Abstract
To investigate determinants of specific transcriptional regulation, we measured factor occupancy and function at a response element, col3A, associated with the collagenase-3 gene in human U2OS osteosarcoma cells; col3A confers activation by phorbol esters, and repression by glucocorticoid and thyroid hormones. The subunit composition and activity of AP-1, which binds col3A, paralleled the intracellular level of cFos, which is modulated by phorbol esters and glucocorticoids. In contrast, a similar AP-1 site at the collagenase-1 gene, not inducible in U2OS cells, was not bound by AP-1. The glucocorticoid receptor (GR) associated with col3A through protein–protein interactions with AP-1, regardless of AP-1 subunit composition, and repressed transcription. TIF2/GRIP1, reportedly a coactivator for GR and the thyroid hormone receptor (TR), was recruited to col3A and potentiated GR-mediated repression in the presence of a GR agonist but not antagonist. GRIP1 mutants deficient in GR binding and coactivator functions were also defective for corepression, and a GRIP1 fragment containing the GR-interacting region functioned as a dominant-negative for repression. In contrast, repression by TR was unaffected by GRIP1. Thus, the composition of regulatory complexes, and the biological activities of the bound factors, are dynamic and dependent on cell and response element contexts. Cofactors such as GRIP1 probably contain distinct surfaces for activation and repression that function in a context-dependent manner.
Keywords: activator protein-1/glucocorticoid receptor/p160 cofactors/thyroid hormone receptor/transcriptional repression
Introduction
Eukaryotic transcriptional regulation is carried out by multiprotein complexes that assemble at genomic response elements close to regulated promoters. Many specific factors and general cofactors capable of participating in such regulatory complexes have been described (Freedman, 1999; Goodman and Smolik, 2000; Lemon and Tjian, 2000; Malik and Roeder, 2000), but the precise determinants of the composition and action of regulatory complexes are not well understood. At a given response element, specific cellular or physiological contexts can promote the assembly of complexes that differ in composition and regulatory activity. Closely related members of a regulator protein family may exert distinct effects on transcription or elicit similar effects by different mechanisms of action. Depending on the context, a single factor may recruit different cofactors or, alternatively, utilize the same cofactors in functionally distinct ways. It is apparent that response elements profoundly influence the activities of the bound components (Lefstin and Yamamoto, 1998); thus, regulatory complexes are fundamentally DNA–protein machines.
Three classes of response elements have been described (Yamamoto et al., 1998): ‘simple’, at which a single regulatory factor binds; ‘composite’, at which two or more different regulators recognize specific DNA sequences; and ‘tethering’, at which one regulator binds to the element, and a second regulatory factor associates with the first through protein–protein interactions. In each case, the prevailing view is that the response element– regulatory factor complexes then recruit cofactors that alter chromatin structure, or contact or modify components of the transcription machinery ultimately leading to enhancement or repression.
Intracellular receptors, such as the glucocorticoid receptor (GR) and the thyroid hormone receptor (TR), function at all three classes of response elements (Chandler et al., 1983; Karin et al., 1984; Diamond et al., 1990; Schule et al., 1990; Yang-Yen et al., 1990; Desbois et al., 1991; Konig et al., 1992). Hormone binding by the ligand binding domains (LBDs) of these receptors triggers changes in their conformations and regulatory activities. In some cases, hormone binding has also been demonstrated to alter the association of receptors with coactivators or corepressors (Fondell et al., 1996; Hanstein et al., 1996; Blanco et al., 1998; Kraus and Kadonaga, 1998; Naar et al., 1999; Rachez et al., 1999). For example, the p160 coactivators (SRC1, TIF2/GRIP1 and ACTR/Rac3/pCIP/AIB1) (Onate et al., 1995; Hong et al., 1996; Voegel et al., 1996; Yao et al., 1996; Anzick et al., 1997; Chen et al., 1997; Li et al., 1997; Torchia et al., 1997) interact with the activation function-2 (AF-2) region within the GR and TR LBDs; the interactions occur in conjunction with agonist, but not antagonist binding. The p160 molecules interact with the receptors through a domain that contains three LxxLL sequence motifs; these sequences, termed NR boxes (Darimont et al., 1998; Ding et al., 1998; Kalkhoven et al., 1998), are differentially recognized by different receptors. Among the three p160 factors, distinct expression patterns and phenotypes of p160-deficient mice (Xu et al., 1998, 2000; Qi et al., 1999; Weiss et al., 1999) suggest selective interactions between particular receptors and individual p160s, but neither the specificity of these interactions in vivo nor the mechanisms of coactivation by the p160s are well understood.
As with activation, transcriptional repression also involves multicomponent protein–DNA complexes. For example, unliganded TR (apoTR) bound to a simple thyroid hormone response element (TRE) can recruit co-repressors N-CoR, SMRT or Alien (Horlein et al., 1995; Chen et al., 1996; Dressel et al., 1999), which in turn bind Sin3A and histone deacetylases (HDACs) (Heinzel et al., 1997; Nagy et al., 1997); the corepressors are released from TR, in favor of coactivators, upon hormone binding at simple TREs. In contrast, hormone-bound TR (holoTR) can repress transcription at tethering response elements, such as certain AP-1 sites (Desbois et al., 1991; Saatcioglu et al., 1993). These complexes are distinct from those at simple TREs but their activity is nevertheless sensitive to HDAC inhibitor trichostatin A (TSA) (M.Cronin and K.R.Yamamoto, unpublished data). GR also represses transcription in an agonist-dependent fashion at tethering response elements such as the AP-1 sites associated with the collagenase-3/MMP13 and collagenase-1/MMP1 genes (Jonat et al., 1990; Yang-Yen et al., 1990; Pendas et al., 1997; Tuckermann et al., 1999) or the NF-κB sites of the IL-8 and ICAM-1 genes (Nissen and Yamamoto, 2000). However, repression by GR appears to require neither HDAC activity (M.Cronin, R.M.Nissen and K.R.Yamamoto, unpublished data) nor corepressors N-CoR or SMRT; indeed, no functional corepressors have been identified for steroid receptors (Chen and Evans, 1995; Horlein et al., 1995; Dressel et al., 1999).
Although activation and repression complexes are clearly distinct, there are several indications that they may be more closely related than appears at first glance: (i) many transcriptional regulators activate transcription in some contexts and repress in others (Roy et al., 1998); (ii) single base pair mutations in composite glucocorticoid response elements (GREs) of the prolactin and proliferin genes (Sakai et al., 1988; Starr et al., 1996) convert them from GR-repressible to GR-inducible elements; conversely, a single amino acid change in the GR DNA-binding domain (DBD) produces a receptor that activates under conditions in which the wild-type GR represses (Starr et al., 1996); (iii) transcriptional repression by GR is agonist dependent and antagonist sensitive, suggesting that repression is mediated by the receptor conformation that binds p160 coactivators (Jonat et al., 1990; Fryer et al., 2000; B.Darimont and K.R.Yamamoto, unpublished data); (iv) overexpression of a p160, TIF2/GRIP1, potentiates estrogen receptor (ER)-mediated repression of tumor necrosis factor-α (TNF) transcription, whereas ER antagonists and ER mutations disrupting the ER–p160 interaction decrease repression (An et al., 1999).
To examine these apparent context-specific effects on regulatory factor assembly and activity, we analyzed the composition and function in vivo of regulatory complexes at a response element, col3A, that governs the expression of the collagenase-3 gene in a human osteosarcoma cell line. The col3A element binds AP-1 and confers phorbol 12-myristate 13-acetate (PMA) inducibility as well as repression by glucocorticoid and thyroid hormones. We monitored factor recruitment and activity at col3A under conditions of activation and repression. We assessed receptor and TIF2/GRIP1 association and function during repression, and determined whether GR and TR, two receptors from the same gene family, repress transcription using similar or distinct components.
Results
AP-1 subunit composition at the collagenase-3 AP-1 element
The collagenase-3 gene is expressed in U2OS human osteosarcoma cells (Jimenez et al., 1999) stably transfected with rat GR [U2OS.G (Rogatsky et al., 1997)]; it can be induced by phorbol esters (PMA) and cytokines IL-6 and IL-1β (Borden et al., 1996; Solis-Herruzo et al., 1999) and repressed by glucocorticoids such as dexamethasone (Pendas et al., 1997; Tuckermann et al., 1999). A single AP-1 binding site positioned at –50/–44 relative to the collagenase-3 transcription start site contributes to basal expression, and is responsible for both PMA inducibility and hormonal repression (Porte et al., 1999; data not shown). Thus, regulation of the collagenase-3 gene emanates from a compact, well defined response element, which we denote col3A.
The cFos component of the AP-1 protein family is critical for collagenase-3 promoter activation: cFos-overexpressing transgenic mice exhibit enhanced expression of collagenase-3, whereas cFos–/– knockout mice are collagenase-3 deficient (Gack et al., 1994; Borden et al., 1996). Furthermore, in fibroblasts cultured from cFos–/–cJun–/– double knockout mice, collagenase-3 is not inducible by PMA and other stimuli (Hu et al., 1994; Schreiber et al., 1995). The expression of the AP-1 family members themselves, particularly cFos, is regulated by AP-1 and GR (Kruijer et al., 1984; Treisman, 1985; Lamph et al., 1988; Subramaniam et al., 1992; Luo and Jackson, 1998; Boudreau et al., 1999). We therefore monitored the effects of PMA and dexamethasone, i.e. inducing and repressing conditions for collagenase-3, on the intracellular levels of cFos and on the subunit composition of AP-1 bound at col3A.
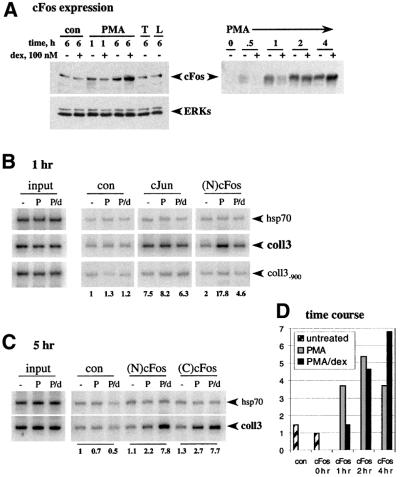
As expected, cFos levels were increased by 1 h, and strongly elevated by 4–6 h of PMA treatment compared with untreated U2OS.G cells (Figure 1A); as controls, TNF and bacterial lipopolysaccharide (LPS) had no effect (left panel). Interestingly, during the first hour of PMA treatment, dexamethasone inhibited PMA-induced cFos accumulation, whereas it potentiated cFos levels at 4 or 6 h of treatment. In contrast, cJun expression was not significantly stimulated by PMA and was mildly inhibited by dexamethasone at all time points tested (data not shown).
