Abstract
The retinoic acid-related orphan receptor β (RORβ) exhibits a highly restricted neuronal-specific expression pattern in brain, retina and pineal gland. So far, neither a natural RORβ target gene nor a functional ligand have been identified, and the physiological role of the receptor is not well understood. We present the crystal structure of the ligand-binding domain (LBD) of RORβ containing a bound stearate ligand and complexed with a coactivator peptide. In the crystal, the monomeric LBD adopts the canonical agonist-bound form. The fatty acid ligand–coactivator peptide combined action stabilizes the transcriptionally active conformation. The large ligand-binding pocket is strictly hydrophobic on the AF-2 side and more polar on the β-sheet side where the carboxylate group of the ligand binds. Site-directed mutagenesis experiments validate the significance of the present structure. Homology modeling of the other isotypes will help to design isotype-selective agonists and antagonists that can be used to characterize the physiological functions of RORs. In addition, our crystallization strategy can be extended to other orphan nuclear receptors, providing a powerful tool to delineate their functions.
Keywords: crystal structure/nuclear receptor/orphan/ROR/RZR
Introduction
Nuclear receptors (NRs) form a superfamily of sequence-specific transcription factors that regulate diverse biological processes including cell growth and differentiation, development, homeostasis and various organ functions in the adult by stimulating or repressing target gene expression (Gronemeyer and Laudet, 1995; Mangelsdorf et al., 1995). NRs all share a common modular structure composed of several domains denoted A–F. Receptor dimerization, ligand binding, repression in the absence of ligand and ligand-dependent transactivation are mediated by the C-terminal region of NRs, termed the ligand-binding domain (LBD), by generating the proper interaction surfaces for multiple partners, including corepressors, coactivators and mediators. The surface to which coactivators and mediators bind is assembled upon ligand binding and comprises H3, H4 and H12; it corresponds to the AF-2, the ligand-dependent transactivation function (reviewed in Renaud and Moras, 2000). In fact, ligand binding appears to trigger a switch in the LBD from a corepressor-binding to a coactivator-binding conformation (Glass and Rosenfeld, 2000; Renaud et al., 2000).
In addition to the ligand-dependent receptors, a vast number of structurally related gene products are described for which no ligands have yet been identified and, therefore, they are referred to as orphan nuclear receptors (Willy and Mangelsdorf, 1998; Giguère, 1999). The retinoic acid-related orphan receptor β [RORβ; NR1F2 (Nuclear Receptors Nomenclature Committee, 1999)], also called retinoid Z receptor β (RZRβ), is an orphan member of family 1, which contains receptors such as RAR or TR. So far, three ROR isotypes, α, β and γ, have been described. Both RORα (NR1F1) and RORγ (NR1F3) are expressed in various tissues (Hirose et al., 1994; Matysiak-Scholze and Nehls, 1997; Koibuchi and Chin, 1998) and seem to be involved in cerebellum development, immune responses (Delerive et al., 2001), lymph node organogenesis and apoptosis during thymopoiesis (Kurebayashi et al., 2000), bone metabolism (Meyer et al., 2000) and adipocyte differenciation (Kurebayashi and Hirose, 1998). In contrast, RORβ is expressed exclusively in areas of the central nervous system (CNS) that are involved in the processing of sensory information, including spinal cord, thalamus and cerebellar cortices, and also the three principal anatomical components of the mammalian timing system, the suprachiasmatic nuclei, the retina and the pineal gland (André et al., 1998). Therefore, it seems that this orphan NR regulates genes whose products play important roles in the context of sensory input integration as well as in the context of the biological clock. RORβ knockout mice exhibit a behavioral phenotype with similarities to a phenotype described some 40 years ago for a spontaneous mouse mutation called vacillans (Sirlin, 1956). These mice display a duck-like gait, transient male incapability to sexually reproduce and a severely disorganized retina that suffers from post-natal degeneration. Biochemical analyses indicated that RORβ can bind as a monomer to hormone response elements formed by the extented half-site sequence motif AnnTAGGTCA and activate reporter genes containing multiple copies of this half-site motif (Greiner et al., 1996). However, in spite of the simplicity of the extented half-site sequence motif, no natural target gene regulated by RORβ could be identified up to now. In addition, RORβ is classified as an orphan receptor, and the lack of a putative ligand has complicated the identification of physiologically relevant targets further. To understand better the role of RORβ in physiology, i.e. regulation of neuronal gene expression, the identification of specific ligands is of utmost importance. To gain insight into the geometry of the potential ligand-binding pocket (LBP) of RORβ and the nature of putative ligands, we concentrated on elucidating the crystal structure of this NR.
Here we present the 1.9 Å crystal structure of a complex between the LBD of the rat RORβ, a fortuitous ligand (stearate) and a peptide from the NR-interacting domain of the coactivator SRC-1 (Onate et al., 1995), a member of the p160 coactivator family (Torchia et al., 1998). The atomic level description of this orphan NR in the active conformation, stabilized by the combined action of the coactivator peptide and the pseudo-ligand, provides an accurate image of the LBP. This information greatly spurs the ability and the rationale for the design of isotype-specific agonists and antagonists that could be used to characterize and modulate the physiological functions of RORβ.
Results
RORβ LBD structure determination
Initially, the RORβ LBD (residues 201–459) was overproduced in Escherichia coli. However, the protein stability was strongly affected by oxidation problems, probably due to the presence of nine cysteines. Homology modeling of the RORβ LBD based on the crystal structure of the RARγ LBD, the closest homolog with known structure (28% identity, 56% similarity), bound to all-trans retinoic acid (ATRA) (Renaud et al., 1995) shows that five cysteines are buried and four are solvent-exposed. Noticeably, the C-terminus contains two cysteines at positions 454 and 458 (after the AF2-AD) that are not conserved in RORα and γ (Figure 1), and thus are probably involved in RORβ-specific regulation but not in the proper folding and ligand-dependent activity of the LBD. As these two cysteines are probably the most accessible ones and thus the most sensitive to oxidation, we chose to remove them by truncation. The minimal LBD (residues 201–452), overproduced in E.coli and purified to homogeneity, proved to be stable towards oxidation. Moreover, it is still transcriptionally active when fused to the GAL4 DNA-binding domain (data not shown).
