Abstract
Yeast spindle pole bodies (SPBs) duplicate once per cell cycle by a conservative mechanism resulting in a pre-existing ‘old’ and a newly formed SPB. The two SPBs of yeast cells are functionally distinct. It is only the SPB that migrates into the daughter cell, the bud, which carries the Bfa1p–Bub2p GTPase-activating protein (GAP) complex, a component of the spindle positioning checkpoint. We investigated whether the functional difference of the two SPBs correlates with the time of their assembly. We describe that in unperturbed cells the ‘old’ SPB always migrates into the bud. However, Bfa1p localization is not determined by SPB inheritance. It is the differential interaction of cytoplasmic microtubules with the mother and bud cortex that directs the Bfa1p–Bub2p GAP to the bud-ward-localized SPB. In response to defects of cytoplasmic microtubules to interact with the cell cortex, the Bfa1p–Bub2p complex binds to both SPBs. This may provide a mechanism to delay cell cycle progression when cytoplasmic microtubules fail to orient the spindle. Thus, SPBs are able to sense cytoplasmic microtubule properties and regulate the Bfa1p–Bub2p GAP accordingly.
Keywords: Bfa1p–Bub2p/mitotic exit network/spindle position checkpoint/SPB inheritance/Tem1p
Introduction
Centrosomes consist of a pair of centrioles surrounded by the pericentriolar material that is involved in microtubule formation. In G1 phase of the cell cycle, the single centrosome of a cell contains a daughter centriole that assembled during the previous S phase and a mother centriole formed in an earlier cell cycle. The mother and daughter centrioles of a centrosome are biochemically, structurally and functionally distinct (Paintrand et al., 1992; Piel et al., 2000, 2001). For example, completion of cytokinesis in animal cells coincides with the migration of the mother but not the daughter centriole towards the cytokinesis site (Piel et al., 2001).
In budding yeast, microtubule organizing functions are provided by the spindle pole body (SPB). The SPB is a multi-layered structure embedded in the nuclear envelope throughout the cell cycle. Like centrioles, the SPB duplicates once per cell cycle and the ‘new’ SPB forms adjacent to the pre-existing ‘old’ one. SPB duplication initiates at the distal end of the half-bridge, a one-sided extension of the central layer of the SPB, with the formation of a precursor named the satellite. The satellite matures into a new SPB around the beginning of S phase. The ‘new’ SPB remains connected to the ‘old’ one by the extended half-bridge, now called the bridge, which is cleaved in S phase. The two SPBs then separate to form the opposite poles of the spindle (Byers and Goetsch, 1975).
Cytoplasmic microtubules directed into the bud orient the spindle along the mother-bud axis through Kar9p, Bim1p and Myo2p, which are associated with the bud cortex (Beach et al., 2000; Korinek et al., 2000; Lee et al., 2000; Pruyne et al., 2000). The short spindle is then situated in the mother cell body with one SPB close to the bud neck and the other opposite towards the pole of the mother cell. Upon the onset of anaphase, cytoplasmic dynein provides the driving force for the nuclear movement into the bud. The Kar9p- and dynein-dependent pathways are to some extent redundant. Loss of one pathway results in nuclear migration defects whereas loss of both is lethal (Miller and Rose, 1998; Heil-Chapdelaine et al., 2000; Farkasovsky and Künzel, 2001).
Bfa1p and Bub2p form a two-component GTPase-activating protein (GAP) complex that is an integral part of the spindle position checkpoint (SPC). The SPC prevents premature cytokinesis when nuclear migration into the bud is delayed, probably by inhibiting the GTPase Tem1p (Bardin et al., 2000; Pereira et al., 2000). Only the SPB that migrates into the bud in anaphase carries Bfa1p and Bub2p (Pereira et al., 2000). Thus, the two SPBs are functionally and biochemically distinct, a property that is here referred to as SPB polarity. SPB polarity is important for SPC regulation. The SPC seems to become inactive when cytoplasmic microtubules organized by the bud-ward-directed SPB fail to interact with the bud neck (Adames et al., 2001). Maintenance of spindle polarity is also important for accurate chromosome segregation (Miller and Rose, 1998).
SPB inheritance describes the mode of partitioning of the ‘old’ and the ‘new’ SPBs between the mother cell body and the bud. ‘Old’ and ‘new’ SPBs may become segregated randomly or preferentially between the two cell bodies. It is unknown whether in budding yeast SPB inheritance and polarity are linked, as this is the case for mammalian centrioles (Piel et al., 2000, 2001). We describe here that SPBs are inherited in a defined manner. The ‘old’ SPB segregates into the bud. Integrity of cytoplasmic microtubules ensures SPB inheritance. However, despite the localization of Bfa1p with the ‘old’ SPB during an unperturbed cell cycle, we found that SPB inheritance does not determine Bfa1p localization. When SPB inheritance is disturbed Bfa1p still localizes to the SPB that enters the bud independently of whether it is the ‘old’ or the ‘new’. Our data suggest that the SPB with which Bfa1p associates is determined by cytoplasmic microtubule–cortex interactions. When these interactions are disturbed, the Bfa1p–Bub2p GAP complex associates with both SPBs. This mechanism may delay cell cycle progression in response to a misaligned spindle.
Results
Spc42p–RFP as a marker for the ‘old’ SPB
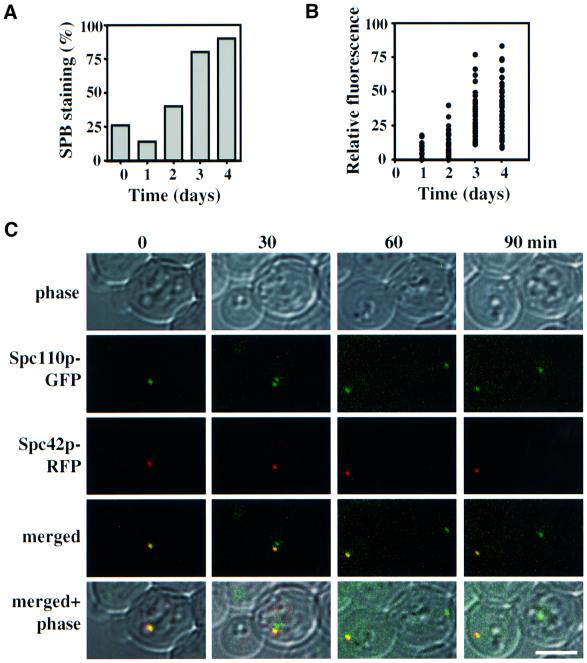
We addressed whether the ‘old’ and ‘new’ SPBs segregate preferentially or randomly into the mother cell body or the bud. For this study, we used cells in which the core SPB components Spc42p and Spc110p were fused to the red (Spc42p–RFP) and the green fluorescent proteins (Spc110p–GFP), respectively. SPC42–RFP SPC110– GFP behaved as wild-type cells and Spc42p–RFP was stably expressed (not shown) suggesting that SPC42–RFP fulfils all essential functions of SPC42. Surprisingly, only 14% of SPC42–RFP SPC110–GFP cells from a logarithmically growing culture displayed a dot-like, red fluorescent SPB signal when compared with the green Spc110p–GFP (Figure 1A, t = 1). We assumed that the rapid division of yeast cells and the slow folding property of the RFP molecule (Baird et al., 2000) prevented the accumulation of fluorescently active Spc42p–RFP at the SPBs. If this is the case, arresting cells in G0 may give the RFP of Spc42p sufficient time to develop into a fluorescent species. Indeed, the numbers of SPC42–RFP SPC110– GFP cells with a red SPB signal increased from 14 to 90% after cells spent 4 days in stationary phase (Figure 1A), as did the relative fluorescence intensity of SPBs (Figure 1B). When stationary SPC42–RFP SPC110–GFP cells were diluted into fresh medium, the SPB duplicated but only one of the two SPBs displayed a red Spc42p–RFP signal (Figure 1C, t = 30) although both SPBs contained Spc42p–RFP as judged by indirect immunofluorescence with anti-RFP antibodies (not shown). A likely explanation of this result is that the just formed ‘new’ SPB incorporated predominantly newly synthesized, fluorescently inactive Spc42p–RFP while the ‘old’ SPB carried fluorescently active Spc42p–RFP molecules.
