Abstract
Geminiviruses have spread worldwide and have become increasingly important in crop plants during recent decades. Recombination among geminiviruses was one major source of new variants. Geminiviruses replicate via rolling circles, confirmed here by electron microscopic visualization and two-dimensional gel analysis of Abutilon mosaic virus (AbMV) DNA. However, only a minority of DNA intermediates are consistent with this model. The majority are compatible with recombination-dependent replication (RDR). During development of naturally infected leaves, viral intermediates compatible with both models appeared simultaneously, whereas agro-infection of leaf discs with AbMV led to an early appearance of RDR forms but no RCR intermediates. Inactivation of viral genes ac2 and ac3 delayed replication, but produced the same DNA types as after wild-type infection, indicating that these genes were not essential for RDR in leaf discs. In conclusion, host factors alone or in combination with the viral AC1 protein are necessary and sufficient for the production of RDR intermediates. The consequences of an inherent geminiviral recombination activity for the use of pathogen-derived resistance traits are discussed.
Keywords: chloroquine/geminivirus/replication/two-dimensional gel electrophoresis
Introduction
Geminiviruses are an increasing major threat to crop plants worldwide, especially in tropical and subtropical countries (Moffat, 1999). One reason for the appearance of epidemics is the spread of the vectors for geminiviruses: a single species of whiteflies, Bemisia tabaci, or several species of leafhoppers (Rybicki and Pietersen, 1999). Another reason has been associated with the recombination of different geminiviruses (Zhou et al., 1997, 1998). Footprints of recombination were found upon sequence comparisons of the DNA of several geminiviruses (Padidam et al., 1999). All this evidence suggests that recombination might be a major driving force for the evolution of geminiviruses and their ability to break resistance in crop plants.
In experiments, recombination occurred quickly if two handicapped virus constructs were co-inoculated (Evans and Jeske, 1993), e.g. with mutated genes for the control of replication or transcription, or if a single construct deviated considerably from viral genome size (Bisaro, 1994). Recombination was also detected between a mutated virus (ΔCP) and its homologous transgene (CP), as shown for African cassava mosaic virus (ACMV) in Nicotiana benthamiana plants (Frischmuth and Stanley, 1998). The latter finding needs attention as many approaches have aimed to create resistance by introducing geminiviral genes into host plant chromosomes, the so-called ‘pathogen derived resistance’ (Frischmuth and Stanley, 1993). Abutilon mosaic virus (AbMV) has been propagated on ornamental Abutilon plants by gardeners (Wege et al., 2000). The single-stranded (ss) circular DNA of geminiviruses is packed into twin-shaped virions (Zhang et al., 2001). They are classified into four genera: Mastrevirus, Curtovirus, Topocuvirus and Begomovirus (Rybicki et al., 2000). Mastreviruses and curtoviruses are transmitted by leafhoppers and possess a monopartite genome, begomoviruses are spread by whiteflies and most of them have a bipartite genome, called DNA A and DNA B. Sequence comparisons between the genera have led to the suggestion that curtoviruses have evolved from a recombination of ancient mastreviruses and begomoviruses (Stanley et al., 1986). Both DNA components of begomoviruses contain different sequences except for a ‘common region’ (CR; Figure 2) of ∼200 bp. The CR is nearly identical in DNA A and B of a single virus. It includes promoters and the origin of replication. A small hairpin–loop-forming sequence in the CR is highly conserved among all geminiviruses (Hanley-Bowdoin et al., 1999). Geminiviruses replicate via double-stranded circular intermediates, which form minichromosomes within the nuclei of infected cells (Abouzid et al., 1988; Pilartz and Jeske, 1992). Analysing the intermediates by two-dimensional gel electrophoresis (Saunders et al., 1991), as well as indirect genetic evidence (Stenger et al., 1991), supported the now generally accepted model that geminiviruses replicate via a rolling circle mechanism (RCR; for review see Hanley-Bowdoin et al., 1999).
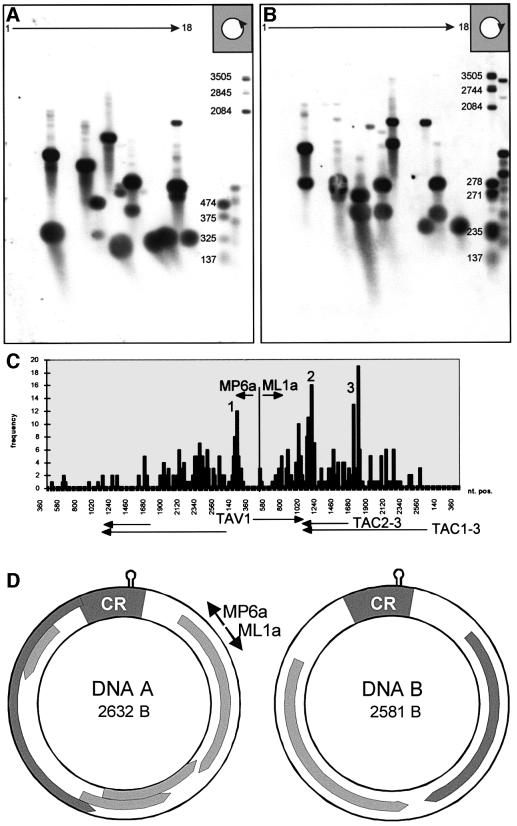
Fig. 2. CsCl-purified DNA from pooled fractions containing lin, oc and hDNA (as seen in Figure 1D) was run on a first agarose gel (1%; TBE); the lane was cut into 18 slices. Eluted DNA was analysed by multiple primer extension, products were run on a second agarose gel (1%; alkaline), blotted and hybridized against DNA A. For primer extension, either primer MP6a (A) or ML1a (B) were used to map DNA ends in the viral (A) or complementary (B) sense. A heterogeneous population of discrete bands resulted for both orientations. The sizes of standards (in bp) are indicated on the right. Several independent primer extension experiments as seen in (A) and (B) have been performed and evaluated. From the size of the primer extension products, the putative genome position of the linDNA ends on AbMV DNA A was deduced. The cumulation of results from 384 samples is represented graphically (C). Most of the ends are scattered across the complete genome length, with higher frequencies around the origin of replication (1) for viral sense DNA, the transcription termination site (2) and the promotor site of the transcript for AC2 and AC3 for complementary sense. AbMV transcripts (TAV1, TAC1-3, TAC2-3) are shown in the linear map for orientation. (D) Genome organization of AbMV including positions of primers (MP6a and ML1a). CR, common region of DNA A and DNA B.
The RCR model explains easily how viral ssDNA is produced from the double-stranded template at the final stage of the multiplication cycle. However, the model is unusual as geminiviruses transcribe bidirectionally (Townsend et al., 1985; Petty et al., 1988; Frischmuth et al., 1991), thus risking collisions between replication and transcription complexes. Brewer (1988) has reviewed the resulting problems and emphasized the evolutionary advantage of replication and transcription proceeding in the same orientation. Being aware of this discrepancy and trying to unravel the recombination mechanism, we asked whether RCR is the only mode of geminivirus replication. The following results, based on two-dimensional gel electrophoresis and electron microscopy, indicate that geminiviruses might multiply by two mechanisms: RCR and recombination-dependent replication (RDR).