Fig. 1. Sequence alignment of ROR LBDs with hRARγ LBD. The LBD sequences of hRARγ (Krust et al., 1989), hRORγ (Hirose et al., 1994), mRORγ, hRORα (Giguère et al., 1994), mRORα (Carlberg et al., 1994), hRORβ (Carlberg et al., 1994), rRORβ (Carlberg et al., 1994) and the Drosophila homolog DHR3 (Koelle et al., 1992) were aligned using the pileup option of the Genetics Computer Group (GCG, 1994). The secondary structure elements of the rRORβ LBD (present study) and the hRARγ LBD (Renaud et al., 1995) are underlined with a plain line (α-helices) or with an arrow (β-strands). The residues involved in the LBP are shown in red; those of rRORβ in close contact with stearate and those of hRARγ in close contact with retinoate are indicated by an asterisk (4 Å cut-off). The residues whose mutation affects the shape of the LBP are colored in yellow (β-specific or β-like), green (α-specific), pink (γ-specific) or blue (DHR3-specific). The amino acid numbering is given for rRORβ.
During the purification process, the presence of a fortuitous ligand was found, which was shown by mass spectrometry to be stearic acid (N.Potier, personal communication). However, crystallization trials in the presence of an excess of stearic acid were unsuccessful. Crystals could only be obtained in the presence of a peptide from the coactivator SRC1 containing the LXXLL motif that was shown to be necessary and sufficient to bind NR LBDs (Heery et al., 1997; Torchia et al., 1997). Crystals of the RORβ LBD–SRC1 peptide complex grew in 1 week from a polyethylene glycol (PEG) 6000 solution without added stearic acid. The structure was solved by molecular replacement using the RARγ holo-LBD (Renaud et al., 1995) as a search model (see Materials and methods for details). The final Rcryst and Rfree were 22.4% and 24.9%, respectively (see Table I). The Ramachandran plot showed only one outlier, a residue located in loop 9–10, which is partially disordered according to the 2Fc – Fo electron density map.
Table I. Data collection and refinement statistics.
| Data collection |
|
| Resolution (Å) | 30.0–1.85 (1.92–1.85) |
| Unique reflections | 28 901 |
| Completeness | 99.9% (99.8%) |
| Rsyma | 3.6% (23.1%) |
| Multiplicity |
4.2 |
| Refinement |
|
| Resolution (Å) | 30.0–1.9 |
| Reflections used | 23 854 |
| Completeness | 100% |
| Rcrystb | 22.4% |
| Rfreec | 24.9% |
| R.m.s.d. on bond length (Å) | 0.008 |
| R.m.s.d. on bond angles (°) |
1.282 |
| Non-hydrogen atoms |
|
| Protein | 1977 |
| Peptide | 91 |
| Ligand | 20 |
| Water molecules |
137 |
| Average B factor for non-hydrogen atoms (Å2) |
|
| Protein | 29.1 |
| Peptide | 37.1 |
| Ligand | 47.9 |
| Water molecules | 39.8 |
In the data collection, the last shell values are presented in parentheses.
aRsym (I) = ΣhklΣi|Ihkl,i – Ihkl>|/ΣhklΣi|Ihkl,i|, where <Ihkl> is the average intensity of the multiple Ihkl,i observations for symmetry-related reflections.
bRcryst = Σhkl|Fobs – Fcalc|/Σhkl|Fobs|.
cRfree = Σhkl∈T|Fobs – Fcalc|/Σhkl∈T|Fobs|, where the T set (10% of reflections) is omitted in the refinement.
Overall structure of the RORβ LBD
The RORβ LBD (Figure 2) presents the canonical fold for the NRs (Wurtz et al., 1996) with two additional α-helices, H2′ and H11′. It is in the agonist-bound state, H12 joining the H3–H4 region to form the proper interaction surface (the complete AF-2) for the coactivator (reviewed in Renaud and Moras, 2000). An additional H2′ helix is also found between H2 and H3 in peroxisome proliferator-activated receptors (PPARs; Nolte et al., 1998). H2 helices in PPAR and ROR are roughly superposed, but H2′ helices are almost perpendicular, pointing directly toward the N-terminus of H3 in the case of ROR and toward the solvent parallel to the β-sheet in the case of PPAR. H11′ is unique to RORβ among known LBD structures; it roughly superposes with the middle part of loop 11–12 of RARγ. PROCHECK analysis (Laskowski et al., 1993) indicates that the dimerization helix H10 is kinked at Ala411– Lys412, which should affect the homo- or heterodimerization of RORβ. Noticeably, there is a one amino acid deletion at the corresponding position in RORγ (Figure 1).
Fig. 2. Schematic representation of the rRORβ LBD in complex with stearate (ball-and-stick) and a SRC-1 peptide (ribbon representation). The kink in H10 has been emphasized by breaking H10 into two segments.
The RARγ and RORβ LBDs were superposed using the LSQ options from the program O (Jones et al., 1991) (Figure 3A). The root mean square deviation (r.m.s.d.) was 1.2 Å on 148 Cαs using a cut-off of 2.5 Å; the aligned regions comprise H1 (second half), H3 (second half)–H5, H7–H9, H10–H11 (first half) and H12. In RORβ, the LBP is shifted laterally toward H7 (Figure 3B). This is due to a different location of H6, which is shifted outwards and tilted downwards, loop 6–7 now lying at the LBD surface; on the other side of the pocket, H3 and s1, the first strand of the β-sheet, are shifted inwards. Secondly, the LBP is larger in the vertical direction, extending toward both the top and the bottom. At the top, the LBP is limited by H5. In RARγ, M272 from this helix points towards the center of the pocket. In RORβ, the side chain of the corresponding residue L304 lies aside. At the bottom, the pocket in RARγ is limited by F230 (H3), F288 (s1) and F304 (loop 6–7). In RORβ, (i) the corresponding C262 in H3 points downwards rather than upwards as F230 in RARγ; (ii) due to the rotation of the plane of the β-turn with strand s1 being more inside the LBP, F320 in RORβ lies ∼5 Å lower than the corresponding F288 in RARγ; and, finally, (iii) loop 6–7 in RORβ is located at the LBD surface and does not contribute to the floor of the pocket, which is now limited on this side by F330 from H6, ∼4 Å lower than F304 from loop 6–7 in RARγ. Globally, the volume of the LBP is much larger in RORβ (766 Å3) than in RARγ (429 Å3).
Fig. 3. (A) Backbone superposition of the structures of the rRORβ LBD (yellow) in complex with stearate (orange) and the SRC1 peptide (green) and of the hRARγ LBD (blue) in complex with retinoate (purple). The superposition was done using the LSQ option of O (Jones et al., 1991). The r.m.s.d. was 1.2 Å for 148 matched Cαs. (B) Superposition of the probe-occupied cavities of rRORβ and hRARγ (view from the top) calculated by MSMS with a probe radius of 1.4 Å. This figure was prepared with DINO (Philippsen, 1999).
