyr et al., 2000). The E1A(PID) fragment has been shown previously to block interactions between p300/CBP and its binding partners, such as RNA pol II complexes (Nakajima et al., 1997b) and TFIIB (Felzien et al., 1999; O’Connor et al., 1999), which it does without inhibiting p300/CBP HAT activity (Kraus et al., 1999). GST fusions of the factor interaction domains (Figure 3A) interacted as expected with their binding partners from the HeLa cell nuclear extract used in the transcription experiments (i.e. p300, CBP and SRC2), as well as with recombinant ERα from an Sf9 cell extract (data not shown).
Fig. 3. Polypeptide inhibitors of interactions among ERα, SRC, p300/CBP and the RNA pol II machinery block ERα-mediated transcription. (A) The indicated fragments of SRC2 and p300 (see Figure 1B), as well as the p300/CBP interaction domain (PID) of the Ad5 E1A protein (residues 1–139), were expressed as GST fusion proteins in E.coli. The purified fusion proteins were analyzed by SDS–PAGE. (B) Chromatin assembly and transcription reactions with ERα + E2 were carried out in the presence of GST-fused SRC, p300 and E1A polypeptides. The polypeptides were added in increasing amounts relative to the concentration of ERα as indicated. (C) Summary of data from multiple transcription experiments like those shown in (B). Errors bars have been omitted for clarity. Each error is <10% of the mean value shown. (D) Experiments similar to those shown in (B) and (C) using Gal4–SRC2(PID) as the activator protein instead of ERα. Line symbols are the same as those used in (C).
Next, we tested the ability of the GST fusions to inhibit ERα-mediated transcription in the context of chromatin using the in vitro transcription system. Note that all of the factors (i.e. p300, CBP, SRC proteins and RNA pol II complexes) except ERα are endogenous components of the HeLa cell nuclear extract. As shown in Figure 3B, GST alone had no effect on ERα-mediated transcription even when used at a 50-fold molar excess over ERα (quantification and summary of multiple experiments are shown in Figure 3C). In contrast, the GST fusions containing SRC2(RID), SRC2(PID), p300(SID) or E1A(PID) were potent inhibitors of ERα-mediated transcription, with all but p300(SID) showing >50% repression at an equimolar amount relative to ERα. As a control, we used the N-terminal region of p300 (NR; residues 1–435), which does not interact strongly with ERα, SRC or RNA pol II (Sheppard et al., 2001; data not shown). Like GST alone, the GST–p300(NR) polypeptide also had no effect on ERα-mediated transcription. Similar results were observed with appropriate inhibitory polypeptides, but not control peptides [i.e. GST and GST–SRC2(RID)], when using Gal4–SRC2(PID) as an activator (Figure 3D).
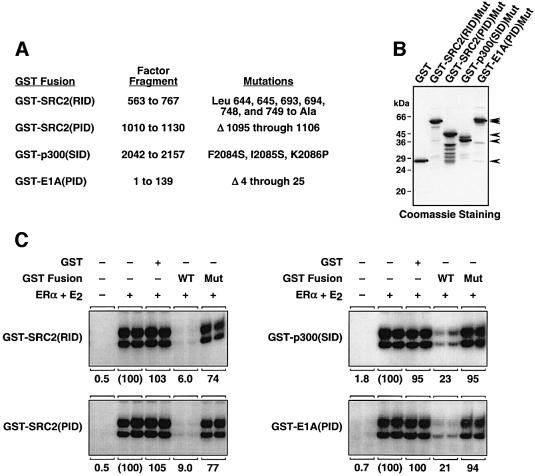
To test the specificity of the inhibitory polypeptides, we introduced mutations into the factor fragments fused to GST (Figure 4A and B). These mutations, which have been characterized previously, disrupt interactions between the factors and their cognate binding partners (Ding et al. 1998; Ma et al., 1999; Nakajima et al., 1997b; Sheppard et al., 2001). The mutant polypeptides were impaired in their ability to inhibit ERα-mediated transcription when added to the in vitro transcription assays (Figure 4C). Together, the experiments in Figures 3 and 4 indicate that polypeptides containing factor interaction domains can function as potent and specific inhibitors of ERα- and coactivator-mediated transcription by blocking critical interactions among the receptor, SRC, p300/CBP and the RNA pol II machinery. Furthermore, they suggest that those interactions are required for ERα-mediated transcription and do not simply serve an enhancing role.
Fig. 4. Mutant coactivator polypeptides have impaired inhibitory activity. (A) List of inhibitory polypeptides and the corresponding mutant versions. (B) The mutant cofactor polypeptides listed in (A) were expressed as GST fusions in E.coli, and the purified proteins were analyzed by SDS–PAGE. (C) The inhibitory activities of the mutant GST-fused SRC, p300 and E1A polypeptides, compared with the wild-type polypeptides, were assessed using chromatin assembly and transcription reactions with ERα + E2 as described in Figure 3B. The polypeptides were added at a 20-fold molar excess relative to ERα, except the GST–E1A(PID) and GST–E1A(PID)mut polypeptides, which were added at a 0.8-fold amount.