Fig. 1. Occupancy of the coll3 AP-1 element by cFos and cJun in vivo. (A) Effect of GR on PMA induction of cFos expression. U2OS.G cells were untreated (con), or treated with PMA, TNF (T) or LPS (L), in the presence or absence of dex, for 1 or 6 h, as indicated, and cFos expression in whole cell extracts was examined by immunoblotting with N-terminal cFos [(N)cFos] polyclonal antibodies (left, upper panel). Equal loading into each lane was verified by blotting with anti-ERK polyclonal antibodies (left, lower panel). cFos expression in U2OS.G cells treated with PMA in the presence or absence of dex for the time indicated was assessed by blotting with C-terminal cFos [(C)cFos] rabbit polyclonal antibodies (right panel). (B and C) Composition of AP-1 subunits within activation and repression complexes at col3A. U2OS.G cells were untreated (–) or cultured in the presence of PMA (P) or PMA+dex (P/d) for 1 (B) or 5 (C) h and chromatin immunoprecipitations were performed with control rabbit serum (con), cJun, (N)cFos and (C)cFos rabbit polyclonal antibodies. In each case, a +153/+423 fragment of hsp70 gene, and the –150/+68 (coll3) and –1179/–897 (coll3–900) fragments of collagenase-3 gene were PCR amplified. Equal amounts of total genomic DNA (input) were used for immunoprecipitations in each treatment condition. 32P incorporation into the coll3 product was quantified on a Storm 860 PhosphorImager and normalized to the signal obtained with control rabbit serum in untreated cells (shown below the gel). (D) Dynamics of cFos recruitment to col3A under conditions of PMA induction and dex repression. U2OS.G cells were untreated, or exposed to PMA or PMA+dex for the times indicated and chromatin immunoprecipitations were performed with control rabbit serum (con) or (N)cFos antibody. coll3 and hsp70 sequences were PCR amplified, quantified and expressed as a coll3:hsp70 ratio. The ratio obtained with (N)cFos antibody in untreated cells was arbitrarily set as 1.
To monitor AP-1 binding to col3A in vivo, we carried out chromatin immunoprecipitation assays using polyclonal antibodies to cFos or cJun (or normal rabbit serum as a negative control), and primers amplifying a 218 bp PCR product from –150 to +68 bp relative to the collagenase-3 transcription start site (coll3), which includes the col3A element. As controls, we amplified a fragment denoted coll3–900, which extends from –897 to –1179 bp upstream of the collagenase-3 transcription unit, and a +153/+423 bp fragment of the hsp70 gene. Cells were untreated, or were incubated for 1 h with PMA or PMA+dex; chromatin fragments containing identical amounts of total genomic DNA (input) were used for the immunoprecipitations (Figure 1B).
Normalized to control serum, the cJun antibodies yielded an ∼8-fold enrichment of the col3A-containing fragment; occupancy by cJun appeared constitutive, unaffected by PMA or PMA+dex treatment (Figure 1B). In contrast, a 1 h exposure to PMA induced a 9-fold enrichment of cFos at col3A relative to untreated cells using antisera against either cFos N-terminal [(N)cFos; Figure 1B] or C-terminal [(C)cFos; data not shown] segments. Strikingly, the PMA-induced reconfiguration of the AP-1 subunits was not observed when the cells were co-treated with PMA+dex. These results suggest that basal AP-1 activity is provided by cJun, that a 1 h exposure to PMA induces cFos expression and binding to the AP-1 site, probably replacing cJun–cJun homodimers with cFos–cJun heterodimers, and that dexamethasone antagonizes this effect. In contrast, when the U2OS.G cells were treated with PMA+dex for 5 h, cFos occupancy of the coll3 fragment was selectively enhanced rather than inhibited, relative to PMA alone or to untreated cells (Figure 1C). The coll3–900 and hsp70 control fragments were not enriched under inducing or repressing conditions.
In a time course experiment, we found that 1, 2 or 4 h of PMA treatment provoked a 3.5- to 5.5-fold increase in cFos occupancy of the coll3 fragment, normalized to the hsp70 control (Figure 1D). Consistent with the hormonal effect on cFos expression (Figure 1A), 1 h with PMA+dex resulted in lower cFos occupancy than with PMA alone, whereas 2 h of hormone treatment had little net effect, and 4 h produced an increase in cFos occupancy (Figure 1D). Thus, cFos and cJun binding at col3A correlated closely with the relative intracellular levels of these proteins in U2OS.G cells, suggesting that cFos occupancy is driven by synthesis and that AP-1 at col3A is dynamic, turning over within the time-frame examined.
GR represses AP-1-dependent transcription independent of the composition of AP-1 subunits
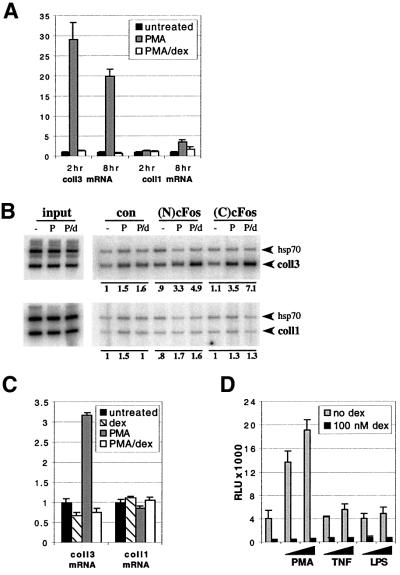
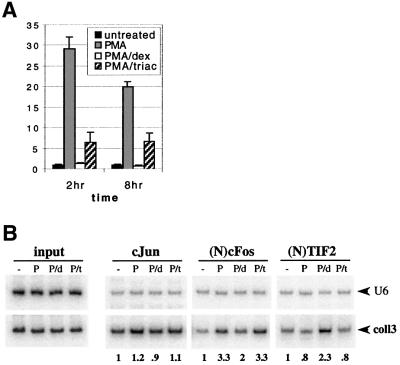
Consistent with the observed PMA-induced increase in cFos expression and col3A occupancy, we found that PMA stimulated the accumulation of collagenase-3 mRNA (measured by quantitative RT–PCR/Taqman) at 1 h (data not shown), and more strongly at 2 and 8 h (Figure 2A) of treatment. In all cases, the induction was completely blocked by dexamethasone, suggesting that collagenase-3 transcription is repressed by GR whether or not cFos is recruited to the repression complex.
Fig. 2. GR represses AP-1 activity independent of the AP-1 subunit composition at col3A. (A) Regulation of collagenase-3 mRNA expression by PMA and dex. U2OS.G cells were treated as shown for the times indicated, total RNA was isolated and reverse-transcribed. Resulting cDNA was subjected to real-time quantitative RT–PCR/Taqman with collagenase-3- and collagenase-1-specific primer/probe sets, in triplicate, using a primer/probe set for G3PDH as an internal control. (B) Binding of cFos to the colA element of the collagenase-1 gene. U2OS.G cells were treated and processed for chromatin immunoprecipitations as described in Figure 1C. A –150/+68 (coll3) region of collagenase-3 gene and a –229/+18 (coll1) region of collagenase-1 gene were co-amplified with +153/+423 and +153/+485 fragments of the hsp70 gene, respectively. 32P incorporation into each PCR product was quantified and expressed as coll3:hsp70 and coll1:hsp70 ratios; in each case, ratios obtained with control serum in untreated cells were arbitrarily set as 1 (shown below the gel). (C) Collagenase-3 induction is not required for repression. U2OS.G cells were treated for 1 h as indicated, total RNA was isolated and analyzed as described in (A). (D) GR-mediated repression of the AP-1-luc reporter activity in U2OS.G cells. Cells were transfected in 24-well plates with 40 ng/well of the AP-1-luc reporter and 50 ng/well of the β-actin-LacZ plasmid expressing β-galactosidase (β-gal). Where indicated, cells were treated with PMA (25 or 50 ng/ml), TNF (5 or 10 ng/ml) or LPS (0.5 or 1 µg/ml), in the absence or presence of 100 nM dex. The AP-1-luc reporter luciferase activity is normalized to β-gal activity and expressed as relative luminescence units (RLU).
Unexpectedly, human fibroblast collagenase-1 (MMP1), which like collagenase-3 is a member of the matrix metalloproteinase family, appeared to be refractory at 2 h and only weakly induced by 8 h of PMA treatment of the U2OS.G cells (Figure 2A). The collagenase-1 gene shares substantial sequence similarity with the collagenase-3 gene, extending through the 5′ flanking region (Vu and Werb, 2000) and including an AP-1 element (termed colA, at –72/–66 bp relative to the transcription start site) that mediates PMA induction and dexamethasone repression in other cell contexts (Angel et al., 1987a; Schule et al., 1990; Yang-Yen et al., 1990). Consistent with the relative inactivity of the collagenase-1 gene in U2OS.G cells, 1 h (data not shown) or 5 h (Figure 2B) of PMA or PMA+dex produced little effect on cFos occupancy of colA. Conceivably, the collagenase-1 gene is packaged in an unfavorable chromatin structure in U2OS.G cells. Regard less, our findings identify collagenase-1 as a negative control for collagenase-3 mRNA expression and AP-1 recruitment in this cell line, and underscore the importance of context differences for gene regulation.
As cJun occupancy of col3A was constitutive (Figure 1B) and cJun–GR interactions have been described (Diamond et al., 1990; Yang-Yen et al., 1990), we next examined whether collagenase-3 expression is dexamethasone-repressible even in the absence of PMA stimulation. As shown in Figure 2C, we found that uninduced collagenase-3 (but not collagenase-1) mRNA accumulation, generated at least in part by cJun homodimer occupancy of col3A (Figure 1B), was reduced in the presence of dex by ∼40%. We also addressed this issue in the context of an AP-1 site as the only regulatory element by monitoring the activity of an AP-1-luc reporter (Rogatsky et al., 1998) transiently transfected into U2OS.G cells. As expected, PMA, but not TNF or LPS, stimulated AP-1-luc reporter activity, and dexamethasone conferred repression (Figure 2D). Importantly, AP-1-luc reporter activity without PMA treatment was also repressed by dexamethasone. Similar effects of PMA and glucocorticoids were observed with several AP-1-containing reporters, including those derived from the bona fide human promoters (data not shown). We conclude that GR represses AP-1-mediated transcription independently of AP-1 subunit composition, consistent with findings that at tethering response elements, GR can repress transcription stimulated by heterologous activation domains (Nissen and Yamamoto, 2000).
