Abstract
The adenomatous polyposis coli (APC) tumor suppressor protein plays a critical role in regulating cellular levels of the oncogene product β-catenin. APC binds to β-catenin through a series of homologous 15 and 20 amino acid repeats. We have determined the crystal structure of a 15 amino acid β-catenin binding repeat from APC bound to the armadillo repeat region of β-catenin. Although it lacks significant sequence homology, the N-terminal half of the repeat binds in a manner similar to portions of E-cadherin and XTcf3, but the remaining interactions are unique to APC. We discuss the implications of this new structure for the design of therapeutics, and present evidence from structural, biochemical and sequence data, which suggest that the 20 amino acid repeats can adopt two modes of binding to β-catenin.
Keywords: adenomatous polyposis coli/β-catenin/crystal structure/Wnt signaling
Introduction
Mutations in the adenomatous polyposis coli (APC) tumor suppressor gene are responsible for the inherited cancer syndrome familial adenomatous polyposis (FAP) (Joslyn et al., 1991; Kinzler et al., 1991), characterized by the development of colorectal polyps at a very young age. APC is also mutated in >85% of sporadic colorectal tumors (Kinzler and Vogelstein, 1996). The role of the APC protein in tumor suppression is believed to be related to its ability to bind to and downregulate the protein β-catenin (Rubinfeld et al., 1993; Su et al., 1993) as part of the Wnt signaling pathway.
β-catenin exists in distinct populations that perform different functions within the cell. Whereas a membrane-associated pool is bound to cadherins and α-catenin in the adherens junction (Yap et al., 1997), cytoplasmic and nuclear β-catenin accumulates only in response to activation of the Wnt/Wingless growth-factor signaling pathway. In the absence of a Wnt signal or pathway mutation, any free β-catenin not in adherens junctions is targeted for degradation by a large, multiprotein complex containing β-catenin, glycogen synthase kinase-3β (GSK3β), APC and another protein known as axin. GSK3β phosphorylation of β-catenin on N-terminal serine and threonine residues targets it for recognition and degradation by the ubiquitin–proteasome system (Aberle et al., 1997; Orford et al., 1997). Mutations in and around these phosphorylated residues result in a stabilized form of β-catenin, and are seen in a wide variety of cancers, including those of the skin, brain, ovaries and uterus (Rubinfeld et al., 1997b; Fukuchi et al., 1998; Palacios and Gamallo, 1998; Zurawel et al., 1998). Axin appears to function as a scaffold for the assembly of the degradation complex, as it contains binding sites for all other complex members, and is the only member of the complex which has been shown to contain a GSK3β binding site (Behrens et al., 1998; Hart et al., 1998).
The precise role of APC in β-catenin targeting and degradation is poorly understood. Cancer cells containing truncations in APC accumulate large quantities of nuclear and cytoplasmic β-catenin, indicating that APC is critical for β-catenin degradation in vivo (Munemitsu et al., 1995). However, phosphorylation of β-catenin can occur in the absence of APC in vitro (Ikeda et al., 1998), and over-expression of axin can override the requirement for APC and promote the down regulation of β-catenin in APC-mutant cells (Hart et al., 1998). APC has been proposed to play a number of roles in β-catenin degradation, including derepression of axin (Hart et al., 1998) and a role in regulating the localization of β-catenin and its degradation complex (Yu et al., 1999; Henderson, 2000; Rosin-Arbesfeld et al., 2000).
In the presence of a Wnt signal, phosphorylation of β-catenin by GSK3β is inhibited through a largely unknown mechanism. This results in the accumulation of free, cytoplasmic β-catenin, which can then bind to proteins of the Lef/Tcf family and function as a transcriptional co-activator (Behrens et al., 1996; Huber et al., 1996; Molenaar et al., 1996). Targets of the β-catenin–Tcf complex include developmentally important genes such as segment polarity genes in Drosophila and axis induction genes in Xenopus, as well as the known proto-oncogenes c-MYC, cyclin D1 and WISP-1 in humans (Brannon et al., 1997; Riese et al., 1997; van de Wetering et al., 1997; Fan et al., 1998; He et al., 1998; Shtutman et al., 1999; Tetsu and McCormick, 1999; Xu et al., 2000).