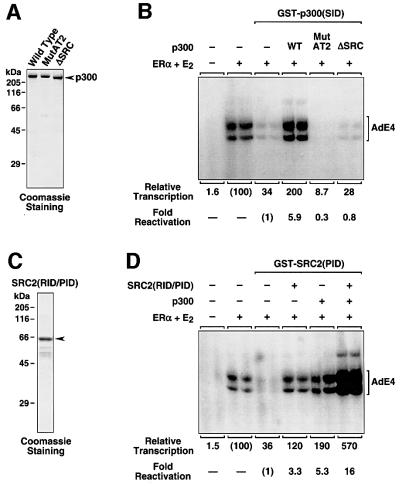
To examine further the specificity of the inhibitory polypeptides, as well as the role of SRC and p300/CBP in ERα-mediated transcription, we asked whether the addition of purified recombinant p300 or SRC2(RID/PID) (see Figure 1B) could relieve the inhibition of ERα-mediated transcription by GST–p300(SID) or GST–SRC2(PID). The addition of purified wild-type p300, but not mutant p300s lacking functional AT activity or an intact SID (MutAT2 and ΔSRC, respectively; Kraus et al., 1999), restored ERα transcriptional activity in the presence of the GST–p300(SID) inhibitor (Figure 5A and B). These results clearly show a requirement for p300 AT activity and SRC–p300 interactions in ERα-mediated transcription. Likewise, wild-type p300, but not a mutant p300 lacking an intact C/H3 (i.e. RNA pol II-binding) domain (ΔC/H3; Kraus et al., 1999), restored ERα transcriptional activity in the presence of the GST–E1A(PID) inhibitor (data not shown). The addition of purified SRC2(RID/PID) or wild-type p300 restored ERα transcriptional activity in the presence of the GST–SRC2(PID) inhibitor (Figure 5C and D). Interestingly, the addition of SRC2(RID/PID) and p300 together had a greater than additive effect (Figure 5D, see ‘Fold reactivation’), indicating a cooperative interaction for these factors in ERα-mediated transcription. The fact that a fragment of SRC2 containing only the RID and PID (see Figure 1B, bottom) was able to relieve transcriptional inhibition by GST–SRC2(PID) and synergize with p300 suggests that other domains of SRC2 are dispensable for ERα-mediated transcription.
Fig. 5. p300 and SRC2(RID/PID) can relieve the repression of ERα-mediated transcription by polypeptide inhibitors. (A and C) SDS–PAGE of purified recombinant His6-tagged p300 and SRC(RID/PID) proteins used in the transcription assays with the chromatin templates shown in (B) and (D). (B) Wild-type p300, but not p300 MutAT2 or p300 ΔSRC, restores ERα-dependent transcriptional activity in GST–p300(SID)-inhibited transcription reactions. GST–p300(SID) was added after chromatin assembly at a 5-fold molar excess relative to ERα to inhibit transcription. Wild-type or mutant p300 proteins were added after chromatin assembly. (D) SRC2(RID/PID) and p300 synergistically restore ERα-dependent transcriptional activity in GST–SRC2(PID)-inhibited transcription reactions. GST–SRC2(PID) was added after chromatin assembly at a 5-fold molar excess relative to ERα to inhibit transcription. SRC2(RID/PID) and p300 were added after chromatin assembly. A lower concentration of p300 (2.5 nM) was used so that synergism with SRC2(RID/PID) could be observed.
SRC2-mediated recruitment of p300 HAT activity to chromatin-bound ERα is sufficient for targeted acetylation of nucleosomal histones
p300/CBP contributes multiple activities to the transcription process (e.g. HAT activity and recruitment of RNA pol II complexes) that are important for ERα-mediated transcription (Figures 3 and 5; Kraus et al., 1999). Thus, the recruitment of p300/CBP by SRC should play an important role in bringing these activities to an estrogen-activated promoter, although it currently is not clear whether recruitment by SRC alone is sufficient. To address more directly the role of SRC in the recruitment of p300/CBP HAT activity to promoters in chromatin, we used a completely biochemically defined targeted histone acetylation assay. The ERE-containing template used for the transcription studies was assembled into chromatin by salt gradient dialysis. ERα, E2, SRC2(RID/PID) and p300 were added in various combinations in the presence of [3H]acetyl CoA, and the targeted acetylation of nucleosomal histones was monitored by fluorography. Note that, for this work, targeted acetylation is defined as acetylation that is dependent on the specific binding of ERα to the EREs in a chromatin template.
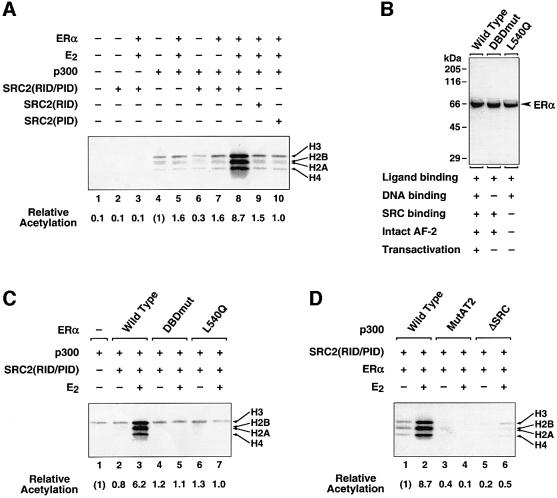
Efficient acetylation of the chromatin template by p300 required ERα, E2 and SRC2(RID/PID) (Figure 6A; compare lanes 4–7 with 8). To explore further the molecular basis for the targeted acetylation of nucleosomal histones by p300, we used two mutant ERα proteins, a DNA-binding mutant (DBDmut; Mader et al., 1989) and an AF-2/helix 12 mutant that exhibits impaired binding to SRC proteins (L540Q; Wrenn and Katzenellenbogen, 1993) (Figure 6B). Neither mutant was able to direct the acetylation of the nucleosomal template by p300 (Figure 6C; compare lanes 5 and 7 with 3), indicating that both the binding of ERα to the EREs in the template and an intact AF-2 (required for the interaction of ERα with SRC2) are necessary for targeted acetylation of nucleosomal histones by p300. We also tested the HAT- and SRC binding-deficient p300 mutants shown in Figure 5A (i.e. MutAT2 and ΔSRC). Note that the ΔSRC mutant retains wild-type HAT activity (Kraus et al., 1999). Neither mutant could support the targeted acetylation of nucleosomal histones in the presence of SRC2 and liganded ERα (Figure 6D, compare lanes 4 and 6 with 2). Together, our results demonstrate that the RID/PID fragment of SRC2 is necessary and sufficient to recruit p300 HAT activity to liganded, chromatin-bound ERα in the absence of ongoing transcription.