GR associates with the AP-1-bound col3A element in vivo in a hormone-dependent manner
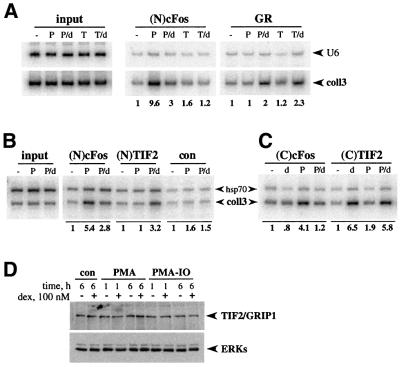
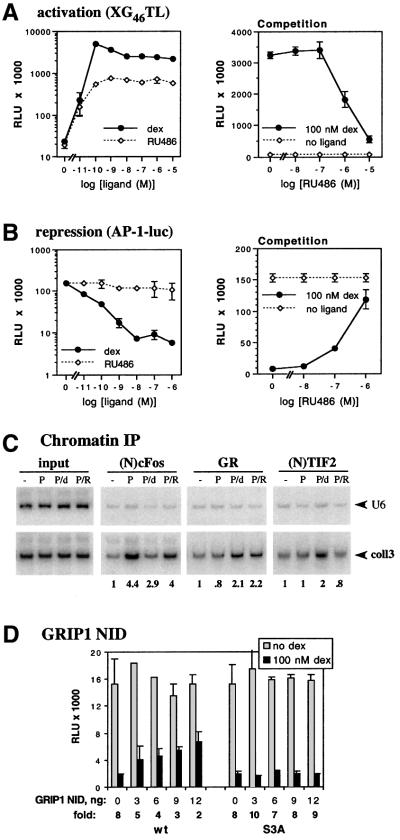
Indirect evidence supports the notion that GR represses AP-1-activated transcription by tethering to the bound AP-1 factor (Konig et al., 1992; M.Cronin and K.R.Yamamoto, unpublished data); AP-1 elements such as col3A do not serve as direct binding sites for GR. As a direct test of tethering, we carried out chromatin immunoprecipitation assays using anti-GR polyclonal antibodies. As shown in Figure 3A, a 1 h co-treatment of U2OS.G cells with PMA+dex produced a modest but reproducible enrichment of the coll3 fragment compared with untreated or PMA-treated cells. We speculate that the rather weak signals obtained in the GR immunoprecipitations may reflect limited epitope accessibility within the AP-1–GR complex or low efficiency of crosslinking. Notably, GR was similarly recruited to col3A in cells co-treated with TNF+dex (Figure 3A). As TNF had little effect on AP-1 activity (Figure 2D), cFos expression (Figure 1A) or recruitment (Figure 3A), this finding is consistent with the notion that GR is recruited to col3A regardless of the AP-1 subunit composition at the site. Under the same treatment conditions, control U6 gene sequences were not enriched. Thus, GR is recruited to the col3A element in a hormone-dependent manner.
Fig. 3. GR and TIF2 are recruited to col3A in a dex-dependent, PMA-independent manner. (A) GR recruitment to col3A. U2OS.G cells were untreated (–) or treated with PMA (P), PMA+dex (P/d), TNF (T) or TNF+dex (T/d) for 1 h and chromatin immunoprecipitations were performed with (N)cFos and GR polyclonal antibodies. coll3 and control U6 sequences were amplified, 32P incorporation into coll3 product was quantified and normalized to the untreated sample. (B) TIF2 occupancy of col3A. U2OS.G cells were treated as in Figure 1B and chromatin immunoprecipitations were performed with (N)cFos, (N)TIF2 polyclonal antibodies or normal rabbit serum (con). coll3 and hsp70 sequences were co-amplified, 32P incorporation into coll3 product was quantified and normalized to the untreated sample in each case. (C) AP-1 activation with PMA is not required for TIF2 recruitment. U2OS.G cells were untreated (–) or treated with dex (d), PMA (P) or PMA+dex (P/d) for 1 h and chromatin immunoprecipitations were performed with (C)cFos polyclonal and (C)TIF2 monoclonal antibodies. Quantitation was performed as described in (B). (D) TIF2 protein expression is unaffected by PMA or dexamethasone. U2OS.G cells were untreated (con) or treated with PMA or PMA/IO, in the presence or absence of dex, for 1 or 6 h, as indicated, and TIF2 expression in whole cell extracts was examined by immunoblotting with anti-TIF2 polyclonal antibodies (upper panel). Equal loading into each lane was verified by blotting with anti-ERK polyclonal antibodies (lower panel).
TIF2/GRIP1 associates with GR at the col3A element in vivo
The most well-documented agonist-dependent protein– protein interaction of intracellular receptors is with the members of the p160 family (Onate et al., 1995; Hong et al., 1996; Voegel et al., 1996; Chen et al., 1997; Li et al., 1997; Torchia et al., 1997; Darimont et al., 1998). Therefore, we tested the possibility that the p160 proteins, originally defined as coactivators, might participate in agonist-dependent repression by GR at col3A. We began by analyzing the expression of the three p160 family members, TIF2/GRIP1, SRC1 and ACTR/Rac3, in U2OS.G cells; TIF2 was readily observed by immunoblotting, whereas SRC1 and ACTR/Rac3 were barely detectable (data not shown). We therefore examined in chromatin immunoprecipitation assays whether TIF2 accumulates in col3A repression complexes under conditions of PMA induction or dexamethasone repression.
As shown in Figure 3B, antibodies to TIF2/GRIP1 selectively precipitated the coll3 fragment but not the hsp70 control when cells were co-treated for 1 h with PMA+dex relative to untreated or PMA-treated cells. Importantly, dexamethasone alone promoted the association of TIF2 with the coll3 fragment (Figure 3C), whereas PMA alone had little or no effect (Figure 3B and C). These results suggest that TIF2 associates with repression complexes at col3A in a GR- and dexamethasone-dependent manner and that this association is independent of AP-1 activation by PMA. The finding that epitopes near the N- and C-terminus of TIF2 were both accessible (Figure 3B and C; see Materials and methods) supported the interpretation that TIF2 is not merely masked by AP-1, but fails to associate with the col3A regulatory complex in the absence of GR.
To examine whether TIF2 recruitment to col3A repression complexes reflects increased TIF2 expression, we analyzed TIF2 protein levels in U2OS.G cells treated (for 1 or 6 h) with PMA or PMA+ionomycin [IO, a calcium ionophore that commonly enhances the effects of PMA (Truneh et al., 1985; Davies et al., 1990)], in the presence or absence of dexamethasone. TIF2 levels in all cases were indistinguishable from those in untreated control cells (Figure 3D) indicating that TIF2 is recruited into the col3A repression complexes from pre-existing intracellular pools.
GRIP1 potentiates GR-dependent transcriptional repression
To determine whether TIF2 plays a role in transcriptional repression, or is merely a non-functional ‘passenger’ recruited to the GR LBD as a result of agonist binding, we examined the effects of introducing GRIP1, the murine homolog of human TIF2, into U2OS.G cells by transient transfection.
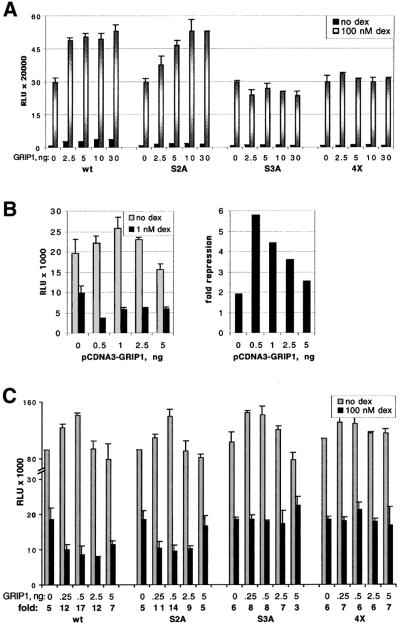
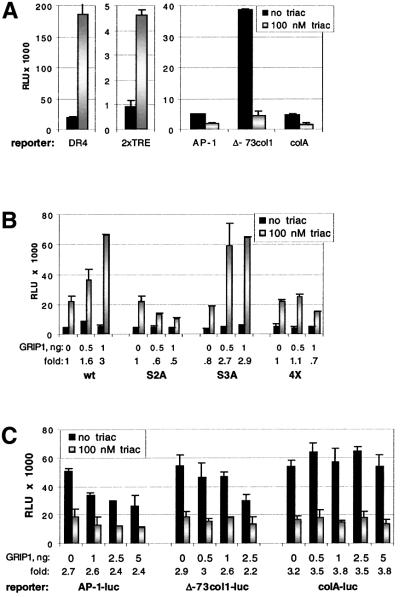
In control experiments, we first confirmed that transfected p160s would enhance the level of glucocorticoid induction of a luciferase reporter gene linked to a simple GRE (XG46TL; Rogatsky et al., 1998). Indeed, SRC1 and ACTR/Rac3 potentiated GR-mediated activation in a dose-dependent manner, maximally 5- to 10-fold (data not shown). GRIP1 enhanced both basal and hormone-dependent expression at low input doses of transfected DNA (Figure 4A), and squelched it at higher doses (data not shown). The modest effect of GRIP1 on GR activation (∼2-fold) probably reflected the high background of endogenous TIF2 protein in U2OS.G cells. Similar results were obtained in two other TIF2-expressing cell lines, A549 and SAOS2.G, whereas GRIP1 had a more robust effect in MCF7 cells, which express relatively low levels of TIF2 (data not shown). Hence, the magnitude of the effect of a given transfected p160 is inversely proportional to the level of that p160 expressed endogenously in the recipient cell line.
Fig. 4. GRIP1 overexpression potentiates GR-mediated activation and repression of AP-1 activity in an NR box 3-dependent manner. U2OS.G cells were transfected in 24-well plates with indicated amounts per well of pCDNA3-GRIP1 (wt, S2A, S3A or 4X). Total amount of transfected DNA was equalized with pCDNA3 empty vector. Transcriptional activation of an XG46TL [(A), 40 ng/well] or repression of AP-1-luc [(B) and (C), 40 ng/well] reporter was measured in the absence or presence of 100 (A and C) or 1 (B) nM dex. Endogenous AP-1 activity in (B) and (C) was stimulated with 25 ng/ml PMA. Luciferase activity is normalized to the β-gal activity of a co-transfected β-actin-LacZ plasmid (50 ng/well) and expressed as RLU. The y-axis in (C) is broken to visualize the effect of GRIP1 on GR-mediated repression at saturating dex concentration. Shown is a representative of two (B) or three (A and C) independent experiments performed in duplicate.
Previous work of others showed that GRIP1–receptor interactions depend upon three NR boxes (Voegel et al., 1998), each bearing a distinct LxxLL motif (Heery et al., 1997), and that different receptors discriminate between the individual NR boxes such that, for instance, GRIP1 associates with GR preferentially through NR box 3 (Darimont et al., 1998; Ding et al., 1998). Consistent with these reports, an NR box 2 mutant (S2A, see Materials and methods) was as effective as wild-type GRIP1 in enhancing GR activity in U2OS.G cells, whereas mutations in NR box 3 (S3A and 4X) abrogated enhancement (Figure 4A); importantly, all four GRIP1 derivatives were expressed at similar levels (data not shown).
We then tested the effects of GRIP1 on GR-dependent repression of AP-1-luc reporter activity. Figure 4B shows that at low doses, GRIP1 increased the magnitude of repression 2- to 3-fold, similar to its effect on GR-mediated enhancement (Figure 4A); as with enhancement, apparent squelching was seen at higher GRIP1 doses. GRIP1 potentiated GR-mediated repression to a similar extent at 1 and 100 nM dexamethasone (Figure 4B and C), suggesting that GRIP1 does not function merely by stabilizing the GR–ligand interaction. The effect of GRIP1 on repression required a functional NR box 3, as mutants with defects in that motif (S3A and 4X), but not in NR box 2 (S3A), failed to confer repression (Figure 4C). We conclude from these experiments that GRIP1 is a GR corepressor in the context of repression complexes formed at col3A, whereas it functions as a GR coactivator at a simple GRE in the same cells.