Fig. 1. The SPB marked by the RFP signal segregates into the bud. (A and B) The number and relative fluorescence intensity of SPBs with Spc42p–RFP staining increases over time. Cells of SPC42–RFP SPC110–GFP were diluted with fresh medium (t = 0). (A) After 1, 2, 3 and 4 days the percentage of cells (n = 50–100) with a red SPB signal and (B) the relative fluorescence intensity of the SPB were determined by fluorescence microscopy. (C) Stationary SPC42–RFP SPC110–GFP cells were diluted into fresh medium. SPB segregation was followed by time-lapse microscopy. Bar, 5 µm.
We studied SPB inheritance using this property of Spc42p–RFP to label the ‘old’ SPB. SPB segregation of SPC42–RFP SPC110–GFP cells was followed by time-lapse microscopy (Figure 1C). In all cells (n = 20) the red-marked SPB migrated into the smaller cell body, the bud (Figure 1C, 60 and 90 min), suggesting that the old SPB is inherited by the daughter cell. To get a more representative picture, stationary SPC42–RFP SPC110–GFP cells were diluted into fresh medium and the logarithmically growing culture was inspected by fluorescence microscopy. The red fluorescent SPB of anaphase cells (n >200) was in 97.8% situated in the bud and in only 2.2% in the mother cell. These results indicate that in the majority of SPC42–RFP SPC110–GFP cells, the RFP-marked SPB segregates into the bud.
The ‘old’ SPB segregates into the bud
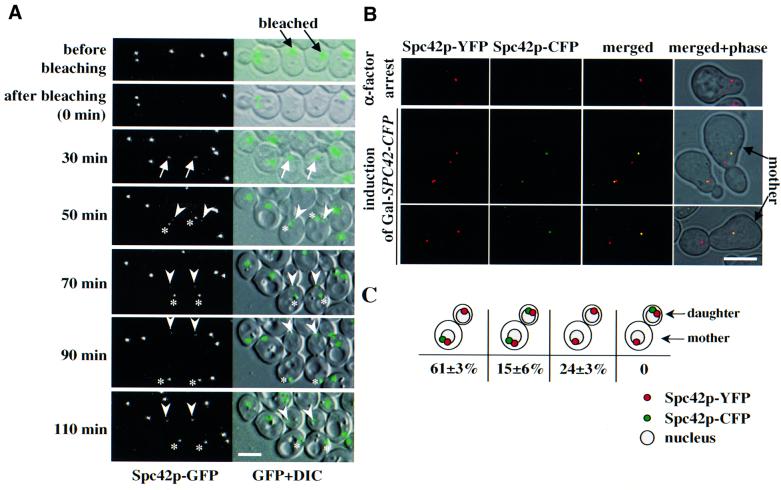
The partitioning of the ‘old’ SPB to the bud was further confirmed by fluorescence recovery after photobleaching (FRAP) and by transient expression of SPC42 fused to the cyan fluorescent protein (SPC42–CFP). For the FRAP experiment, a laser beam was used to photobleach Spc42p–GFP of the single SPB in selected G1 cells (Figure 2A, t = 0 min, middle two cells). Cells were then followed by time-lapse microscopy. With SPB duplication at the time of bud emergence, the GFP signal recovered (Figure 2A, t = 30 min, arrow). When the SPBs became separated, one weak (Figure 2A, t = 50 min, arrowhead) and one strong (asterisk) GFP signal was detected. We reasoned that the new SPB is marked by the strong GFP signal, because de novo synthesized and unbleached cytoplasmic Spc42p–GFP was incorporated into this SPB during duplication. The weak GFP signal at the ‘old’ SPB can be explained by the slow turnover of Spc42p subunits. Upon anaphase, the weakly labelled SPB entered the bud while the strongly labelled ‘new’ SPB remained in the mother cell body (t = 90 min). All together, 11 G1 phase cells were followed after photobleaching. In all cases, the SPB with the stronger GFP signal stayed in the mother cell during anaphase.
Fig. 2. The ‘old’ SPB migrates into the bud. (A) SPB inheritance determined by FRAP. Spc42p–GFP signals in unbudded cells (indicated by black arrows) were photobleached (t = 0) using a confocal microscope. Fluorescent and differential interference contrast microscopy (DIC) images were taken at the times indicated. The white arrows mark the Spc42p–GFP signal re-appearing after 30 min. The strongly labelled ‘new’ and the weakly labelled ‘old’ SPBs (50–110 min) are indicated by asterisks and arrowheads, respectively. (B) Selective expression of SPC42–CFP. Ga1S–SPC42–CFP SPC42–YFP cells grown in glucose medium were arrested in G1 with α-factor. α-factor was removed by washing the cells with raffinose medium. Galactose (2%) was added (GalS induction) for 20 min followed by the addition of 4% glucose (GalS repression). Samples were analysed by fluorescence microscopy every 10 min over 1.5 h. Note that the YFP signal is shown in red and the CFP signal in green. (C) Quantification of (B) (n = 120; two experiments). Nuclei were added in an illustrative form as they were not stained in the samples.
In the second experiment, SPC42–YFP cells (chromosomal SPC42 fused to the yellow fluorescent protein) carrying SPC42–CFP under control of the regulatory GalS promoter (GalS–SPC42–CFP) were arrested with α-factor in glucose medium to repress GalS expression. The single SPB of α-factor-arrested SPC42–YFP GalS–SPC42–CFP cells only displayed Spc42p–YFP fluorescence (Figure 2B, top). After release of the cell cycle block, at the time of SPB duplication, GalS–SPC42–CFP expression was shortly induced. This ensured, in the majority of cells, selective incorporation of Spc42p–CFP into only the newly formed SPB (Figure 2B, bottom and C). Since SPC42–YFP was constitutively expressed, both SPBs contained Spc42p–YFP. In 61% of the cells, only the new SPB was marked by CFP. This SPB stayed in the mother cell body (Figure 2B and C). In ∼15% of the cells, both SPBs displayed Spc42p–CFP labelling (Figure 2C). Incorporation of Spc42p–CFP into the ‘old’ SPB is probably caused by the extension of the Spc42p layer (Bullitt et al., 1997). Cells lacking a CFP signal (24%) may have failed to induce the GalS promoter (Figure 2C). Together, these experiments demonstrate that in wild-type cells the ‘old’ SPB segregates into the daughter cell.