Fig. 6. SRC2(RID/PID) is necessary and sufficient for the recruitment of p300 HAT activity to chromatin-bound ERα. (A, C and D) Targeted histone acetylation assays. ERα (wild-type or mutant), E2, p300 (wild-type or mutant), SRC2(RID-PID), SRC2(RID) and/or SRC2(PID) were added as indicated to reactions containing purified salt-dialyzed chromatin and [3H]acetyl CoA. After incubation, the reactions were subjected to electrophoresis on 15% polyacrylamide–SDS gels with subsequent fluorography. The 3H-labeled histone bands were excised from the gel and quantified by liquid scintillation counting. The core histones (H2A, H2B, H3 and H4) and relative acetylation levels are indicated. (B) SDS–PAGE of purified wild-type and mutant ERα proteins used in the HAT assay shown in (C). The various biochemical activities retained by the mutant ER proteins are indicated in the chart below the gel.
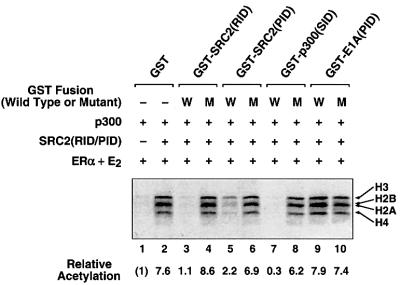
To explore further the recruitment and assembly of the ERα–SRC–p300 complex leading to targeted histone acetylation, we added the wild-type inhibitory polypeptides (shown in Figure 3), as well as the corresponding mutant versions (shown in Figure 4), to the HAT assays. The inhibitory polypeptides that target interactions among ERα, SRC and p300 [i.e. GST–SRC2(RID), GST– SRC2(PID) and GST–p300(SID)], but not the mutant versions, inhibited the targeted acetylation of nucleosomal histones (Figure 7, lanes 1–8). In contrast, GST– E1A(PID), which blocks interactions between p300 and RNA pol II but should not prevent the assembly of the ERα–SRC–p300 complex, did not inhibit histone acetylation (Figure 7, compare lanes 2, 9 and 10). Using the inhibitory polypeptides in this completely defined assay further helps to establish the composition of the ERα-containing complex that assembles at the promoter in response to ligand, leading to targeted histone acetylation. Furthermore, these experiments demonstrate clearly that the protein–protein interactions required for targeted histone acetylation constitute a subset of the interactions required for transcriptional activation.
Fig. 7. Polypeptide inhibitors of interactions among ERα, SRC and p300/CBP block receptor-mediated targeted histone acetylation. Targeted histone acetylation assays were set up as described in Figure 6 in the presence or absence of a 20-fold excess of wild-type (W) or mutant (M) GST-fused SRC, p300 and E1A polypeptides (relative to the amount of ERα), as indicated.
SRC-mediated recruitment of p300/CBP stimulates transcription initiation, but not reinitiation, with chromatin templates
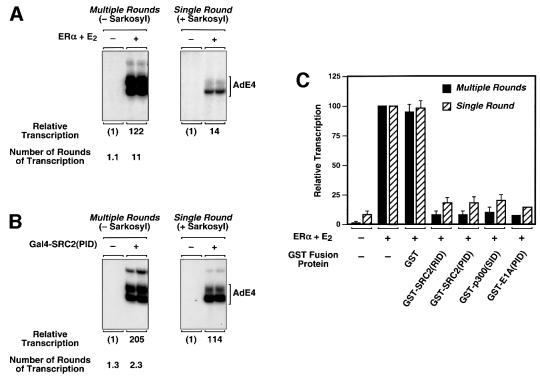
A fundamental question in transcriptional regulation is at which steps in the transcription process do transcriptional activators and coactivators exert their regulatory effects? In previous experiments, ERα was shown to have a dual role in the transcription process with chromatin templates, enhancing the efficiency of both transcription initiation and reinitiation (Kraus and Kadonaga, 1998). FurtherKraus and Kadonaga, 1998). These previous mechanistic experiments, considered in the context of the model shown in Figure 1A, suggested to us that SRC proteins might play a role in transcription initiation by recruiting p300/CBP. To test this possibility, we examined the activity of Gal4– SRC2(PID) in the presence or absence of the detergent Sarkosyl. Sarkosyl limits transcription to a single round (i.e. it blocks reinitiation), allowing the examination of events associated with transcription initiation (Hawley and Roeder, 1985, 1987). For comparison, we examined the activity of ERα under the same conditions.
Unlike ERα, which stimulated an 11-fold increase in the number of rounds of transcription (compared with no activator) (Figure 8A), Gal4–SRC2(PID) stimulated only a 2-fold increase in the number of rounds of transcription (Figure 8B). Thus, Gal4–SRC2(PID), which functions by recruiting p300/CBP, stimulates transcription initiation without affecting reinitiation substantially. Together, these results suggest a specific role for the recruitment of p300/CBP by the SRC2(PID), namely to enhance the formation of a stable transcription pre-initiation complex and subsequent transcription initiation. This conclusion is supported by experiments comparing the activities of the inhibitory GST-fused polypeptides in single versus multiple round transcription experiments (Figure 8C). The polypeptides, which block SRC or p300/CBP function, inhibited ERα-mediated transcription in a single round of transcription, indicating that they target events related to transcription initiation.
