Abstract
Dynamins form a family of multidomain GTPases involved in endocytosis, vesicle trafficking and maintenance of mitochondrial morphology. In contrast to the classical switch GTPases, a force-generating function has been suggested for dynamins. Here we report the 2.3 Å crystal structure of the nucleotide-free and GDP-bound GTPase domain of Dictyostelium discoideum dynamin A. The GTPase domain is the most highly conserved region among dynamins. The globular structure contains the G-protein core fold, which is extended from a six-stranded β-sheet to an eight-stranded one by a 55 amino acid insertion. This topologically unique insertion distinguishes dynamins from other subfamilies of GTP-binding proteins. An additional N-terminal helix interacts with the C-terminal helix of the GTPase domain, forming a hydrophobic groove, which could be occupied by C-terminal parts of dynamin not present in our construct. The lack of major conformational changes between the nucleotide-free and the GDP-bound state suggests that mechanochemical rearrangements in dynamin occur during GTP binding, GTP hydrolysis or phosphate release and are not linked to loss of GDP.
Keywords: crystal structure/Dictyostelium discoideum/dynamin/GTPase
Introduction
The large GTPases of the dynamin family function in endocytosis, vesicle trafficking, maintenance of mitochondrial morphology, and viral resistance and cell plate formation in plants (reviewed in van der Bliek, 1999a). Dynamin 1, the founding and best characterized member, plays an important role in synaptic vesicle recycling (Kosaka and Ikeda, 1983; Poodry, 1990). It can form collars around the neck of clathrin-coated buds (Takei et al., 1995) and is required for pinching off vesicles from the plasma membrane. Overexpression of dynamin mutants defective in GTP binding or hydrolysis leads to an inhibition of receptor-mediated endocytosis because coated pits do not become constricted and coated vesicles fail to bud (Herskovits et al., 1993; Damke et al., 1994). However, the exact role of dynamin in vesicle budding is still a matter of debate (van der Bliek, 1999b; Yang and Cerione, 1999; Marks et al., 2001). It has been proposed to work like a classical switch GTPase, signaling to downstream effectors in the GTP-bound state (Sever et al., 1999). In contrast, accumulating evidence points towards dynamin being a mechanochemical enzyme that uses GTP hydrolysis and a resulting conformational change actively to drive membrane fission. Such a role as a force-generating GTPase would set dynamin apart from other GTPases, which function as switches.
Dynamin A, with a molecular mass of 96 kDa, is one of at least three dynamin-related proteins from the lower eukaryote Dictyostelium discoideum (Wienke et al., 1999). Dynamin A knockout cells show a pleiotropic phenotype, hinting at a role in membrane severing. The observed effects include alterations of mitochondrial and endosomal morphology and a defect in cytokinesis (Wienke et al., 1999). Dynamin A contains three domains common to all dynamins: (i) an N-terminal highly conserved GTPase domain; (ii) a middle domain of unknown function; and (iii) a C-terminal GTPase effector domain (GED). Based on sequence similarity and domain organization, dynamin A belongs to the Vps1p subfamily, which includes the yeast proteins Vps1p and Dnm1p (Rothman et al., 1990; Gammie et al., 1995) as well as mammalian DLP1 (Yoon et al., 1998). Proteins from this subfamily lack a pleckstrin homology (PH) and a proline-rich domain. Instead they have an additional segment inserted between the middle domain and the GED. In dynamin A, this is a region, unique among dynamins, rich in asparagine, glutamine and serine. Sequence similarity between dynamin A and dynamin 1 is highest in the GTPase domain (60%) and in the GED (43%), with the middle domain being more divergent.
Dynamin A, like dynamin 1, forms oligomeric rings and spirals in vitro, which constrict upon GTP binding (Hinshaw and Schmid, 1995; B.Klockow, W.Tichelaar, D.Madden, H.H.Niemann, T.Akiba, K.Hirose and D.J.Manstein, submitted). In vitro purified dynamin 1 is able to tubulate protein-free liposomes, forming a protein coat around the lipid tubules (Takei et al., 1998), and can vesiculate lipid tubes by constricting upon GTP hydrolysis (Sweitzer and Hinshaw, 1998). Other in vitro studies have shown that the pitch of dynamin spirals assembled on lipid nanotubes increased upon GTP hydrolysis, suggesting that dynamin might act in a spring-like manner and ‘pop’ off vesicles (Stowell et al., 1999).
Common biochemical properties of dynamin family members are a low affinity for GTP (10–25 µM) and a high basal and activated turnover rate when compared with small GTPases (Sever et al., 2000). It has been shown that the GED of dynamin 1 binds to its GTPase domain, thereby activating its GTPase activity (Muhlberg et al., 1997). Using the yeast two-hybrid system, the GED of dynamin 1 was shown to interact with itself, with the GTPase domain and with the middle domain. The strength of the interaction of the GTPase domain and GED is influenced by the nucleotide state of the GTPase domain (Smirnova et al., 1999). Recently, a mutant that is impaired in GTP hydrolysis but normal in GTP binding was shown to inhibit endocytosis, underlining the importance of GTP hydrolysis as opposed to GTP binding (Marks et al., 2001).
In order to understand more fully the mechanism by which dynamins operate, we set out to determine the atomic structure of a dynamin family member. Although we were successful in crystallizing full-length dynamin A, the poorly ordered paracrystals, probably composed of stacked dynamin rings, were not suitable for further crystallographic analysis (unpublished data). As the highest degree of sequence identity among dynamin family members is in the GTPase domain, we then set out to obtain the structure of this critical region. Here we present the crystal structures of the GTPase domain of D.discoideum dynamin A in the nucleotide-free and in the GDP-bound form at 2.3 Å resolution. The structures were determined using a myosin fusion approach similar to the one used to solve the structure of repeats 1 and 2 of α-actinin (Kliche et al., 2001).