In order to perform its various roles within the cell, β-catenin must bind to a large number of protein partners. The majority of these ligands, including cadherin, APC, axin and Lef/Tcf, bind to the central armadillo repeat region of β-catenin (Hülsken et al., 1994; Rubinfeld et al., 1995; Behrens et al., 1996, 1998; van de Wetering et al., 1997; Ikeda et al., 1998), which consists of 12 imperfect repeats of an ∼42 amino acid sequence motif (Peifer et al., 1994). The crystal structure of this region (Huber et al., 1997) revealed that each of the repeats consists of three α-helices, designated H1, H2 and H3. The 12 repeats form a continuous superhelix of helices, with a long, positively charged groove running along the H3 helices from repeats 1–10. The crystal structures of this region of β-catenin in complex with the catenin-binding regions of Xenopus Tcf3 (XTcf3) (Graham et al., 2000) and E-cadherin (Huber and Weis, 2001) revealed that this groove constitutes the binding site for both ligands. Although no sequence similarity had been previously detected between the two proteins, their interactions with β-catenin are surprisingly similar. This homology is particularly striking in a nine amino acid stretch designated the ‘extended region’ for its bound conformation.
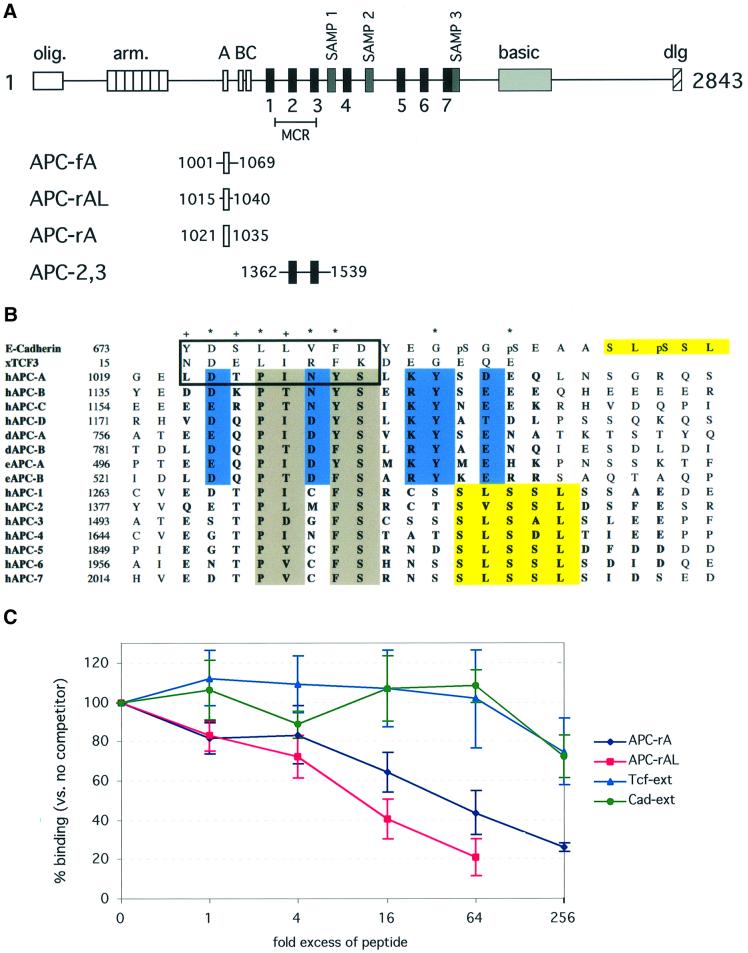
APC binds to β-catenin using series of three 15 amino acid repeat sequences, designated repeats A, B and C, and seven 20 amino acid repeats, designated repeats 1–7 (Figure 1A). Fragments of APC containing as little as one repeat have been shown to have β-catenin binding activity (Rubinfeld et al., 1997a). The 15 and 20 amino acid repeats share a conserved ‘core homology region’ in their N-terminal regions, but they diverge in their C-terminal regions (Figure 1B).