A transcriptionally active LBD conformation
The LBD is in the canonical, transcriptionally active conformation, and the coactivator peptide binds the AF-2 surface as described previously for other NR–coactivator peptide complexes (Darimont et al., 1998; Nolte et al., 1998; Shiau et al., 1998). The LXXLL motif-containing peptide used for crystallization corresponds to the second NR-box from the p160 coactivator SRC-1 (residues 686–700) (Onate et al., 1995; Heery et al, 1997; Torchia et al., 1997). The following residues of the peptide are seen in the crystal structure: HKILHRLLQE. The LXXLL motif forms the hydrophobic face of an amphipathic α-helix interacting with a hydrophobic cleft on the LBD surface. In particularly, the side chains of L313 and L314 make van der Waals contacts with V274 (H3), and I292 and L295 (H4) from the RORβ LBD.
The side chain carboxylate of the conserved E448 (H12) known to be important for transactivation (see mutant studies below) forms hydrogen bonds with the backbone amide nitrogens of I309 and L310 at the N-terminus of the peptide helix. At the other end, the main chain oxygen atoms of L313 and L314 are hydrogen-bonded to K278 (H3) from the RORβ LBD. These ‘capping interactions’ are similar to those already described for other NRs (Darimont et al., 1998; Nolte et al., 1998; Shiau et al., 1998).
The ligand-binding pocket
Compared with the human ortholog, the rat RORβ LBD sequence differs at only three amino acid positions: S210T (beginning of H1), L376I (loop H7–H8) and D436E (H11′). A456T and V457A are located at the C-terminus after H12 and are not present in our construct. None of these residues are part of the LBP; therefore, the present analysis can be extrapolated to the human receptor.
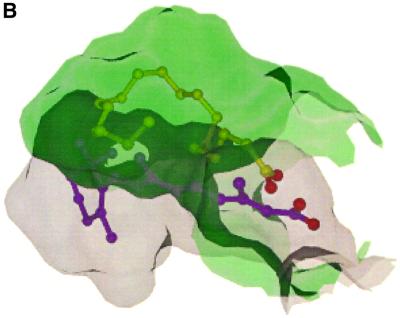
The probe-occupied volume of the LBP (766 Å3) is larger than that of the vitamin D receptor (VDR) (660 Å3; Rochel et al., 2000) and ranks third among the LBDs of known structure, after PPAR (1400 Å3; Nolte et al., 1998) and PXR (1150 Å3; Watkins et al., 2001). The LBP is essentially hydrophobic on the AF-2 side (H5 N-terminus, H6, H7, H10, H12) and more polar on the H3 side (L1–2, H3, H5 C-terminus). Stearic acid was co-purified fortuitously from E.coli and co-crystallized with the heterologously expressed RORβ LBD. The hydrophobic side of the pocket is partially filled up with the aliphatic chain of stearic acid, while the polar side is occupied by the carboxylate group of the fatty acid and 11 ordered water molecules (Figure 4). Upon binding, stearate adopts a U-shaped conformation similar to that observed for oleate in the mutant RXRαF318A LBD (Bourguet et al., 2000) and also for several fatty acids in fatty acid-binding proteins (Young et al., 1994; Thompson et al., 1997; Balendiran et al., 2000). One oxygen atom of the carboxylate group forms a hydrogen bond with Q265 NE2. This residue differs in RORα and β. The other oxygen atom of the carboxylate group is hydrogen-bonded through water molecules to other conserved residues among RORα and β of the LBP, namely Q228 and R306, and to the carbonyl group of V303 (Figure 4). The higher average B-value for the ligand (48 Å2 compared with 29 Å2 for the protein) suggests that the fatty acid thus probably adopts multiple low-energy conformations inside the pocket, in good agreement with the few distant van der Waals contacts between the aliphatic chain and the pocket. Indeed, the electron density map is less well defined in the middle portion of the chain.
Fig. 4. (A) Detailed view of the LBP showing the stearate ligand and the residues in close van der Waals contact (4.0 Å cut-off) or inter acting through hydrogen bonds. (B) Another view, emphasizing the second layer of water molecules forming a channel.
Isotype variability
The amino acid sequence conservation among ROR LBDs is not very high, RORβ being 61% identical (74% similar) to RORα and 49% identical (67% similar) to RORγ; it is also 33% identical (49% similar) to DHR3 (NR1F4), the Drosophila homolog of RORs. Moreover, only 17 out of the 32 residues whose side chain contributes to the LBP are strictly conserved in the three isotypes (Figure 1), raising the possibility of different ligands.
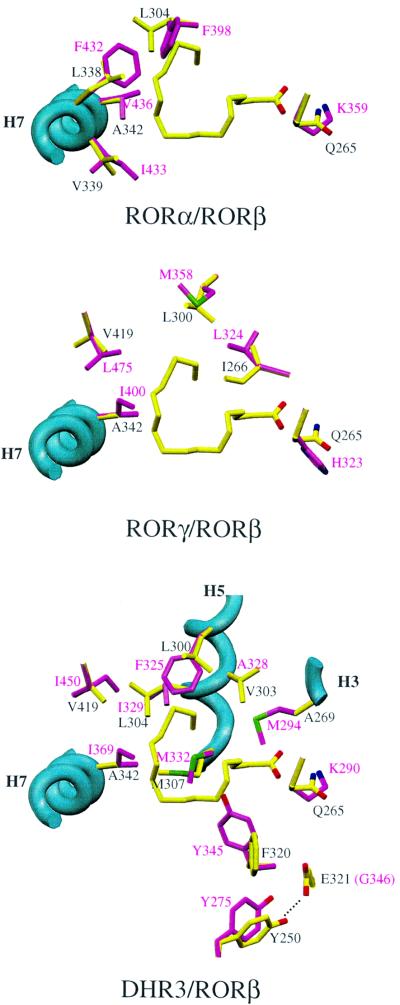
Homology models for the hRORα, hRORγ and DHR3 LBDs were built using the present rRORβ structure as a template. The hRORα LBP is smaller on the hydrophobic side (toward H7) due to a cluster of four substitutions for bulkier side chains: hRORα F398/rRORβ L304, F432/L338, I433/V339 and V436/A342 (Figure 5A). Accordingly, the volume of the pocket is 568 Å3, compared with 766 Å3 for rRORβ. This suggests that the natural ligand for the α and β isotypes could be different. The hRORγ LBP is more similar in size (705 Å3) but exhibits a slightly different shape due to four conservative substitutions—hRORγ L324/rRORβ I266 and M358/L300—which slightly enlarge the cavity on one side, and I400/A342 and L475/V419 that decrease the size of the cavity on another side (Figure 5B). Strikingly, the homology model of the DHR3 LBD displays a much smaller pocket (221 Å3) due to: (i) a few substitutions for bulkier side chains: DHR3 M294/rRORβ A269, F325/L300, I329/L304 and I369/A342, which makes M332 protrude more into the pocket than the corresponding M307, and I450/V419; and (ii) a reorientation of Y275 and Y345 (Figure 5C).