Results
The dynamin A GTPase domain (residues 2–316) was fused to the C-terminus of the Dictyostelium myosin II catalytic domain. The myosin tag led to enhanced expression, facilitated a standard purification and simplified phase determination by molecular replacement. The fusion protein was purified using the established procedure for myosin head fragments, with minor modifications (Manstein and Hunt, 1995). Briefly, the fusion protein was separated from the bulk cytosolic proteins by pelleting it with actin. The acto-myosin complex was dissociated by addition of ATP and the solubilized protein was purified over an Ni-NTA column. Crystallization occurred under conditions very close to those used both for the myosin motor domain alone and for the myosin–α-actinin fusion. Crystals grew equally well in the absence and presence of GDP. The crystals of the nucleotide-free and the GDP-bound form are isomorphous and belong to space group P21 with one molecule in the asymmetric unit (Table I). The structure was determined by molecular replacement using the myosin motor domain as search model. The current model of the GTPase domain contains residues 2–307, of which three surface loops are disordered (Table II). As the myosin region of the fusion protein mediates most crystal contacts, the dynamin region seems to be more flexible and has somewhat higher B-factors.
Table I. Crystallographic data.
| Nucleotide-free | GDP | |
|---|---|---|
| Crystal space group | P21 | P21 |
| Unit cell parameters | ||
| a (Å) | 54.45 | 54.33 |
| b (Å) | 62.04 | 61.95 |
| c (Å) | 181.2 | 181.07 |
| β (°) | 94.79 | 94.52 |
| Parameter | ||
| Resolution (Å)a | 15–2.3 (2.4–2.3) | 29–2.3 (2.4–2.3) |
| Wavelength (Å) | 1.000 | 0.920 |
| Completeness (%) | 97.9 (94.6) | 94.0 (53.6) |
| Unique reflections | 52 742 | 50 492 |
| Redundancy | 3.7 (3.0) | 3.4 (1.6) |
| Rsym (%)b | 5.0 (21.5) | 5.4 (18.6) |
| I/σ | 19.30 (4.98) | 15.66 (4.13) |
aValues in parentheses correspond to the highest resolution shell.
bRsym = ΣhΣi|I(h) – Ii(h)|/ΣhΣiIi(h), where Ii(h) and I(h) are the ith and mean measurements of the intensity of reflection h.
Table II. Refinement statistics.
| Nucleotide-free | GDP | |
|---|---|---|
| Resolution (Å) | 15–2.3 | 29–2.3 |
| Reflections (work set/test set) | 49 050/3692 | 47 022/3470 |
| Protein atoms | 8297 | 8297 |
| Ligand atoms | 40 | 69 |
| Water molecules | 376 | 369 |
| Average B-factor overall (dynamin) (Å2) | 38.9 (51.8) | 46.7 (64.6) |
| R.m.s.d. bond length (Å)/angles (°) | 0.008/1.2 | 0.008/1.1 |
| Rwork (%)a/Rfree (%)b | 19.6/25.5 | 20.3/25.8 |
aRwork = Σh|Fo – Fc|/ΣhFo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection h.
bRfree is the same as Rwork, but calculated on the 7% of the data excluded from refinement.
Overall structure
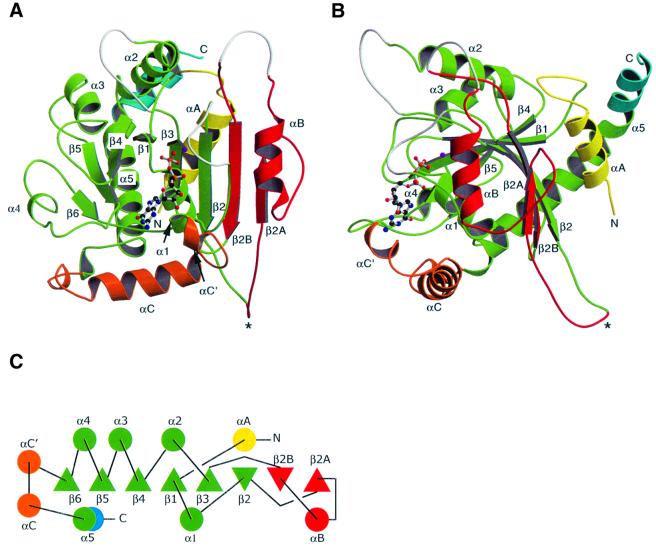
The GTPase domain consists of an eight-stranded β-sheet with six parallel and two antiparallel strands surrounded by nine helices (Figure 1). The G domain core motif, a six-stranded β-sheet with one antiparallel strand surrounded by five α-helices (Kjeldgaard et al., 1996), is present within our structure and closely resembles that found in Ras (Pai et al., 1990). Corresponding secondary structure elements of dynamin A and Ras, which share 10% sequence identity, can be superposed with an r.m.s.d. of 1.83 Å for 90 common Cα atoms.
Fig. 1. Structure and topology of the GTPase domain of dynamin A. The G-protein core fold is shown in green; αA in yellow; the insertion comprising β2A, αB, β2B in red; αC′ and αC in orange; and the extension of α5 after the kink in blue. The asterisk marks the tip of the variable loop connecting β2 and β2A. Missing loops are in white. (A) Front view. The β-sheet is extended beyond β2 to a total of eight strands. (B) Side view. αA and αB pack against the sheet from different sides, while αC runs perpendicular to the β-strands. αA and the extension of α5 make contacts at the back of the β-sheet. β3 and β6 are not labeled as they are hidden behind β2 and α1, respectively. (C) Topology diagram. β-strands coming out of the plane of the paper are triangles with tip up, while those running into the plane are tip down. Figures 1A and B, 4 and 5 were produced using Molscript (Kraulis, 1991) and Raster3d (Merritt and Bacon, 1997).