Fig. 1. The β-catenin-binding sites of APC. (A) Schematic of the APC primary structure. The conserved axin binding (SAMP1-3), oligomerization (olig.), armadillo repeat (arm.), basic and discs large interaction (dlg) regions are indicated. The 15 amino acid β-catenin-binding repeats are labeled A, B and C (white boxes). The 20 amino acid β-catenin-binding repeats are labeled 1–7 (black boxes). Truncations in the midpoint cluster region (MCR), which eliminate all of the axin-binding and most of the β-catenin-binding repeats, account for >60% of oncogenic mutations in APC (Miyoshi et al., 1992). The APC constructs used in binding experiments and crystallization are shown, with the beginning and end residue numbers in human APC indicated. (B) Alignment of the APC 15 and 20 amino acid repeats with E-cadherin and XTcf3. The alignment of the 15mers with E-cadherin and XTcf3 was performed based on the homologous regions of the E-cadherin–β-catenin, XTcf3–β-catenin and APC-rA–β-catenin structures (boxed). The 20mers were aligned with the 15mers based on alignment of the core homology regions. For an alternative alignment using the SLSSL sequences of E-cadherin and the 20mers, see Figure 4A, bottom panel. The residues that constitute the 15 and 20 amino acid repeat sequences are in bold. The homologous residues of the 15 and 20mer ‘core homology region’ are shaded gray; those conserved only in the 15mers are blue. The phosphorylation-specific binding motif of E-cadherin and the homologous APC 20mer sequences are highlighted in yellow. Beginning residue numbers based on the full-length proteins are indicated before the alignment. Residues from APC-rA that form contacts with β-catenin are indicated by asterisks (contacts by side chain only or main chain and side chain atoms) or plus signs (contacts by mainchain atoms only) above the alignment. hAPC-A, hAPC-B, hAPC-C: human APC 15mer repeats A, B and C. hAPC-D: hypothesized fourth human 15mer. dAPC-A, dAPC-B: Drosophila APC 15mers. eAPC-A, eAPC-B: Drosophila APC2 15mers. hAPC-1, hAPC-2, etc.: human APC 20mers. (C) Competition experiments to test the relative affinities of several β-catenin-binding peptides. GST-pulldown assays were performed using GST–β-catenin (full length) in the presence of a 5-fold excess of APC-fA. Increasing quantities of the APC-rA, APC-rAL, Tcf-ext or Cad-ext peptides were tested for their ability to compete with APC-fA for binding to limiting β-catenin. Fold molar excess of peptide (as compared with APC-fA) is plotted on the x-axis, as a pseudo log base-4 plot. APC-fA band intensities were quantified using the NIH Image program and are shown on the y-axis as percent of binding relative to that with no peptide competitor. Each point is plotted as mean ± SD of three experiments, except for the 256-fold excess of APC-rA, for which only two data points were obtained. APC-fA did not bind to GST alone (data not shown). See Materials and methods for details.
Here we present the structure of 15mer repeat A from APC (termed APC-rA) in complex with the armadillo repeat region of β-catenin. We discuss the similarities and differences of the β-catenin–APC-rA complex with the two previously determined complexes, and the implications for binding of the APC 20mer repeats. An examination of the structure in light of the existing sequence, biochemical and mutagenesis data suggests that the 20mers can bind in two modes to β-catenin, which may be important in the function of the degradation complex. In addition, we present evidence for a fourth, previously unidentified 15 amino acid repeat in mammalian APCs.