Fig. 5. Homology modeling of RORα, RORγ and DHR3 LBDs, showing the non-conserved residues affecting the pocket’s shape. Figures 2, 3A, 4 and 5 were prepared with SETOR (Evans, 1993).
In vivo mutational analysis of the LBD of RORβ
Prior to the determination of the crystal structure, we chose to model the RORβ LBD from the hRARγ LBD structure in the agonist-bound form even though no natural or synthetic agonist was known. Homology modeling allowed us to emphasize some important residues in the activation helix H12 and in the putative LBP that were subjected to mutational analysis. The crystal structure now validates this choice.
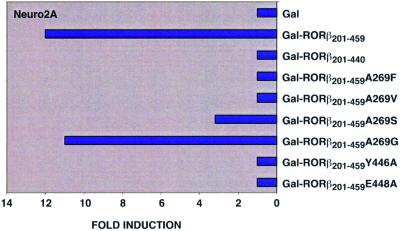
To test whether RORβ might be a ligand-dependent NR and thus support the existence of an endogenous ligand, a first series of site-directed mutagenesis experiments was designed. A269 was chosen because its side chain points into the pocket. We replaced it by bulkier residues in order to hinder the binding of a putative ligand. Increasing the size of the side chain decreased transactivation by Gal-RORβ201–459 in Neuro2A cells (Figure 6) and other cell lines (HT22 and NIH 3T3). All mutants were expressed to similar levels in the tested cells (data not shown). In the A269S mutant, a partial transcriptional activity of RORβ is still observed, whereas both mutants with bulkier side chains, A269V and A269F, failed to transactivate the reporter gene at all. Additional reporter gene assays showed that the transcriptional activity of Gal-RORβ201–459A269G in various cell lines does not differ from that of Gal-RORβ201–459. Altogether, these data suggest that transactivation by RORβ is ligand dependent and that the binding of the endogenous ligand is hampered by bulky side chains at position 269. Indeed, according to the crystal structure, mutation of A269 to a phenylalanine residue prevents the binding of stearate but should not affect the correct folding of the LBD.
Fig. 6. Histogram showing the results of the transactivation assays.
Deletion of the AF2-AD domain in both the full-length receptor and the fusion protein Gal-RORβ201–440 resulted in a complete loss of transcriptional activation by RORβ (Figure 6). Previous mutational analyses also demonstrated that the activation domains AF2-AD of RORβ and RARγ are functionally interchangeable (Greiner et al. 1996). To test the functional importance of the RORβ AF2-AD, two residues located in the activation helix H12 were specifically mutated. Both mutants Gal-RORβ201–459E448A and Gal-RORβ201–459Y446A are transcriptionally inactive on a (GALp)3-TK luciferase reporter in Neuro2A (Figure 6) and all other cell lines analyzed (data not shown). In the crystal structure, the side chain carboxylate of E448 is hydrogen-bonded to the coactivator peptide residues L310 and I309. The maintenance of this interaction is thus important for transactivation, most probably by acting in the recruitment of the coactivator. Y446 OH makes a hydrogen bond with H423 NE2 from H11. Moreover, the Y446 side chain is at the core of a highly conserved hydrophobic cluster comprising W259, A263 and I266 (H3), L427(H11), F442 (L11′–12) and F450 (H12). The Y446A mutation directly affects this cluster, probably causing a local collapse perturbing the coactivator-binding surface.
Discussion
The strategy developed in the present study in order to crystallize RORβ LBD without prior knowledge of a natural or synthetic ligand is applicable to other orphan NR LBDs. In our study, three major problems were overcome: (i) oxidation due to the presence of accessible cysteines not involved in the LBP or other critical contacts (in principle, exposed cysteines should be mutated unless located at potentially critical functional sites such as the LBP or its vicinity or interfaces; Gangloff et al., 2001); (ii) stabilization of the LBD fold by a pseudo-ligand coming from the expression host or from a parallel search; and (iii) use of a coactivator peptide to stabilize the agonist-bound, transcriptionally active conformation.
LBD stabilization through the presence of a fortuitous ligand
Several structures of NR LBDs in complex with natural or synthetic ligands have been reported. In most cases (RAR, TR, ER and VDR), the natural ligand was known and was co-crystallized with the LBD (Renaud et al., 1995; Wagner et al., 1995; Brzozowski et al., 1997; Rochel et al., 2000). In the case of PPARs, the natural ligands are not known with certainty, though it has been proposed that PPARs act as lipid sensors since they bind to and are activated by many low-affinity fatty acids and fatty acid derivatives; indeed, the PPARβ LBD was co-crystallized with eicosapentaenoic acid (Xu et al., 1999). Besides, many PPAR isotype-specific ligands have been synthetized (Willson, 2000). Apo-LBDs are rather unstable in the absence of cofactors; indeed, up to now, crystals of the apo-LBD could only be obtained in the case of RXRα (Bourguet et al., 1995), PPARβ (Xu et al., 1999), PPARγ (Nolte et al., 1998) and PXR (Watkins et al., 2001).
Two independent structural observations pointed to the unexpected ability of lipids to stabilize mainly hydrophobic LBPs. The first was the discovery of the presence of oleate in RXRα within the heterodimer RXRα–RARα (Bourguet et al., 2000). More recently, the crystal structure of an insect USP LBD was reported (Billas et al., 2001), showing a very large, fortuitous ligand that co-purifies with the protein, a phospholipid of mass 745. In the present study, we observe a similar situation, where the pseudo-ligand stearate is co-purified from the expression host.