Results
hDNA characterization
When DNA samples of AbMV-infected leaves were analysed by electrophoresis in the presence of ethidium bromide, blotting and virus-specific hybridization, several bands appeared, as seen in Figure 1A. The ssDNA was well separated from covalently closed (ccc), linear (lin) and open circular (oc) double-stranded (ds) molecules. This is the general DNA picture after geminivirus multiplication. When different samples were compared, originating from all leaves of an Abutilon shoot (Figure 1A), beginning with the apex (A) and the smallest leaf (#1), additional hybridizing material accumulated transiently during leaf development. Most prominent in leaf #5, heterogeneous high-molecular-weight DNA (hDNA), co-migrating with chromosomal host DNA merged into a downward smear of hybridizing DNA. In 72 samples, each of infected or uninfected leaves, the smear appeared only in the infected ones, excluding unspecific binding of the probe to chromosomal DNA or to DNA of sporadically appearing bacterial contaminants on leaf surfaces (data not shown). Two further reasons for an artificial smear were excluded by the results shown in Figure 1B. A smear might result from gel overloading or from contaminating proteins and RNA. In addition, with Abutilon plants it is crucial to remove all the mucilage, which is present in tremendous amounts in the sap. In order to control these possible artefacts, DNA was extracted from a first gel and re-run on a second gel in the presence of SDS, either directly or after treatment with RNase or proteinase (Figure 1B). The results confirmed that a substantial part of the smear represents true hDNA molecules migrating at the same position as in the first electrophoresis. When DNA of virus-infected plants was fractionated on CsCl gradients either in the presence or absence of ethidium bromide, the hDNA co-migrated with dsDNA (Figure 1C) and was present in the pool of oc and lin-dsDNA (Figure 1D), but not of cccDNA. These results further exclude an interaction of hDNA with proteins or other molecules and are consistent with the interpretation that hDNA is predominantly dsDNA without superhelicity.
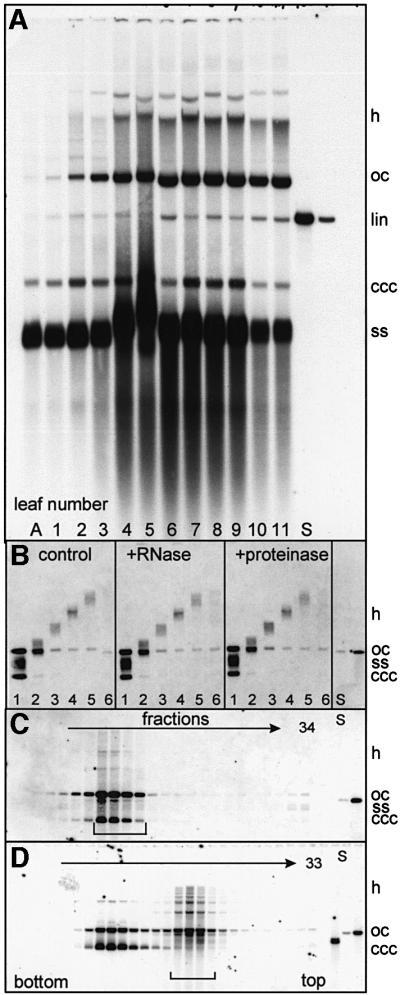
Fig. 1. Detection of gel-separated AbMV DNA from naturally infected Abutilon following Southern blotting and hybridization against a DNA A probe. Gels (1% agarose) were run in the presence of ethidium bromide (A) or SDS (B–D). (A) Samples (100 ng of DNA/lane) of different leaves from a single Abutilon shoot (A to 11) were loaded. (B) An agarose lane [as in #5 of (A)] from a separate gel was cut into six pieces, #1 being the fraction of highest mobility. DNA was extracted and one-third was re-run untreated (control), or pre-treated with RNase or proteinase before. After re-electrophoresis, the smear was still present and unaffected by RNase or proteinase treatment. The only form that was separated from the smear was ocDNA in all fractions. This behaviour of ocDNA was often observed under several conditions and is possibly the result of a release of ocDNA from linDNA molecules that had been threaded through the ring of ocDNA. Such a type of mechanical interaction is mostly disrupted by the addition of SDS to the gel buffer as seen here. (C and D) Viral DNA intermediates of infected Abutilon plants analysed by two methods of isopycnic CsCl gradient centrifugation in the absence (C) and presence (D) of ethidium bromide. Ethidium bromide intercalates differentially into supercoiled and linDNA or ocDNA, thereby separating both DNA types. From 34 or 33 fractions collected from the bottom of the tube after centrifugation, DNAs were extracted and analysed on an SDS-containing agarose gel, blotted and hybridized against a DNA A probe. (C) hDNA was found in the fractions of dsDNA (bracket). Its amount is somewhat lower in comparison with (A) as heterogeneous ssDNA components had been removed by the separation. (D) hDNA co-migrated with linDNA and ocDNA (bracket), but not with cccDNA. Some ocDNA appeared in the cccDNA pool, presumably due to nicking during handling of the samples after centrifugation. hDNA migrated up to the position of host chromosomal DNA, as deduced from comparing the blots with the ethidium bromide-stained gels (data not shown). S, standards (1 or 10 pg of DNA).
Mapping the ends of hDNA
DNA of virus-infected plants was purified by CsCl gradient centrifugation in the presence of ethidium bromide (as seen in Figure 1D); the hDNA pool was separated on a neutral agarose gel, which was cut into slices after electrophoresis. DNA was eluted from the gel pieces to perform multiple primer extensions in order to map the ends of the different DNA forms. Figure 2 shows two representative blots for virion sense (Figure 2A) and for complementary sense (Figure 2B) extension products. Discrete bands appeared after hybridization, excluding random nicking activity during preparation as the source of heterogeneity. However, various bands were scattered across the gel in different samples without any regular pattern. If the hDNA was produced by RCR, a defined end at the origin of replication would be expected for virus sense primer extension, resulting in the same band in all fractions upward from the ocDNA position. The complementary sense strand might be heterogeneous in principle as it is assumed that complementary sense replication is carried out by host primase, but Saunders et al. (1992) mapped the complementary sense priming site on ACMV to a discrete region in the vicinity of the CR. Using different samples of DNA, the bands resulting from primer extension varied in mobility in an unpredictable manner, but the bands were always discrete (as in Figure 2). A summary of 384 primer extension products is given in Figure 2C. The frequencies of the calculated ends of DNA were drawn to the respective nucleotide positions of the genomic map of AbMV. The ends of virion sense and complementary sense products were scattered over the whole genome of AbMV, with some higher frequencies at the origin of replication (Figure 2C, 1), the transcription termination region (Figure 2C, 2) and the promotor region of the transcript TAC2-3 (Figure 2C, 3; Frischmuth et al., 1991). It must be kept in mind that ocDNA just nicked by the Rep protein during RCR was included in the samples and presumably increased the relative amount of primer extension products with single-stranded tails starting at the origin of replication. Longer RCR products should have been removed by the chosen CsCl centrifugations. To summarize, these results are consistent with the idea that hDNA is not only heterogeneous in length, but also according to the 5′- and 3′-ends in reference to the genomic map.
Two-dimensional gel electrophoresis
Two-dimensional electrophoresis has been useful in a variety of versions to analyse replicative intermediates of prokaryotic and eukaryotic organisms (Friedman and Brewer, 1995). The first detection of RCR intermediates for geminiviruses was also based on two-dimensional analysis in a combination of neutral and alkaline gels (Saunders et al., 1991). To analyse AbMV intermediates, several variations of two-dimensional gels were tested and the one described below (neutral/chloroquine; Figure 3) was best for the following reasons. Chloroquine intercalates into dsDNA, reducing its mobility in comparison to ssDNA, and adds positive superhelical turns into negatively supercoiled circular DNA (for detailed discussion see Snapka et al., 1991). Depending on the concentration of chloroquine and the size of the circular DNA, several topoisomers of cccDNA can be visualized.