Results and discussion
The structure of the β-catenin–APC-rA complex
A complex between a 15mer peptide corresponding to repeat A (Figures 1A and B) with blocked N- and C-termini (APC-rA), and the arm repeat region of β-catenin (henceforth referred to as ‘β-catenin’) was crystallized. The structure was determined by molecular replacement, and the model refined to a maximum resolution of 3.1 Å. Although the crystals diffract to only moderate resolution, the asymmetric unit of the crystal contains two independent copies of the complex, and the maps were readily interpretable after refinement imposing non-crystallographic symmetry (NCS) restraints on the two β-catenin molecules. NCS restraints were not applied to the APC-rA peptide in the early rounds of refinement (see Materials and methods), but the peptide was found to form the same contacts with β-catenin in both copies. In copy 1, amino acids 151–546 and 561–663 of β-catenin and 1021–1031 of APC are visible. In copy 2, amino acids 150–545 and 558–663 of β-catenin and 1021–1034 of APC are visible. The only significant difference between the two β-catenin–APC-rA complexes is that residue 1021 of APC adopts different backbone and side chain conformations in the two copies. The following description of the complex is based on copy 2, as more of the APC-rA peptide is ordered.
Leu 1021 is seen in two distinct conformations and no density is seen for the last residue (Gln 1035), making it likely that residues 1022–1034 constitute the full binding region of the APC 15mers. As the peptide was synthesized with blocked N- and C-termini, no charges are present at the ends of the peptide, and the first and last residues would be expected to bind essentially as they would in the full-length protein.
The APC-rA peptide competes for binding with a fragment of APC containing 15mer repeat A plus 25–30 residues of flanking sequence on each end of the repeat (APC-fA) (Figure 1A and C). An ∼30-fold excess of the APC-rA peptide was required to reduce binding of the longer fragment by half (Figure 1C). This corresponds to a difference of ∼2 kcal/mol in binding energy between the two proteins. An energetic difference this small could result from missing direct contacts with β-catenin on the order of one to two hydrogen bonds. Alternatively, this energy difference could be the result of non-specific enthalpic (e.g. electrostatic) interactions missing in the shorter peptide, or of a difference in the conformational equilibria of the shorter and longer proteins, resulting in somewhat different entropic penalties for binding of the two constructs. In order to distinguish between these possibilities, we tested the binding of the longer APC-rAL peptide, corresponding to APC residues 1015–1040 (Figure 1A). This peptide, which extends far beyond the region of 15 amino acid repeat homology, would be expected to contain any contacts that may be missing in the APC-rA peptide. However, the longer peptide competes with APC-fA only slightly more efficiently than APC-rA, requiring an ∼10-fold excess of peptide to reduce APC-fA binding by half (Figure 1C). These data support the hypothesis that the observed binding differences are not due to specific contacts with β-catenin that are missing in the APC-rA peptide.
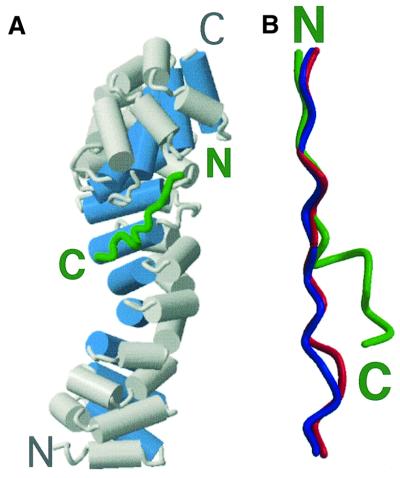
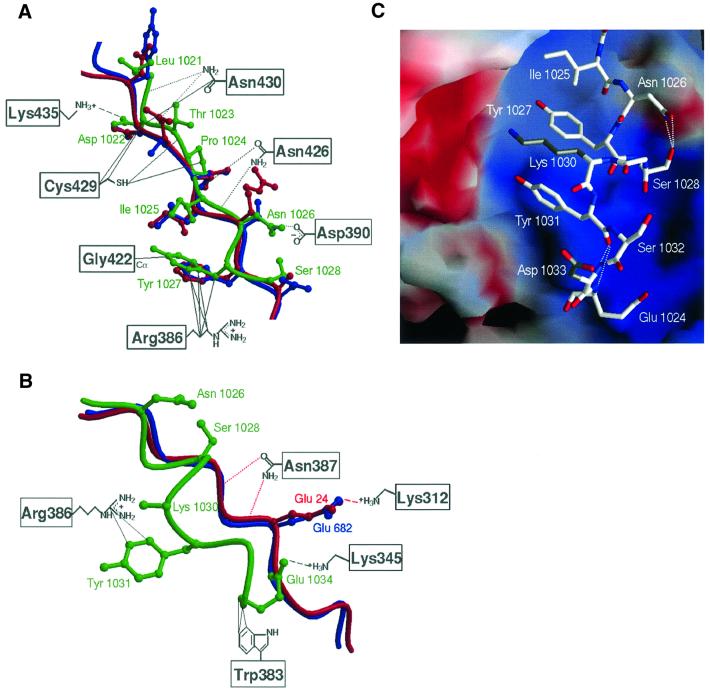