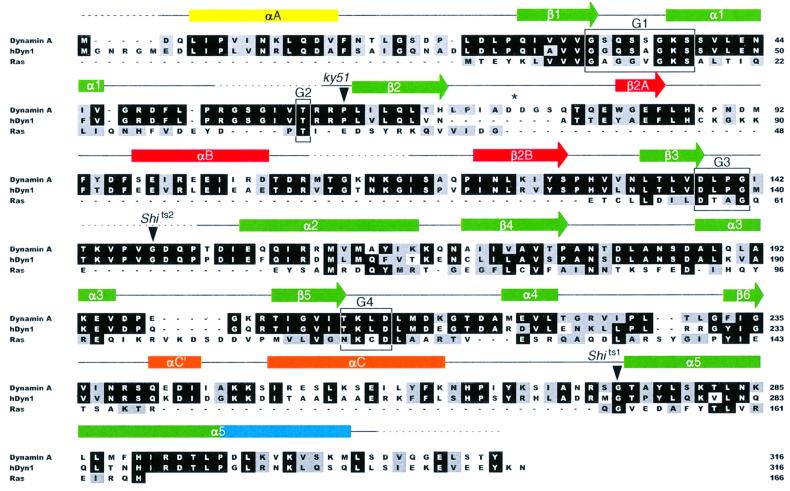
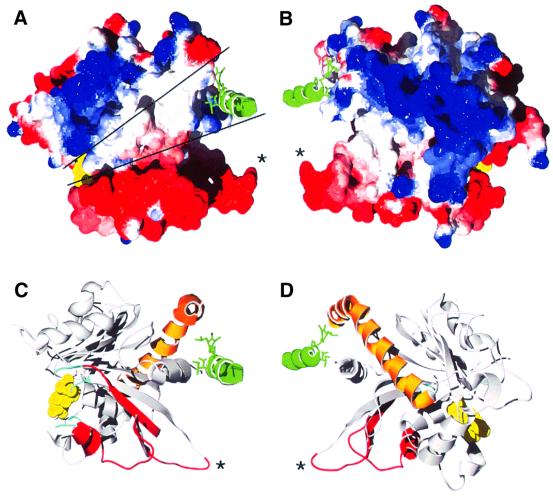
The central β-sheet of dynamin A contains eight strands instead of the six strands found in the common core fold of GTPases. It is extended beyond β2 by strands β2A and β2B, which are linked by helix αB (Figure 1). Strands β2 and β3, which are connected by a short β-turn in most other GTP-binding proteins, are linked by a 55 amino acid insertion in dynamin A (Figure 2). The start of this unique insertion coincides with the only intron in the Dictyostelium dymA gene coding for dynamin A. The Drosophila shibire (van der Bliek and Meyerowitz, 1991) and the Caenorhabditis elegans dyn-1 (Clark et al., 1997) genes also have splice sites close to the start or end of the insertion. In fact, the connection between β2 and β2A is the most diverse region within the dynamin GTPase domain. In dynamin A, it is eight residues longer than in dynamin 1 (Figure 2), and forms a long protruding loop with two aspartate residues at its tip (Figure 1A and B). In the Saccharomyces cerevisiae dynamin homologs Vps1p (Rothman et al., 1990) and Dnm1p (Gammie et al., 1995), the connection is >30 amino acids longer than that of dynamin A and is predicted to contain two additional β-strands. In rat DLP1, there are three alternatively spliced forms containing between eight and 21 residues more than dynamin A (Yoon et al., 1998). In Vps1p, Dnm1p and DLP1, this region is rich in charged amino acids. The variability, coupled with its extended conformation and highly charged nature (Figure 3A and B), suggests that the loop connecting β2 and β2A is a site for interaction with other proteins. The high degree of divergence between different dynamin family members and the alternative splicing of DLP1 in this region would then reflect the ability to interact with different binding partners.
Fig. 2. Structure-based sequence alignment of dynamin A from D.discoideum, human dynamin 1 and Ras. Rectangles indicate helices, arrows β-sheets and dashed lines disordered regions. Colors of secondary structure elements correspond to those in Figure 1. The consensus elements for GTP binding are boxed and labeled G1–G4. The position of the residues mutated in the temperature-sensitive alleles of Drosophila (shits1,2) and C.elegans (ky51) is marked. The asterisk marks the same residues as in Figure 1. Residues identical in at least two of the proteins are boxed in black; conservative substitutions are shaded in gray.
Fig. 3. Possible interaction site of the GTPase domain with the GED. The C-terminal helix of the myosin tag is shown in green with three hydrophobic side chains as a stick model. It binds into a groove of hydrophobic residues formed by helices αA and α5 of dynamin. The nucleotide is shown in yellow, and the asterisks mark the position of Asp74 and Asp75 in the loop connecting β2 and β2A. (A) Electrostatic surface potential of the dynamin A GTPase domain. Electrostatic potential is shown in red for negative charge, ramping through white at neutral charge to blue at positive charge. The two lines mark an exposed hydrophobic patch that runs from the groove formed by αA and α5 to the nucleotide-binding site. Disordered loops that might need stabilization by the C-terminal part of dynamin cluster around the nucleotide. The GED could occupy a position equivalent to that of the myosin helix and extend to the nucleotide, thereby covering the exposed hydrophobic patch. The insertion comprising β2A, αB and β2B is highly negatively charged. (B) The view from (A) rotated by 180°. The surface of helices αC and α5 is positively charged. (C and D) Ribbon diagrams of the same orientation as in (A) and (B). The topologically unique dynamin insertion is colored red, the C-terminal helix α5 is orange, and the ends of disordered loops are in cyan. The figure was produced with the Swiss-PDBViewer (Guex and Peitsch, 1997) and POV-Ray.
Another feature of the dynamin GTPase domain is the additional N-terminal helix αA and the extension of the C-terminal helix α5, which contact each other. Helices αA and αB pack against the β-sheet from opposite sides (Figure 1B). Helix αA forms hydrogen bonds with polar side chains of the β-sheet, which are solvent exposed in Ras. An N-terminal helix preceding the GTPase core domain is also found in other GTPases, e.g. the Ras-like ARFs. However, the position occupied by the N-terminal helix in dynamins is different from that in ARFs (Amor et al., 1994), with the helices packing against opposite sides of the β-sheet in these two GTPase families. The C-terminal helix α5 is considerably longer in dynamin than in other G-proteins, forming a continuous helix, kinked at Pro296 (Figure 1B). The part of helix α5 C-terminal to Pro296 makes hydrophobic contacts only to the N-terminal helix αA and the loop connecting αA and β1, suggesting that they form a stable structural unit. Surface potential analysis shows that αA and the C-terminal end of α5 form a hydrophobic groove. In our structure, this groove is occupied by hydrophobic residues from the C-terminus of myosin and the linker between the fusion partners (Figure 3). In the full-length protein, this groove might be occupied by a part of dynamin’s C-terminus. For dynamin 1, it was shown by yeast two-hybrid analysis that the GED is able to interact with a fragment corresponding to residues 1–322 of dynamin A, while a shorter fragment extending only to residue 300 did not interact with the GED (Smirnova et al., 1999). These observations are consistent with our suggestion that the groove formed by αA and α5 might be a site for interaction between the GTPase domain and the GED.