RU486 or dominant-negative GRIP1 relieves GR-dependent repression of AP-1 activity
To assess whether inhibiting the interaction of GR with endogenous TIF2 would compromise GR-mediated repression, we used RU486, which competes with dexamethasone for binding to the GR LBD. RU486 is typically a partial agonist, disabling the LBD-associated AF-2 by generating an LBD conformation incompatible with p160 binding (Darimont et al., 1998; Williams and Sigler, 1998; B.Darimont and K.R.Yamamoto, unpublished data), while permitting enhancement by activation function-1 (AF-1) in the GR N-terminal domain. In U2OS.G cells, saturating concentrations of RU486 induced a 30-fold activation of the XG46TL luciferase reporter, whereas dexamethasone stimulated the same reporter 110-fold (Figure 5A, left). In contrast, RU486-bound GR triggered little repression from the AP-1-luc reporter (Figure 5B, left). Importantly, RU486 competitively inhibited both dexamethasone-mediated activation and repression (Figure 5A and B, right panels), demonstrating that GR binds RU486 in both reporter contexts.
Fig. 5. RU486 or GRIP1 NID antagonizes GR-mediated activation and repression of AP-1 activity. U2OS.G cells were transfected in 24-well plates with (A) GR-inducible XG46TL or (B) GR-repressible AP-1-luc (40 ng/well) reporter and a β-actin-LacZ plasmid (50 ng/well). GR-dependent activation and repression was assayed across a range of dex (solid line) and RU486 (dashed line) concentrations (left panels). Activation and repression conferred by 100 nM dex were competitively inhibited by increasing concentrations of RU486 (right panels, solid line); activity of XG46TL (A) and AP-1-luc (B) reporter in the absence of GR ligand is shown by a dashed line. Endogenous AP-1 activity in (B) was stimulated with 25 ng/ml PMA. Luciferase activity is normalized to β-gal activity and expressed as RLU. Shown is a representative of two independent experiments performed in triplicate. (C) coll3 occupancy in the presence of RU486. U2OS.G cells were either untreated (–) or treated with PMA (P), PMA+dex (P/d) or PMA+RU486 (P/R) for 1 h, and chromatin immunoprecipitation was performed as described in Figure 3. An equal amount of genomic DNA (input) was used in each treatment condition; coll3 and control U6 sequences were amplified, 32P incorporation into each PCR product was quantified and expressed as a coll3:U6 ratio; the coll3:U6 ratio in untreated cells were set as 1. (D) The GRIP1 NID is an NR box 3-dependent dominant-negative for GR-mediated repression of AP-1 activity. U2OS.G cells were transfected in 24-well plates with indicated amounts per well of pCDNA3-GRIP1 NID (wt or S3A) or pCDNA3 empty vector. GR transcriptional repression of AP-1-luc reporter was assayed as described in Figure 4C. Shown is a representative of three independent experiments performed in duplicate.
In principle, the result obtained with RU486 might reflect an inhibition of GR tethering to the AP-1 bound at coll3. We therefore performed chromatin immunoprecipitations, scoring for tethered GR in U2OS.G cells treated with PMA, PMA+dex or PMA+RU486. As shown in Figure 5C, both GR ligands resulted in comparable GR occupancy of coll3, whereas TIF2 occupancy was strictly dexamethasone dependent. Thus, RU486 enables the GR–AP-1 tethering interaction, but unlike dexamethasone, RU486 precludes GRIP1 binding by GR.
As an independent approach, we constructed a plasmid expressing a GRIP1 fragment, amino acid residues 563–765, encompassing the GRIP1 NR interaction domain (NID). As shown in Figure 5D, the GRIP1 NID functions as a dominant-negative for GR-mediated repression when transiently overexpressed in U2OS.G cells. The effect is GRIP1 NID dose dependent and requires an intact NR box 3, suggesting that the fragment acts by displacing endogenous p160s. Taken together, the RU486 and GRIP1 NID experiments reinforce the notion that GRIP1 serves as a functional corepressor in these regulatory complexes.
GRIP1 overexpression does not affect TR-mediated repression of AP-1 activity
Hormone-bound thyroid hormone receptor (holoTR), like holoGR, can repress transcription from various tethering response elements, including AP-1 elements (Desbois et al., 1991; Saatcioglu et al., 1993, 1997). However, TR-mediated repression of AP-1 is sensitive to the HDAC inhibitor TSA and can be restored by HDAC overexpression (M.Cronin and K.R.Yamamoto, unpublished data), suggesting that these repression complexes may be similar to apoTR complexes at simple TREs (Heinzel et al., 1997; Nagy et al., 1997), and functionally distinct from the holoGR complexes at col3A.
To investigate the role of p160 factors in regulation by TR, we generated U2OS.GT, a derivative of U2OS.G cells stably expressing human TRβ-1 (see Materials and methods). In these cells, transiently transfected reporters bearing simple TREs (DR4 or 2× TRE) were induced by triac, a TR synthetic agonist (Figure 6A, left and center). In contrast, three different AP-1-containing reporters [denoted AP-1-luc, Δ-73col1-luc (Angel et al., 1987b) and colA-luc (Starr et al., 1996); see Materials and methods] were repressed by triac (Figure 6A, right). As expected, none of the reporters was affected by triac in parental U2OS.G cells (data not shown), confirming that the observed regulation was TR dependent.
Fig. 6. GRIP1 increases TRβ-mediated transcriptional activation but not repression of AP-1 activity. (A) TRβ-1 ectopically expressed in U2OS.GT cells is competent for activation and repression. U2OS.GT cells were transfected in 24-well plates with 40 ng/well of TRβ-responsive reporter (DR4 or 2× TRE, left and middle panel), or AP-1 reporters (AP-1-luc, Δ-73col1-luc and colA-luc, right panel), and 50 ng/well of the β-actin-LacZ plasmid. Luciferase activity was measured in the presence and absence of 100 nM triac, normalized to β-gal activity and expressed as RLU; endogenous AP-1 activity was stimulated with 25 ng/ml PMA. (B) Transiently overexpressed GRIP1 coactivates TRβ in an NR box 2-dependent manner. U2OS.GT cells were transfected with amounts indicated per well of pCDNA3-GRIP1 (wt, S2A, S3A and 4X), pCDNA3 to equalize total amount of DNA, 2× TRE reporter and β-actin-LacZ. TRβ-dependent activation of 2× TRE reporter was assayed in the presence or absence of 100 nM triac, normalized to β-gal activity and expressed as RLU. (C) GRIP1 does not potentiate TRβ-dependent repression of AP-1 activity. U2OS.GT cells were transfected with amounts indicated per well of pCDNA3-GRIP1 (or pCDNA3 vector), indicated AP-1 reporters and β-actin-LacZ. AP-1 activity in 25 ng/ml PMA was assayed in the absence and presence of 100 nM triac; luciferase activity of AP-1-luc and colA-luc reporters was multiplied by 10 for uniformity with the activity of Δ-73col1-luc reporter.
Transiently transfected GRIP1 enhanced TRβ-mediated activation from the 2× TRE (Figure 6B); consistent with previous observations (Darimont et al., 1998), this enhancement required GRIP1 NR box 2 rather than NR box 3 (Figure 6B). In contrast to the results with GR, however, overexpression of GRIP1 did not alter TRβ-dependent repression from the AP-1-luc, Δ-73col1-luc and colA-luc reporters (Figure 6C). Thus, within a single cell line, GRIP1 is a coactivator for both GR and TRβ, and a corepressor for GR but not TRβ. We conclude that different receptors utilize distinct regulatory complexes to effect transcriptional repression from common response elements.
GRIP1 is not recruited to col3A by TRβ
As with the transiently introduced AP-1-reporters, transcription of the chromosomal collagenase-3 gene in U2OS.GT cells was also repressed by TRβ. Figure 7A shows the relative levels of collagenase-3 mRNA as assessed by RT–PCR/Taqman at 2 or 8 h of PMA or PMA+triac treatment; the repression was robust albeit not as strong as that provoked by holoGR in the PMA+dex condition.
Fig. 7. TRβ repression complex at the col3A is different from that formed by GR. (A) TRβ represses collagenase-3 mRNA expression in U2OS.GT cells. Cells were treated as shown, for times indicated, total RNA was isolated and analyzed as described in Figure 2A. (B) GR and TRβ transcriptional repression complexes at col3A. U2OS.GT cells were untreated (–) or exposed to PMA (P), PMA+dex (P/d) or PMA+triac (P/t) for 1 h and chromatin immunoprecipitations were performed with cJun, (N)cFos and (N)TIF2 polyclonal antibodies. An equal amount of genomic DNA (input) was used in each treatment condition. coll3 and control U6 sequences were amplified and quantified as described in Figure 5C.
We used chromatin immunoprecipitation to compare the repression complexes formed by TRβ and GR at col3A in U2OS.GT cells. We found that 1 h exposure of the cells to PMA, PMA+dex or PMA+triac had no significant effect on cJun occupancy of the coll3 fragment, and that unlike dexamethasone, triac did not affect the PMA-induced level of cFos occupancy (Figure 7B). Furthermore, dexamethasone but not triac stimulated col3A occupancy by TIF2 (Figure 7B), consistent with the results of transient GRIP1 overexpression on GR- and TRβ-dependent repression of AP-1 reporters (Figures 4B and 6C). Thus, at a single response element, GR and TRβ promote the assembly of distinct repression complexes.
Discussion
The revelation that transcription can be controlled by combinations of regulatory factors rather than by a single factor emerged first from studies of the Escherichia coli lac operon, where full expression requires both relief from inhibition by the lac repressor and activation by the cyclic AMP receptor (Emmer et al., 1970). Other studies in prokaryotes showed that one factor can function as either a positive or a negative regulator (Meyer and Ptashne, 1980), and that distinct ‘functional surfaces’ mediate the activation or repression (Guarente et al., 1982). Later work in metazoans revealed multicomponent regulatory complexes specified in part by the sequence and organization of composite response elements (Yamamoto et al., 1992; Giese et al., 1995; Thanos and Maniatis, 1995; Lefstin and Yamamoto, 1998), which can extend over tens of kilobases of DNA (Small and Levine, 1991). Whereas certain components in the complexes bind directly to response element sequences, others associate through protein–protein contacts (Yuan et al., 1993; Yamamoto et al., 1998). Remarkably, point mutations in factors or response elements can ‘invert’ positive and negative regulation (Sakai et al., 1988; Starr et al., 1996), implying that the core components of activation and repression complexes may employ functional surfaces that are more similar than their divergent activities might otherwise suggest.
Despite these extensive analyses, the determinants of combinatorial specificity are not known. Cell-specific regulation is commonly a consequence of unique combinations of components that are themselves less specifically expressed (Bruhn et al., 1997; Goodman and Smolik, 2000). Even knowing a response element, a promoter sequence and the regulatory factors within a cell, the precise composition of the regulatory complex and even the direction of regulation are unpredictable. Given this uncertainty, fundamental questions remain. What components comprise a given regulatory complex? Are distinct surfaces of these factors active in different functional contexts? Are the mechanisms of activation and repression context specific?