Use of a coactivator peptide as a tool for crystallization
The idea of masking the hydrophobic cleft between H3 and H4 in order to make the LBD more soluble and more amenable to crystallization originated from the observation that in all reported LBD crystal structures, the cleft is always occupied by an amphipilic helix or molecule: (i) H12 of the same molecule in the known antagonist-bound complexes; (ii) H12 of a neighboring LBD in RXR and PPAR apo-LBDs; (iii) a detergent molecule in the case of RAR holo-LBD (Klaholz and Moras, 2000); or (iv) a coactivator peptide when added to the agonist-bound LBD complex (Darimont et al., 1998; Nolte et al., 1998; Shiau et al., 1998). In fact, the addition of a high-affinity, cognate coactivator peptide removes a major constraint on crystallization. Another reason for using the coactivator peptide is the low affinity of stearate and its lack of agonistic capability in transactivation assays (data not shown), suggesting that this fortuitous ligand alone is not able to stabilize the active conformation. This reasoning is consistent with the equilibrium model where a coactivator can stabilize the active conformation of an NR LBD by shifting the equilibrium between the ligand-bound and unbound forms (Gangloff et al., 2001; Steinmetz et al., 2001). In addition, the present structure, showing that an SRC1 peptide can bind the RORβ LBD coactivator-binding site in the classical conformation, confirms the assumption of multiple possible combinations between NRs and coactivators. Any minimal LXXLL-containing helix should bind to the hydrophobic coactivator-binding site, the specific pairwise interaction being modulated by the flanking regions. In summary, the present structure argues strongly in favor of a classical coactivation mechanism for RORβ.
RORβ probably functions as a monomer
RORα and RORβ have been proposed to function as both monomers and homodimers (Carlberg et al., 1994; Giguère et al., 1994). However, according to in vitro and in vivo data, RORβ is unable to form homodimers (Greiner et al., 1996). RORβ binds to monomeric response elements formed by the extented half-site sequence motif AnnTAGGTCA but cannot transactivate reporter genes containing only a single copy of this motif (Greiner et al., 1996). On the other hand, transactivation of RORβ-dependent reporter genes is only achieved with direct repeat binding sites (DR6–DR9) or by two binding sites oriented as inverted palindromes (P0). However, even for the transcriptionally active response elements, no cooperative binding is detected, which indicates that RORβ occupies both sites independently. Furthermore, in contrast to most members of family 1, RORβ does not heterodimerize with RXR (Greiner et al., 1996). In all NR LBD homo- and heterodimer structures, the dimerization interface is topologically conserved, and for the residues in contact no significant conformational change upon dimerization is observed. A homodimer built upon this assumption generates important clashes at the level of H10: a steric one between the 2-fold related I410 and a repulsive interaction involving E386 from one LBD and E404 from the other. These contacts would be sufficient to destabilize a canonical homodimer and explain the biochemical observations.
Is stearate close to the human physiological ligand?
Over the past 10 years, it has been shown that fatty acids can act as signaling molecules in regulating gene expression (Duplus et al., 2000). The mechanism by which fatty acids modulate gene transcription still remains largely unknown. The CNS contains large amounts of polyunsaturated fatty acids such as arachidonic acid (20:4 n-6) and docosahexaenoic acid (DHA) (22:6 n-3). The total synthesis of these two polyunsaturated fatty acids is not possible in mammals so that their precursors, linoleic acid (18:2 n-6) and α-linolenic acid (18:3 n-3), must be provided in the diet (Contreras et al., 2000). DHA has been shown to be a ligand for RXR in mouse brain (de Urquiza et al., 2000). However, two facts argue against stearate being close to the real ligand: (i) stearate does not activate RORβ in a cell reporter assay; and (ii) the low percentage of pocket occupancy by stearate (33%) and its partially disorded conformation. Thus, stearate most probably acts as a LBD stabilizer by filling the pocket, but it is unable on its own to generate sufficient interactions with the pocket to stabilize the active conformation, which is achieved by addition of the coactivator peptide. This conformational heterogeneity is reflected by the fact that LBD crystals could not be obtained in the absence of the peptide.
The low sequence conservation among the residues mapping the LBP and the large volume of the cavity (766 Å3) could suggest that the three isotypes do not share the same physiological ligand. Indeed, homology models display important differences at the LBP level, especially between RORα and RORβ/γ, the RORα pocket being 25% smaller. RORβ and RORγ LBPs have similar volumes but their shape is slightly different.
Among the four polar residues interacting directly or indirectly with the stearate carboxylate group, only Gln265 varies. This amino acid is replaced by a lysine residue in RORα and by a histidine residue in RORγ. Both residues could bind the carboxylate group of stearate with slightly different geometries. Thus, two possibilities may exist: (i) the natural ligand is the same for the three isotypes and residue 265 is not involved in the anchoring of the ligand, explaining its lack of conservation; or (ii) different ligands exist and the difference of side chain at position 265 is crucial in the ligand specificity for the three isotypes. However, one cannot exclude the possibility of a common ligand accommodated with a subtle rearrangement of the side chains at position 265 to maintain the interaction, although this would affect the binding affinity.
Conclusion
Escherichia coli allows the production of large amounts of heterologous proteins but also contains biomolecules that can be trapped by the heterologously produced proteins. In the present study, stearate was found in the LBP of the orphan NR RORβ LBD. Both the presence of the fortuitous ligand and the addition of a peptide containing the LXXLL motif stabilize the active holo-LBD conformation, the low affinity of stearate for RORβ being compensated by the high-affinity binding of the coactivator peptide, which prevents alternative conformations. Despite the fact that stearate does not function as an RORβ agonist in transactivation assays, a longer fatty acid or related lipids endogenously present in neuronal cells may bind with higher affinity to the RORβ LBP. Such a ligand might explain the neuronal-specific activity of RORβ. The high concentration of fatty acids in neuronal tissues and their role in brain development support this idea. The question remains open of whether there is a unique ligand or a family of compounds that bind to the RORβ LBP and regulate transcription. Nevertheless, the structure of the RORβ LBD provides a detailed picture of its LBP. This knowledge will greatly enhance the ability to design agonist and antagonist molecules that can be used to characterize in detail the physiological functions of RORs. Such synthetic compounds will be powerful tools to investigate ROR signaling pathways without prior knowledge of the natural ligands. Taken together, we have developed an efficient strategy to crystallize the RORβ LBD. This strategy can be extended to orphan NRs in general and will thus provide a very useful tool to delineate novel functions of orphan NRs.