Two other prominent insertions are present in dynamin A when compared with Ras. First, the loop connecting β3 and α2 is 13 residues longer (Figure 2). Although several of the additional residues extend helix α2 N-terminally, making it longer than in Ras, most of these residues are disordered in our structure. This is not surprising, as in other GTP-binding proteins, α2 and the preceding loop have been shown to move between different nucleotide states and are often flexible in the absence of γ-phosphate. A second insertion is located between β6 and α5 (Figure 2). It contains helix αC, which does not pack against either side of the β-sheet, but rather runs perpendicular to the strands (Figure 1).
Nucleotide-binding site
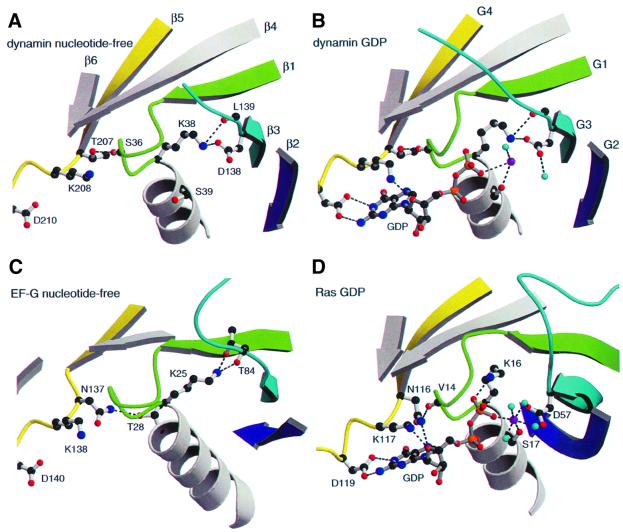
Four consensus elements, G1–G4, are involved in GTP binding in all GTPases (Bourne et al., 1991). The G1 motif (GxxxxGKS/T) or P-loop binds the phosphates of the nucleotide. The interactions of P-loop residues (32GSQSSGKS39) with the nucleotide found in dynamin A are similar to those found in Ras, with a few exceptions (Figure 4B and C). Lys38 does not bind to the β-phosphate, but instead makes hydrogen bonds to the side chain of Asp138 and the backbone carbonyl of Leu139, a situation similar to that found in GDP-bound EF-G (Czworkowski et al., 1994). In the position equivalent to Ser36 and Ser40 of dynamin A, Ras has hydrophobic residues. Ser36 and Ser40 make hydrogen bonds to the oxygens of the β- and α-phosphate, respectively. In many GTPases, a serine or threonine is found in the corresponding positions, forming hydrogen bonds similar to those observed in dynamin A. The G2 motif (a conserved threonine) is involved in coordination of the Mg2+ in the GTP-bound state. It is part of the so-called switch I region, which moves significantly between the GDP- and the GTP-bound state, and is formed by Thr59 in dynamin A. This threonine is also part of the dynamin family signature, a highly conserved region of dynamins, (51LPRGSGIVTR60 in dynamin A) as defined by PROSITE (Hofmann et al., 1999). Thr59 is not resolved in our structures, because, in the absence of γ-phosphate, the switch I region can be disordered, as in some other GTPase, e.g. in EF-G (Ævarsson et al., 1994; Czworkowski et al., 1994). The G3 motif DxxG (138DLPG141 in dynamin A) contains two invariant residues, an aspartate, which coordinates the Mg2+ via a bridging water molecule, and a glycine whose amide nitrogen forms a hydrogen bond to the nucleotide γ-phosphate. In our structures, Asp138 binds the ε-amino group of the P-loop lysine (Figure 4A and B), and Gly141 is close to where the γ-phosphate would be, occupying an intermediate position between that found in the Ras-GDP and Ras-GTP structures. The G4 motif (consensus sequence N/TKxD, 207TKLD210 in dynamin A) is involved in base binding. The side chain of Thr207 does not interact with the base but makes a hydrogen bond to the carbonyl of Ser36 in the P-loop (Figure 4B). Lys208 interacts with the endocyclic ribose oxygen as expected. The side chain of Asp210 also makes two hydrogen bonds to the nucleotide (Figure 4B).
Fig. 4. Comparison of the nucleotide-binding site of empty and GDP-bound dynamin A with those of empty EF-G and GDP-bound Ras. (A) In nucleotide-free dynamin A, the side chain of Thr207 from the TKLD motif makes a hydrogen bond (dashed lines) to the carbonyl of Ser36 in the P-loop. Lys38 binds to residues from switch II (Asp138 and Leu139). (B) In GDP-bound dynamin A, Lys38 preserves its interactions with residues from switch II and does not bind to the β-phosphate. Lys208 binds the endocyclic oxygen of the ribose and Asp210 makes two hydrogen bonds with the base, while Thr207 does not bind the base. The coordination of the Mg2+ (magenta), which is usually octahedral in G-proteins [see (D)], is non-standard due to the disorder of the structural elements and water molecules (cyan) in this region. (C) In nucleotide-free EF-G, the P-loop Lys25 binds to residues from switch II, as in dynamin A. Asn137, equivalent to dynamin A Thr207, interacts with the side chain of Thr28 in the helix following the P-loop. (D) The canonical nucleotide-binding site of Ras-GDP. Lys16 binds to the β-phosphate. The interactions of Lys117 and Asp119 with the nucleotide correspond to those of Lys208 and Asp210 in dynamin A. Asn116 in Ras makes two hydrogen bonds, one to the carbonyl of Val14 in the P-loop and one to the base.
Additional contacts with the nucleotide are mediated by the backbone of residues Asn238 and Arg239, which do not belong to one of the consensus elements for GTP binding. The carbonyl of Arg239 hydrogen-bonds to 2′O of the ribose, while the amide of Asn238 binds the exocyclic oxygen of the base. Asn238 and Arg239 occupy positions roughly equivalent to those of the G5 motif (SAK/L), which is found in small GTPases, but not conserved between all GTP-binding proteins (Figure 5). In Ras, residues from G5 hydrogen-bond to the base oxygen via backbone amides, as does Asn238 of dynamin A.