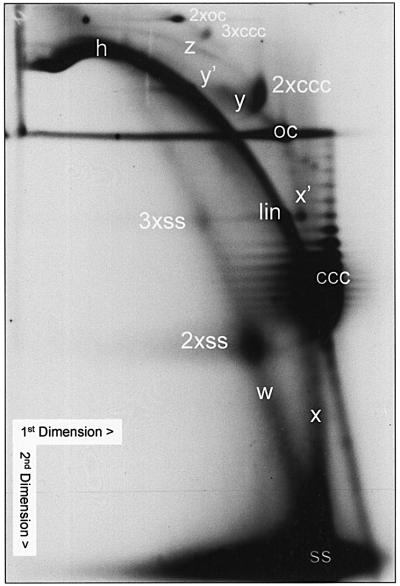
Fig. 3. AbMV DNA A intermediates as detected after two-dimensional gel electrophoresis, blotting and hybridization against a DNA A probe. The sample (100 ng of DNA) was first run in an SDS gel (0.5% agarose; first dimension), and subsequently in a chloroquine gel (1.4% agarose; second dimension). 2× and 3× indicate dimer and trimer molecules, respectively. For detailed description and interpretation see the text.
The two-dimensional system was relatively insensitive to nicking during preparation in comparison with alkaline gels. ssDNA, forming nearly straight lines, was well separated from dsDNA, forming arcs. cccDNA was easily recognized by the separation of topoisomers or the resulting comma-like appearance in molecules of lower mobility, whereas ocDNA appeared as oval flecks. The assignment of the respective DNA species was confirmed by the comparison of blots after neutral or alkaline transfer (data not shown). There was no great difference in the appearance of signals whether DNA A- or DNA B-specific probes were used or whether the DNA was heated to 65°C under high stringency and in the presence of SDS before loading or not (data not shown). The latter comparison excluded the possibility that some DNA forms remain attached to each other just by unspecific adsorption. All the DNA forms discussed in the following paragraph (Figure 3) were found to be reproduced in >50 two-dimensional gels whenever the respective blot exhibited replication intermediates. Under these conditions all the forms appeared simultaneously.
The most prominent fleck (Figure 3; ss) represents ssDNA that was run to the border of the gel to allow optimal resolution of the other forms. Starting from this fleck, a straight line (w) indicates heterogeneous ssDNA that crosses dimeric (2×ss) and trimeric ssDNA (3×ss). The second prominent fleck is cccDNA, resolved into a set of bands corresponding to its topoisomers (ccc). The number and the maximum intensity of the topoisomers changed with different samples. The most prominent diagonal arc represents heterogeneous lin-dsDNA (h) overprinting spots of monomeric linDNA and multimeric linDNA. The identity of this arc with dsDNA was confirmed by co-electrophoresis of AbMV intermediates with bacteriophage λ DNA cut by HindIII restriction endonuclease and successive hybridization with DNA A and a λ-specific probe. The λ DNA spots migrated on the arc of hDNA (data not shown). ocDNA forms a round fleck and tends to smear horizontally for reasons discussed in the legend to Figure 1.
The intermediate DNA forms are interpreted as follows (Figure 3). A vertical straight line (x) starts at the ssDNA, crosses an unidentified spot (x′), which is only present in DNA A, and ends at the position of ocDNA (oc). This type of line is expected for complementary strand synthesis on circular single-stranded monomeric matrices, as analysed in greater detail by Saunders et al. (1991). Another straight line (y) starts from the position of ocDNA (oc) in a northwest direction and ends abruptly (y′) at a position of a roughly estimated size of one additional monomeric ssDNA. The y complexes are compatible with the behaviour of RCR intermediates in the case where one monomeric genome length of ssDNA has been synthesized in one round of replication. We did not observe an extension of this form to dimeric or trimeric ssDNA. If it had occurred, it must have been present in amounts less than the detection limit of this technique. An additional arc (z) points to the dimeric cccDNA (2×ccc). This feature is indicative of an association of cccDNA with heterogeneous lin-dsDNA. The z complexes might have been formed by accidental homologous recombination, either between cccDNA and dsDNA with protruding 3′-ends, or between cccDNA and lin-ssDNA, which has subsequently been copied by de novo second-strand synthesis. Alternatively, they are consistent with RDR intermediates, as discussed below.
The y as well as the z complexes appeared only transiently in those samples where an increase in the amount of AbMV DNA, compared with the sample from the next younger leaf at the shoot, was observed as an indication for replication. Therefore, it is improbable that they were generated by the purification procedure.
Electron microscopy
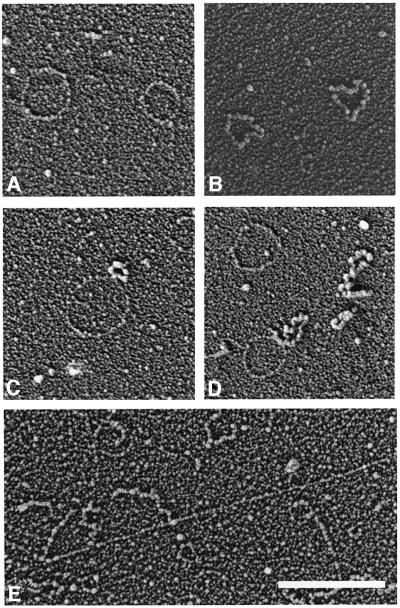
To visualize the DNA intermediates, different versions of gel electrophoresis were combined with spreading techniques in order to differentiate ssDNA and dsDNA or ocDNA and cccDNA. After one-dimensional gel separation (Figure 1B), viral ocDNA (Figure 4A), circular ssDNA (Figure 4B) and intermediates expected for RCR (Figure 4C and D) are readily discriminated by the differential binding of T4 ssDNA-binding protein. The postulated RDR intermediates have not been resolved under these conditions, presumably because they are obscured by the high amount of hDNA and chromosomal DNA.
Fig. 4. Electron microscope visualization of AbMV DNA intermediates after spreading in the presence of T4 gene 32 protein and ethidium bromide. ssDNA appears thicker in the protein complex than the thin naked dsDNA. Representative examples after one-dimensional gel electrophoresis and elution (Figure 1B) of circular dsDNA (A), ssDNA of genomic length (B) and RCR intermediates (C and D). In all fractions of the gel (Figure 1B), heterogeneous linDNA was observed, but without in situ hybridization it is impossible to discriminate between host and viral DNA. Nevertheless, it is remarkable to note that a high number of these heterogeneous dsDNA molecules possess lin-ssDNA extensions (E), which are not seen in DNA of uninfected plants. Bar = 500 nm. The contour length of circular molecules appears somewhat smaller than expected from classical Kleinschmidt preparations, especially after interaction with ssDNA-binding protein. This effect has been observed for AbMV molecules as well as for the standard (bacteriophage ΦX174 DNA) in parallel preparations (data not shown).