The APC-rA peptide binds within the groove formed by the H3 helices of β-catenin, where XTcf3 and E-cadherin were previously observed to bind (Figure 2A) (Graham et al., 2000; Huber and Weis, 2001). The structure of β-catenin is essentially the same as has been observed previously (Huber et al., 1997; Graham et al., 2000; Huber and Weis, 2001). A comparison with the β-catenin–XTcf3 and β-catenin–E-cadherin complexes reveals that the core homology region of the APC-rA peptide binds to β-catenin in a similar manner to the extended regions of the other two ligands (Figures 2B and 3A). In this region, each residue of E-cadherin adopts the same backbone and side chain conformations as its XTcf3 counterpart (Figure 2B), and makes nearly identical contacts with β-catenin. At either end of the extended region, lysine residues 435 and 312 of β-catenin form salt bridges, designated ‘charged buttons’, with acidic residues from E-cadherin or XTcf3 (Asp16 and Glu24 of XTcf3, Asp674 and Glu682 of E-cadherin). Over its first eight residues (Leu 1021– Ser1028), APC-rA adopts nearly identical backbone and side chain conformations as XTcf3 (Asn15–Lys22) and E-cadherin (Tyr673–Asp680) (Figure 3A). Most of the contacts observed in the corresponding region of the other two ligands are preserved in the APC-rA complex. This includes the first charged button (between APC-rA Asp1022 and β-catenin Lys435), hydrogen bonding between β-catenin Asn residues 430 and 426 and the APC-rA backbone, and hydrophobic contacts of several residues with β-catenin Cys429 and Arg386 (Figure 3A). Additionally, the side chain of Asn1026 forms a hydrogen bond with β-catenin Asp390 that is not present in the E-cadherin and XTcf3 complex structures (Figure 3A).
Fig. 2. Overall structure of the β-catenin–APC-rA complex. (A) Overall view of the β-catenin–APC-rA complex. Cylinders represent α-helices. β-catenin is gray, except for the H3 helices, which are blue. The Cα backbone trace of APC-rA is shown in green. The N- and C-terminus of each protein is labeled. (B) Comparison of the Cα backbone positions of β-catenin-bound APC-rA (green, APC residues 1021–1034), E-cadherin (blue, extended region residues 673–686) and XTcf3 (red, extended region residues 15–28). The figure was made by superimposing β-catenin in all three complexes using the LSQ function in O (Jones et al., 1991), and observing the relative positions of the three ligands. The N- and C-termini of APC-rA are indicated; E-cadherin and XTcf-3 are parallel to APC-rA. The figure was generated using Molscript and Raster3D (Kraulis, 1991; Merrit and Murphy, 1994).
Fig. 3. Interactions in the β-catenin–APC-rA complex. (A) Comparison of β-catenin-bound APC-rA, XTcf3 and E-cadherin in the core homology region of APC-rA. β-catenin residues are labeled in gray boxes. Other colors are as in Figure 2B. Contacts between APC-rA and β-catenin are drawn as solid lines (non-polar interactions), dotted lines (hydrogen bonds) or dashed lines (salt bridges). APC-rA residue numbers are indicated in green. (B) Comparison of β-catenin bound APC-rA, XTcf3 and E-cadherin in the region of the APC-rA bulge. Coloring and labeling is as in (A). Contacts of β-catenin with APC-rA are drawn in gray, and those with XTcf3 and E-cadherin in red. (C) Stabilizing forces in the APC-rA C-terminal bulge. β-catenin is drawn in a surface representation, colored blue for positive and red for negative electrostatic potential at the 10 kT/e level. The APC-rA peptide is colored by atom type with carbon white, oxygen red and nitrogen blue. Although no density is seen for the APC-rA Lys1030 or Asp1033 side chains in the structure, they are modeled (gray side chains) to demonstrate their likely interactions with regions of electrostatic potential on the surface of β-catenin. Hydrogen bonds between backbone and side chain atoms within the peptide are drawn as dotted lines. The Leu 1029 side chain is not shown for clarity. (A) and (B) were generated using Molscript and Raster3D (Kraulis, 1991; Merrit and Murphy, 1994).