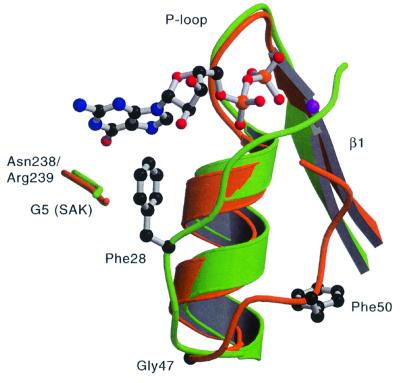
Fig. 5. Comparison of dynamin A Phe50 and Ras Phe28. Dynamin A is colored in orange and Ras in green. GDP is only shown for Ras. Gly47 at the end of helix α1 allows the switch I loop to take off at a different angle in dynamin A than in Ras, which has no glycine at the equivalent position. In dynamin A, Phe50 is buried by hydrophobic residues of the β-sheet (not shown) instead of stabilizing the base as Ras Phe28 does. The position of the Ras G5 motif (145SAK147) and of dynamin A Asn238 and Arg 239 is also shown.
In Ras, Phe28 from the switch I region forms the bottom of the binding pocket, stabilizing the guanine base by aromatic–aromatic interactions (Pai et al., 1990). Dynamin A and many other dynamin family members have a phenylalnine in the equivalent sequence position (Phe50; Figure 2). In dynamin A, Phe50 is pointing in a direction opposite to that of Phe28 in Ras, leaving the guanine ring more solvent accessible (Figure 5). Instead of stabilizing the base, Phe50 is buried in a hydrophobic pocket formed by residues mainly from helix αB and strands β2A and β3. This results in the Cα and Cζ atoms of Ras Phe28 and dynamin A Phe50 being 9.6 and 13.8 Å apart. The cause of this difference is that the switch I loops take off at different angles from the C-terminal end of helix α1 (Figure 5). This flexibility in trajectory is probably due to Gly47, which is located at the position at which the dynamins deviate from that of Ras. This glycine is highly conserved among dynamins and is only absent in some proteins from the Mgm family, which show greater divergence in the dynamin family motif forming the switch I region (Figure 4 in van der Bliek, 1999a).
The nucleotide-binding site of nucleotide-free dynamin A looks very similar to that of the GDP-bound form (Figure 4A and B). The interaction of Lys38 with Asp138 and Leu139 is the same as for the GDP-bound form (Figure 4A and C). While this position of the lysine side chain is uncommon for GDP-bound GTPases, interactions of the P-loop lysine with residues from or close to the G3 motif are usually found in nucleotide-free structures [e.g. FtsY (Montoya et al., 1997), EF-G (Ævarsson et al., 1994) or the EF-Tu–EF-Ts complex (Kawashima et al., 1996)] and is most probably a general feature stabilizing empty P-loops. A minor difference found in the nucleotide-free form is that the carbonyl of Gln34 is flipped. In this conformation, the hydrogen bond from the amide of Ser35 to the β-phosphate cannot form.
Discussion
Three loops, all clustered in one region of the molecule, are not defined in our structures (top middle and top right in Figure 1). These are the tip of the loop connecting α1 and β2 (switch I), the connection between αB and β2B, and the loop between β3 and α2 (following switch II). It is not without precedent that the switch regions are flexible in the absence of a γ-phosphate, as the switch I region is not defined in either nucleotide-free or GDP-bound EF-G (Ævarsson et al., 1994; Czworkowski et al., 1994). A possible explanation for the observed clustered disorder is that, in the full-length molecule, this region might interact with parts from the C-terminus not present in our construct. Therefore, in the absence of C-terminal structures, this region may be destabilized. A patch of hydrophobic, surface-exposed residues leads from this region to the hydrophobic groove, a predicted C-terminal interaction area (Figure 3). In the full-length molecule, a part of the C-terminus of dynamin A could replace the myosin helix in the hydrophobic groove, cover the hydrophobic patch leading to the nucleotide-binding site and stabilize the disordered loops.
This work provides a framework to understand some of the mutations within the dynamin GTPase domain that have been described to impair function of the molecule. Two temperature-sensitive mutations were identified in the D.shibire gene (van der Bliek and Meyerowitz, 1991). In the Shits1 allele, the glycine corresponding to Gly275 at the start of helix α5 is mutated to aspartate. This glycine has backbone angles forbidden for non-glycine residues and the conformation of this region would probably be disrupted by a mutation to a non-glycine residue, which would have limited conformational flexibility. However, the presence of the aspartate side chain would not be sterically blocked, and could stabilize the helix by interacting favorably with backbone amides that are otherwise unable to form hydrogen bonds (Wan and Milner-White, 1999). As this compensating stabilization is predicted to become weaker with increasing temperature, distortion of the natural protein conformation would be greater at higher temperatures. The glycine mutated to Ser in Shits2 corresponds to Gly148 in the loop before helix α2. This helix is part of the switch II, a region essential for mechanochemical coupling that is not defined in our structure, but would be very sensitive to mutation away from a flexible glycine residue. In the C.elegans dyn-1 gene, a temperature-sensitive mutation [dyn-1 (ky51)] changes Pro62 at the start of β2 to serine (Clark et al., 1997). This residue is located in a hydrophobic environment. The change to a polar residue might be tolerated at lower, but not at higher temperature, where hydrophobic forces become stronger. Another mutant has been described in which the P-loop Ser45 in dynamin 1, or Ser17 in Ras, has been mutated to asparagine, resulting in a dominant-negative phenotype having reduced GTP binding in both dynamin and Ras (Feig and Cooper, 1988; Herskovits et al., 1993; Marks et al., 2001). In dynamin A, the corresponding Ser39 makes the same interactions with the nucleotide as Ser17 in Ras, namely a hydrogen bond to an oxygen of the β-phosphate and to the Mg2+ ion. It is likely that the observed effects in dynamin have the same structural basis as in Ras, for which an altered Mg2+ binding in the triphosphate state is assumed (Farnsworth and Feig, 1991). Recently, several mutations that impair the GTP hydrolysis of dynamin 1, its function in endocytosis and the affinity for nucleotide have been reported (Marks et al., 2001). All of the residues affected are conserved between dynamin 1 and dynamin A and all are located in the critical switch elements, not well resolved in our structures.