Fig. 8. Recruitment of p300/CBP by SRC2 enhances ER-mediated transcription initiation, but not reinitiation, with chromatin templates. The transcriptional activity of ERα and Gal4–SRC2(PID) (A and B), as well as the inhibitory activity of the GST–coactivator polypeptide fusions shown in Figure 3 (C), were analyzed in single round (+Sarkosyl) versus multiple round (–Sarkosyl) transcription experiments with chromatin templates. The number of rounds of transcription obtained was determined by dividing the amount of transcription in the absence of Sarkosyl by the amount of transcription in the corresponding sample in the presence of Sarkosyl. In (C), the GST polypeptides were added at a 50-fold molar excess relative to ERα. ‘ERα + E2’ for each set of samples (±Sarkosyl) was set to 100%. Each bar represents the mean plus the range from two separate determinations.
Discussion
Previous studies have defined a set of interactions among nuclear receptors, the SRC proteins, p300/CBP and the RNA pol II machinery (see Figure 1A) (McKenna et al., 1999; Leo and Chen, 2000; Robyr et al., 2000). In the studies described herein, we have explored the functional contribution of these interactions to ERα-mediated transcription in the context of chromatin using a biochemical approach. Our results indicate that the assembly of complexes containing these factors is required for the targeted acetylation of nucleosomal histones and subsequent transcription initiation. Given the high level of structural and functional conservation among members of the nuclear receptor superfamily, it is likely that these mechanisms are conserved with other nuclear receptors.
Polypeptides containing factor interaction domains function as potent dominant-negative inhibitors of ERα-mediated transcription
Polypeptides that bind specifically to protein factors can block protein–protein interactions, thereby inhibiting the functional activity of the factors. For example, small peptides related to the NR boxes found in many nuclear receptor-interacting proteins can bind to and inhibit the transcriptional activity of ERs in cell-based assays (Norris et al., 1999). In our studies, we have used polypeptides containing previously defined factor interaction domains from p300, SRC2 and the adenovirus E1A protein, and show that they can function as potent inhibitors of ERα-mediated targeted acetylation and transcription in vitro. Our results indicate that the inhibitory polypeptides, in the context of in vitro assays with chromatin templates, are useful tools for exploring the biochemistry of ERα-mediated transcription.
Previous binding studies, as well as the assays shown herein, suggest that the specificity of the p300(SID), SRC2(RID) and SRC2(PID) inhibitors is very high (McKenna et al., 1999; Leo and Chen, 2000). Although the specificity of the E1A(PID) fragment for p300/CBP is likely to be good, the E1A protein also interacts with and regulates the activity of other transcription-related factors (Miller et al., 1996; Boyer et al., 1999; Li et al., 1999), as well as the HAT activities of p300/CBP and PCAF (see e.g. Ait-Si-Ali et al., 1998; Reid et al., 1998; Chakravarti et al., 1999). Furthermore, the C/H3 region of p300/CBP, to which E1A(PID) binds, is also the site of interaction of other factors in addition to components of the RNA pol II machinery (Goodman and Smolik, 2000; Vo and Goodman, 2001). Although these facts might reduce the utility of E1A as a specific inhibitor of p300/CBP–RNA pol II machinery interactions and activity, this is not necessarily the case. For example, the E1A(PID) fragment that we used in our assays does not inhibit the HAT activity of p300/CBP (Kraus et al., 1999). In addition, the interaction of RNA pol II complexes with the C/H3 region, as well as the inhibitory effects of E1A on the binding of RNA pol II to that region, have been well characterized using a variety of biochemical assays (Nakajima et al., 1997a,b). Likewise, deletional analyses of the p300 C/H3 region have demonstrated a critical role for that region in ERα-mediated transcription (Kraus et al., 1999). Finally, we have found that wild-type p300, but not a mutant p300 lacking an intact C/H3 (i.e. E1A- and RNA pol II-binding) domain (ΔC/H3; Kraus et al., 1999), restores ERα transcriptional activity in the presence of the GST– E1A(PID) inhibitor (data not shown). Thus, although we cannot completely exclude the possibility that other p300/CBP–factor interactions are being blocked or the activities of other factors are being altered, the results with the E1A(PID) fragment are consistent with the inhibition of functional interactions between the C/H3 region of p300/CBP and components of the RNA pol II machinery.
The inhibitory polypeptides have provided new insights into the biochemistry of ERα-mediated transcription. For example, using the polypeptides, we have found that interactions among ERα, SRC, p300/CBP and the RNA pol II machinery are required for ERα-mediated transcription and do not simply serve an enhancing role (Figure 3). Together, these results indicate that the intrinsic transcriptional activity of ERα is low and that ERα functions primarily as a nucleation site for the assembly of coactivator complexes at promoters assembled into chromatin. In addition, our experiments using the inhibitory polypeptides suggest that interactions among SRC, p300/CBP and RNA pol II complexes are dynamic because, to function as inhibitors, the polypeptides must disrupt interactions between the native complexes present in the HeLa cell nuclear extract. Also, the inhibitory polypeptide complementation studies (Figure 5D) have provided clear biochemical evidence for synergism between SRC and p300/CBP in ERα- mediated transcription. Furthermore, the complementation assays using the p300 mutants (Figure 5B) have helped to define the activities of p300 that are required for ERα-mediated transcription, demonstrating a critical role for p300–SRC interactions and p300 AT activity. In future studies, it will be interesting and informative to use these inhibitory polypeptides to explore receptor–coactivator interactions in vivo. Recent studies using dominant-negative coactivator fragments in cell-based assays suggest that such an approach will be feasible (Voegel et al., 1998; Li et al., 2000).