We addressed some of these issues by examining regulatory complexes and mechanisms associated with the col3A response element of the collagenase-3 gene, which specifies basal expression, induction by PMA and repression by glucocorticoid or thyroid hormones. Others had shown that cFos is required for induced expression of collagenase-3 in mice (Gack et al., 1994; Hu et al., 1994; Schreiber et al., 1995; Borden et al., 1996). Furthermore, cFos expression correlates with collagenase-3 mRNA expression in proliferating osteoblasts, and cFos overexpression increases collagenase-3 promoter activity; in contrast, another Fos family member, Fra-2, inhibits collagenase-3 transcription (Winchester et al., 2000). Extending earlier observations, we provide direct evidence that AP-1 subunit occupancy at col3A is dynamic, changing over time and as a function of synthesis; thus, we suggest that cJun–cJun homodimers bound at col3A in uninduced cells are displaced during PMA induction by cJun–cFos heterodimers. Based on these results, one simple model is that glucocorticoid or thyroid hormones antagonize PMA effects by inhibiting cFos expression or its binding to col3A. Such a scheme would be reminiscent of the reported effects of PMA or TNF on occupancy of the NF-κB site proximal to the IκB promoter (Algarte et al., 1999); brief treatment with these agents leads to p50–RelA binding and increased transcription, whereas prolonged treatment results in p50–cRel occupancy and a relative decrease in transcription.
Our findings, however, revealed that glucocorticoid- and thyroid hormone-mediated repression at col3A were distinguishable processes, but that neither depended upon inhibiting cFos occupancy. Thus, TRβ repressed collagenase-3 without altering the cJun–cFos binding to col3A induced by PMA (Figure 7B). In contrast, brief treatment with dexamethasone led to lower levels of cFos occupancy and collagenase-3 expression, whereas longer treatments maintained the repression despite increased cFos occupancy relative to PMA alone (Figures 1 and 2A); the glucocorticoid-specific temporal shifts in cFos occupancy of col3A reflected glucocorticoid-regulated changes in cFos protein levels (Figure 1A). GR repressed transcription (Figure 2C and D) from the col3A element by tethering to the bound AP-1 (Figure 3A) independent of its subunit composition or activation state, suggesting that repression occurs downstream of AP-1 binding.
One example of such late-stage repression has been described for various members of the intracellular receptor family, and appears to involve deacetylation of histones or other protein targets. These HDAC-mediated mechanisms have been implicated for both aporeceptors (Nagy et al., 1997; Wong et al., 1998) and holoreceptors (Desbois et al., 1991; Saatcioglu et al., 1993; M.Cronin and K.R.Yamamoto, unpublished data), in different response element contexts. Whether GR and other vertebrate steroid receptors repress by this mechanism has not been firmly established. That is, under repressing conditions, N-CoR, SMRT and Alien do not associate with steroid receptors, GR-mediated repression of AP-1 at colA and col3A is insensitive to TSA (M.Cronin, I.Rogatsky and K.R.Yamamoto, unpublished data), and we have failed to detect a decline in histone H4 and H3 acetylation at col3A (I.Rogatsky and K.R.Yamamoto, unpublished data). On the other hand, Ito et al. (2000) reported that GR represses interleukin-1β induction of granulocyte-macrophage colony-stimulating factor expression by recruiting HDAC2 and reducing the level of histone H4 acetylation. It is apparent that modulation of histone acetylation is not a universal mechanism of metazoan transcriptional regulation (Eberhardy et al., 2000; Koipally and Georgopoulos, 2000), and that additional studies are required to understand its context dependence.
Although corepressors for the steroid receptors have not been identified, various reports have suggested a role for the LBD in repression. For example, RIP140, which can bind LBDs and displace bound p160 when overexpressed, abrogates both activation and repression by GR (Subramaniam et al., 1999); ER antagonists and ER LBD mutations that disrupt p160 interactions decrease ER-mediated repression of the TNF promoter (An et al., 1999). We now demonstrate directly that TIF2/GRIP1 is recruited to col3A in a GR- and hormone-dependent manner to facilitate GR-mediated repression. Thus, like GR itself, TIF2/GRIP1 can mediate either positive or negative transcriptional regulation.
What determines whether TIF2/GRIP1 will facilitate activation or repression? Conceivably, GR might transduce signals from response elements into conformational alterations in the LBD, thereby producing different GR–p160 interfaces for activation and repression. However, disruption of the GRIP1 NR box 3 eliminated both coactivator and corepressor activities (Figure 4), as well as the ability of the GRIP1 NID to function as a dominant-negative for repression (Figure 5D). More detailed analyses may yet reveal distinctions. In this regard, it is intriguing that the GR N-terminal domain, as well as the LBD, is essential for repression from colA (Schule et al., 1990; M.Cronin and K.R.Yamamoto, unpublished data). Thus, molecular surfaces outside of the GR LBD and/or the GRIP1 NR box 3 may specify interfaces that function selectively in repression.
Alternatively, activation or repression by GR–p160 complexes may be determined by context parameters that are independent of the GR–p160 interaction per se. Other components of the regulatory complexes, recruited either by DNA binding or protein–protein interactions, may produce different regulatory outcomes. For example, regulatory complexes in which intracellular receptors activate transcription appear to contain CBP/p300 or Mediator components perhaps recruited in ordered fashion (Shang et al., 2000; D.Burakov and L.P.Freedman, personal communication). Our failure to detect changes in histone acetylation at col3A does not exclude the possibility that CBP serves as a component of GR repression complexes independent of its HAT activity or its ability to function as a coactivator for AP-1. However, preliminary experiments have failed to reveal p300 at col3A under inducing or repressing conditions (I.Rogatsky and K.R.Yamamoto, unpublished data). Clearly, further studies are required to identify functional components of GR repression complexes. Repression, for example, might result from recruitment of a novel enzymatic activity that modifies GRIP1, GR or the transcription machinery, or precludes pre-initiation complex formation. In the case of GR-mediated repression of NF-κB-induced IL-8 expression, for instance, GR tethers to NF-κB bound at the operative response element, and interferes with phosphorylation of Ser2 in the RNA polymerase II C-terminal domain (Nissen and Yamamoto, 2000), a modification essential for polymerase activity. It will be interesting to investigate the potential role of TIF2/GRIP1 in such a mechanism.
In contrast to our findings with GR, it appears that TRβ represses collagenase-3 transcription utilizing a different regulatory complex. We failed to detect a TRβ-dependent increase in GRIP1 occupancy of col3A, and overexpressed GRIP1 did not affect TRβ-mediated repression of AP-1-responsive reporters (Figures 6C and 7B), although it functioned as a TRβ coactivator in the same cell line (Figure 6B). Our results are consistent with the finding that TR mutations disrupting GRIP1 binding compromise its activation but not its repression functions (Saatcioglu et al., 1997). Thus, TR-mediated repression of AP-1-activated transcription is GRIP1 independent.
These studies reinforce the notion that the selectivity of transcriptional regulation is determined by multifactor complexes that are specified by the availability and activities of regulatory factors, and by the sequence, organization and accessibility of response elements. The actions of the resultant complexes cannot be deduced from the identity of their components, as the binding and activity of a given factor, such as TIF2/GRIP1, depends on context effects imposed by response elements and by other factors. Functionally, such regulatory factors are ‘chameleon-like’ [a term suggested for certain RNA-binding proteins (Smith et al., 2000)], their behavior fashioned by their surrounding environment.
Viewed in isolation, transcriptional regulation seems baroque, decorated with multifunctional factors, extensive factor families, multicomponent complexes, branched pathways and linked networks. However, it is now clear that organismal complexity correlates with the complexity of gene regulation (Wilson et al., 1977), including the location, timing and magnitude of transcription, rather than the number of genes per se; thus, humans carry only ∼50% more genes than fruit flies (Myers et al., 2000; Venter et al., 2001). This implies that the acquisition of new regulatory interactions, which enable greater sophistication of organismal form and function, is accomplished by using pre-existing genes and mechanisms in slightly different ways or combinations. Only occasionally would new genes be added, as with the emergence of the intracellular receptors in metazoans, and the small subfamily of steroid receptors only in the vertebrates.
Given the complexity of transcriptional regulation, what must we do to understand it? Our studies and those of others imply that it may be informative to characterize in detail small sets of related response elements and the associated regulatory complexes in several functional states (inactive, basal, induced, repressed) in different cells or tissues. A complementary approach would include analyses of a factor that performs distinct functions at unrelated response elements. From such investigations, testable predictions for context-specific determinants of regulatory specificity may emerge.
Materials and methods
Cell lines and treatments
U2OS.G human osteosarcoma cells expressing wild-type rat GR (Rogatsky et al., 1997) were cultured in Dulbecco’s modified Eagle medium (DMEM, Life Technologies) supplemented with 10% fetal bovine serum (FBS, HyClone laboratories) and 350 µg/ml G418 (Life Technologies). U2OS.GT cells were generated by transfecting a full-length human TRβ-1 cDNA in the pRep4 expression plasmid containing hygromycinR gene [2.5 µg DNA/100 mm dish using Lipofectamine-PLUS reagent (Life Technologies) as per the manufacturer’s instructions] and selecting for resistant colonies at 400 µg/ml hygromycin B (Life Technologies). Multiple hygromycin-resistant clones were assayed for TRβ expression by indirect immunofluorescence and western blotting with TRβ-specific rabbit polyclonal antibody. Clones homogeneously expressing TRβ at levels comparable to endogenous levels of TR expression in mouse p19 cells were further maintained at 200 µg/ml hygromycin B.
All dexamethasone, RU486, triac and PMA (Life Technologies) dilutions were made in 100% ethanol; ‘untreated’ cells received an equivalent amount of the ethanol vehicle. Human TNFα and bacterial LPS (derived from E.coli J5 strain) stock solutions were purchased from Sigma. Ionomycin was purchased from Calbiochem.
Plasmids
Mammalian reporters including XG46TL containing two consensus GREs upstream of (–109) thymidine kinase (TK) promoter, AP-1-luc containing a single consensus AP-1 site upstream of the TK promoter (Rogatsky et al., 1998), Δ-73col1-luc containing –73/+63 sequences of the human collagenase-1 gene (Angel et al., 1987b), colA-luc containing a collagenase-1-derived AP-1 site upstream of the Drosophila alcohol dehydrogenase (Adh) minimal promoter (–33/+53) (Starr et al., 1996), pGL3-DR4 (Vivanco Ruiz et al., 1991) and ps2ΔxTREpal (Sharif and Privalsky, 1992) were previously described. A β-actin-LacZ plasmid expressed β-galactosidase (β-gal) under the control of human β-actin promoter. Full-length wt GRIP1, GRIP1 4X (Ding et al., 1998) with mutated NR boxes 2 and 3, GRIP1 S2A and GRIP1 S3A (Darimont et al., 1998) with individually mutated NR boxes 2 and 3, respectively, were excised from the pSG5 plasmid and subcloned into the EcoRI site of pCDNA3 plasmid (Invitrogen), which enables expression in mammalian cells from the cytomegalovirus promoter. GRIP1 NID (wt or S3A) fragment was excised from pGex2TK-GRIP1563–765 and pGEX4T1-GRIP1563–765His6, respectively (Darimont et al., 1998), using BamHI–XhoI and subcloned into BamHI–XhoI sites of pCDNA3. A full-length hTRβ-1 cDNA was subcloned into XhoI–Klenow–HindIII sites of the pRep4 plasmid (Invitrogen).