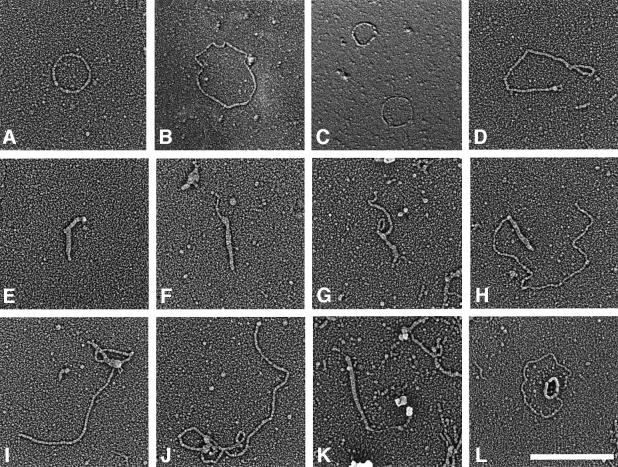
A better distinction was possible after extraction of DNA intermediates from two-dimensional gels (as shown in Figure 3). To allow assignment of the electron microscopic images to certain fractions of the gel, two gels were cast and loaded with the same sample in parallel. One gel was cut into regular pieces of 1 cm2, the slices were stored frozen, and the second gel was processed for hybridization as in Figure 3. With reference to this blot, interesting pieces of the first gel were chosen, their DNA extracted and one-tenth of it run on an agarose gel, followed by hybridization to make sure that the correct gel pieces were collected. Eluates from 13 pieces, covering all the DNA forms above the cccDNA (see Figure 3), were analysed by electron microscopy. To make the images more comparable with the in-gel situation, DNA was spread in the presence of the same amount of chloroquine as in the gel, instead of ethidium bromide as for the preparation shown in Figure 4. Chloroquine gave dsDNA a thicker appearance, making electron microscopic detection easier, but reducing the difference in appearance between ssDNA and dsDNA (see discussion in the legend to Figure 5). However, the cccDNA was better visualized and recognizable. As predicted from the two-dimensional gels, cccDNA was found attached to the end of heterogeneous lin-dsDNA frequently for dimeric cccDNA (Figure 5I–K) in the respective fractions of the gel. Moreover, monomeric cccDNA (Figure 5F–H) was also detected in a similar linkage, which is not resolved in the two-dimensional gels, presumably because it was hidden under the strong arc of heterogeneous lin-dsDNA (Figure 5H). Forms predicted for RCR (Figure 5L) were observed as in the previous experiments (Figure 4). In Figure 5L, the rare case is documented that RCR can also start from dimeric ocDNA.
Fig. 5. Electron microscope visualization of AbMV DNA intermediates after spreading in the presence of T4 gene 32 protein and chloroquine. Representative examples purified from two-dimension gels (Figure 3) are shown for monomeric oc-dsDNA (A), dimeric oc-dsDNA (B), oc-dsDNA (C, lower molecule) in comparison with circular ssDNA, multimeric cccDNA (D), monomeric cccDNA (E), monomeric (F–H) and dimeric (I and J) cccDNA linked with lin-dsDNA. Intermediates shown in (E–H) were from two-dimensional fractions of monomeric cccDNA, and in (I) and (J) of dimeric cccDNA and the adjacent gel blocks in the respective northwest directions (see Figure 3). A rare curiosity is seen in (K), where cccDNA is connected to lin-dsDNA continued by ssDNA that ends in a structure similar in size and shape to a geminivirion. (L) An RCR intermediate as seen in Figure 4C and D. Bar = 500 nm.
Apart from the DNA species described so far, which were classified easily due to their conformation and size, all fractions in a northeast direction from the hDNA diagonal arc harboured large networks of poorly resolved lin-dsDNA with ssDNA extensions as shown in Figure 4E. It remains to be shown by in situ hybridization whether they result from host DNA or viral intermediates. If they really represent viral heterogeneous lin-dsDNA that replicates either by an RCR or RDR mechanism, they should appear as a field of hybridizing material northeast of the hDNA arc (Figure 3) rather than in discrete arcs or spots.
The presence of z intermediates is independent of most viral genes
Homologous recombination could occur due to host or viral gene products’ activities, or a combination of both. As homologous recombination might be especially useful for begomoviruses to combine DNA A and DNA B via their homologous regions (Figure 2D, CR) we wondered whether certain genes (ac2/ac3) present only in the genus Begomovirus (and Curtovirus) are necessary for the generation of z intermediates. The third interesting candidate gene (ac1), which might also be a participant in such a process, cannot be mutated without destroying the basis of replication (Evans and Jeske, 1993; for a review see Hanley-Bowdoin et al., 1999). To test the role of viral gene products, leaf discs were agro-inoculated with DNA A or a mutated DNA A (ΔAC2/3) that is impaired in long-distance spread through plants (Evans and Jeske, 1993). Both were applied alone or together with DNA B.
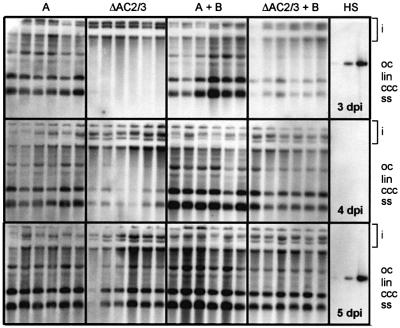
Figure 6 shows the differential appearance of the wild-type and mutated version of AbMV DNA A in single leaf discs from 3 to 5 d.p.i. The accumulation of viral DNA is generally slower for the mutant, but can be accelerated by co-inoculation of DNA B. At 3 and 4 d.p.i., the amount of viral ssDNA in relation to dsDNA is reduced in the mutant, but at 5 d.p.i. some samples had reached the same amount of viral total DNA and ssDNA as in the wild-type samples. Therefore, the phenotype of this AbMV mutant is due to a replication delay rather than a general prevalence for certain DNA forms as it has been described for other geminiviruses defective in ac2 (Hayes and Buck, 1989; Sunter et al., 1990). An inefficient replication of the AbMV mutant is, however, consistent with the function of AC3 as replication enhancer (for review see Hanley-Bowdoin et al., 1999).
Fig. 6. Viral DNA A from leaf discs agro-inoculated with DNA A (A), the mutant (ΔAC2/3), DNA A and DNA B, ΔAC2/3 and DNA B. DNA forms are indicated as in Figure 1. ‘i’ marks input DNA of plasmids. HS, hybridization standard (1, 10 and 100 pg).
The analysis of selected samples (DNA A plus DNA B at 3, 4 and 5 d.p.i.; DNA A, DNA ΔAC2/3 and DNA ΔAC2/3 plus DNA B at 5 d.p.i.) by two-dimensional gel electrophoresis showed no significant differences in the pattern of DNA intermediates. As representative examples, two-dimensional blots of DNA A and DNA ΔAC2/3 are shown in Figure 7. In contrast to samples from naturally infected plants (Figure 3), no x or y intermediates (indicative for RCR) were detectable in any of the two-dimensional blots after leaf disc infection, even if the blot was overexposed (data not shown). Note that under these conditions, 3 d.p.i. is the first time point when newly replicated viral DNA was found reproducibly. ssDNA, however, might have been produced before by RCR, but escaped detection by hybridization. Alternatively, z intermediates and hDNA were clearly represented in each of the two-dimensional blots, irrespective of whether wild-type or mutant DNA constructs were inoculated. We conclude that neither ac2, ac3 genes nor DNA B are necessary to produce z intermediates and hDNA. Either host factors alone or in combination with AC1 are sufficient. Whether AC2 and/or AC3 (as replication enhancer) display a helper function in this process remains to be shown.