Order for the assembly and function of an ERα–SRC–p300/CBP complex
The fact that artificial recruitment of p300/CBP by Gal4– SRC2(PID), but not artificial recruitment of SRC by Gal4–p300(SID), is sufficient for potent transcriptional activation (Figure 2C) indicates an order for the assembly and function of SRC–p300/CBP complexes that was suggested by previous interaction assays, i.e. SRC recruits p300/CBP and not vice versa. Recent studies indicate, in contrast to earlier reports, that p300/CBP does not interact directly with some nuclear receptors, including ERα (Li et al., 2000; Sheppard et al., 2001). Thus, one important function served by the SRC proteins is to recruit p300/CBP to liganded receptors bound at the promoter. In fact, our results indicate that this may be the most important function served by the SRC proteins since a minimal fragment of SRC2 containing the RID and PID domains, but lacking the putative HAT domain, was sufficient to stimulate ERα-mediated transcription and synergize with p300 (Figure 5D). Similar effects with SRC(RID/PID) fragments have been observed using cell-based assays (Voegel et al., 1998; Sheppard et al., 2001). Additional support for the primary role of SRC proteins as bridging factors was provided by a recent report using a genetic approach with cell-based assays (Mak and Parker, 2001). Although p300/CBP may stabilize interactions involving SRC at the promoter, our results suggest that p300/CBP does not play a significant role in recruiting SRC to promoters assembled into chromatin.
ERα–SRC–p300/CBP interactions leading to the targeted acetylation of nucleosomal histones and the activation of transcription
Our results indicate that the SRC-mediated recruitment of two p300/CBP activities, namely HAT activity and transcriptional intermediary activity (i.e. interactions with the RNA pol II machinery), is required for ERα- mediated transcription. In the biochemically defined targeted HAT assays (Figures 6 and 7), we show that the recruitment of p300 to liganded, chromatin-bound ERα via SRC is necessary and sufficient for the efficient acetylation of nucleosomal histones. The studies with the ERα and p300 mutants in the acetylation assay emphasize the requirement for the simultaneous binding of SRC to both p300 and ERα for acetylation. In addition, our studies indicate that p300 is a relatively poor nucleosomal HAT in the absence of tethering to a chromatin-bound factor, such as ERα.
The results from the acetylation assays show a good correlation between the targeted acetylation of nucleosomal histones by p300 and the transcriptional activity of ERα. For example, both the acetylation of nucleosomal histones by p300 (Figure 6A) and the transcriptional activity of ERα with chromatin templates (Figure 2A) are ligand dependent. Likewise, the AF-2- and DNA-binding mutant ERαs, as well as the MutAT2 and ΔSRC mutant p300s, are unable to support the acetylation of nucleosomal histones (Figure 6C and D) or estrogen-stimulated transcription (Figure 5B; data not shown; Kraus and Kadonaga, 1998; Kraus et al., 1999). Note, however, that the protein–protein interactions required for histone acetylation constitute a subset of the interactions required for transcriptional activation, since p300–RNA pol II interactions are required for transcription (Figure 3) but not targeted histone acetylation (Figure 7). The results from our biochemical assays are in good agreement with recent chromatin immunoprecipitation (ChIP) assays showing ligand-dependent localization of acetylated histones, SRC proteins and p300/CBP at the promoters of estrogen-regulated genes in vivo (Chen et al., 1999; Shang et al., 2000). The question of whether p300/CBP-mediated histone acetylation is localized to the promoter or extends to regions upstream or downstream has not been resolved clearly (Dilworth et al., 2000; Kundu et al., 2000).
Interestingly, the acetylation of nucleosomal histones in our assays was mediated efficiently by the SRC2(RID/PID) fragment, which lacks the putative C-terminal HAT domain (Chen et al., 1997; Spencer et al., 1997). As mentioned above, this same fragment also supported ERα-mediated transcription and synergism with p300. Thus, the SRC2(RID/PID) fragment is sufficient to support both targeted histone acetylation and transcription with chromatin templates. These results support the conclusion that the most important role for the SRC proteins in ERα-mediated transcription is to recruit p300/CBP to estrogen-regulated promoters in a ligand-dependent manner.
A specific role for ERα–SRC–p300/CBP interactions in the transcription process
The transcription process can be divided into a number of distinct steps including: (i) the assembly of a pre-initiation complex; (ii) initiation and promoter clearance; (iii) elongation; and (iv) reinitiation (Hawley and Roeder, 1985, 1987). In the context of a chromatin template, chromatin remodeling and modification events also occur during the transcription process (Kingston and Narlikar, 1999; Kornberg and Lorch, 1999). Different transcription-related factors play distinct roles in one or more of these processes. Herein, we demonstrate that SRC stimulates transcription initiation by recruiting p300/CBP, without substantially affecting transcription reinitiation. p300/CBP, once recruited, can contribute various activities toward transcription initiation, including the recruitment of the RNA pol II machinery and the acetylation of nucleosomal histones. These results are in good agreement with previous studies showing a role for p300 in ERα- mediated transcription initiation (Kraus and Kadonaga, 1998) and a requirement for p300 AT activity in transcription initiation (Kraus et al., 1999). Furthermore, we find that targeted histone acetylation occurs in the absence of ongoing transcription (Figures 6 and 7), which suggests that it precedes and is required for a subsequent step, such as the assembly of a stable pre-initiation complex. This fits well with recent demonstrations of an acetyl-coenzyme A-dependent event required for the stimulation of transcription initiation by ERα and retinoic acid receptors with chromatin templates (Dilworth et al., 2000; Jiang et al., 2000).
What might be the role of activator-targeted nucleosomal histone acetylation by p300/CBP in transcription initiation? Although histone acetylation is thought to facilitate ATP-dependent chromatin remodeling, recent studies suggest that the AT activity of p300/CBP is not required for chromatin remodeling, but rather for a subsequent step in the transcription process (Li et al., 1999; Dilworth et al., 2000; Ito et al., 2000; Kundu et al., 2000). In fact, the post-remodeling acetylation of nucleosomal histones by p300 has been shown to facilitate the transfer of histone H2A–H2B dimers from nucleosomes to NAP-1, a histone chaperone (Ito et al., 2000). The resulting altered nucleosome structure may help to maintain an open chromatin conformation that is conducive to the formation of a stable transcription pre-initiation complex and transcription initiation. This model for the timing of events is in agreement with recent in vitro order-of-addition experiments (Dilworth et al., 2000; Kundu et al., 2000), as well as ChIP assays showing ligand-dependent association of p300 with estrogen-regulated promoters in vivo prior to the recruitment of RNA pol II (Shang et al., 2000).