Beginning at Leu 1029, the APC-rA peptide adopts a conformation strikingly different from that of XTcf3 or E-cadherin (Figure 3B). The C-terminal half of the peptide, from Leu1029 to Glu1034, bulges out of the β-catenin groove, away from the paths of XTcf3 and E-cadherin (Figures 2B and 3B). This difference in conformation is probably due to the presence of a lysine residue at amino acid 1030 of APC-rA. In XTcf3 and E-cadherin this position is occupied by an acidic residue (Glu24 of XTcf3, Glu682 of E-cadherin), which forms a salt bridge with β-catenin Lys312 to form the second charged button of the extended region (Figure 3B). The lysine at this position in APC-rA probably causes charge repulsion with β-catenin Lys312, leading to a deviation from the conformations of the other two ligands. The conformation of the backbone at Lys1030 suggests that the side chain of this residue is in the vicinity of β-catenin Glu462 and several other acidic residues. Although APC-rA Lys1030 is disordered and thus does not form a direct interaction with any of these residues, the bulge in APC-rA is likely stabilized by an electrostatic attraction between the basic lysine of APC-rA and an acidic patch on β-catenin (Figure 3C). A positively charged residue is present at the position equivalent to Lys1030 in all 15mer repeats, suggesting that this feature is important for β-catenin binding (Figure 1B). Several internal structural features also appear to contribute to the stability of the APC-rA bulge (Figure 3C). The side chain of Asn1026 forms hydrogen bonds with the side chain of Ser1028, and the backbone of several other residues within this region form hydrogen bonds with one another.
At the end of the bulge, APC-rA Glu1034 points back into the positively charged β-catenin groove (Figure 3C). There it forms what may be regarded as an alternative second charged button: a salt bridge with β-catenin Lys345 (Figure 3B). In addition, the aliphatic portion of the Glu1034 side chain forms non-polar contacts with β-catenin Trp383. The only other bulge residue to make direct interactions with β-catenin is APC-rA Tyr1031, which makes several contacts with β-catenin Arg386 (Figure 3B).
The structure of the β-catenin–APC-rA complex is consistent with site-directed mutagenesis data on the interactions of the APC 15mers with β-catenin (von Kries et al., 2000). Mutation of β-catenin Lys435 or Arg386 to alanine was found to eliminate binding to the 15mers. In the structure, Lys435 interacts with APC-rA Asp1022 to form the first charged button, and Arg386 forms numerous contacts with Tyr1027 and Tyr1031 of APC-rA. Interest ingly, mutation of the alternative second charged button lysine of β-catenin, Lys345, to Ala had almost no effect on 15mer binding (von Kries et al., 2000). These results, together with the observation that the C-terminus of APC-rA is disordered in one of the two independent copies of the complex, suggest that the N-terminal core homology region of APC-rA is more important for binding than the C-terminal bulge.