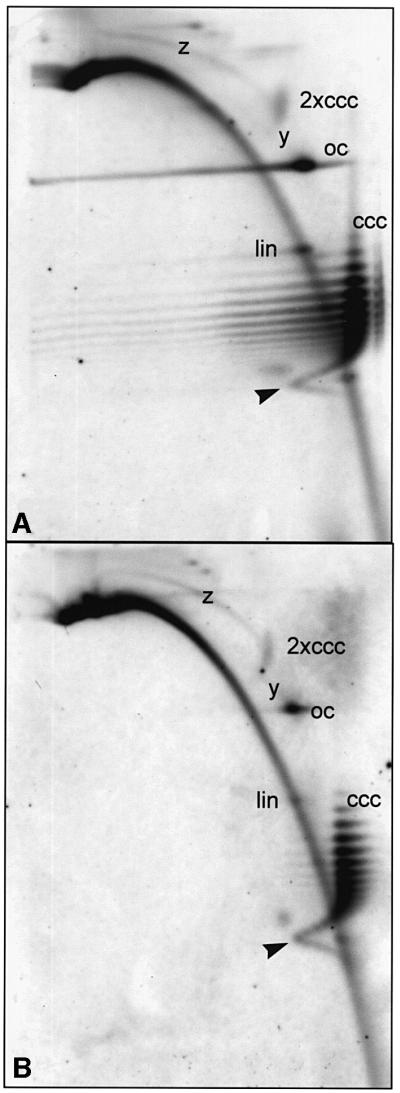
Fig. 7. Two-dimensional gel analysis of selected samples shown in Figure 6. Samples of leaf discs agro-inoculated with DNA A (A) or ΔAC2/3 (B) were compared at 5 d.p.i. Note that z intermediates were present in all samples, whereas x and y intermediates were not detectable. A so far unprecedented arc of cccDNA (arrowheads) was observed in all samples during these experiments. These forms presumably represent extremely open chromatin structures with less negative or even positive supercoiling.
To ensure that the mutants were stable during the time course of agro-infection and did not revert to wild type, all samples of Figure 6 were analysed by PCR and diagnostic restriction as described (Evans and Jeske, 1993). Selected PCR products with the most abundant amount of replicated viral DNA were re-cloned and sequenced. No reversion to wild-type sequences was present in these clones.
Discussion
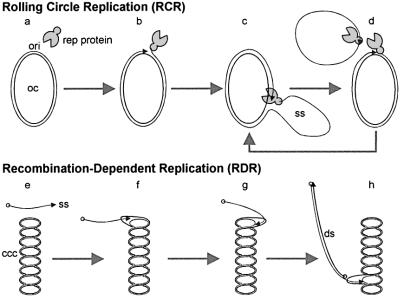
The presence of lin-dsDNA with heterogeneous length and ends (hDNA) as a major component of AbMV DNA intermediates, and the linkage of such DNA to cccDNA (z intermediates), as seen in two-dimensional gel electrophoresis and after electron microscopy, require an explanation extending our current model of geminivirus replication. As observed for geminiviruses in general, AbMV produces the typical forms of RCR, which have been visualized by electron microscopy here for the first time. One simple explanation for the high amount of hDNA and for the z intermediates is that AbMV has developed an additional capacity to multiply via an alternative route, RDR (Figure 8). Preliminary in vitro experiments to show that z intermediates are indeed substrates for run-on replication have failed so far. This was not unexpected as the isolated naked DNA might impose topological constraints on the process, whereas in the cell, viral dsDNA is wrapped around nucleosomes and supplemented with other proteins, which might be necessary to elongate the recombined DNA molecules. Further experiments using reconstituted chromatin and nuclear extracts should prove the model unequivocally.
Fig. 8. Models of RCR and RDR. Step a: binding of a replication-associated protein (Rep; corresponding to ORF AC1, C1 or C1-2 for geminiviruses), respectively, to the origin of replication (ori). Step b: nicking of DNA and covalently binding of Rep to the 5′-end of DNA. Step c: ssDNA displacement and replication. Step d: new nicking, ssDNA closing and Rep release. Step e: incomplete ssDNA interacts with cccDNA at homologous sites. Step f: homologous recombination. Step g: loop migration and ssDNA elongation. Step h: ssDNA elongation and complementary strand synthesis resulting in dsDNA.
hDNA might result from different processes. Depending on the particular virus or the host plant, it might be produced by nicking enzymes from multimeric RCR intermediates, which are complemented by second strand synthesis, or directly by RDR as proposed in Figure 8. In the case of AbMV, we did not detect multimeric RCR products (Figure 3), but instead very long z intermediates in the absence of any indication of RCR (Figure 7). Therefore, this is interpretated, at least for AbMV under the chosen conditions, as hDNA being predominantly formed by RDR. Moreover, if hDNA were the product of RCR and cDNA synthesis we would have expected a smear after primer extension rather than discrete bands (Figure 2). In contrast, RDR provides the possibility to amplify hDNA, leading to discrete populations of DNA molecules. With this view, incomplete replicated or nicked DNA from RCR could be repaired by RDR connecting both processes.
An RDR model can explain a series of biological phenomena, first of all the rapid or frequent recombinations observed. Moreover, an RDR mechanism could provide several advantages for geminiviruses. Whenever normal viral replication is hindered, either due to shortage of nucleotides, DNA digestion by host enzymes or conflicts between replication and transcription, incomplete DNA molecules can be recovered for productive infection by homologous recombination and converted into full-size genomic DNA. The process normally involves proteins of host or viral origin, which work in a recA-like manner (Kowalczykowski and Eggleston, 1994). Invasion of disrupted strands into cccDNA compared with hDNA would increase the fidelity of repair by recruiting a full-length endless template. For this aim, cccDNA has to be opened by a helicase, but topoisomerases are not necessary because torsional stress remains equal during loop migration (Figure 8). As a consequence, the master sequence is left intact.
AbMV might be an exceptional case among geminiviruses as it has adapted for >100 years to its host Abutilon, which has been propagated only by vegetative means (Wege et al., 2000). However, unexplained forms of viral DNA have also been observed in two-dimensional gels in the case of the distantly related ACMV (Saunders et al., 1991). They correspond well to the hDNA presented here. As recombination is a widespread biological phenomenon among geminiviruses, we assume that RDR is rather a general replication process in this virus group, which has escaped our notice so far.
The combination of two different replication strategies is not without precedence among viruses. The best analysed example is the replication of bacteriophage T4 (Formosa and Alberts, 1986). Heterogeneous T4 DNA molecules had indicated a recombination-dependent mechanism, and the viral protein UvsX was shown to promote recombination in a recA-like manner. Template switching to homologous nucleic acids is even an obligatory step for the replication of retroviruses and pararetroviruses (Hohn and Fütterer, 1997). Moreover, the recombination–replication–repair connection has been an emerging field for research of eukaryotes too (Beernink and Morrical, 1999; Haber, 1999). In contrast to bacteria and yeast, homologous recombination is rare in higher plants (Swoboda et al., 1994; Puchta and Hohn, 1996) and animals.
In addition to homologous recombination, illegitimate recombination has also been observed for geminiviruses. Cloned viral constructs with large defects in the coat-protein gene tend to reconstitute genomic size DNA for unknown reasons (for review see Bisaro, 1994). A detailed sequence analysis of the size revertants revealed footprints of an illegitimate recombination (Etessami et al., 1989). Indirect evidence for recombination between geminivirus and host DNA has been obtained by Bejarano et al. (1996) and Kenton et al. (1995), who detected geminiviral DNA sequences as evolutionary footprints in Nicotiana tabacum genomic DNA. Currently, no information is available on the frequency of illegitimate recombination during geminiviral multiplication.