Materials and methods
Expression, purification and crystallization
The rat RORβ LBD (residues 201–459) was overproduced as a histidine-tagged protein in E.coli BL21(DE3) by using pET-15b vector (Novagen). Due to aggregation problems, rRORβ was recloned as a truncated version (residues 201–452). Two non-conserved cysteines are deleted while maintaining the integrity of H12 (Figure 1). The purified monomer was concentrated to 6 mg/ml. A native gel reveals that the protein of interest migrates as a single species. The cells were grown in LBM at 37°C to an OD 0.6 and induced with 0.8 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The incubation was continued at 16°C overnight. Cells from 1 l of culture were resuspended in 50 ml of 20 mM Tris–HCl pH 8.5, 100 mM NaCl, 10% glycerol, 2 mM CHAPS and 2 mM β-mercaptoethanol (buffer A), and sonicated. The lysate was centrifuged at 50 000 r.p.m. for 2 h at 4°C, and the supernatant was loaded on a 2 ml cobalt affinity column. The column was washed with 10 ml of buffer A and the protein was eluted with a gradient of 0–1 M imidazole in buffer A. Subsequent gel filtration was performed on a Superdex S-200 HiLoad 16/60 from Pharmacia using as elution buffer 20 mM Tris–HCl pH 8.5, 100 mM NaCl, 2 mM CHAPS and 5 mM dithiothreitol (DTT) (at 1 ml/min). The protein eluted at 87 ml and was estimated to be >95% pure and homogeneous by SDS–PAGE. Co-crystallization with a 3 M excess of SRC-1 NR-interacting peptide (686-RHKILHRLLQEGSPS-700) was carried out with the hanging drop vapor diffusion method (2 µl of LBD–peptide complex solution + 2 µl of reservoir solution against 500 µl of reservoir solution). A proprietary NR LBD screening kit (D.Zeyer, S.Duclaud, D.Moras and J.P.Renaud, unpublished results) allowed us to find preliminary crystallization conditions. In the refined conditions, crystals grow within 1 week at 22°C to a size of ∼110 × 60 × 30 µm with a reservoir of 100 mM Tris–HCl pH 8.0, 15% PEG 6000. Crystallization trials without the SRC-1 peptide were unsuccessful, even in the presence of an excess of stearic acid.
Data collection, structure determination and refinement
Crystals were cryoprotected by equilibration in 15% PEG 6000 at pH 8.0 containing 15% glycerol and then flash-frozen in liquid ethane at liquid nitrogen temperature. X-ray diffraction data were collected at liquid nitrogen temperature from a single frozen crystal at the ID14-3 beamline at the ESRF Grenoble, France. Crystals diffracted X-rays to a resolution limit of 1.9 Å. All data were integrated and scaled using DENZO and SCALEPACK (Otwinowski and Minor, 1997) (Table I). The space group is P212121, with unit cell parameters a = 52.302 Å, b = 58.490 Å, c = 106.036 Å, α = β = γ = 90°. There is one monomer per asymmetric unit and a solvent content of 52%. The estimated B-factor by Wilson plot is 29 Å2. The structure was solved by molecular replacement using the program AMoRe (Navaza, 1994) and the RARγ holo-LBD [Protein Data Bank (PDB) accession code 2lbd] as search model. The top solution had a correlation coefficient of 27.8% and an R-factor of 52.7% after AMoRe rigid-body refinement. Automated model building using ARP/wARP (Perrakis et al., 1999) yielded three chains (243 residues, connectivity index 0.98). The partial model was subjected to alternating rounds of manual building using O (Jones et al., 1991) and refinement using CNS (Brünger et al., 1998). The final model, refined at 1.9 Å (Rcryst = 22.4%, Rfree = 24.9%), comprises 244 residues (208–451), the ligand, 10 residues of the SRC1 peptide (687–696) and 137 water molecules. According to PROCHECK (Laskowski et al., 1993), 99.2% of peptide φ/ψ angle pairs lie in allowed regions of the Ramachandran plot, 0.4% in generously allowed regions and 0.4% in disallowed regions. This last percentage corresponds to one residue (D403) from loop 9–10 for which the electron density is not well defined. The probe-occupied volume of the cavity was calculated with VOIDOO (Kleywegt, 1994) using a probe radius of 1.4 Å, and the volume of stearate with GRASP (Nicholls, 1993). In the RORβ LBD homodimer modeling study, the buried surface was also calculated with GRASP. The atomic coordinates have been deposited with the PDB (accession code: 1K4W; Berman et al., 2000).
Wild-type and mutant expression vectors and reporter plasmids
The luciferase reporter plasmid (GALp)3-TKLuc containing three copies of the GAL4-binding site upstream of a thymidine kinase (TK) promoter and the expression vectors for pCMXGal-RORβ201–459 and pCMXGal-RORβ201–440 were described previously (Greiner et al., 2000). The prokaryotic expression vector pRSETB-RORβ201–459 was generated by subcloning RORβ201–459 from pCMXGal-RORβ201–459 into pRSETB purchased from Invitrogen (Carlsbad, CA). Point mutations in the ROR LBD were generated by using an overlapping PCR approach. PCR products were cloned in-frame into pCMXGal at EcoRI and SalI restriction sites (Umesono et al., 1991). Detailed descriptions on the cloning of all expression vectors for Gal-RORβ fusion proteins described in the manuscript are available on request. All generated plasmids were verified by sequencing.
Cell culture and transfection assays
HT22 and NIH 3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM). Neuro2A cells were cultured in Earl’s modified Eagle’s medium (EMEM). Both media were supplemented with 10% fetal calf serum, penicillin, streptomycin and glutamine. Transient transfection assays were carried out in 12-well plates (4 × 104 cells per well) using the standard calcium phosphate co-precipitation technique (Pfitzner et al., 1995) or DOTAP lipofection (Roche Molecular Biochemicals) according to the manufacturer’s protocol. Luciferase activity was assayed as recommended by the manufacturer (Promega) in a Luminometer ML 3000 (Dynatech). Relative light units were normalized according to Pfitzner et al. (1995). All experiments were repeated at least five times.
Acknowledgments
Acknowledgements
We are grateful to Vincent Desserich for the construction of the pET-15b/rRORβ(201–452) plasmid and to Denis Zeyer and André Mitschler for help in data collection. We thank Steffi Arzt for support on ESRF beamline ID14-3. We are also thankful to Luc Moulinier and Isabelle Billas for help with ARP-wARP, to Pascal Eberling for peptide synthesis, and to Pascal Egea for help with DINO. This work was supported by the post-genomic program of the Ministère de la Recherche and by a grant from the Deutsche Forschungsgemeinschaft to R.S. (Schu 688/5-1).