Antibodies
Anti-p160 antibodies included anti-SRC1, anti-Rac3/ACTR and anti-TIF2 [(N)TIF2, residues 206–217] rabbit polyclonal (Affinity Bioreagents, PA1-840, PA1-845 and PA1-846, respectively). Anti-TIF2 mouse monoclonal antibody 3Ti3F1 directed against TIF2 amino acid residues 940–1010 [(C)TIF2] was kindly provided by P.Chambon (Voegel et al., 1998). Anti-cFos (SC-52 and SC-7202), anti-ERK (SC-94) and anti-cJun (SC-1694) rabbit polyclonal and anti-cJun mouse monoclonal (SC-7481) antibodies were purchased from Santa Cruz Biotechnology, Santa Cruz, CA. Anti-GR rabbit polyclonal antibodies N499 were generated against a purified polypeptide corresponding to amino acid residues 1–499 of the human GR (R.M.Nissen, B.Darimont and K.R.Yamamoto, unpublished data). Anti-TRβ mouse monoclonal antibody (MA1-216; Affinity Bioreagents) was used for the immunoblotting and indirect immunofluorescence.
Chromatin immunoprecipitations
Protocols described in Nissen and Yamamoto (2000) were modified for chromatin immunoprecipitations. U2OS.G and U2OS.GT cells grown in 150-mm dishes were treated as indicated in figure legends, chilled at 4°C for 10 min and 11× formaldehyde solution (50 mM HEPES–KOH pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, 11% formaldehyde) was added to a final concentration of 1×. After a 30 min incubation at 4°C, formaldehyde was quenched with 125 mM glycine for 5 min at 4°C, cells were rinsed in ice-cold phosphate-buffered saline (PBS), scraped off the dishes and harvested by centrifugation (600 g, 10 min at 4°C). Cell pellets were lysed in 10 ml of ice-cold lysis buffer (50 mM HEPES–KOH pH 7.4, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100) supplemented with protease inhibitors [1 mM phenymethylsulfonyl fluoride (PMSF) and 1 µg/ml each of aprotinin, leupeptin and pepstatin A] by nutating at 4°C for 10 min and crude nuclei were collected by centrifugation (600 g, for 5 min at 4°C). Nuclei were nutated in 8 ml of wash buffer (10 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 200 mM NaCl, supplemented with protease inhibitors) for 10 min at room temperature, collected as above and resuspended in 2 ml of ice-cold RIPA buffer [10 mM Tris–HCl pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 140 mM NaCl, 5% glycerol, 0.1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), 1% Triton X-100, supplemented with protease inhibitors]. Samples were sonicated with Branson Sonifier 250, with a microtip at power setting 5, in 20 s bursts separated by 1 min incubation on ice for total sonication time 3 min per sample (nine cycles). Average DNA fragment size of 200–800 bp was assessed by agarose gel electrophoresis. Lysates were then cleared by centrifugation (16 000 g, 15 min at 4°C) and 20 µl of each sample were saved as ‘total genomic input’.
The rest of the lysate was used for immunoprecipitation with (N)cFos, (C)cFos, cJun, (N)TIF2 (2–3 µg/sample each), N499 (24 µg total IgG/sample), (C)TIF2 (8 µg/sample) antibodies or equivalent amount of normal mouse or normal rabbit serum. Immunoprecipitations were performed for 6–16 h, at 4°C, and immune complexes were collected by nutating the lysates for 1 h at 4°C with 30 µl/sample of 50% slurry protein A/G agarose beads (Santa Cruz Biotechnology) pre-incubated with 100 µg/ml salmon sperm DNA (Life Technologies). The beads were washed once with 0.5 ml of ice-cold RIPA buffer and then five times for 5 min with 1 ml of ice-cold RIPA buffer supplemented with 1 mM PMSF and 100 µg/ml yeast tRNA (Life Technologies). The beads were then incubated in 100 µl of TE, 0.5% SDS, 200 µg/ml proteinase K (Sigma) for 3 h at 55°C, and cross-links were reversed for 6 h at 65°C. The DNA was extracted twice with phenol–chloroform, once with chloroform, ethanol-precipitated in the presence of 20 µg of glycogen at –20°C overnight and resuspended in 40 µl of TE buffer. PCR was performed exactly as described (Nissen and Yamamoto, 2000) using 4 µl (10%) of DNA sample.
PCR primers used for chromatin immunoprecipitations
A –150/+68 fragment of the human collagenase-3 gene (coll3) was amplified using a primer pair 5′-CTACCACAAACCACACTCGG and 5′-AGCTCAAGAAGAGGAAGGCAG. An upstream promoter region –1179/–897 (coll3–900) was amplified with a 5′-CTACCCATTTACTTCTGCAGGG and 5′-AATGTTCTTGACCACCGTCC primer set. A –229/+18 fragment of the human collagenase-1 gene (coll1) was amplified with 5′-CCACTGTTTACATGGCAGAGTG and 5′-TCTTGCTGCTCCAATATCCC primer pair. A primer set that amplified a +153/+423 fragment of the human hsp70 gene (5′-GGATCCAGTGTTCCGTTTCC and 5′-GTCAAACACGGTGTTCTGCG) was used as a control for collagenase-3 amplification; an hsp70 +154/+485 fragment amplified with the forward primer described above and a reverse primer 5′-AGTGCTTCATGTCCGACTGC was used as a control for collagenase-1 amplification. A primer pair used to amplify U6 snRNA –245/+85 region was described previously (Nissen and Yamamoto, 2000).
RNA isolation, RT and Taqman
Subconfluent U2OS.G or U2OS.GT cells were treated in 100-mm dishes as described in the figure legends, trypsinized, resuspended in DMEM–10% FBS, and collected by centrifugation (1500 r.p.m. for 5 min, Beckman GS-6KR centrifuge). Cell pellets were washed once with PBS and total RNA was isolated using QIAshredder and RNeasy-Mini kits (Qiagen) as per the manufacturer’s instructions. Random hexamer-primed cDNA was prepared from 1 µg of total RNA using the Clontech Advantage RT-for-PCR kit following manufacturer’s instructions. Taqman analysis was performed on an ABI Prism 7700 Sequence Detection System using standard protocols. Collagenase-3 cDNA fragment was amplified with the 5′-ATGATCTCTTTTGGAATTAAGGAGCAT and 5′-CATAATTTGGCCCAGGAGGAA primer pair using 5′-dT-FAM-CTACCCATTTGATGGGCCCTCTGGC-TAMRA doubly-modified oligonucleotide (Synthegen) as a fluorogenic probe. Collagenase-1 cDNA fragment was amplified with the 5′-CATGCGCACAAATCCCTTCT and 5′-CATCTCTGTCGGCAAATTCGT primer pair using 5′-dT-FAM-AGCAGCTTCAAGCCCATTTGGCAGT-TAMRA doubly-modified oligonucleotide (Biosearch Technologies) as a fluorogenic probe. Samples were analyzed in triplicate and normalized using a G3PDH primer/probe set (PE Biosystems) run in parallel. Data presented are transformed using the ∂∂Ct method as described (PE Biosystems User Bulletin 2, 1997).
Immunoblotting
Subconfluent U2OS.G cells were treated in 60-mm dishes as described in the figure legends, chilled on ice, washed with PBS, and lysed in 200 µl of RIPA lysis buffer (see above). The lysates were cleared by centrifugation (16 000 g, 15 min at 4°C), total protein concentration of the extracts was measured using Bradford Reagent (Bio-Rad), equalized with RIPA buffer and the extracts were boiled for 2 min in 5× SDS sample buffer. Protein extracts were fractionated by SDS–PAGE, transferred to Immobilon paper (Millipore Corp.) and probed with antibodies of interest. Blots were developed using horseradish peroxidase-conjugated sheep anti-mouse or donkey anti-rabbit secondary antibodies and the enhanced chemiluminescence (ECL) substrate (Amersham Pharmacia) according to the manufacturer’s instructions.
Transient transfections
U2OS.G cells were split in DMEM–10% FBS into 24-well plates at 30 000 cells/well and transfected the following day in FBS-free DMEM with Lipofectamine-PLUS reagent (Life Technologies) using 0.8 µl/well Lipofectamine and 1.6 µl/well PLUS as per the manufacturer’s instructions. Three hours post-transfection, cells were re-fed with DMEM–10% FBS, allowed to recover for 3 h and re-fed with DMEM–10% FBS containing appropriate hormone dilutions. The following day (12–14 h later) cells were lysed in 100 µl/well of 1× lysis buffer (Pharmingen) and assayed for luciferase and β-gal activity as previously described (Iniguez-Lluhi et al., 1997).
Acknowledgments
Acknowledgements
We thank M.Stallcup, B.Darimont and R.Uht for providing expression plasmids and M.Garabedian for providing U2OS.G cells. We are grateful to P.Chambon for providing anti-TIF2 antibodies and R.Nissen and B.Darimont for anti-GR N499 antibodies. We thank B.Darimont, B.Freeman, C.Jamieson, H.Luecke and R.Nissen for helpful discussions and B.Black, N.Freedman, H.Luecke and M.Van Gilst for critical comments on the manuscript. I.R. is supported by the Parker B.Francis Fellowship for Pulmonary Research. Research support was from the National Institutes of Health and the National Science Foundation.