Recombination has been considered a major concern in safety assessments for genetically modified crops (for review see Hammond et al., 1999). Kiraly et al. (1998) have demonstrated that recombination of a challenging virus with a transgene can change symptom expression and host range specificity. The capability of whitefly to transmit geminiviruses was localized on the coat protein (Höfer et al., 1997), and rapid recovery of a coat-protein gene deletion mutant by uptake of a transgene has been found for ACMV if the gene was integrated together with the origin of replication (Frischmuth and Stanley, 1998). If RDR is a major process in geminivirus multiplication, as proposed in this paper, all transgenic constructs that provide information for symptom expression, host range, tissue and vector specificities should be avoided. It might be advisable to choose defective constructs with the aim of excluding such coding sequences. In this way, it is a fortunate coincidence that the resistance strategy for geminiviruses utilizing defective interfering DNA as control elements was successful (Frischmuth and Stanley, 1993).
Materials and methods
Plants and virus
AbMV-infected, vegetatively propagated Abutilon sellovianum (Jeske et al., 1977), infectious clones of DNA A and DNA B (Frischmuth et al., 1990; Wege et al., 2000) and a mutated version of AbMV DNA A with inactive open reading frames AC2/AC3 (Evans and Jeske, 1993) have been described.
Leaf disc infection
Nicotiana benthamiana DOMIN leaves, 5 cm in length, were harvested and surface sterilized with 10% Klorix (a household chlorine bleach) supplemented with 0.01% SDS for 10 min, followed by short washes in 70% ethanol and two in sterile H2O. Leaf discs of 9 mm in diameter were punched out and laid upside upon sterile cultivation media [4.38 g/l Murashige-Skoog salts (Sigma M5524), 30 g/l sucrose, 0.5 mg/ml nicotinic acid, 0.5 mg/l pyridoxine–HCl, 0.5 mg/l thiamine–HCl, 2 mg/l glycine, 1 mg/ml 6-benzylaminopurine, 0.1 mg/l 1-naphthaleneacetic acid, 3 mM 2-[N-morpholino]ethanesulfonic acid pH 5.7, 0.8% agar; Gibco] in Petri dishes. After pre-incubation (28°C, 1500 lx light for 16 h, 25°C at night) for 1 day, leaf discs were dipped into suspensions of Agrobacterium tumefaciens carrying the appropriate plasmids. For co-inoculation experiments, different A.tumefaciens clones were mixed just before use. The leaf discs were placed onto cultivation media and incubated as before for 3, 4 or 5 days.
DNA isolation
The following DNA isolation procedure enriched extrachromosomal DNA according to Hirt (1967), with the modifications of Givens et al. (1996) to preserve topoisomers. Leaf discs or single leaves from 30- to 50-cm-tall shoots of AbMV-infected plants were shock frozen in liquid nitrogen, ground to powder in a mortar, transferred frozen to 80% acetone and stored at –20°C overnight. After centrifugation (Variofuge, Heraeus; 4000 r.p.m. for 10 min at 4°C), the supernatant was removed, the pellet resuspended in lysis buffer [100 mM Tris–HCl pH 7.5, 10 mM EDTA, 1% SDS, 10 mM N-ethylmaleimide (NEM), 5% β-mercaptoethanol], the suspension rolled for 20 min at room temperature and NaCl added to 1 M final concentration with gentle mixing. Incubation overnight at 0°C and centrifugation (SS34 rotor, Sorvall; 15 000 r.p.m. for 30 min at 4°C) yielded a supernatant that was poured onto solid polyethylene glycol (PEG) 6000 to a final PEG concentration of 12% (w/w). PEG was dissolved slowly by rolling the samples at 4°C for a minimum of 2 h. The suspension was centrifuged (Variofuge, Heraeus; 4000 r.p.m. for 40 min at 4°C), the pellet resuspended in TE (10 mM Tris–HCl pH 7.5, 1 mM EDTA), digested with RNase A, adjusted to 1% SDS, and extracted twice with phenol–chloroform and once with chloroform. DNA in the aqueous phase was precipitated by isopropanol, washed with 70% ethanol and dissolved in TE. DNA amounts were quantified using 4′,6-diamidino-2-phenylindole (DAPI; Tanious et al., 1992) with the fluorescence normalized against a standard of CsCl gradient-purified total plant DNA of known concentration.
Standard gel electrophoresis and hybridization
Gels with agarose concentrations as indicated in the Results were cast in TBE (89 mM Tris–HCl, 89 mM boric acid, 2 mM EDTA) adding either SDS to 0.03%, 0.5 µg/ml ethidium bromide or 50 µg/ml chloroquine, called in the following ‘SDS’, ‘ethidium bromide’ or ‘chloroquine gels’. Samples in loading buffer (TE, 1% SDS, 0.025% bromophenol blue, 2.5% Ficoll) with or without heating at 65°C for 10 min prior to loading were electrophoresed with various field strengths as indicated in Results. For elution from the gel, DNA was recovered by glass milk adsorption (Prep-A-gene kit; Bio-Rad). For the experiments shown in Figure 1, eluted DNA was digested with 10 µg/ml RNase A or 100 µg/ml proteinase K (Boehringer) for 30 min at 20°C. Southern blotting using alkaline (Chomczynski and Qasba, 1984) or neutral transfer, and subsequent hybridization, were performed as described (Song and Jeske, 1994). Probes were digoxigenin-labelled (Boehringer kit #158606), gel-purified fragments either of cloned full-length AbMV DNA A (Frischmuth et al., 1990) excised with PstI, of the AC1 region amplified by PCR from the full-length DNA A clone, or of the BV1 gene (Wege and Jeske, 1998). Hybridization standards were 1, 10 or 100 pg PstI fragments of DNA A.
CsCl gradient centrifugation
Solutions of DNA in TE were adjusted to CsCl densities of 1.69 g/ml for gradients without ethidium bromide, or of 1.55 g/ml for gradients containing 0.8 mg/ml ethidium bromide. Gradients were generated by centrifugation (VTi 65.1 rotor, Beckman; 50 000 r.p.m. for 16 h at 20°C). Fractions were collected from the bottom. Ethidium bromide was removed by extraction with water-saturated 1-butanol. Carrier tRNA (to 50 µg/ml) and 3 vols of TE were added, and nucleic acids precipitated by adding 2 vols of ethanol. After washing with 70% ethanol, nucleic acids were dissolved in TE.
Multiple primer extension
Primers MP6a (5′-CGCGCTTAGGCATTTTGGGTTAAAGC-3′; nucleotides 370–345 in AbMV DNA A) and ML1a (5′-ATCTCCCATGGCGATCGATGCCTGGA-3′; nucleotides 371–396; for genome position see Frischmuth et al., 1990; Figure 2) were used to map the ends of linDNA in viral sense (MP6a) or complementary sense (ML1a) orientation, respectively. linDNA and ocDNA, pooled from fractions after CsCl/ethidium bromide gradient centrifugation (as prepared for Figure 1D), were run on 1% agarose gels. Each lane was cut into 18 pieces, DNA was glass-milk extracted and analysed by multiple primer extensions: 50 pmol of a single primer (either MP6a or ML1a) were added to the extracted DNA of each fraction in a volume of 50 µl containing 1.5 mM MgCl2, 67 mM Tris–HCl pH 8.8, 16 mM (NH4)2SO4, 0.01% Tween-20, 0.25 mM each dNTP and 1 U of Eurobiotaq polymerase and cycled in a thermoblock (94°C for 2 min, 60°C for 1 min, 72°C for 90 s for one cycle; 94°C for 30 s, 60°C for 1 min, 72°C for 90 s for 41 cycles). The products were separated on alkaline gels (2% agarose, 50 mM NaOH, 1 mM EDTA at 64 V/354 mA and 4°C under buffer recirculation for 20 h). Gels were blotted using alkaline transfer solution and hybridized against digoxigenin-labelled AbMV DNA A probes. For size standards, cloned DNA fragments of AbMV DNA A were cut with various restriction enzymes giving fragments as indicated in the figures.