It is interesting to compare the GTPase domains of dynamin A and human guanylate-binding protein 1 (hGBP1). Although hGBP1 and dynamins share no obvious sequence similarity, biochemical and anticipated structural similarities have led to the suggestion that hGBP1 may belong to the dynamin family of large GTPases (Prakash et al., 2000a). hGBP1 is a large interferon-induced GTPase with antiviral activity that, unlike dynamins, hydrolyses GTP to GDP and GMP. Like dynamins, hGBP1 has low nucleotide affinity, displays a high GTP turnover rate and shows nucleotide-dependent oligomerization, though it has not been shown to form spirals or tubes (Praefcke et al., 1999). The crystal structure of the full-length protein was solved and shows hGBP1 to consist of two domains, a globular GTPase domain and a purely helical domain (Prakash et al., 2000a). The helical domain consists of two three-helix bundles extending >100 Å away from the GTPase and a long helix looping back to the GTPase. It has been proposed that the two three-helix bundles correspond to the middle domain of dynamin and the long helix corresponds to the GED of dynamin. There are also structural similarities between the two proteins within the GTPase domain. For example, the loop connecting α1 and β2, which contains the switch I region, varies considerably between GTPases in terms of length, sequence and structure. In dynamin A, the trajectory and length of this loop are very similar to those in hGBP1, but different from other GTPases. Like all dynamins except Mgm, hGBP1 has a glycine at the end of helix α1 (Gly60) which has almost the same φ- and ψ-angles as found in dynamin A. In the triphosphate form of hGBP1, this loop forms the so-called phosphate cap (Prakash et al., 2000b). It seems possible that in dynamin, these residues, which are flexible in the absence of a γ-phosphate, may have a similar function. Two of the insertions found in dynamin A are present in hGBP1 as well. In both proteins, α2 and the loop connecting β3 and α2 are longer than in Ras and β6 does not connect to α5 directly, but goes through additional helices.
The similarity of these structural elements, however, contrasts with differences in the overall topology and in the nucleotide-binding region of the two GTPase domains. In hGBP1, the central β-sheet is also extended to an eight-stranded sheet with additional strands next to β2, as in dynamin A, but these two β-strands are topologically different. In hGBP1, they are formed by an N-terminal extension, while in dynamin they are inserted between β2 and β3. Furthermore, the direction of the additional strands is reversed between hGBP1 and dynamin, and there is no spatial overlap between β2A of dynamin A and β0 of hGBP1. These differences make a direct evolutionary conversion from one topology into the other very unlikely. Regarding the recognition of the guanine base, dynamins are related much more closely to virtually any other GTPase than to hGBP1, which does not have a classical N/TKxD motif. Additionally, while the so-called guanine cap in hGBP1 takes over the function of Phe28 in Ras and completly covers the base (Prakash et al., 2000b), in dynamin A there is no real substitute for Ras Phe28 and the base is even more accessible than in hGBP1. The conclusion we draw from the differing fold topologies and nucleotide-binding characteristics of the GTPase domains is that dynamin and hGBP1 are significantly different and represent distinct subfamilies of GTP-binding proteins. The presence of several shared structural features is, most probably, a result of evolutionarily convergent events.
The crystal structure of the dynamin A GTPase domain provides the first atomic-level look at a critical catalytic domain of a dynamin family protein and places dynamins into a new subfamily of GTP-binding proteins. The structure indicates that interactions between the GTPase domain and C-terminal regions of the molecule (or a neighboring molecule) are likely to be necessary for correct folding and placement of the catalytic loops surrounding the nucleotide-binding site. Furthermore, the similarity of the nucleotide-free and GDP-bound structures suggests that dynamin’s major conformational changes occur, not upon GDP release, but rather upon GTP binding, hydrolysis or phosphate release. Further elucidation of dynamin’s mechanism awaits the determination of GTP and transition state structures, as well as a high-resolution structure of the full-length protein.
Materials and methods
Protein expression and purification
Amino acids 2–316 of dynamin A were fused to the C-terminus of the Dictyostelium myosin II motor domain (residue 3–765) carrying an N-terminal His tag. The protein domains are separated by a thrombin cleavage site (Gly-Leu-Val-Pro-Arg-Gly-Ser), which acts as a linker. The protein was expressed in Dictyostelium AX3-ORF+ and purified essentially as described for the myosin part alone (Manstein and Hunt, 1995; Manstein et al., 1995). Briefly, the cells were lysed with 0.5% Triton X-100 in 20 cell volumes of Tris, pH 8.0, 2 mM EDTA, 0.2 mM EGTA, 1 mM dithiothreitol (DTT) containing protease inhibitors. The fusion protein was pelleted with actin for 1 h at 30 000 g and then solubilized by homogenizing the pellet in two cell volumes of 50 mM HEPES pH 7.3, 30 mM K-acetate, 10 mM Mg-acetate, 7 mM mercaptoethanol and 10 mM ATP. After centrifugation at 20 000 g for 30 min, the supernatant was applied to an Ni-NTA column. The column was washed extensively with 50 mM HEPES, pH 7.3, 300 mM K-acetate and with 50 mM HEPES, pH 7.3, 30 mM K-acetate and 40 mM imidazole, followed by elution with a gradient from 40 to 500 mM imidazole. Peak fractions were dialyzed against 50 mM Tris pH 8.0, 1 mM MgCl2, 1 mM DTT, 3% sucrose. The fusion protein was concentrated to 5 mg/ml, flash-frozen in liquid nitrogen and stored at –80°C.
Crystallization and data collection
For crystallization, 2 mM ADP/MgCl2 was added to the protein. Crystals were grown by the hanging drop vapor diffusion method at 4°C by mixing 2 µl of protein and 2 µl of reservoir solution. In order to obtain better crystals, microseeding was employed. The reservoir solution was 11% PEG-8000, 50 mM Tris, pH 8.5, 200 mM KCl, 5 mM MgCl2, 10% glucose, 2% methyl-propane-diol, 1 mM EGTA and 5 mM DTT. Crystals grew within 2 weeks to a size of ∼300 × 300 × 15 µm. The crystals complexed with GDP were grown under identical conditions but with an additional 2 mM GDP/MgCl2 added to the protein. Crystals were cryo-protected in reservoir solution with 12.5% PEG-8000 and 20% glycerol, and flash-frozen in liquid nitrogen. Before data collection, all crystals had to be annealed several (usually three) times by briefly blocking the cryo stream. Annealing reduced the mosaicity and improved the diffraction limit. For the nucleotide-free form, a native data set was collected at Elettra beamline 5.2R in Trieste at 100 K using a MAR image plate. For the GDP-bound form, a data set was collected at beamline ID14-4 in Grenoble at 100 K on an ADSC CCD detector.