References
- André E., Conquet,F., Steinmayr,M., Stratton,S.C., Porciatti,V. and Becker-André,M. (1998) Disruption of retinoid-related orphan receptor β changes circadian behavior, causes retinal degeneration and leads to vacillans phenotype in mice. EMBO J., 17, 3867–3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balendiran K.G., Schnugten,F., Scapin,G., Borchers,T., Xhong,N., Lim,K., Godbout,R., Spener,F. and Sacchettini,J.C. (2000) Crystal structure and thermodynamic analysis of human brain fatty acid-binding protein. J. Biol. Chem., 275, 27045–27054. [DOI] [PubMed] [Google Scholar]
- Berman H.M., Westbrook,J., Feng,Z., Gilliland,G., Bhat,T.N., Weissig,H., Shindyalov,I.N. and Bourne,P.E. (2000) The Protein Data Bank. Nucleic Acids Res., 28, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billas I.M.L., Moulinier,L., Rochel,N. and Moras,D. (2001) Crystal structure of the ligand-binding domain of the ultraspiracle protein USP, the ortholog of retinoid X receptors in insects. J. Biol. Chem., 276, 7465–7474. [DOI] [PubMed] [Google Scholar]
- Bourguet W., Ruff,M., Chambon,P., Gronemeyer,H. and Moras,D. (1995) Crystal structure of the ligand-binding domain of the human nuclear receptor RXR-α. Nature, 375, 377–382. [DOI] [PubMed] [Google Scholar]
- Bourguet W., Vivat,V., Wurtz,J.M., Chambon,P., Gronemeyer,H. and Moras,D. (2000) Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol. Cell, 5, 289–298. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Brzozowski A.M. et al. (1997) Molecular basis of agonism and antagonism in the oestrogen receptor. Nature, 389, 753–758. [DOI] [PubMed] [Google Scholar]
- Carlberg C., Hooft van Huijsduijnen,R., Staple,J.K., DeLamarter,J.F. and Becker-André,M. (1994) RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol. Endocrinol., 8, 757–770. [DOI] [PubMed] [Google Scholar]
- Contreras M.A., Greiner,R.S., Chang,M.C., Myers,C.S., Salem,N.,Jr and Rapoport,S.I. (2000) Nutritional deprivation of α-linolenic acid decreases but does not abolish turnover and availability of unacylated docosahexaenoic acid and docosahexaenoyl-CoA in rat brain. J. Neurochem., 75, 2392–2400. [DOI] [PubMed] [Google Scholar]
- Darimont B.D. et al. (1998) Structure and specificity of nuclear receptor–coactivator interactions. Genes Dev., 12, 3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delerive P., Monte,D., Dubois,G., Trottein,F., Fruchart-Najib,J., Mariani,J., Fruchart,J.C. and Staels,B. (2001) The orphan nuclear receptor RORα is a negative regulator of the inflammatory response. EMBO Rep., 2, 42–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Urquiza A.M., Liu,S., Sjoberg,M., Zetterstrom,R.H., Griffiths,W., Sjovall,J. and Perlmann,T. (2000) Docosahexaenoic acid, a ligand for the retinoid X receptor in mouse brain. Science, 290, 2140–2144. [DOI] [PubMed] [Google Scholar]
- Duplus E., Glorian,M. and Forest,C. (2000) Fatty acid regulation of gene transcription. J. Biol. Chem., 275, 30749–30752. [DOI] [PubMed] [Google Scholar]
- Evans S.V. (1993) SETOR: hardware-lighted three dimensional solid model representations of macromolecules. J. Mol. Graph., 11, 134–138. [DOI] [PubMed] [Google Scholar]
- Gangloff M., Ruff,M., Eiler,S., Duclaud,S., Wurtz,J.M. and Moras,D. (2001) Crystal structure of a mutant hERα ligand-binding domain reveals key structural features for the mechanism of partial agonism. J. Biol. Chem., 276, 15059–15065. [DOI] [PubMed] [Google Scholar]
- Genetics Computer Group (1994) Program Manual for the Wisconsin Package, Version 8. GCG, Madison, WI.
- Giguère V. (1999) Orphan nuclear receptors: from gene to function. Endocr. Rev., 20, 689–725. [DOI] [PubMed] [Google Scholar]
- Giguère V., Tini,M., Flock,G., Ong,E., Evans,R.M. and Otulakowski,G. (1994) Isoform-specific amino-terminal domains dictate DNA-binding properties of RORα, a novel family of orphan hormone nuclear receptors. Genes Dev., 8, 538–553. [DOI] [PubMed] [Google Scholar]
- Glass C.K. and Rosenfeld,M.G. (2000) The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev., 14, 121–141. [PubMed] [Google Scholar]
- Greiner E.F., Kirfel,J., Greschik,H., Dorflinger,U., Becker,P., Mercep,A. and Schüle,R. (1996) Functional analysis of retinoid Z receptor β, a brain-specific nuclear orphan receptor. Proc. Natl Acad. Sci. USA, 93, 10105–10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner E.F., Kirfel,J., Greschik,H., Huang,D., Becker,P., Kapfhammer,J.P. and Schüle,R. (2000) Differential ligand-dependent protein– protein intteractions between nuclear receptors and a neuronal-specific cofactor. Proc. Natl Acad. Sci. USA, 97, 7160–7165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gronemeyer H. and Laudet,V. (1995) Nuclear receptors. Protein Profile, 2, 1173–1308. [PubMed] [Google Scholar]
- Heery D.M., Kalkhoven,E., Hoare,S. and Parker,M.G. (1997) A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature, 387, 733–736. [DOI] [PubMed] [Google Scholar]
- Hirose T., Smith,R.J. and Jetten,A.M. (1994) RORγ: the third member of ROR/RZR orphan receptor subfamily that is highly expressed in skeletal muscle. Biochem. Biophys. Res. Commun., 205, 1976–1983. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr., A47, 110–119. [DOI] [PubMed] [Google Scholar]
- Klaholz B.P. and Moras,D. (2000) Structural role of a detergent molecule in retinoic acid nuclear receptor crystals. Acta Crystallogr., D56, 933–935. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1994) Detection, delineation, measure ment and display of cavities in macromolecular structures. Acta Crystallogr., D50, 178–185. [DOI] [PubMed] [Google Scholar]
- Koelle M.R., Segraves,W.A. and Hogness,D.S. (1992) DHR3: a Drosophila steroid receptor homolog. Proc. Natl Acad. Sci. USA, 89, 6167–6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koibuchi N. and Chin,W. (1998) RORα gene expression in the perinatal rat cerebellum: ontogeny and thyroid hormone regulation. Endocrinology, 139, 2335–2341. [DOI] [PubMed] [Google Scholar]
- Krust A., Kastner,P., Petkovich,M., Zelent,A. and Chambon,P. (1989) A third human retinoic acid receptor, hRAR-γ. Proc. Natl Acad. Sci. USA, 86, 5310–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurebayashi S. and Hirose,T. (1998) Novel orphan receptor: RORγ expressed during adipocyte differentiation. Nippon Rinsho, 56, 1729–1733. [PubMed] [Google Scholar]
- Kurebayashi S., Ueda,E., Sakaue,M., Patel,D.D., Medvedev,A., Zhang,F. and Jetten,A.M. (2000) Retinoid-related orphan receptor γ (RORγ) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc. Natl Acad. Sci. USA, 97, 10132–10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., Mc Arthur,M.W., Moss,D.S. and Thornton,J.M. (1993) Procheck: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Mangelsdorf D.J. et al. (1995) The nuclear receptor superfamily: the second decade. Cell, 83, 835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matysiak-Scholze U. and Nehls,M. (1997) The structural integrity of RORα isoforms is mutated in staggerer mice: cerebellar coexpression of RORα1 and RORα4. Genomics, 43, 78–84. [DOI] [PubMed] [Google Scholar]
- Meyer T., Kneissel,M., Mariani,J. and Fournier,B. (2000) In vitro and in vivo evidence for orphan nuclear receptor RORα function in bone metabolism. Proc. Natl Acad. Sci. USA, 97, 9197–9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated package for molecular replacement. Acta Crystallogr., A50, 157–163. [Google Scholar]
- Nicholls A. (1993) GRASP: Graphical Representation and Analysis of Surface Properties. Columbia University, New York, NY.