Two-dimensional gel electrophoresis
First-dimension gel electrophoresis (0.5% agarose in TBE supplemented with 0.03% SDS) was performed in 3-mm-wide and 12-cm-long tubes at 10 V for 19 h at room temperature. The gel cylinders were layed into the preformed slot of a second-dimension slab gel (12 × 14 cm, 1.4% agarose, TBE, 50 µg/ml chloroquine) and electrophoresed for 19 h at 45 V and room temperature. Gels were incubated for 20 min in 0.25 M HCl before alkaline blotting and hybridization. Gels run in parallel were cut into squares of 1 cm edge length to extract the DNA.
Electron microscopy
Gel-purified DNA was spread using the on-drop technique described by Coggins (1987). DNA (from several picograms to a maximum of 100 ng, as estimated from hybridization signals) was incubated with a 40-fold amount of T4 gene 32 protein (Boehringer) for 10 min at 37°C. Glutaraldehyde was added to a final concentration of 0.2% and fixed the sample for 10 min at 37°C. With the supplement of ethidium bromide (final concentration 100 µg/ml) or chloroquine (final concentration 50 µg/ml), the solution was pipetted onto parafilm as a drop of 50 or 100 µl. The drops were allowed to stand in a humid chamber for a minimum of 2 h or overnight in the case of two-dimension gel experiments in order to spread the DNA–protein complexes on the surface. The complexes were quickly attached to a carbon-coated 300-mesh copper grid, washed for 30 s in H2O, stained for 30 s with uranyl acetate (1 µl of a 50 mM stock solution in 50 mM HCl was added to 999 µl of 90% ethanol just before use), washed with 90% ethanol and air-dried. Samples were shaded with platinum–carbon at an angle of 8° in a Balzers coating system (MED 020). Electron microscopy was carried out with an EM 10 (Zeiss) using 60 or 20 kV acceleration voltage. Photographs were taken at 10 000× or 20 000× magnification on Agfa or Kodak films. Negatives were scanned at 2000 l.p.i. and processed for printing using Paintshop-Pro 5 (Jasc Software). Magnification standards were circular dsDNA or ssDNA of bacteriophage ΦX174 (Boehringer) processed on separate grids. Contour length measurements were performed from scanned negatives using ScanPro 4 (Jandel Scientific).
Acknowledgments
Acknowledgements
The authors thank Dr Christina Wege, Professor Dr Robin Ghosh for helpful discussions, Mrs Cornelia Kocher and Mrs Alexandra Schwierzok for skillful technical assistance, Mr Diether Gotthardt, Mrs Sabine Weller and Mrs Sigrid Kober for taking care of the plants. We are very much obliged to Professor Dr Dieter Hülser, who provided the facilities for electron microscopy and encouraged our work by friendly discussions. This work was supported by Land Baden-Württemberg providing a sabbatical to H.J.
References
- Abouzid A.M., Frischmuth,T. and Jeske,H. (1988) A putative replicative form of the Abutilon mosaic virus (gemini group) in a chromatin-like structure. Mol. Gen. Genet., 212, 252–258. [Google Scholar]
- Beernink H.T.H. and Morrical,S.W. (1999) RMPs: recombination/replication mediator proteins. Trends Biochem. Sci., 24, 385–389. [DOI] [PubMed] [Google Scholar]
- Bejarano E.R., Khashoggi,A., Witty,M. and Lichtenstein,C. (1996) Integration of multiple repeats of geminiviral DNA into the nuclear genome of tobacco during evolution. Proc. Natl Acad. Sci. USA, 93, 759–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisaro D.M. (1994) Recombination in geminiviruses: mechanisms for maintaining genome size and generating genomic diversity. In Paszkowski,J. (ed.), Homologous Recombination and Gene Silencing in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp. 39–60.
- Brewer B. (1988) When polymerases collide: replication and the transcriptional organization of the E. coli chromosome. Cell, 53, 679–686. [DOI] [PubMed] [Google Scholar]
- Chomczynski P. and Qasba,P.K. (1984) Alkaline transfer of DNA to plastic membrane. Biochem. Biophys. Res. Commun., 122, 340–344. [DOI] [PubMed] [Google Scholar]
- Coggins L.W. (1987) Preparation of nucleic acids for electron microscopy. In Sommerville,J. and Scheer,U. (eds), Electron Microscopy in Molecular Biology—A Practical Approach. IRL Press, Oxford, UK, pp. 1–29.
- Etessami P., Watts,J. and Stanley,J. (1989) Size reversion of African cassava mosaic virus coat protein gene deletion mutants during infection of Nicotiana benthamiana. J. Gen. Virol., 70, 277–289. [DOI] [PubMed] [Google Scholar]
- Evans D. and Jeske,H. (1993) Complementation and recombination between mutants of complementary sense genes of DNA A of Abutilon mosaic virus. Virology, 197, 492–496. [DOI] [PubMed] [Google Scholar]
- Formosa T. and Alberts,B.M. (1986) DNA synthesis dependent on genetic recombination: characterization of a reaction catalyzed by purified bacteriophage T4 proteins. Cell, 47, 793–806. [DOI] [PubMed] [Google Scholar]
- Friedman K.L. and Brewer,B.J. (1995) Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. In Colowick,S.P. and Kaplan,N.O. (eds), Methods Enzymol., 262, 613–627. [DOI] [PubMed] [Google Scholar]
- Frischmuth S., Frischmuth,T. and Jeske,H. (1991) Transcript mapping of Abutilon mosaic virus, a geminivirus. Virology, 185, 596–604. [DOI] [PubMed] [Google Scholar]
- Frischmuth T. and Stanley,J. (1993) Strategies for the control of geminivirus disease. Semin. Virol., 4, 329–337. [Google Scholar]
- Frischmuth T. and Stanley,J. (1998) Recombination between viral DNA and the transgenic coat protein gene of African casava mosaic geminivirus. J. Gen. Virol., 79, 1265–1271. [DOI] [PubMed] [Google Scholar]
- Frischmuth T., Zimmat,G. and Jeske,H. (1990) The nucleotide sequence of Abutilon mosaic virus reveals prokaryotic as well as eukaryotic features. Virology, 178, 461–468. [DOI] [PubMed] [Google Scholar]
- Givens R.M., Saavedra,R.A. and Huberman,J.A. (1996) Topological complexity of SV40 minichromosomes. J. Mol. Biol., 257, 53–65. [DOI] [PubMed] [Google Scholar]
- Haber J.E. (1999) DNA recombination: the replication connection. Trends Biochem. Sci., 24, 271–275. [DOI] [PubMed] [Google Scholar]
- Hammond J., Lecoq,H. and Raccah,B. (1999) Epidemiological risks from mixed virus infections and transgenic plants expressing viral genes. Adv. Virus Res., 54, 189–314. [DOI] [PubMed] [Google Scholar]