The sequence conservation observed in APC 15 and 20mers (Figure 1B) can be rationalized from the structure. Several residues that form specific side chain contacts with β-catenin (Asp1022, Pro1024, Tyr1027, Tyr1031) are highly conserved in 15mers or both 15 and 20mers. Asn1026, Ser1028 and Lys1030 appear to contribute to the stability of the C-terminal bulge, and are highly conserved in 15mers. However, the reasons for conservation of some residues are less obvious. The second ‘charged button’ is formed not by the conserved acid (Asp1033), but rather by the residue that follows it (Glu1034). As the APC-rA peptide makes no interaction with β-catenin between Tyr1031 and Glu1034, there may be some flexibility as to which of these acidic positions forms the second charged button. Another residue for which the reason for conservation is not immediately obvious is APC-rA Pro1024. This residue makes hydrophobic contacts with β-catenin Cys429, as does the leucine residue present at the equivalent position in XTcf3 and E-cadherin. However, a proline is invariant at this position in all APC 15 and 20mers. As proline residues are very restricted in their allowed backbone conformations, this residue may be responsible for reducing the entropic cost of binding of the APC repeats.
E-cadherin and Tcf4 are unstructured in the absence of β-catenin (Huber et al., 2001; Knapp et al., 2001). A circular dichroism spectrum of the 15 amino acid repeat region of APC shows no regular secondary structure, suggesting that it too may be disordered (data not shown). The XTcf3 and E-cadherin complexes bury 4820 and 6100 Å2 of surface area, respectively (Graham et al., 2000; Huber and Weis, 2001), as compared with 1220 Å2 for the APC-rA complex. Thus, in order to bind with reasonable affinity to β-catenin, it is likely that the APC 15mers bind more efficiently within their limited interaction area. Experiments comparing the binding affinities of APC-rA and the corresponding regions of XTcf3 (residues 15–25) and E-cadherin (residues 673–683) (Figure 1C) reveal that APC-rA does in fact bind to the extended region binding site of β-catenin with higher affinity than the other two ligands. This may be a result of more favorable contacts by the bulge region of APC-rA as compared with the C-terminal residues of the XTcf3 and E-cadherin extended regions, or of the entropic advantage provided by the conserved proline in the APC repeats.
A fourth 15mer in mammalian APCs
While studying the conservation of APC 15mer repeats, we noticed a previously unreported fourth sequence in human APC that is similar to the known 15mers. This sequence (residues 1173–1187), which we refer to as repeat D, lies just C-terminal to repeat C (residues 1156–1170). Similar fourth 15mer sequences are found in the equivalent position in rat (residues 1171–1185) and mouse (residues 1172–1186) APCs. Each of these sequences scores as highly significant in a MAST search for 15mer related sequences in the non-redundant protein database (Henikoff et al., 1995; Bailey and Gribskov, 1998) (see Materials and methods). All residues observed to make contacts with β-catenin in APC-rA are conserved in human, mouse and rat repeats D (Figure 1B).
Insights into binding of the APC 20mers
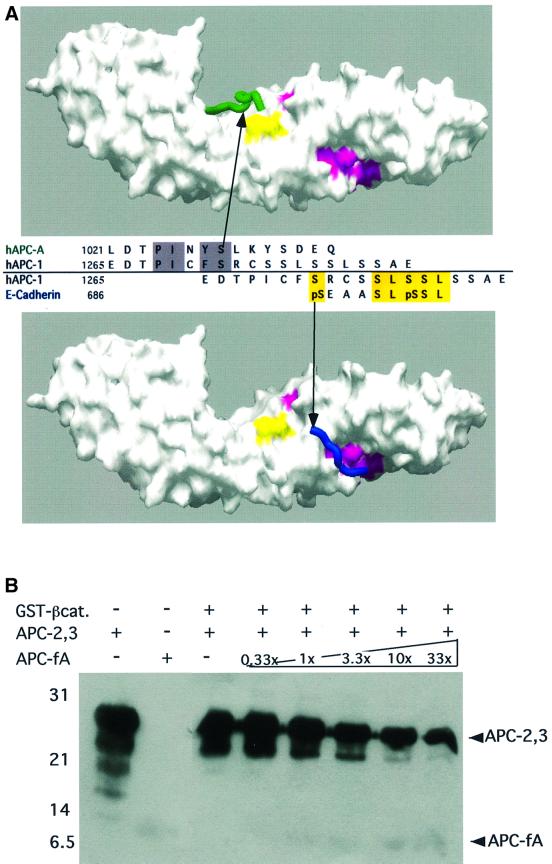
The homology between the N-terminal regions of the APC 15 and 20mer repeats (Figure 1B) suggests that the APC 20mers may bind to β-catenin by a mechanism similar to that of APC-rA binding (Figure 4A, top panel). Alignment of the core homology region of the 20mers with that of the 15mers results in a reasonable alignment with E-cadherin and XTcf3, even outside of the core homology region. Serines from the 20mer C-terminus (e.g. hAPC-1 Ser1276 and 1278) align with the first two phosphoserines seen in the E-cadherin structure (pSer684 and 686) and two glutamic acid residues from XTcf3 (Glu26 and 28) (Figure 1B). This alignment is consistent with data on the binding of an APC 20mer construct to a series of β-catenin point mutants (von Kries et al., 2000). The two mutations that eliminate APC 20mer binding (Lys345Ala and Trp383Ala) are in residues that interact with Glu1034 of APC-rA, at the C-terminus of the 15mer peptide (Figure 4A).