Structure determination and refinement
Data were processed with XDS (Kabsch, 1993). Initial phases for the nucleotide-free crystals were obtained by molecular replacement in CNS (Brünger et al., 1998) using the myosin catalytic domain (Kliche et al., 2001) as search model and data from 15 to 3.5 Å. After rigid body refinement, the myosin converter domain (residues 686–761) was repositioned manually into density using the program O (Jones et al., 1991), as it had moved relative to the rest of the molecule. The initial density for the dynamin GTPase was used to build six β-strands and two helices as polyalanine. Repetitive rounds of refinement using simulated annealing and gradient energy minimization in CNS followed by manual rebuilding using O added more residues and side chains as they became visible. When the R-factor had dropped to 25%, waters were picked automatically as implemented in CNS and checked manually. B-factors were refined individually. The current models of the nucleotide-free and the GDP-bound form contain in the myosin part residues 2–19, 25–201, 208–770, an ADP, an Mg2+ and a glucose. The dynamin part contains residues 2–52, 61–108, 117–142 and 152–307. In the nucleotide-free structure, 89% of the residues are in the most favored region of the Ramachandran plot, and no non-glycine residues are in disallowed regions. The crystals containing GDP were completely isomorphous with the nucleotide-free crystals. After a rigid body refinement and simulated annealing using the nucleotide-free structure, several rounds of manual rebuilding followed by simulated annealing were carried out. Eighty-eight percent of the residues are in the most favored region of the Ramachandran plot for the structure with GDP. The PDB accession code is 1JX2 for the nucleotide-free structure, and 1JWY for the GDP structure.
Acknowledgments
Acknowledgements
We would like to thank M.Degano and the staff at beamline 5.2R, Elettra, Trieste; the staff at the ESRF beamline ID14-4; B.Klockow for helpful discussions; and K.C.Holmes for continuous support. H.H.N. is supported by the Boehringer Ingelheim Fonds.
References
- Ævarsson A., Brazhnikov,E., Garber,M., Zheltonosova,J., Chirgadze,Y., Al-Karadaghi,S., Svensson,L.A. and Liljas,A. (1994) Three-dimensional structure of the ribosomal translocase: elongation factor G from Thermus thermophilus. EMBO J., 13, 3669–3677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor J.C., Harrison,D.H., Kahn,R.A. and Ringe,D. (1994) Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature, 372, 704–708. [DOI] [PubMed] [Google Scholar]
- Bourne H.R., Sanders,D.A. and McCormick,F. (1991) The GTPase superfamily: conserved structure and molecular mechanism. Nature, 349, 117–127. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., D54, 905–921. [DOI] [PubMed] [Google Scholar]
- Clark S.G., Shurland,D.L., Meyerowitz,E.M., Bargmann,C.I. and van der Bliek,A.M. (1997) A dynamin GTPase mutation causes a rapid and reversible temperature-inducible locomotion defect in C.elegans. Proc. Natl Acad. Sci. USA, 94, 10438–10443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czworkowski J., Wang,J., Steitz,T.A. and Moore,P.B. (1994) The crystal structure of elongation factor G complexed with GDP, at 2.7 Å resolution. EMBO J., 13, 3661–3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damke H., Baba,T., Warnock,D.E. and Schmid,S.L. (1994) Induction of mutant dynamin specifically blocks endocytic coated vesicle formation. J. Cell Biol., 127, 915–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie A.E., Kurihara,L.J., Vallee,R.B. and Rose,M.D. (1995) DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J. Cell Biol., 130, 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N. and Peitsch,M.C. (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis, 18, 2714–2723. [DOI] [PubMed] [Google Scholar]
- Farnsworth C.L. and Feig,L.A. (1991) Dominant inhibitory mutations in the Mg2+-binding site of rasH prevent its activation by GTP. Mol. Cell. Biol., 11, 4822–4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig L.A. and Cooper,G.M. (1988) Inhibition of NIH 3T3 cell proliferation by mutant ras protein with preferential affinity for GDP. Mol. Cell. Biol., 8, 3235–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskovits J.S., Burgess,C.C., Obar,R.A. and Vallee,R.B. (1993) Effects of mutant rat dynamin on endocytosis. J. Cell Biol., 122, 565–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinshaw J.E. and Schmid,S.L. (1995) Dynamin self-assembles into rings suggesting a mechanism for coated vesicle budding. Nature, 374, 190–192. [DOI] [PubMed] [Google Scholar]
- Hofmann K., Bucher,P., Falquet,L. and Bairoch,A. (1999) The PROSITE database, its status in 1999. Nucleic Acids Res., 27, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Kabsch W. (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J. Appl. Crystallogr., 26, 795–800. [Google Scholar]
- Kawashima T., Berthet-Colominas,C., Wulff,M., Cusack,S. and Leberman,R. (1996) The structure of the Escherichia coli EF-Tu·EF-Ts complex at 2.5 Å resolution. Nature, 379, 511–518. [DOI] [PubMed] [Google Scholar]
- Kjeldgaard M., Nyborg,J. and Clark,B.F. (1996) The GTP binding motif: variations on a theme. FASEB J., 10, 1347–1368. [PubMed] [Google Scholar]