Integrity of cytoplasmic microtubules ensures inheritance of the ‘old’ SPB into the bud
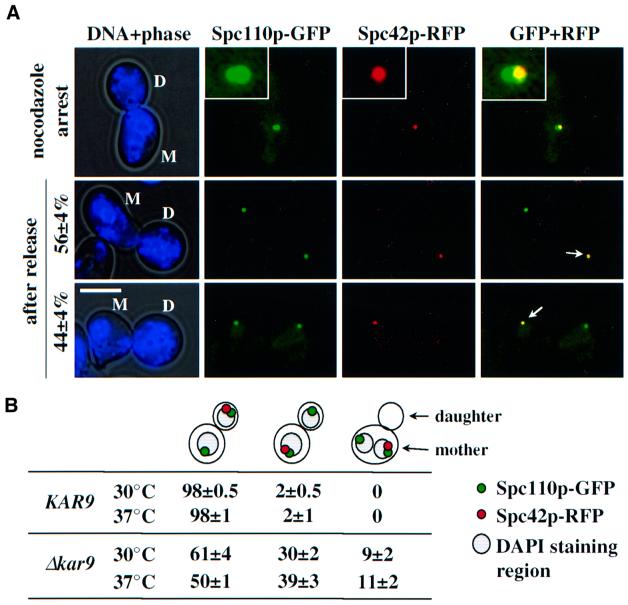
Using SPC42–RFP cells, we investigated whether microtubules are required to ensure SPB inheritance. Stationary SPC42–RFP SPC110–GFP cells were diluted into medium containing α-factor to mark the mother cell body with a mating projection. Cells were released from the cell cycle block into medium with the microtubule depolymerizing drug nocodazole, resulting in spindle collapse and mitotic arrest (Jacobs et al., 1988; Figure 3A, top). When nocodazole was removed, the microtubules reformed, thus re-establishing a bipolar spindle. In these cells the ‘old’ SPB, marked by the RFP signal, segregated with an almost equal likelihood to the mother cell body as to the bud (Figure 3A, bottom; the arrow marks the ‘old’ SPB). In conclusion, transient microtubule depolymerization disrupts the inheritance of the ‘old’ SPB to the bud.
Fig. 3. The continuous presence of cytoplasmic microtubules ensures migration of the ‘old’ SPB into the bud. (A) Transient microtubule depolymerization disrupts SPB inheritance. α-factor-synchronized SPC42–RFP SPC110–GFP cells were incubated with nocodazole to depolymerize microtubules resulting in spindle collapse with two SPBs close together. Nocodazole was then removed by washing. After 1 h samples were analysed for Spc42p–RFP and Spc110p–GFP fluorescence. The indicated percentages are based on three independent experiments (n >80). The arrows point towards the ‘old’ RFP-labelled SPBs. The insets are magnifications of the signals at the top. Abbreviations: D, daughter cell; M, mother cell body. Bar, 5 µm. (B) Interaction of cytoplasmic microtubules with the cell cortex ensures segregation of the ‘old’ SPB into the bud. SPB inheritance was determined in KAR9 SPC42–RFP SPC110–GFP and Δkar9 SPC42–RFP SPC110–GFP cells, after release from α-factor arrest at 30 and 37°C (n = 100; two experiments).
To address whether cytoplasmic microtubules play a role in SPB inheritance, we used Δkar9 cells. The nuclear microtubules are not affected by Δkar9 while cytoplasmic microtubules fail to interact with the bud cell cortex resulting in ∼10–20% of cells in anaphase with both 4′,6-diamino-2-phenylindole (DAPI) staining regions situated within the mother cell body. Eventually, the cytoplasmic microtubules interact with the bud cortex by an alternative dynein-dependent mechanism orientating the spindle along the mother bud axis (Miller and Rose, 1998; Heil-Chapdelaine et al., 2000; Korinek et al., 2000; Lee et al., 2000; Farkasovsky and Künzel, 2001). Cells of Δkar9 SPC42–RFP SPC110–GFP were arrested with α-factor and then released at 30°C. The ‘old’ SPB of anaphase cells segregated into the bud in 61% of cells (Figure 3B) and stayed in the mother cell body in ∼30% of cells, indicating an SPB inheritance defect. In the residual 9%, both SPBs were found in the mother cell. Mis-segregation of the ‘old’ SPB into the mother cell increased to 39% (2% in wild type) at 37°C. In summary, functional cytoplasmic microtubules are required to ensure the inheritance of the ‘old’ SPB into the bud.
Bfa1p SPB polarity is not determined by SPB inheritance
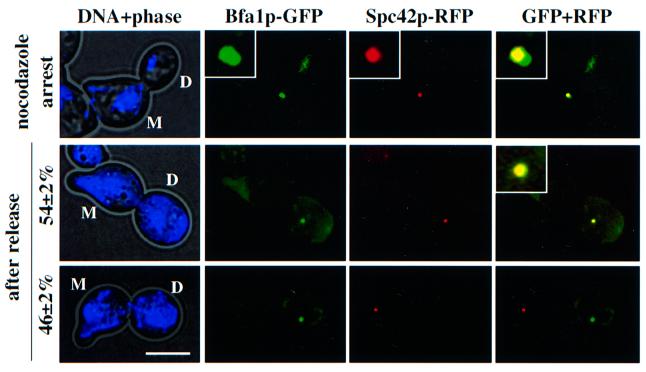
Bfa1p always associates with the SPB that migrates into the bud (Pereira et al., 2000), suggesting that it is linked to the ‘old’ SPB. Since the ‘old’ but not the ‘new’ SPB has experienced at least one cell cycle, a modification of the ‘old’ SPB in the previous cell cycle may determine its preferential association with Bfa1p. If this is the case, Bfa1p should always associate with the ‘old’ SPB, even when SPB inheritance is disturbed. To test this hypothesis, we performed the experiment shown in Figure 3A but now using BFA1–GFP SPC42–RFP cells. BFA1–GFP SPC42– RFP cells were first treated with α-factor to mark the mother cell body with a mating projection. These cells were then diluted into medium with nocodazole to depolymerize microtubules and to arrest cells in metaphase. As previously reported (Pereira et al., 2000), both SPBs of nocodazole-treated cells carry a Bfa1p–GFP signal (Figure 4, top). When nocodazole was removed, the spindle reformed along the mother bud axis and finally elongated into the bud in anaphase. Furthermore, Bfa1p SPB polarity was re-established such that in anaphase cells Bfa1p–GFP was always (n >100; 100%) associated with the SPB in the bud independently, whether it was the ‘old’ or the ‘new’ SPB (Figure 4, bottom; the ‘old’ SPB is marked by the RFP signal). Identical results were obtained for Bub2p, the other component of the GAP and for the GTPase Tem1p, which is regulated by the Bfa1p–Bub2p GAP (not shown). We conclude that Bfa1p associates with the bud-ward-directed SPB even when the ‘old’ SPB stays in the mother cell body.