Materials and methods
Synthesis and purification of recombinant proteins
Wild-type and mutant FLAG-tagged ER proteins and His6-tagged p300 proteins were expressed and purified as described previously (Kraus and Kadonaga, 1999). The ER mutants [L540Q and DBDmut (previously referred to as HE82)] have been described elsewhere (Mader et al., 1989; Wrenn and Katzenellenbogen, 1993). The p300 mutants (MutAT2 and ΔSRC) have also been described elsewhere (Kraus et al., 1999). GST–E1A(PID) (amino acids 1–139 of E1A) and Gal4-VP16 were expressed in E.coli and purified as described previously (Chasman et al., 1989; Nakajima et al., 1997b). His6-tagged coactivator fragments, His6-tagged Gal4(DBD) fusions and GST–coactivator fragment fusions were expressed in E.coli and purified using standard nickel-NTA or glutathione–agarose affinity chromatography. The coactivator fragments contained the following amino acid residues of mouse SRC2 or human p300: SRC2(RID), 563–767; SRC2(PID), 1010–1130; SRC2(RID/PID), 624–1130; p300(SID), 2042–2157; p300(NR), 1–435 (see Figure 1B). The mutations in the factor fragments listed in Figure 4A are based on previously characterized mutations (Nakajima et al., 1997b; Ding et al., 1998; Ma et al., 1999; Sheppard et al., 2001) and were generated by PCR.
In vitro chromatin assembly and transcription reactions
The pERE and pGIE0 plasmid templates have been described elsewhere (Pazin et al., 1994; Kraus and Kadonaga, 1998). Chromatin assembly reactions were performed as described previously (Kraus and Kadonaga, 1999) using a chromatin assembly extract derived from Drosophila embryos (Bulger and Kadonaga, 1994). Transcriptional activator proteins [e.g. ER ± E2 and the Gal4(DBD) fusions] were added during the chromatin assembly reactions, whereas coactivators [e.g. p300 and SRC2(RID/PID)] and the GST fusion proteins were added after chromatin assembly was complete. The final concentrations of the factors in the transcription reactions were as follows, unless noted otherwise: ERα (3 nM), E2 (30 nM), Gal4(DBD) fusions (7.5 nM), p300 (30 nM) and SRC2(RID/PID) (50 nM). Multiple and single round in vitro transcription reactions with the chromatin templates were performed as described previously (Kraus and Kadonaga, 1999). All reactions were performed in duplicate, and each experiment was performed a minimum of three separate times to ensure reproducibility. The data were analyzed and quantified using a PhosphorImager (Molecular Dynamics).
Histone acetylation assays with reconstituted chromatin templates
Salt-dialyzed chromatin, prepared essentially as described previously (Jeong et al., 1991) using the pERE plasmid template, was purified on a linear 15–40% (w/v) sucrose gradient. For the acetylation assays, a 20 µl aliquot of chromatin containing ∼1.2 µg of plasmid DNA and 0.9 µg of histones was mixed with [3H]acetyl CoA (5 µM) and various combinations of factors [i.e. ERα (200 nM), p300 (40 nM), SRC2(RID/ PID) (40 nM) and E2 (1 µM)] as indicated in a final volume of 35 µl under reaction conditions described previously (Kraus et al., 1999; Manning et al., 2001). GST-fused SRC, p300 and E1A polypeptides were added at a 20-fold excess relative to the amount of ERα, as indicated. The reactions were incubated at 27°C for 30 min and then analyzed on 15% polyacrylamide–SDS gels with subsequent fluorography. The 3H-labeled histones were excised from the gel and quantified by liquid scintillation counting.
Acknowledgments
Acknowledgements
We thank Edwin Cheung, John Lis and Erik Andrulis for critical reading of this manuscript. We are grateful to Mike Stallcup for the wild-type and mutant SRC2 cDNAs, Marc Montminy for the GST–E1A constructs, Beatrice Darimont for the GST–SRC2(RID) construct, Edwin Cheung for the mutant ERα proteins, and Tory Manning for the p300 proteins. This work was supported by a Career Award in the Biomedical Sciences from the Burroughs Wellcome Fund and a grant from the National Institutes of Health (DK58110) to W.L.K.