- Kliche W., Fujita-Becker,S., Kollmar,M., Manstein,D.J. and Kull,F.J. (2001) Structure of a genetically engineered molecular motor. EMBO J., 20, 40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka T. and Ikeda,K. (1983) Possible temperature-dependent blockage of synaptic vesicle recycling induced by a single gene mutation in Drosophila. J. Neurobiol., 14, 207–225. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Manstein D.J. and Hunt,D.M. (1995) Overexpression of myosin motor domains in Dictyostelium: screening of transformants and purification of the affinity tagged protein. J. Muscle Res. Cell Motil., 16, 325–332. [DOI] [PubMed] [Google Scholar]
- Manstein D.J., Schuster,H.P., Morandini,P. and Hunt,D.M. (1995) Cloning vectors for the production of proteins in Dictyostelium discoideum. Gene, 162, 129–134. [DOI] [PubMed] [Google Scholar]
- Marks B., Stowell,M.H., Vallis,Y., Mills,I.G., Gibson,A., Hopkins,C.R. and McMahon,H.T. (2001) GTPase activity of dynamin and resulting conformation change are essential for endocytosis. Nature, 410, 231–235. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Montoya G., Svensson,C., Luirink,J. and Sinning,I. (1997) Crystal structure of the NG domain from the signal-recognition particle receptor FtsY. Nature, 385, 365–368. [DOI] [PubMed] [Google Scholar]
- Muhlberg A.B., Warnock,D.E. and Schmid,S.L. (1997) Domain structure and intramolecular regulation of dynamin GTPase. EMBO J., 16, 6676–6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai E.F., Krengel,U., Petsko,G.A., Goody,R.S., Kabsch,W. and Wittinghofer,A. (1990) Refined crystal structure of the triphosphate conformation of H-ras p21 at 1.35 Å resolution: implications for the mechanism of GTP hydrolysis. EMBO J., 9, 2351–2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poodry C.A. (1990) shibire, a neurogenic mutant of Drosophila. Dev. Biol., 138, 464–472. [DOI] [PubMed] [Google Scholar]
- Praefcke G.J., Geyer,M., Schwemmle,M., Robert Kalbitzer,H. and Herrmann,C. (1999) Nucleotide-binding characteristics of human guanylate-binding protein 1 (hGBP1) and identification of the third GTP-binding motif. J. Mol. Biol., 292, 321–332. [DOI] [PubMed] [Google Scholar]
- Prakash B., Praefcke,G.J., Renault,L., Wittinghofer,A. and Herrmann,C. (2000a) Structure of human guanylate-binding protein 1 representing a unique class of GTP-binding proteins. Nature, 403, 567–571. [DOI] [PubMed] [Google Scholar]
- Prakash B., Renault,L., Praefcke,G.J., Herrmann,C. and Wittinghofer,A. (2000b) Triphosphate structure of guanylate-binding protein 1 and implications for nucleotide binding and GTPase mechanism. EMBO J., 19, 4555–4564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman J.H., Raymond,C.K., Gilbert,T., O’Hara,P.J. and Stevens,T.H. (1990) A putative GTP binding protein homologous to interferon-inducible Mx proteins performs an essential function in yeast protein sorting. Cell, 61, 1063–1074. [DOI] [PubMed] [Google Scholar]
- Sever S., Muhlberg,A.B. and Schmid,S.L. (1999) Impairment of dynamin’s GAP domain stimulates receptor-mediated endocytosis. Nature, 398, 481–486. [DOI] [PubMed] [Google Scholar]
- Sever S., Damke,H. and Schmid,S.L. (2000) Garrotes, springs, ratchets and whips: putting dynamin models to the test. Traffic, 1, 385–392. [DOI] [PubMed] [Google Scholar]
- Smirnova E., Shurland,D.L., Newman-Smith,E.D., Pishvaee,B. and van der Bliek,A.M. (1999) A model for dynamin self-assembly based on binding between three different protein domains. J. Biol. Chem., 274, 14942–14947. [DOI] [PubMed] [Google Scholar]
- Stowell M.H., Marks,B., Wigge,P. and McMahon,H.T. (1999) Nucleotide-dependent conformational changes in dynamin: evidence for a mechanochemical molecular spring. Nature Cell Biol., 1, 27–32. [DOI] [PubMed] [Google Scholar]
- Sweitzer S.M. and Hinshaw,J.E. (1998) Dynamin undergoes a GTP-dependent conformational change causing vesiculation. Cell, 93, 1021–1029. [DOI] [PubMed] [Google Scholar]
- Takei K., McPherson,P.S., Schmid,S.L. and De Camilli,P. (1995) Tubular membrane invaginations coated by dynamin rings are induced by GTP-γS in nerve terminals. Nature, 374, 186–190. [DOI] [PubMed] [Google Scholar]
- Takei K., Haucke,V., Slepnev,V., Farsad,K., Salazar,M., Chen,H. and De Camilli,P. (1998) Generation of coated intermediates of clathrin-mediated endocytosis on protein-free liposomes. Cell, 94, 131–141. [DOI] [PubMed] [Google Scholar]
- van der Bliek A.M. (1999a) Functional diversity in the dynamin family. Trends Cell Biol., 9, 96–102. [DOI] [PubMed] [Google Scholar]
- van der Bliek A.M. (1999b) Is dynamin a regular motor or a master regulator? Trends Cell Biol., 9, 253–254. [DOI] [PubMed] [Google Scholar]
- van der Bliek A.M. and Meyerowitz,E.M. (1991) Dynamin-like protein encoded by the Drosophila shibire gene associated with vesicular traffic. Nature, 351, 411–414. [DOI] [PubMed] [Google Scholar]
- Wan W.Y. and Milner-White,E.J. (1999) A natural grouping of motifs with an aspartate or asparagine residue forming two hydrogen bonds to residues ahead in sequence: their occurrence at α-helical N termini and in other situations. J. Mol. Biol., 286, 1633–1649. [DOI] [PubMed] [Google Scholar]
- Wienke D.C., Knetsch,M.L., Neuhaus,E.M., Reedy,M.C. and Manstein,D.J. (1999) Disruption of a dynamin homologue affects endocytosis, organelle morphology and cytokinesis in Dictyostelium discoideum. Mol. Biol. Cell, 10, 225–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. and Cerione,R.A. (1999) Endocytosis: is dynamin a ‘blue collar’ or ‘white collar’ worker? Curr. Biol., 9, R511–R514. [DOI] [PubMed] [Google Scholar]
- Yoon Y., Pitts,K.R., Dahan,S. and McNiven,M.A. (1998) A novel dynamin-like protein associates with cytoplasmic vesicles and tubules of the endoplasmic reticulum in mammalian cells. J. Cell Biol., 140, 779–793. [DOI] [PMC free article] [PubMed] [Google Scholar]