Fig. 4. Bfa1p SPB polarity is not determined by SPB inheritance. BFA1–GFP SPC42–RFP cells were synchronized with α-factor, released into nocodazole medium, and then washed free of nocodazole. GFP and RFP signals were determined by fluorescence microscopy (n >100, two independent experiments). DNA was stained with DAPI. The insets are magnifications of the signals. Abbreviations: D, daughter cell; M, mother cell. Bar, 5 µm.
Microtubules and not factors associated with the bud or bud neck determine Bfa1p SPB localization
Factors associated with the bud neck or bud may facilitate Bfa1p binding to the bud-ward-oriented SPB. Alternat ively, signals transmitted from the cell cortex to the SPB along cytoplasmic microtubules may determine preferential association of Bfa1p with one of the two SPBs. To discriminate between both possibilities, we investigated whether the cellular position of SPBs and cytoplasmic microtubule functions influence Bfa1p SPB localization.
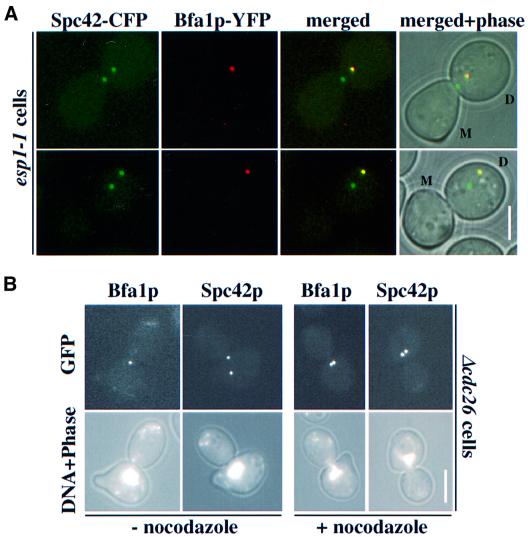
In esp1-1 cells, the entire nucleus migrates into the bud (McGrew et al., 1992). To test whether factors in the bud or bud neck are determining binding of Bfa1p to SPBs, Bfa1p SPB localization was studied in esp1-1 BFA1–YFP SPC42–CFP cells. We found that in 98% of esp1-1 cells, with one SPB in the bud neck and the other in the bud (Figure 5A, top) or both SPBs in the bud (Figure 5A, bottom), only the SPB closest to the bud tip carried a Bfa1p–YFP signal. In 2% of these esp1-1 cells, both SPBs were associated with Bfa1p–YFP (not shown). The fact that the bud-neck-oriented SPB hardly showed Bfa1p–YFP staining in esp1-1 cells with both SPBs in the bud argues against a model in which proteins associated with the bud neck or bud stabilize Bfa1p SPB localization.
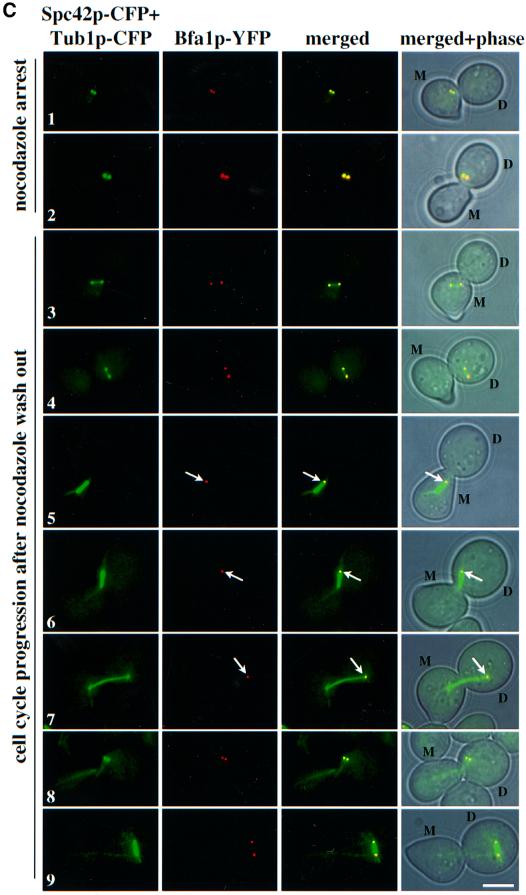
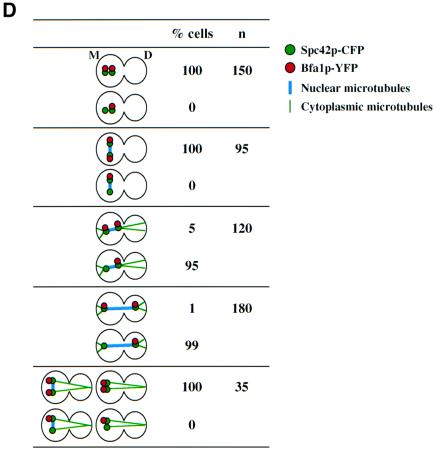
Fig. 5. Microtubules and not factors associated with the bud neck or bud are determinants of Bfa1p SPB polarity. (A) Bfa1p SPB localization in esp1-1 cells. Cells of esp1-1 BFA1–YFP SPC42–CFP were synchronized by α-factor block and released at 37°C. Shown is the Bfa1p–YFP SPB localization of cells in which both SPBs are located in the bud. (B) Bfa1p binds to the bud-ward-oriented SPB in arrested Δcdc26 cells. α-factor synchronized Δcdc26 BFA1–GFP and Δcdc26 SPC42–GFP cells were shifted to 37°C for 3 h. Nocodazole was then added for 1 h. Spc42p–GFP and Bfa1p–GFP SPB localization were determined by fluorescence microscopy. DNA was stained with DAPI. (C) Bfa1p SPB localization after nocodazole wash out. α-factor-synchronized BFA1–YFP SPC42–CFP CFP–TUB1 cells were incubated with nocodazole for 2 h at 30°C (rows 1 and 2). Nocodazole was removed by washing the cells. Spindle formation and Bfa1p–YFP at SPBs (marked by Spc42p–CFP) were followed over time (rows 3–9). (D) Quantification of (C). Only cells in which the spindle assembled in the mother cell body were counted. Abbreviations: D, daughter cell; M, mother cell. Bars, 5 µm.