References
- Ait-Si-Ali S. et al. (1998) Histone acetyltransferase activity of CBP is controlled by cycle-dependent kinases and oncoprotein E1A. Nature, 396, 184–186. [DOI] [PubMed] [Google Scholar]
- Bannister A.J. and Kouzarides,T. (1996) The CBP co-activator is a histone acetyltransferase. Nature, 384, 641–643. [DOI] [PubMed] [Google Scholar]
- Boyer T.G., Martin,M.E., Lees,E., Ricciardi,R.P. and Berk,A.J. (1999) Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature, 399, 276–279. [DOI] [PubMed] [Google Scholar]
- Bulger M. and Kadonaga,J.T. (1994) Biochemical reconstitution of chromatin with physiological nucleosome spacing. Methods Mol. Genet., 5, 241–262. [Google Scholar]
- Chakravarti D., Ogryzko,V., Kao,H.Y., Nash,A., Chen,H., Nakatani,Y. and Evans,R.M. (1999) A viral mechanism for inhibition of p300 and PCAF acetyltransferase activity. Cell, 96, 393–403. [DOI] [PubMed] [Google Scholar]
- Chasman D.I., Leatherwood,J., Carey,M., Ptashne,M. and Kornberg,R.D. (1989) Activation of yeast polymerase II transcription by herpesvirus VP16 and GAL4 derivatives in vitro. Mol. Cell. Biol., 9, 4746–4749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Schiltz,R.L., Chakravarti,D., Nash,A., Nagy,L., Privalsky,M.L., Nakatani,Y. and Evans,R.M. (1997) Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell, 90, 569–580. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Xie,W., Wilpitz,D. and Evans,R.M. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- Couse J.F. and Korach,K.S. (1999) Estrogen receptor null mice: what have we learned and where will they lead us? Endocr. Rev., 20, 358–417. [DOI] [PubMed] [Google Scholar]
- Darimont B.D., Wagner,R.L., Apriletti,J.W., Stallcup,M.R., Kushner,P.J., Baxter,J.D., Fletterick,R.J. and Yamamoto,K.R. (1998) Structure and specificity of nuclear receptor–coactivator interactions. Genes Dev., 12, 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth F.J., Fromental-Ramain,C., Yamamoto,K. and Chambon,P. (2000) ATP-driven chromatin remodeling activity and histone acetyltransferases act sequentially during transactivation by RAR/RXR in vitro. Mol. Cell, 6, 1049–1058. [DOI] [PubMed] [Google Scholar]
- Ding X.F., Anderson,C.M., Ma,H., Hong,H., Uht,R.M., Kushner,P.J. and Stallcup,M.R. (1998) Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol. Endocrinol., 12, 302–313. [DOI] [PubMed] [Google Scholar]
- Felzien L.K., Farrell,S., Betts,J.C., Mosavin,R. and Nabel,G.J. (1999) Specificity of cyclin E–Cdk2, TFIIB and E1A interactions with a common domain of the p300 coactivator. Mol. Cell. Biol., 19, 4241–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Hawley D.K. and Roeder,R.G. (1985) Separation and partial characterization of three functional steps in transcription initiation by human RNA polymerase II. J. Biol. Chem., 260, 8163–8172. [PubMed] [Google Scholar]
- Hawley D.K. and Roeder,R.G. (1987) Functional steps in transcription initiation and reinitiation from the major late promoter in a HeLa nuclear extract. J. Biol. Chem., 262, 3452–3461. [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven,E., Hoare,S. and Parker,M.G. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature, 387, 733–736. [DOI] [PubMed] [Google Scholar]
- Ito T., Ikehara,T., Nakagawa,T., Kraus,W.L. and Muramatsu,M. (2000) p300-mediated acetylation facilitates the transfer of histone H2A–H2B dimers from nucleosomes to a histone chaperone. Genes Dev., 14, 1899–1907. [PMC free article] [PubMed] [Google Scholar]
- Jeong S.W., Lauderdale,J.D. and Stein,A. (1991) Chromatin assembly on plasmid DNA in vitro. Apparent spreading of nucleosome alignment from one region of pBR327 by histone H5. J. Mol. Biol., 222, 1131–1147. [DOI] [PubMed] [Google Scholar]
- Jiang W., Nordeen,S.K. and Kadonaga,J.T. (2000) Transcriptional analysis of chromatin assembled with purified ACF and dNAP1 reveals that acetyl CoA is required for preinitiation complex assembly. J. Biol. Chem., 275, 39819–39822. [DOI] [PubMed] [Google Scholar]
- Kamei Y. et al. (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell, 85, 403–414. [DOI] [PubMed] [Google Scholar]
- Kingston R.E. and Narlikar,G.J. (1999) ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev., 13, 2339–2352. [DOI] [PubMed] [Google Scholar]
- Kornberg R.D. and Lorch,Y. (1999) Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev., 9, 148–151. [DOI] [PubMed] [Google Scholar]
- Kraus W.L. and Kadonaga,J.T. (1998) p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev., 12, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus W.L. and Kadonaga,J.T. (1999) Ligand- and cofactor-regulated transcription with chromatin templates. In Picard,D. (ed.), Steroid/Nuclear Receptor Superfamily: A Practical Approach. Oxford University Press, Oxford, pp. 167–189.
- Kraus W.L., Manning,E.T. and Kadonaga,J.T. (1999) Biochemical analysis of distinct activation functions in p300 that enhance transcription initiation with chromatin templates. Mol. Cell. Biol., 19, 8123–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kundu T.K., Palhan,V.B., Wang,Z., An,W., Cole,P.A. and Roeder,R.G. (2000) Activator-dependent transcription from chromatin in vitro involving targeted histone acetylation by p300. Mol. Cell, 6, 551–561. [DOI] [PubMed] [Google Scholar]
- Leo C. and Chen,J.D. (2000) The SRC family of nuclear receptor coactivators. Gene, 245, 1–11. [DOI] [PubMed] [Google Scholar]
- Li H. and Chen,J.D. (1998) The receptor-associated coactivator 3 activates transcription through CREB-binding protein recruitment and autoregulation. J. Biol. Chem., 273, 5948–5954. [DOI] [PubMed] [Google Scholar]