References
- Algarte M., Kwon,H., Genin,P. and Hiscott,J. (1999) Identification by in vivo genomic footprinting of a transcriptional switch containing NF-κB and Sp1 that regulates the IκBα promoter. Mol. Cell. Biol., 19, 6140–6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An J., Ribeiro,R.C., Webb,P., Gustafsson,J.A., Kushner,P.J., Baxter,J.D. and Leitman,D.C. (1999) Estradiol repression of tumor necrosis factor-α transcription requires estrogen receptor activation function-2 and is enhanced by coactivators. Proc. Natl Acad. Sci. USA, 96, 15161–15166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P., Baumann,I., Stein,B., Delius,H., Rahmsdorf,H.J. and Herrlich,P. (1987a) 12-O-tetradecanoyl-phorbol-13-acetate induction of the human collagenase gene is mediated by an inducible enhancer element located in the 5′-flanking region. Mol. Cell. Biol., 7, 2256–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angel P., Imagawa,M., Chiu,R., Stein,B., Imbra,R.J., Rahmsdorf,H.J., Jonat,C., Herrlich,P. and Karin,M. (1987b) Phorbol ester-inducible genes contain a common cis element recognized by a TPA-modulated trans-acting factor. Cell, 49, 729–739. [DOI] [PubMed] [Google Scholar]
- Anzick S.L. et al. (1997) AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science, 277, 965–968. [DOI] [PubMed] [Google Scholar]
- Blanco J.C., Minucci,S., Lu,J., Yang,X.J., Walker,K.K., Chen,H., Evans,R.M., Nakatani,Y. and Ozato,K. (1998) The histone acetylase PCAF is a nuclear receptor coactivator. Genes Dev., 12, 1638–1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden P., Solymar,D., Sucharczuk,A., Lindman,B., Cannon,P. and Heller,R.A. (1996) Cytokine control of interstitial collagenase and collagenase-3 gene expression in human chondrocytes. J. Biol. Chem., 271, 23577–23581. [DOI] [PubMed] [Google Scholar]
- Boudreau F., Zannoni,S., Pelletier,N., Bardati,T., Yu,S.J. and Asselin,C. (1999) Negative regulation of glucocorticoid-dependent induction of c-fos by ras in intestinal epithelial cells. Mol. Cell. Biochem., 195, 99–111. [DOI] [PubMed] [Google Scholar]
- Bruhn L., Munnerlyn,A. and Grosschedl,R. (1997) ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRα enhancer function. Genes Dev., 11, 640–653. [DOI] [PubMed] [Google Scholar]
- Chandler V.L., Maler,B.A. and Yamamoto,K.R. (1983) DNA sequences bound specifically by glucocorticoid receptor in vitro render a heterologous promoter hormone responsive in vivo. Cell, 33, 489–499. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Schiltz,R.L., Chakravarti,D., Nash,A., Nagy,L., Privalsky,M.L., Nakatani,Y. and Evans,R.M. (1997) Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell, 90, 569–580. [DOI] [PubMed] [Google Scholar]
- Chen J.D. and Evans,R.M. (1995) A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature, 377, 454–457. [DOI] [PubMed] [Google Scholar]
- Chen J.D., Umesono,K. and Evans,R.M. (1996) SMRT isoforms mediate repression and anti-repression of nuclear receptor heterodimers. Proc. Natl Acad. Sci. USA, 93, 7567–7571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darimont B.D., Wagner,R.L., Apriletti,J.W., Stallcup,M.R., Kushner,P.J., Baxter,J.D., Fletterick,R.J. and Yamamoto,K.R. (1998) Structure and specificity of nuclear receptor–coactivator interactions. Genes Dev., 12, 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies M.P., Hallam,T.J. and Merritt,J.E. (1990) A role for calcium and protein kinase C in agonist-stimulated adhesion of human neutrophils. Biochem. J., 267, 13–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois C., Aubert,D., Legrand,C., Pain,B. and Samarut,J. (1991) A novel mechanism of action for v-ErbA: abrogation of the inactivation of transcription factor AP-1 by retinoic acid and thyroid hormone receptors. Cell, 67, 731–740. [DOI] [PubMed] [Google Scholar]
- Diamond M.I., Miner,J.N., Yoshinaga,S.K. and Yamamoto,K.R. (1990) Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science, 249, 1266–1272. [DOI] [PubMed] [Google Scholar]
- Ding X.F., Anderson,C.M., Ma,H., Hong,H., Uht,R.M., Kushner,P.J. and Stallcup,M.R. (1998) Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol. Endocrinol., 12, 302–313. [DOI] [PubMed] [Google Scholar]
- Dressel U., Thormeyer,D., Altincicek,B., Paululat,A., Eggert,M., Schneider,S., Tenbaum,S.P., Renkawitz,R. and Baniahmad,A. (1999) Alien, a highly conserved protein with characteristics of a corepressor for members of the nuclear hormone receptor superfamily. Mol. Cell. Biol., 19, 3383–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberhardy S.R., D’Cunha,C.A. and Farnham,P.J. (2000) Direct examination of histone acetylation on myc target genes using chromatin immunoprecipitation. J. Biol. Chem., 275, 33798–33805. [DOI] [PubMed] [Google Scholar]
- Emmer M., de Crombrugghe,B., Pastan,I. and Perlman,R. (1970) Cyclic AMP receptor protein of E.coli: its role in the synthesis of inducible enzymes. Proc. Natl Acad. Sci. USA, 66, 480–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fondell J.D., Ge,H. and Roeder,R.G. (1996) Ligand induction of a transcriptionally active thyroid hormone receptor coactivator complex. Proc. Natl Acad. Sci. USA, 93, 8329–8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedman L.P. (1999) Increasing the complexity of coactivation in nuclear receptor signaling. Cell, 97, 5–8. [DOI] [PubMed] [Google Scholar]
- Fryer C.J., Kinyamu,H.K., Rogatsky,I., Garabedian,M.J. and Archer,T.K. (2000) Selective activation of the glucocorticoid receptor by steroid antagonists in human breast cancer and osteosarcoma cells. J. Biol. Chem., 275, 17771–17777. [DOI] [PubMed] [Google Scholar]
- Gack S., Vallon,R., Schaper,J., Ruther,U. and Angel,P. (1994) Phenotypic alterations in fos-transgenic mice correlate with changes in Fos/Jun-dependent collagenase type I expression. Regulation of mouse metalloproteinases by carcinogens, tumor promoters, cAMP, and Fos oncoprotein. J. Biol. Chem., 269, 10363–10369. [PubMed] [Google Scholar]
- Giese K., Kingsley,C., Kirshner,J.R. and Grosschedl,R. (1995) Assembly and function of a TCRα enhancer complex is dependent on LEF-1-induced DNA bending and multiple protein–protein interactions. Genes Dev., 9, 995–1008. [DOI] [PubMed] [Google Scholar]
- Goodman R.H. and Smolik,S. (2000) CBP/p300 in cell growth, transformation, and development. Genes Dev., 14, 1553–1577. [PubMed] [Google Scholar]
- Guarente L., Yocum,R.R. and Gifford,P. (1982) A GAL10–CYC1 hybrid yeast promoter identifies the GAL4 regulatory region as an upstream site. Proc. Natl Acad. Sci. USA, 79, 7410–7414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanstein B., Eckner,R., DiRenzo,J., Halachmi,S., Liu,H., Searcy,B., Kurokawa,R. and Brown,M. (1996) p300 is a component of an estrogen receptor coactivator complex. Proc. Natl Acad. Sci. USA, 93, 11540–11545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven,E., Hoare,S. and Parker,M.G. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature, 387, 733–736. [DOI] [PubMed] [Google Scholar]
- Heinzel T. et al. (1997) A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature, 387, 43–48. [DOI] [PubMed] [Google Scholar]
- Hong H., Kohli,K., Trivedi,A., Johnson,D.L. and Stallcup,M.R. (1996) GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc. Natl Acad. Sci. USA, 93, 4948–4952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horlein A.J. et al. (1995) Ligand-independent repression by the thyroid hormone receptor mediated by a nuclear receptor co-repressor. Nature, 377, 397–404. [DOI] [PubMed] [Google Scholar]
- Hu E., Mueller,E., Oliviero,S., Papaioannou,V.E., Johnson,R. and Spiegelman,B.M. (1994) Targeted disruption of the c-fos gene demonstrates c-fos-dependent and -independent pathways for gene expression stimulated by growth factors or oncogenes. EMBO J., 13, 3094–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iniguez-Lluhi J.A., Lou,D.Y. and Yamamoto,K.R. (1997) Three amino acid substitutions selectively disrupt the activation but not the repression function of the glucocorticoid receptor N terminus. J. Biol. Chem., 272, 4149–4156. [DOI] [PubMed] [Google Scholar]
- Ito K., Barnes,P.J. and Adcock,I.M. (2000) Glucocorticoid receptor recruitment of histone deacetylase 2 inhibits interleukin-1β-induced histone H4 acetylation on lysines 8 and 12. Mol. Cell. Biol., 20, 6891–6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez M.J., Balbin,M., Lopez,J.M., Alvarez,J., Komori,T. and Lopez-Otin,C. (1999) Collagenase 3 is a target of Cbfa1, a transcription factor of the runt gene family involved in bone formation. Mol. Cell. Biol., 19, 4431–4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonat C., Rahmsdorf,H.J., Park,K.K., Cato,A.C., Gebel,S., Ponta,H. and Herrlich,P. (1990) Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell, 62, 1189–1204. [DOI] [PubMed] [Google Scholar]
- Kalkhoven E., Valentine,J.E., Heery,D.M. and Parker,M.G. (1998) Isoforms of steroid receptor co-activator 1 differ in their ability to potentiate transcription by the oestrogen receptor. EMBO J., 17, 232–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karin M., Haslinger,A., Holtgreve,H., Richards,R.I., Krauter,P., Westphal,H.M. and Beato,M. (1984) Characterization of DNA sequences through which cadmium and glucocorticoid hormones induce human metallothionein-IIA gene. Nature, 308, 513–519. [DOI] [PubMed] [Google Scholar]
- Koipally J. and Georgopoulos,K. (2000) Ikaros interactions with CtBP reveal a repression mechanism that is independent of histone deacetylase activity. J. Biol. Chem., 275, 19594–19602. [DOI] [PubMed] [Google Scholar]
- Konig H., Ponta,H., Rahmsdorf,H.J. and Herrlich,P. (1992) Interference between pathway-specific transcription factors: glucocorticoids antagonize phorbol ester-induced AP-1 activity without altering AP-1 site occupation in vivo. EMBO J., 11, 2241–2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus W.L. and Kadonaga,J.T. (1998) p300 and estrogen receptor cooperatively activate transcription via differential enhancement of initiation and reinitiation. Genes Dev., 12, 331–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruijer W., Cooper,J.A., Hunter,T. and Verma,I.M. (1984) Platelet-derived growth factor induces rapid but transient expression of the c-fos gene and protein. Nature, 312, 711–716. [DOI] [PubMed] [Google Scholar]
- Lamph W.W., Wamsley,P., Sassone-Corsi,P. and Verma,I.M. (1988) Induction of proto-oncogene JUN/AP-1 by serum and TPA. Nature, 334, 629–631. [DOI] [PubMed] [Google Scholar]
- Lefstin J.A. and Yamamoto,K.R. (1998) Allosteric effects of DNA on transcriptional regulators. Nature, 392, 885–888. [DOI] [PubMed] [Google Scholar]
- Lemon B. and Tjian,R. (2000) Orchestrated response: a symphony of transcription factors for gene control. Genes Dev., 14, 2551–2569. [DOI] [PubMed] [Google Scholar]