Bfa1p is targeted to both SPBs when cells are treated with nocodazole (Figure 4). In response to nocodazole treatment, microtubules depolymerize, cells arrest in metaphase and the mitotic checkpoint is activated. Thus, any of these events may direct Bfa1p to both SPBs. Cells without CDC26 (Δcdc26) were used to investigate what regulates Bfa1p SPB binding. Cdc26p is a component of the anaphase-promoting complex and it becomes essential at 37°C for the onset of anaphase (Zachariae et al., 1996). Therefore, Δcdc26 cells, like nocodazole-treated cells, arrest in metaphase. However, in contrast to nocodazole-treated cells, Δcdc26 cells contain a short spindle positioned at the mother bud neck. We found that in arrested Δcdc26 cells only the bud-ward-oriented SPB was associated with Bfa1p–GFP (Figure 5B, compare Bfa1p–GFP signal with the SPB marker Spc42p–GFP). However, when Δcdc26 (Figure 5B) or Δcdc26 Δmad2 cells (not shown) were treated with nocodazole both SPBs showed Bfa1p–GFP staining. This suggests that microtubule depolymerization and not cell cycle arrest or activation of the MAD2-dependent mitotic checkpoint directs Bfa1p to both SPBs.
The role played by microtubules in regulating Bfa1p SPB localization was dissected by following the re-establishment of Bfa1p SPB polarity after nocodazole was washed out. For this experiment we used BFA1–YFP SPC42–CFP CFP–TUB1 cells, in which the nuclear and cytoplasmic microtubules were labelled by CFP–Tub1p and the SPB by Spc42p–CFP. α-factor-synchronized cells were treated with nocodazole. Both SPBs carried Bfa1p– YFP and Spc42p–CFP signals and no nuclear or cytoplasmic microtubules were detectable (Figure 5C, rows 1 and 2). In ∼71% of the cells, the two SPBs localized in the mother cell body (Figure 5C, row 1) while in 29% of cells, both SPBs resided in the bud (Figure 5C, row 2), probably depending on which set of cytoplasmic microtubules depolymerized first. Upon removal of nocodazole, the SPBs became separated with the reformation of a nuclear spindle. At this point, no cytoplasmic microtubules were observed and Bfa1p localized to both SPBs (Figure 5C, rows 3 and 4). Bfa1p–YFP SPB polarity became apparent with the reformation of cytoplasmic microtubules that positioned the spindle towards the mother bud axis (Figure 5C, rows 5 and 6). In cells with the two SPBs in the mother cell body, the SPB closest to the bud neck was associated with Bfa1p–YFP (Figure 5C, row 5). However, in cells with the SPBs in the bud, the SPB opposite to the bud neck carried the Bfa1p–YFP signal (Figure 5C, row 6). Eventually the spindle elongated and Bfa1p was associated with the SPB in the bud (Figure 5C, row 7). In conclusion, SPB polarity is re-established as soon as the cytoplasmic microtubules position the spindle towards the mother bud axis. The SPB closest to the bud tip retains Bfa1p independently of the position of the SPBs within the cell.
A closer inspection of BFA1–YFP SPC42–CFP CFP– TUB1 cells after nocodazole wash-out revealed that when cytoplasmic microtubules were organized before nuclear microtubules, Bfa1p was still associated with both SPBs (Figure 5C, row 8 and D). We also observed cells with a misaligned nuclear spindle associated with cytoplasmic microtubules that carried two Bfa1p SPB signals (Figure 5C, row 9 and D). In both cell types, the two cytoplasmic microtubule bundles were directed into the same cell body suggesting that no forces were applied onto the SPBs. This suggests that not simply the presence of cytoplasmic microtubules but the way they interact with the cell cortex regulates SPB association of Bfa1p. In summary, cytoplasmic microtubule–cortex interactions rather than the position of the SPBs may determine SPB localization of Bfa1p.
Interactions of cytoplasmic microtubules with the cell cortex determine Bfa1p SPB localization
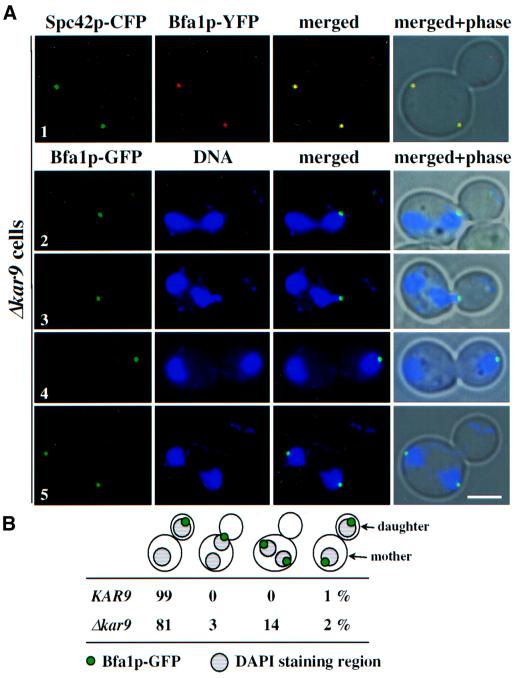
Using Δkar9 cells, we investigated whether defects in the interaction of cytoplasmic microtubules with the bud cell cortex affect Bfa1p SPB localization. Cytoplasmic microtubule defects become apparent in Δkar9 cells through the misalignment of the two separated DAPI-staining regions in the mother cell (Miller et al., 1999; Korinek et al., 2000; Lee et al., 2000). We confirmed with Δkar9 BFA1–YFP SPC42–CFP cells that Bfa1p is at SPBs even when the spindle is misaligned in the mother cell body (Figure 6A, row 1). Bfa1p was then analysed in Δkar9 BFA1–GFP cells in which the DNA was stained with DAPI. In cells with two DAPI-staining regions orientated along the mother bud axis, Bfa1p–GFP was associated with the SPB closest to the bud tip (Figure 6A, rows 2–4 and B). This was regardless of whether the chromosomes were already segregated into the bud (Figure 6A, row 4) or in the process of being segregated (Figure 6A, rows 2 and 3). Only in a few cells with correctly positioned DAPI regions did both SPBs carry Bfa1p (Figure 6B). In contrast, in Δkar9 cells, with the two DAPI regions misaligned in the mother cell body, both SPBs were always associated with a Bfa1p–GFP signal (Figure 6A, row 5 and B). This result indicates that Bfa1p is probably directed to both SPBs in response to the failure of cytoplasmic microtubules to interact with the bud cortex.