- Nolte R.T. et al. (1998) Ligand binding and co-activator assembly of the peroxisome proliferator-activated receptor-γ. Nature, 395, 137–143. [DOI] [PubMed] [Google Scholar]
- Nuclear Receptors Nomenclature Committee (1999) A unified nomenclature system for the nuclear receptor superfamily. Cell, 97, 161–163. [DOI] [PubMed] [Google Scholar]
- Onate S.A., Tsai,S.Y., Tsai,M.J., and O’Malley,B.W. (1995) Sequence and characterization of a co-activator for the steroid hormone receptor superfamily. Science, 270, 1354–1357. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin,V.S. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Pfitzner E., Becker,P., Rolke,A. and Schüle,R. (1995) Functional antagonism between the retinoic acid receptor and the viral transactivator BZLF1 is mediated by protein–protein interactions. Proc. Natl Acad. Sci. USA, 92, 12265–12269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen A. (1999) DINO: visualizing structural biology. http://www.bioz.unibas.ch/∼xray/dino.
- Renaud J.P. and Moras,D. (2000) Structural studies on nuclear receptors. Cell. Mol. Life Sci., 57, 1748–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud J.P., Rochel,N., Ruff,M., Vivat,V., Chambon,P., Gronemeyer,H. and Moras,D. (1995) Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature, 378, 681–689. [DOI] [PubMed] [Google Scholar]
- Renaud J.P., Harris,J.M., Downes,M., Burke,L.J. and Muscat,G.E.O. (2000) Structure–function analysis of the Rev-erbA and RVR ligand-binding domains reveals a large hydrophobic surface that mediates corepressor binding and a ligand cavity occupied by side chains. Mol. Endocrinol., 14, 700–717. [DOI] [PubMed] [Google Scholar]
- Rochel N., Wurtz,J.M., Mitschler,A., Klaholz,B. and Moras,D. (2000) The crystal structure of the nuclear receptor for vitamin D bound to its natural ligand. Mol. Cell, 5, 173–179. [DOI] [PubMed] [Google Scholar]
- Shiau A.K., Barstad,D., Loria,P.M., Cheng,L., Agard,D.A. and Greene,G.L. (1998) The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction with tamoxifen. Cell, 95, 927–937. [DOI] [PubMed] [Google Scholar]
- Sirlin J.L. (1956) Vacillans, a neurological mutant in the house mouse linked with brown. J. Genet., 54, 42–48. [Google Scholar]
- Steinmetz A., Renaud,J.P. and Moras,D. (2001) Binding of ligands and activation of transcription by nuclear receptors. Annu. Rev. Biophys. Biomol. Struct., 30, 329–359. [DOI] [PubMed]
- Torchia J. et al. (1997) The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature, 387, 677–684. [DOI] [PubMed] [Google Scholar]
- Torchia J., Glass,C. and Rosenfeld,M.G. (1998) Co-activators and co-repressors in the integration of transcriptional responses. Curr. Opin. Cell Biol., 10, 373–383. [DOI] [PubMed] [Google Scholar]
- Thompson J., Winters,N., Terwey,D., Bratt,J. and Banaszak,L. (1997) The crystal structure of the liver fatty acid-binding protein. J. Biol. Chem., 272, 7140–7150. [DOI] [PubMed] [Google Scholar]
- Umesono K., Murakami,K.K., Thompson,C.C. and Evans,R.M. (1991) Direct repeats as selective response elements for the thyroid hormone, retinoic acid, and vitamin D3 receptors. Cell, 65, 1255–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner R.L., Apriletti,J.W., McGrath,M.E., West,B.L., Baxter,J.D. and Fletterick,R.J. (1995) A structural role for hormone in the thyroid hormone receptor. Nature, 378, 690–697. [DOI] [PubMed] [Google Scholar]
- Watkins R.E., Wisely,G.B., Moore,L.B., Collins,J.L., Lambert,M.H., Williams,S.P., Willson,T.M., Kliewer,S.A. and Redinbo,M.R. (2001) The human nuclear xenobiotic receptor PXR: structural determinants of directed promiscuity. Science, 292, 2329–2333. [DOI] [PubMed] [Google Scholar]
- Willson T.M., Brown,P.J., Sternbach,D.D. and Henke,B.R. (2000) The PPARs: from orphan receptors to drug discovery. J. Med. Chem., 43, 527–550. [DOI] [PubMed] [Google Scholar]
- Willy P.J. and Mangelsdorf,D.J. (1998) Nuclear orphan receptors: the search for novel ligands and signaling pathways. In O’Malley,B.W. (ed.), Hormones and Signaling. Vol. 1. Academic Press, pp. 307–358.
- Wurtz J.M., Bourguet,W., Renaud,J.P., Vivat,V., Chambon,P., Moras,D. and Gronemeyer,H. (1996) A canonical structure for the ligand-binding domain of nuclear receptors. Nature Struct. Biol., 3, 87–94. [DOI] [PubMed] [Google Scholar]
- Xu H.E. et al. (1999) Molecular recognition of fatty acids by peroxisome proliferator-activated receptors. Mol. Cell, 3, 397–403. [DOI] [PubMed] [Google Scholar]
- Young A.C., Scapin,G., Kromminga,A., Patel,S.B., Veerkamp,J.H. and Sacchettini,J.C. (1994) Structural studies on human muscle fatty acid binding protein at 1.4 Å resolution: binding interactions with three C18 fatty acids. Structure, 2, 523–534. [DOI] [PubMed] [Google Scholar]