- Li H., Gomes,P.J. and Chen,J.D. (1997) RAC3, a steroid/nuclear receptor-associated coactivator that is related to SRC-1 and TIF2. Proc. Natl Acad. Sci. USA, 94, 8479–8484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L.G. and Jackson,I.M. (1998) Glucocorticoids stimulate TRH and c-fos/c-jun gene co-expression in cultured hypothalamic neurons. Brain Res., 791, 56–62. [DOI] [PubMed] [Google Scholar]
- Malik S. and Roeder,R.G. (2000) Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem. Sci., 25, 277–283. [DOI] [PubMed] [Google Scholar]
- Meyer B.J. and Ptashne,M. (1980) Gene regulation at the right operator (OR) of bacteriophage λ. III. λ repressor directly activates gene transcription. J. Mol. Biol., 139, 195–205. [DOI] [PubMed] [Google Scholar]
- Myers E.W. et al. (2000) A whole-genome assembly of Drosophila. Science, 287, 2196–2204. [DOI] [PubMed] [Google Scholar]
- Naar A.M., Beaurang,P.A., Zhou,S., Abraham,S., Solomon,W. and Tjian,R. (1999) Composite co-activator ARC mediates chromatin-directed transcriptional activation. Nature, 398, 828–832. [DOI] [PubMed] [Google Scholar]
- Nagy L., Kao,H.Y., Chakravarti,D., Lin,R.J., Hassig,C.A., Ayer,D.E., Schreiber,S.L. and Evans,R.M. (1997) Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell, 89, 373–380. [DOI] [PubMed] [Google Scholar]
- Nissen R.M. and Yamamoto,K.R. (2000) The glucocorticoid receptor inhibits NFκB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev., 14, 2314–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onate S.A., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (1995) Sequence and characterization of a coactivator for the steroid hormone receptor superfamily. Science, 270, 1354–1357. [DOI] [PubMed] [Google Scholar]
- Pendas A.M., Balbin,M., Llano,E., Jimenez,M.G. and Lopez-Otin,C. (1997) Structural analysis and promoter characterization of the human collagenase-3 gene (MMP13). Genomics, 40, 222–233. [DOI] [PubMed] [Google Scholar]
- Porte D., Tuckermann,J., Becker,M., Baumann,B., Teurich,S., Higgins,T., Owen,M.J., Schorpp-Kistner,M. and Angel,P. (1999) Both AP-1 and Cbfa1-like factors are required for the induction of interstitial collagenase by parathyroid hormone. Oncogene, 18, 667–678. [DOI] [PubMed] [Google Scholar]
- Qi C. et al. (1999) Mouse steroid receptor coactivator-1 is not essential for peroxisome proliferator-activated receptor α-regulated gene expression. Proc. Natl Acad. Sci. USA, 96, 1585–1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachez C., Lemon,B.D., Suldan,Z., Bromleigh,V., Gamble,M., Naar,A.M., Erdjument-Bromage,H., Tempst,P. and Freedman,L.P. (1999) Ligand-dependent transcription activation by nuclear receptors requires the DRIP complex. Nature, 398, 824–828. [DOI] [PubMed] [Google Scholar]
- Rogatsky I., Trowbridge,J.M. and Garabedian,M.J. (1997) Gluco corticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol. Cell. Biol., 17, 3181–3193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I., Waase,C.L. and Garabedian,M.J. (1998) Phosphorylation and inhibition of rat glucocorticoid receptor transcriptional activation by glycogen synthase kinase-3 (GSK-3). Species-specific differences between human and rat glucocorticoid receptor signaling as revealed through GSK-3 phosphorylation. J. Biol. Chem., 273, 14315–14321. [DOI] [PubMed] [Google Scholar]
- Roy S., Garges,S. and Adhya,S. (1998) Activation and repression of transcription by differential contact: two sides of a coin. J. Biol. Chem., 273, 14059–14062. [DOI] [PubMed] [Google Scholar]
- Saatcioglu F., Bartunek,P., Deng,T., Zenke,M. and Karin,M. (1993) A conserved C-terminal sequence that is deleted in v-ErbA is essential for the biological activities of c-ErbA (the thyroid hormone receptor). Mol. Cell. Biol., 13, 3675–3685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saatcioglu F. et al. (1997) Mutations in the conserved C-terminal sequence in thyroid hormone receptor dissociate hormone-dependent activation from interference with AP-1 activity. Mol. Cell. Biol., 17, 4687–4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai D.D., Helms,S., Carlstedt-Duke,J., Gustafsson,J.A., Rottman,F.M. and Yamamoto,K.R. (1988) Hormone-mediated repression: a negative glucocorticoid response element from the bovine prolactin gene. Genes Dev., 2, 1144–1154. [DOI] [PubMed] [Google Scholar]
- Schreiber M., Baumann,B., Cotten,M., Angel,P. and Wagner,E.F. (1995) Fos is an essential component of the mammalian UV response. EMBO J., 14, 5338–5349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schule R., Rangarajan,P., Kliewer,S., Ransone,L.J., Bolado,J., Yang,N., Verma,I.M. and Evans,R.M. (1990) Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell, 62, 1217–1226. [DOI] [PubMed] [Google Scholar]
- Shang Y., Hu,X., DiRenzo,J., Lazar,M.A. and Brown,M. (2000) Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell, 103, 843–852. [DOI] [PubMed] [Google Scholar]
- Sharif M. and Privalsky,M.L. (1992) V-erbA and c-erbA proteins enhance transcriptional activation by c-jun. Oncogene, 7, 953–960. [PubMed] [Google Scholar]
- Small S. and Levine,M. (1991) The initiation of pair-rule stripes in the Drosophila blastoderm. Curr. Opin. Genet. Dev., 1, 255–260. [DOI] [PubMed] [Google Scholar]
- Smith C.A., Calabro,V.V. and Frankel,A.D. (2000) An RNA-binding chameleon. Mol. Cell, 6, 1067–1076. [DOI] [PubMed] [Google Scholar]
- Solis-Herruzo J.A., Rippe,R.A., Schrum,L.W., de La Torre,P., Garcia,I., Jeffrey,J.J., Munoz-Yague,T. and Brenner,D.A. (1999) Interleukin-6 increases rat metalloproteinase-13 gene expression through stimulation of activator protein 1 transcription factor in cultured fibroblasts. J. Biol. Chem., 274, 30919–30926. [DOI] [PubMed] [Google Scholar]
- Starr D.B., Matsui,W., Thomas,J.R. and Yamamoto,K.R. (1996) Intracellular receptors use a common mechanism to interpret signaling information at response elements. Genes Dev., 10, 1271–1283. [DOI] [PubMed] [Google Scholar]
- Subramaniam M., Colvard,D., Keeting,P.E., Rasmussen,K., Riggs,B.L. and Spelsberg,T.C. (1992) Glucocorticoid regulation of alkaline phosphatase, osteocalcin, and proto-oncogenes in normal human osteoblast-like cells. J. Cell. Biochem., 50, 411–424. [DOI] [PubMed] [Google Scholar]
- Subramaniam N., Treuter,E. and Okret,S. (1999) Receptor interacting protein RIP140 inhibits both positive and negative gene regulation by glucocorticoids. J. Biol. Chem., 274, 18121–18127. [DOI] [PubMed] [Google Scholar]
- Thanos D. and Maniatis,T. (1995) Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell, 83, 1091–1100. [DOI] [PubMed] [Google Scholar]
- Torchia J., Rose,D.W., Inostroza,J., Kamei,Y., Westin,S., Glass,C.K. and Rosenfeld,M.G. (1997) The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature, 387, 677–684. [DOI] [PubMed] [Google Scholar]
- Treisman R. (1985) Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5′ element and c-fos 3′ sequences. Cell, 42, 889–902. [DOI] [PubMed] [Google Scholar]
- Truneh A., Albert,F., Golstein,P. and Schmitt-Verhulst,A.M. (1985) Early steps of lymphocyte activation bypassed by synergy between calcium ionophores and phorbol ester. Nature, 313, 318–320. [DOI] [PubMed] [Google Scholar]
- Tuckermann J.P., Reichardt,H.M., Arribas,R., Richter,K.H., Schutz,G. and Angel,P. (1999) The DNA binding-independent function of the glucocorticoid receptor mediates repression of AP-1-dependent genes in skin. J. Cell Biol., 147, 1365–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter J.C. et al. (2001) The sequence of the human genome. Science, 291, 1304–1351. [DOI] [PubMed] [Google Scholar]
- Vivanco Ruiz M.M., Bugge,T.H., Hirschmann,P. and Stunnenberg,H.G. (1991) Functional characterization of a natural retinoic acid responsive element. EMBO J., 10, 3829–3838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegel J.J., Heine,M.J., Zechel,C., Chambon,P. and Gronemeyer,H. (1996) TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J., 15, 3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Voegel J.J., Heine,M.J., Tini,M., Vivat,V., Chambon,P. and Gronemeyer,H. (1998) The coactivator TIF2 contains three nuclear receptor-binding motifs and mediates transactivation through CBP binding-dependent and -independent pathways. EMBO J., 17, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vu T.H. and Werb,Z. (2000) Matrix metalloproteinases: effectors of development and normal physiology. Genes Dev., 14, 2123–2133. [DOI] [PubMed] [Google Scholar]
- Weiss R.E., Xu,J., Ning,G., Pohlenz,J., O’Malley,B.W. and Refetoff,S. (1999) Mice deficient in the steroid receptor co-activator 1 (SRC-1) are resistant to thyroid hormone. EMBO J., 18, 1900–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.P. and Sigler,P.B. (1998) Atomic structure of progesterone complexed with its receptor. Nature, 393, 392–396. [DOI] [PubMed] [Google Scholar]
- Wilson A.C., Carlson,S.S. and White,T.J. (1977) Biochemical evolution. Annu. Rev. Biochem., 46, 573–639. [DOI] [PubMed] [Google Scholar]
- Winchester S.K., Selvamurugan,N., D’Alonzo,R.C. and Partridge,N.C. (2000) Developmental regulation of collagenase-3 mRNA in normal, differentiating osteoblasts through the activator protein-1 and the runt domain binding sites. J. Biol. Chem., 275, 23310–23318. [DOI] [PubMed] [Google Scholar]
- Wong J., Patterton,D., Imhof,A., Guschin,D., Shi,Y.B. and Wolffe,A.P. (1998) Distinct requirements for chromatin assembly in transcriptional repression by thyroid hormone receptor and histone deacetylase. EMBO J., 17, 520–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J., Qiu,Y., DeMayo,F.J., Tsai,S.Y., Tsai,M.J. and O’Malley,B.W. (1998) Partial hormone resistance in mice with disruption of the steroid receptor coactivator-1 (SRC-1) gene. Science, 279, 1922–1925. [DOI] [PubMed] [Google Scholar]
- Xu J., Liao,L., Ning,G., Yoshida-Komiya,H., Deng,C. and O’Malley,B.W. (2000) The steroid receptor coactivator SRC-3 (p/CIP/RAC3/AIB1/ACTR/TRAM-1) is required for normal growth, puberty, female reproductive function, and mammary gland development. Proc. Natl Acad. Sci. USA, 97, 6379–6384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto K.R., Pearce,D., Thomas,J. and Miner,J.N. (1992) Com binatorial regulation at mammalian composite response elements. In McKnight,S.L. and Yamamoto,K.R. (eds), Transcriptional Regulation. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 2, pp. 1169–1192.
- Yamamoto K.R., Darimont,B.D., Wagner,R.L. and Iniguez-Lluhi,J.A. (1998) Building transcriptional regulatory complexes: signals and surfaces. Cold Spring Harb. Symp. Quant. Biol., 63, 587–598. [DOI] [PubMed] [Google Scholar]
- Yang-Yen H.-F., Chambard,J.-C., Sun,Y.-L., Smeal,T., Schmidt,T.J., Drouin,J. and Karin,M. (1990) Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein–protein interaction. Cell, 62, 1205–1215. [DOI] [PubMed] [Google Scholar]
- Yao T.P., Ku,G., Zhou,N., Scully,R. and Livingston,D.M. (1996) The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl Acad. Sci. USA, 93, 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y.O., Stroke,I.L. and Fields,S. (1993) Coupling of cell identity to signal response in yeast: interaction between the α1 and STE12 proteins. Genes Dev., 7, 1584–1597. [DOI] [PubMed] [Google Scholar]