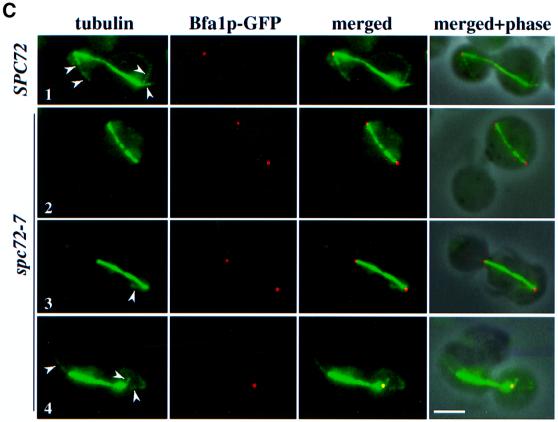
Fig. 6. Interactions of cytoplasmic microtubules with the cell cortex determine Bfa1p SPB localization. (A) Cytoplasmic microtubule–cortex interactions influence Bfa1p SPB localization. Δkar9 BFA1–YFP SPC42–CFP and Δkar9 BFA1–GFP cells were analysed by fluorescence microscopy. DNA was stained with DAPI. Note that the CFP signal is shown in green and the YFP signal in red. (B) Quantification of (A) (n = 200). Only cells with two separated DAPI-staining regions were counted. (C) SPB localization of Bfa1p is dependent on functional cytoplasmic microtubules. α-factor-synchronized SPC72 BFA1–GFP and spc72-7 BFA1–GFP cells were incubated for 1.5 h at 37°C. Microtubules were visualized by indirect immunofluorescence. The arrowheads mark cytoplasmic microtubules. Note that the Bfa1p–GFP signal is shown in red and the microtubules in green. Bars, 5 µm.
Spc72p tethers the γ-tubulin complex to the cytoplasmic side of the SPB. The SPBs of temperature-sensitive spc72(ts) cells frequently fail to organize cytoplasmic microtubules resulting in a mixed population of cells with and without cytoplasmic microtubules (Knop and Schiebel, 1998). Cells of spc72-7 BFA1–GFP were used to correlate cytoplasmic microtubule defects with Bfa1p SPB association. In spc72-7 BFA1–GFP cells with a misaligned spindle in the mother cell body, cytoplasmic microtubules were not detectable and Bfa1p was associated with both SPBs (Figure 6C, row 2). Similarly, in cells (n = 21) with a correctly positioned anaphase spindle but with at least one missing cytoplasmic microtubule bundle, Bfa1p–GFP was associated with both SPBs (Figure 6C, row 3), suggesting a link between defective cytoplasmic microtubules and Bfa1p association with both SPBs. In support of this notion, cells (n = 23) with a correctly positioned anaphase spindle and two cytoplasmic microtubule bundles had Bfa1p–GFP located only at the bud-ward-orientated SPB (Figure 6C, row 4), as was observed in wild-type cells (Figure 6C, row 1). Together, these data suggest a mechanism controlling Bfa1p localization through cytoplasmic microtubule–cortex interactions.
Discussion
Based on the binding of a Kar1p–LacZ fusion protein to the SPB that migrates into the bud in wild-type cells and to the ‘new’ but defective SPB in ndc1-1 cells, it has been suggested that the new SPB segregates into the bud (Vallen et al., 1992). However, because the Kar1p–LacZ protein is non-functional and over-produced and SPB duplication is defective in ndc1-1 cells, these experiments are difficult to evaluate.
Here, we show by three different approaches that SPBs have a defined mode of inheritance (Figures 1 and 2). In 98% of yeast cells, the ‘old’ SPB was found to migrate into the bud dependent upon functional cytoplasmic microtubules (Figures 1–3). In G1–S phase of the cell cycle, the cytoplasmic microtubules originate from the bridge between the duplicated SPBs and are directed into the growing bud (Byers and Goetsch, 1975). Segregation of the ‘old’ SPB into the bud is probably only ensured through the bridge-associated cytoplasmic microtubules staying associated with the ‘old’ SPB. Support for this model comes from the preferential segregation of the Tub1p–RFP signal, marking the already assembled microtubules with the SPB migrating into the bud, while the SPB in the mother cell was associated with newly assembled, non-fluorescent microtubules (G.Pereira, unpublished data). We propose that cytoplasmic microtubules of the bridge move to the cytoplasmic side of the ‘old’ SPB in S phase. This movement may be driven by the directed re-localization of the γ-tubulin complex-binding protein Spc72p from the bridge to the cytoplasmic side of the SPB (Pereira et al., 1999). The cytoplasmic microtubules at the ‘new’ SPB probably form de novo after SPB separation (Segal et al., 2000).
The property of the Spc42p–RFP molecule to mark the ‘old’ SPB now allows the study of features of the ‘new’ and the ‘old’ SPB in more detail. For example, evidence was obtained that sister chromatids attach in the absence of the cohesin Scc1p or in mutants of the aurora-like kinase IPL1 preferentially to one spindle pole (Biggins et al., 1999; Tanaka et al., 2000), raising the question as to whether there is a preferential binding to the ‘old’ or the ‘new’ SPB.
The mitotic exit network (MEN) is a GTPase-driven signal transduction cascade that controls inactivation of cyclin-dependent kinases and thereby the timing of cytokinesis at the end of anaphase. The Bfa1p–Bub2p GAP complex is part of the SPC that inhibits the MEN until the nucleus has migrated into the bud, making cytokinesis dependent on successful chromosome segregation (Bardin et al., 2000; Pereira et al., 2000). The fact that SPC and MEN components are associated with the yeast SPB suggests that SPBs are involved in the regulation of late mitotic events including cytokinesis. Also, in animal cells, centrioles have a function in regulating cytokinesis. Completion of cytokinesis coincides with the migration of the mother centriole towards the cytokinesis site, possibly through its capability to bind microtubules stably (Piel et al., 2000, 2001). Thus, it seems that the daughter centriole has to go through an entire cell cycle to mature into a form that allows regulation of cytokinesis. In contrast, polar SPB localization of the Bfa1p–Bub2p complex in yeast is not determined by the age of the SPB (Figure 4). Instead, SPB localization of the Bfa1p–Bub2p GAP is dependent on cytoplasmic microtubule–cortex interactions that position the spindle along the mother bud axis (Figures 5 and 6). We propose that the differential interactions of cytoplasmic microtubules with the mother cell and bud cortex determine Bfa1p–Bub2p GAP association with SPBs. The fact that the bud-ward-directed SPB migrates into the bud in anaphase through cytoplasmic microtubule-transmitted forces while the other SPB stays in the mother cell (Miller and Rose, 1998; Heil-Chapdelaine et al., 2000; Farkasovsky and Künzel, 2001) indicates that the two cytoplasmic microtubule sets are different. Their distinct properties may modulate the SPBs differently, retaining the Bfa1p–Bub2p GAP complex at one SPB but preventing its binding to the other (Figures 5 and 6). In this respect, it is interesting that the Bfa1p–Bub2p GAP complex binds to the core SPB protein, Nud1p, which also has a function in cytoplasmic microtubule organization (Gruneberg et al., 2000). It is possible that cytoplasmic microtubules regulate Nud1p, allowing Bfa1p–Bub2p binding accordingly.