- Hanley-Bowdoin L., Settlage,S.B., Orozco,B.M., Nagar,S. and Robertson,D. (1999) Geminiviruses: models for plant DNA replication, transcription and cell cycle regulation. Crit. Rev. Plant Sci., 18, 71–106. [PubMed] [Google Scholar]
- Hayes R.J. and Buck,K.W. (1989) Replication of tomato golden mosaic virus DNA B in transgenic plants expressing open reading frames (ORFs) of DNA A: requirement of ORF AL2 for production of single-stranded DNA. Nucleic Acids Res., 17, 10213–10222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn T. and Fütterer,J. (1997) The proteins and functions of plant pararetroviruses: knowns and unknowns. Crit. Rev. Plant Sci., 16, 133–161. [Google Scholar]
- Hirt B. (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol., 26, 365–369. [DOI] [PubMed] [Google Scholar]
- Höfer P., Bedford,I.D., Markham,P.G., Jeske,H. and Frischmuth,T. (1997) Coat protein gene replacement results in whitefly transmission of an insect nontransmissible geminivirus isolate. Virology, 236, 288–295. [DOI] [PubMed] [Google Scholar]
- Jeske H., Menzel,D. and Werz,G. (1977) Electron microscopic studies on intranuclear virus-like inclusions in mosaic-diseased Abutilon sellowianum Reg. Phytopathol. Z., 89, 289–295. [Google Scholar]
- Kenton A., Khashoggi,A., Parokonny,A., Bennett,M.D. and Lichtenstein,C. (1995) Chromosomal location of endogenous geminivirus-related DNA sequences in Nicotiana tabacum L. Chromosome Res., 3, 346–350. [DOI] [PubMed] [Google Scholar]
- Kiraly L., Bourque,J.E. and Schoelz,J.E. (1998) Temporal and spatial appearance of recombinant viruses formed between cauliflower mosaic virus (CaMV) and CaMV sequences present in transgenic Nicotiana bigelovii. Mol. Plant–Microbe Interact., 11, 309–316. [Google Scholar]
- Kowalczykowski S.C. and Eggleston,A.K. (1994) Homologous pairing and DNA strand-exchange proteins. Annu. Rev. Biochem., 63, 991–1043. [DOI] [PubMed] [Google Scholar]
- Moffat A. (1999) Geminiviruses emerge as serious crop threat. Science, 286, 1835. [Google Scholar]
- Padidam M., Sawyer,S. and Fauquet,C.M. (1999) Possible emergence of new geminiviruses by frequent recombination. Virology, 285, 218–225. [DOI] [PubMed] [Google Scholar]
- Petty I.T.D., Coutts,R.H.A. and Buck,K.W. (1988) Transcriptional mapping of the coat protein gene of tomato golden mosaic virus. J. Gen. Virol., 69, 1359–1365. [Google Scholar]
- Pilartz M. and Jeske,H. (1992) Abutilon mosaic geminivirus double-stranded DNA is packed into minichromosomes. Virology, 189, 800–802. [DOI] [PubMed] [Google Scholar]
- Puchta H. and Hohn,B. (1996) From centiMorgans to base pairs: homologous recombination in plants. Trends Plant Sci., 10, 340–348. [Google Scholar]
- Rybicki E.P. et al. (2000) Family Geminiviridae. In van Regenmortel,M.H.V., Fauquet,C.M. and Bishop,D.H.L. (eds), Virus Taxonomy—Classification and Nomenclature of Viruses. Academic Press, San Diego, CA, pp. 285–297.
- Rybicki E.P. and Pietersen,G. (1999) Plant virus disease problems in the developing world. Adv. Virus Res., 53, 127–175. [DOI] [PubMed] [Google Scholar]
- Saunders K., Lucy,A. and Stanley,J. (1991) DNA forms of the geminivirus African cassava mosaic virus consistent with a rolling circle mechanism of replication. Nucleic Acids Res., 19, 2325–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders K., Lucy,A. and Stanley,J. (1992) RNA-primed complementary-sense DNA synthesis of the geminivirus African cassava mosaic virus. Nucleic Acids Res., 20, 6311–6315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapka R.M., Permana,P.A., Marquit,G. and Shin,C.-G. (1991) Two-dimensional agarose gel analysis of simian virus 40 DNA replication intermediates. Methods, 3, 73–82. [Google Scholar]
- Song J.-Y. and Jeske,H. (1994) The level of Abutilon mosaic geminivirus in leaf discs and wound callus. J. Phytopathol., 140, 45–52. [Google Scholar]
- Stanley J., Markham,P.G., Callis,R.J. and Pinner,M.S. (1986) The nucleotide sequence of an infectious clone of the gemini virus beet curly top virus. EMBO J., 5, 1761–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenger D.C., Revington,G.N., Stevenson,M.C. and Bisaro,D.M. (1991) Replication release of geminivirus genomes from tandemly repeated copies: evidence for a rolling circle replication of a plant viral DNA. Proc. Natl Acad. Sci. USA, 88, 8029–8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunter G., Hartitz,M.D., Hormuzdi,S.G., Brough,C.L. and Bisaro,D.M. (1990) Genetic analysis of tomato golden mosaic virus: ORF AL2 is required for coat protein accumulation while ORF AL3 is necessary for efficient DNA replication. Virology, 179, 69–77. [DOI] [PubMed] [Google Scholar]
- Swoboda P., Gal,S., Hohn,B. and Puchta,H. (1994) Intrachromosomal homologous recombination in whole plants. EMBO J., 13, 484–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanious F.A., Veal,J.M., Buczak,H., Ratmeyer,L.S. and Wilson,W.D. (1992) DAPI (4′,6-diamidino-2-phenylindole) binds differently to DNA and RNA: minor-groove binding at AT sites and intercalation at AU sites. Biochemistry, 31, 3103–3112. [DOI] [PubMed] [Google Scholar]
- Townsend R., Stanley,J., Curson,S.J. and Short,M.N. (1985) Major polyadenylated transcripts of cassava latent virus and location of the gene encoding coat protein. EMBO J., 4, 33–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege C. and Jeske,H. (1998) Abutilon mosaic geminivirus proteins expressed and phosphorylated in Escherichia coli. J. Phytopathol., 146, 613–621. [Google Scholar]
- Wege C., Gotthardt,R.-D., Frischmuth,T. and Jeske,H. (2000) Fulfilling Koch’s postulates for abutilon mosaic virus. Arch. Virol., 145, 2217–2225. [DOI] [PubMed] [Google Scholar]
- Zhang W., Olson,N.H., Baker,T.S., Faulkner, L, Agbandje-McKenna,M., Boulton,M.I., Davies,J.W. and McKenna,R. (2001) Structure of the maize streak virus geminate particle. Virology, 279, 471–477. [DOI] [PubMed] [Google Scholar]
- Zhou X., Liu,Y., Calvert,L., Munoz,C., Otim-Nape,G.W., Robinson,D.J. and Harrison,B.D. (1997) Evidence that DNA-A of a geminivirus associated with severe cassava mosaic disease in Uganda has arisen by interspecific recombination. J. Gen. Virol., 78, 2101–2111. [DOI] [PubMed] [Google Scholar]
- Zhou X.P., Liu,Y.L., Robinson,D.J. and Harrison,B.D. (1998) Four DNA-A variants among Pakistani isolates of cotton leaf curl virus and their affinities to DNA-A of geminivirus isolates from okra. J. Gen. Virol., 79, 915–923. [DOI] [PubMed] [Google Scholar]