- Li Q., Imhof,A., Collingwood,T.N., Urnov,F.D. and Wolffe,A.P. (1999) p300 stimulates transcription instigated by ligand-bound thyroid hormone receptor at a step subsequent to chromatin disruption. EMBO J., 18, 5634–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., O’Malley,B.W. and Wong,J. (2000) p300 requires its histone acetyltransferase activity and SRC-1 interaction domain to facilitate thyroid hormone receptor activation in chromatin. Mol. Cell. Biol., 20, 2031–2042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H., Hong,H., Huang,S.M., Irvine,R.A., Webb,P., Kushner,P.J., Coetzee,G.A. and Stallcup,M.R. (1999) Multiple signal input and output domains of the 160-kilodalton nuclear receptor coactivator proteins. Mol. Cell. Biol., 19, 6164–6173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mader S., Kumar,V., de Verneuil,H. and Chambon,P. (1989) Three amino acids of the oestrogen receptor are essential to its ability to distinguish an oestrogen from a glucocorticoid-responsive element. Nature, 338, 271–274. [DOI] [PubMed] [Google Scholar]
- Mak H.Y. and Parker,M.G. (2001) Use of suppressor mutants to probe the function of estrogen receptor–p160 coactivator interactions. Mol. Cell. Biol., 21, 4379–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf D.J. et al. (1995) The nuclear receptor superfamily: the second decade. Cell, 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning E.T., Ikehara,T., Ito,T., Kadonaga,J.T. and Kraus,W.L. (2001) p300 forms a stable, template-committed complex with chromatin: role for the bromodomain. Mol. Cell. Biol., 21, 3876–3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna N.J., Lanz,R.B. and O’Malley,B.W. (1999) Nuclear receptor coregulators: cellular and molecular biology. Endocr. Rev., 20, 321–344. [DOI] [PubMed] [Google Scholar]
- Miller M.E., Cairns,B.R., Levinson,R.S., Yamamoto,K.R., Engel,D.A. and Smith,M.M. (1996) Adenovirus E1A specifically blocks SWI/SNF-dependent transcriptional activation. Mol. Cell. Biol., 16, 5737–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizzen C.A. and Allis,C.D. (1998) Linking histone acetylation to transcriptional regulation. Cell. Mol. Life Sci., 54, 6–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima T., Uchida,C., Anderson,S.F., Lee,C.G., Hurwitz,J., Parvin,J.D. and Montminy,M. (1997a) RNA helicase A mediates association of CBP with RNA polymerase II. Cell, 90, 1107–1112. [DOI] [PubMed] [Google Scholar]
- Nakajima T., Uchida,C., Anderson,S.F., Parvin,J.D. and Montminy,M. (1997b) Analysis of a cAMP-responsive activator reveals a two-component mechanism for transcriptional induction via signal-dependent factors. Genes Dev., 11, 738–747. [DOI] [PubMed] [Google Scholar]
- Nolte R.T. et al. (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature, 395, 137–143. [DOI] [PubMed] [Google Scholar]
- Norris J.D., Paige,L.A., Christensen,D.J., Chang,C.Y., Huacani,M.R., Fan,D., Hamilton,P.T., Fowlkes,D.M. and McDonnell,D.P. (1999) Peptide antagonists of the human estrogen receptor. Science, 285, 744–746. [DOI] [PubMed] [Google Scholar]
- O’Connor M.J., Zimmermann,H., Nielsen,S., Bernard,H.U. and Kouzarides,T. (1999) Characterization of an E1A–CBP interaction defines a novel transcriptional adapter motif (TRAM) in CBP/p300. J. Virol., 73, 3574–3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko V.V., Schiltz,R.L., Russanova,V., Howard,B.H. and Nakatani,Y. (1996) The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell, 87, 953–959. [DOI] [PubMed] [Google Scholar]
- Pazin M.J., Kamakaka,R.T. and Kadonaga,J.T. (1994) ATP-dependent nucleosome reconfiguration and transcriptional activation from preassembled chromatin templates. Science, 266, 2007–2011. [DOI] [PubMed] [Google Scholar]
- Reid J.L., Bannister,A.J., Zegerman,P., Martinez-Balbas,M.A. and Kouzarides,T. (1998) E1A directly binds and regulates the P/CAF acetyltransferase. EMBO J., 17, 4469–4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robyr D., Wolffe,A.P. and Wahli,W. (2000) Nuclear hormone receptor coregulators in action: diversity for shared tasks. Mol. Endocrinol., 14, 329–347. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Sheppard H.M., Harries,J.C., Hussain,S., Bevan,C. and Heery,D.M. (2001) Analysis of the steroid receptor coactivator 1 (SRC1)–CREB binding protein interaction interface and its importance for the function of SRC1. Mol. Cell. Biol., 21, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiau A.K., Barstad,D., Loria,P.M., Cheng,L., Kushner,P.J., Agard,D.A. and Greene,G.L. (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell, 95, 927–937. [DOI] [PubMed] [Google Scholar]
- Spencer T.E. et al. (1997) Steroid receptor coactivator-1 is a histone acetyltransferase. Nature, 389, 194–198. [DOI] [PubMed] [Google Scholar]
- Torchia J., Rose,D.W., Inostroza,J., Kamei,Y., Westin,S., Glass,C.K. and Rosenfeld,M.G. (1997) The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature, 387, 677–684. [DOI] [PubMed] [Google Scholar]
- Vo N. and Goodman,R.H. (2001) CREB-binding protein and p300 in transcriptional regulation. J. Biol. Chem., 276, 13505–13508. [DOI] [PubMed] [Google Scholar]
- Voegel J.J., Heine,M.J., Tini,M., Vivat,V., Chambon,P. and Gronemeyer,H. (1998) The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J., 17, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner M., Nilsson,S. and Gustafsson,J.A. (1999) The estrogen receptor family. Curr. Opin. Obstet. Gynecol., 11, 249–254. [DOI] [PubMed] [Google Scholar]
- Winston F. and Allis,C.D. (1999) The bromodomain: a chromatin-targeting module? Nature Struct. Biol., 6, 601–604. [DOI] [PubMed] [Google Scholar]
- Wolffe A.P. and Kurumizaka,H. (1998) The nucleosome: a powerful regulator of transcription. Prog. Nucleic Acid Res. Mol. Biol., 61, 379–422. [DOI] [PubMed] [Google Scholar]
- Wrenn C.K. and Katzenellenbogen,B.S. (1993) Structure–function analysis of the hormone binding domain of the human estrogen receptor by region-specific mutagenesis and phenotypic screening in yeast. J. Biol. Chem., 268, 24089–24098. [PubMed] [Google Scholar]
- Yang X.J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]