Bfa1p associates with both SPBs if the binding of cytoplasmic microtubules to the cell cortex is disturbed, as in the case of Δkar9 and spc72-7 cells (Figures 5 and 6), or when microtubules are completely depolymerized by treating cells with nocodazole (Figure 5C and D). It is likely that only the SPB-associated Bfa1p–Bub2p GAP complex is able to inhibit the MEN (Bardin et al., 2000; Pereira et al., 2000; Adames et al., 2001). Therefore, directing Bfa1p and Bub2p from a cytoplasmic pool to both SPBs in response to microtubule defects will increase the number of active Bfa1p–Bub2p GAP complexes, thus blocking cell cycle progression more efficiently. This is consistent with the observation that in nocodazole-treated cells, the GTPase Tem1p, which is inhibited by the Bfa1p–Bub2p GAP complex, associates with both SPBs dependent on Bub2p (Pereira et al., 2000). Furthermore, evidence was obtained for an increase in Bfa1p–Bub2p activity when metaphase-arrested cells were treated with nocodazole (Fesquet et al., 1999). Finally, when cytoplasmic microtubules regain their function and start to align the spindle towards the mother bud axis, Bfa1p is released from the SPB that stays in the mother cell body (Figure 6). Reformation of Bfa1p SPB polarity may be important for the proper function of the MEN at the end of anaphase since many components of this pathway are positioned within the cell in a polar manner (Bardin et al., 2000; Pereira et al., 2000; Menssen et al., 2001).
A misaligned spindle also delays cell cycle progression in mammalian and Schizosaccharomyces pombe cells (O’Connell and Wang, 2000; Gachet et al., 2001), pointing to a similar control of cell cycle regulators at the centrosome or SPB in these organisms. Delaying cytokinesis in response to a misaligned spindle will ensure accurate chromosome segregation and prevent the deleterious consequences of chromosome instability, which are associated with many types of cancer cells. A centrosome-based checkpoint may also explain the link between centrosome abnormalities and cancer (Marx, 2001).
Materials and methods
Plasmids, yeast strains and growth conditions
Strains and plasmids used in this study are described in Table I. An RFP–KanMX6 cassette for the tagging of genes was constructed by amplifying RFP of plasmid DSRed1 (Clontech) by PCR. RFP was then subcloned into pYM12 (Knop et al., 1999). CFP (from T.Davis), GFP (Knop et al., 1999), RFP and YFP cassettes (from T.Davis) were PCR amplified, and strains were constructed and evaluated as described (Knop et al., 1999). The URA3-based CFP–TUB1 integration plasmid has been described (Jensen et al., 2001). The GalS–SPC42–CFP plasmid was constructed by cloning SPC42–CFP into p416–GalS (Mumberg et al., 1995). To arrest yeast cells in stationary phase, cells were grown for 3–5 days in YPAD medium at 23°C. Cells were allowed to recover in fresh medium for 2 h before the start of an experiment. Synthetic α-factor (10 µg/ml) was used to arrest cells in G1. Microtubules were depolymerized with 15 µg/ml nocodazole.
Table I. Yeast strains and plasmids.
| Name | Genotype, construction | Source or reference |
|---|---|---|
| Yeast strains | ||
| ESM356-1 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 | this study |
| ESM616 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 SPC42–GFP-KanMX6 | this study |
| ESM1431 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 BFA1–GFP-His3MX6 | this study |
| ESM1505 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 Δcdc26::klTRP BFA1–GFP-His3MX6 | this study |
| ESM1508 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 spc72-7 BFA1–GFP-His3MX6 | this study |
| ESM1509 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::pSM784 Δcdc26::klTRP | this study |
| ESM1596 | MATa ura3-52 leu2Δ1::pSM976 trp1Δ63 his3Δ200 SPC42–RFP-KanMX6 | this study |
| ESM1597 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 SPC42–RFP-KanMX6 SPC110–GFP-klTRP Δkar9::His3MX6 | this study |
| ESM1598 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 SPC42–YFP-His3MX6 pSM1024 | this study |
| ESM1611 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1::pSM976 Δkar9::klTRP | this study |
| GPY499 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 SPC42–RFP-KanMX6 SPC110–GFP-klTRP | this study |
| GPY572 | MATa ura3-52::pCFP-TUB1 leu2Δ1 trp1Δ63 his3Δ200 SPC42–CFP-KanMX6 BFA1–YFP-His3MX6 | this study |
| GPY598 | MATa esp1-1 ura3 leu2 lys2 trp1ade2 BFA1–YFP-His3MX6 SPC42::pXHO144 | this study |
| GPY599 | MATa ura3-52 leu2Δ1 trp1Δ63 his3Δ200 BFA1–YFP-His3MX6 SPC42–CFP-KanMX6 Δkar9::klTRP | this study |
| YPH499 | MATa ura3-52 lys2-801 ade2-101 trp1Δ63 his3Δ200 leu2Δ1 | Sikorski and Hieter (1989) |
| Plasmids | ||
| p416-GalS | pRS416 containing the GalS promotor | Mumberg et al. (1995) |
| pCFP-TUB1 | pRS306 carrying CFP–TUB1 | Jensen et al. (2001) |
| pRS304 | TRP1-based yeast integration vector | Sikorski and Hieter (1989) |
| pRS305 | LEU2-based yeast integration vector | Sikorski and Hieter (1989) |
| pSM784 | pRS305 carrying SPC42–GFP | this study |
| pSM976 | pRS305 containing BFA1–GFP | this study |
| pSM1024 | p416-GalS carrying SPC42–CFP | this study |
| pXHO144 | pRS304 carrying SPC42 (codons 194–363) fused to CFP | He et al. (2000) |
Time-lapse observation, FRAP, indirect immunofluorescence and fluorescence microscopy
For time-lapse microscopy, stationary SPC42–RFP SPC110–GFP cells were mounted onto a glass slide with a 25% gelatin pad containing complete synthetic medium and 2% glucose. For the FRAP experiment, the Spc42p–GFP signal of G1 cells was photobleached by a laser (488 nm) using a confocal microscope (Zeiss LSM510). At each time point, GFP images were collected from 12 Z sections (each 0.3 µm apart), which were converted into a single two-dimensional image by taking the maximum signal at each pixel (Tanaka et al., 2000). Indirect immunofluorescence was performed using a standard protocol (Pereira et al., 2000). CFP-, GFP-, RFP- and YFP-labelled cells were analysed by fluorescence microscopy after fixing the cells with paraformaldehyde (Pereira et al., 2000). DNA was stained with DAPI. The relative fluorescence intensity of RFP-labelled SPBs was measured with the NIH image program as the difference between the maximal fluorescence at the SPB and the background fluorescence in the immediate proximity of the SPB.
Acknowledgments
Acknowledgements
We especially thank K.Paiha and P.Steinlein for their help with the time-lapse microscopy. We are also grateful to T.Davis and S.Jensen for plasmids. We thank C.Manson for reading the manuscript. The work of E.S. is supported by the Cancer Research Campaign and Human Frontiers Science Program Organization and the work of T.U.T. and K.N. by The Wellcome Trust and Human Frontiers Science Program Organization.
References
- Adames N.R., Oberle,J.R. and Cooper,J.A. (2001) The surveillance mechanism of the spindle position checkpoint in yeast. J. Cell Biol., 153, 159–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird G.S., Zacharias,D.A. and Tsien,R.Y. (2000) Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc. Natl Acad. Sci. USA, 97, 11984–11989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A.J., Visintin,R. and Amon,A. (2000) A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell, 102, 21–31. [DOI] [PubMed] [Google Scholar]
- Beach D.L., Thibodeaux,J., Maddox,P., Yeh,E. and Bloom,K. (2000) The role of the proteins Kar9 and Myo2 in orienting the mitotic spindle of budding yeast. Curr. Biol., 10, 1497–1506. [DOI] [PubMed] [Google Scholar]
- Biggins S., Severin,F.F., Bhalla,N., Sassoon,I., Hyman,A.A. and Murray,A.W. (1999) The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev., 13, 532–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullitt E., Rout,M.P., Kilmartin,J.V. and Akey,C.W. (1997) The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell, 89, 1077–1086. [DOI] [PubMed] [Google Scholar]
- Byers B. and Goetsch,L. (1975) Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae.J. Bacteriol., 124, 511–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkasovsky M. and Künzel,H. (2001) Cortical Num1p interacts with dynein intermediate chain Pac11p and cytoplasmic microtubules in budding yeast. J. Cell Biol., 152, 251–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesquet D., Fitzpatrick,P.J., Johnson,A.L., Kramer,K.M., Toyn,J.H. and Johnston,L.H. (1999) A Bub2p-dependent spindle checkpoint pathway regulates the Dbf2p kinase in budding yeast. EMBO J., 18, 2424–2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y., Tournier,S., Millar,J.B.A. and Hyams,J. (2001) A MAP kinase-dependent actin checkpoint ensures proper spindle orientation in fission yeast. Nature, 412, 352–355. [DOI] [PubMed] [Google Scholar]
- Gruneberg U., Campbell,K., Simpson,C., Grindlay,J. and Schiebel,E. (2000) Nud1p links cytoplasmic microtubule organization and the control of exit from mitosis. EMBO J., 19, 6475–6488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Asthana,S. and Sorger,P.K. (2000) Transient sister chromatid separation and elastic deformation of chromosomes during mitosis in budding yeast. Cell, 101, 763–775. [DOI] [PubMed] [Google Scholar]
- Heil-Chapdelaine R.A., Oberle,J.R. and Cooper,J.A. (2000) The cortical Num1p is essential for dynein-dependent interactions of microtubules with the cortex. J. Cell Biol., 151, 1337–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs C.W., Adams,A.E.M., Szaniszlo,P.J. and Pringle,J.R. (1988) Functions of microtubules in the Saccharomyces cerevisiae cell cycle. J. Cell Biol., 107, 1409–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S., Segal,M., Clarke,D.J. and Reed,S.I. (2001) A novel role of the budding yeast separin Esp1 in anaphase spindle elongation: evidence that proper spindle association of Esp1p is regulated by Pds1p. J. Cell Biol., 152, 27–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M. and Schiebel,E. (1998) Receptors determine the cellular localization of a γ-tubulin complex and thereby the site of microtubule formation. EMBO J., 17, 3952–3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Siegers,K., Pereira,G., Zachariae,W., Winsor,B., Nasmyth,K. and Schiebel,E. (1999) Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast, 15, 963–972. [DOI] [PubMed] [Google Scholar]
- Korinek W.S., Copeland,M.J., Chaudhuri,A. and Chant,J. (2000) Molecular linkage underlying microtubule orientation toward cortical sites in yeast. Science, 287, 2257–2259. [DOI] [PubMed] [Google Scholar]
- Lee L., Tirnauer,J.S., Li,J., Schuyler,S.C., Liu,J.Y. and Pellman,D. (2000) Positioning of the mitotic spindle by a cortical-microtubule capture mechanism. Science, 287, 2260–2262. [DOI] [PubMed] [Google Scholar]
- Marx J. (2001) Do centrosome abnormalities lead to cancer? Science, 292, 426–429. [DOI] [PubMed] [Google Scholar]
- McGrew J.T., Goetsch,L., Byers,B. and Baum,P. (1992) Requirement for ESP1 in the nuclear division of Saccharomyces cerevisiae. Mol. Biol. Cell, 3, 1443–1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen R., Neutzner,A. and Seufert,W. (2001) Asymmetric spindle pole localization of yeast Cdc15 kinase links mitotic exit and cytokinesis. Curr. Biol., 11, 345–350. [DOI] [PubMed] [Google Scholar]
- Miller R.K. and Rose,M.D. (1998) Kar9p is a novel cortical protein required for cytoplasmic microtubule orientation in yeast. J. Cell Biol., 140, 377–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller R.K., Matheos,D. and Rose,M.D. (1999) The cortical localization of the microtubule orientation protein, Kar9p, is dependent upon actin and proteins required for polarization. J. Cell Biol., 144, 963–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D., Müller,R. and Funk,M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- O’Connell C.B. and Wang,Y. (2000) Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol. Biol. Cell, 11, 1765–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintrand M., Moudjou,M., Delacroix,H. and Bornens,M. (1992) Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol., 108, 107–128. [DOI] [PubMed] [Google Scholar]
- Pereira G., Grueneberg,U., Knop,M. and Schiebel,E. (1999) Interaction of the yeast γ-tubulin complex binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J., 18, 4180–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Höfken,T., Grindlay,J., Manson,C. and Schiebel,E. (2000) The Bub2p spindle checkpoint links nuclear migration with mitotic exit. Mol. Cell, 6, 1–10. [PubMed] [Google Scholar]
- Piel M., Meyer,P., Khodjakow,A., Rieder,C.L. and Bornens,M. (2000) The respective contributions of the mother and daugther centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol., 149, 317–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M., Nordberg,J., Euteneuer,U. and Bornens,M. (2001) Centrosome-dependent exit of cytokinesis in animal cells. Science, 291, 1550–1553. [DOI] [PubMed] [Google Scholar]
- Pruyne Y.H., Huffaker,D. and Bretschner,A. (2000) Myosin V orientates the mitotic spindle in yeast. Nature, 406, 1013–1015. [DOI] [PubMed] [Google Scholar]
- Segal M., Clarke,D.J., Maddox,P., Salmon,E.D., Bloom,K. and Reed,S.I. (2000) Coordinated spindle assembly and orientation requires Clb5-dependent kinase in budding yeast. J. Cell Biol., 148, 441–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae.Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T., Fuchs,J., Loidl,J. and Nasmyth,K. (2000) Cohesin ensures bipolar attachment of microtubules to sister centromeres and resists their precocious separation. Nature Cell Biol., 2, 492–499. [DOI] [PubMed] [Google Scholar]
- Vallen E.A., Scherson,T.Y., Roberts,T., Van Zee,K. and Rose,M.D. (1992) Asymmetric mitotic segregation of the yeast spindle pole body. Cell, 69, 505–515. [DOI] [PubMed] [Google Scholar]
- Zachariae W., Shin,T.H., Galova,M., Obermaier,B. and Nasmyth,K. (1996) Identification of subunits of the anaphase-promoting complex of Saccharomyces cerevisiae. Science, 274, 1201–1204. [DOI] [PubMed] [Google Scholar]