Fig. 4. Implications for APC 20mer binding. (A) Two potential modes for APC 20mer binding. β-catenin is shown as a surface representation. β-catenin residues whose mutation affected APC binding in previous experiments are colored. Those mutations that affected APC 20mer binding (Lys345, Trp383) (von Kries et al., 2000) are shown in yellow. Residues whose mutation affected binding of a construct containing both 15 and 20mer repeats (Lys312, Phe253, Phe293, Ala295, Ile296) are magenta. (Top panel) The β-catenin–APC-rA structure is used as a model for core homology region binding. The APC-rA Cα trace is drawn as a green tube. (Bottom panel) The SLSSL region of E-cadherin (residues 686–694) is shown in its binding site, as a model for SLSSL-region binding. The E-cadherin Cα trace is drawn as a blue tube. The sequence alignment between the panels shows human APC 20mer repeat 1 (hAPC-1) aligned with human APC 15mer repeat A (hAPC-A) and E-cadherin as would be predicted by each of the models. The relative positions of hAPC-A and E-cadherin have been preserved as in Figure 1B, and the hAPC-1 sequence is moved to align with each, demonstrating the two binding modes. Conserved core homology region residues are highlighted in gray, conserved SLSSL region residues in yellow. Arrows indicate the predicted position of hAPC-1 Ser1274 in each structural model. The binding sites for this residue in the two models are separated by more than 18 Å. (B) A construct containing a single 15mer repeat competes with a 20mer construct for binding to β-catenin. See Figure 1A for identification of the constructs used. Lane 1, 0.5 nmol APC-2,3; lane 2, 0.5 nmol APC-fA; lanes 3–8, GST-pulldown assays with constant GST-β-catenin and APC-2,3 and increasing amounts of competing APC-fA. Note: the maximum level of competitor contains 33 times more APC-fA than APC-2,3, or 16.5 times more 15mer repeats than 20mers. The antibody used for western blotting reacted poorly with the APC-fA construct, as it was generated to a fragment of APC containing only a 2 amino acid overlap with APC-fA. See Materials and methods for details.
In order to assess whether the 15 and 20mer repeats of APC bind at the same site on β-catenin, we tested whether the two classes of repeats could compete for binding to β-catenin (Figure 4B). A construct containing 15mer repeat A (APC-fA) could compete with binding of a construct containing two 20mer repeats (APC-2,3). The efficiency of the observed competition (with significant reduction of APC-2,3 binding even at a 1:1 ratio with APC-fA), suggests an extensive overlap between the binding sites of the two proteins. However, it remains possible that the 15 and 20mer binding sites are distinct but overlapping, or that presentation of the binding repeats within the larger constructs may result in steric clashes, even if their binding sites do not overlap.
The theory that the core homology regions of the 15 and 20mer repeats bind at the same site differs from a proposed mechanism for 20mer binding derived from the β-catenin– E-cadherin complex structure (Huber and Weis, 2001). In that structure, a Ser-Leu-Ser-Ser-Leu (SLSSL) sequence from E-cadherin bound to β-catenin in a phosphorylation-dependent manner, with phosphoserines at consensus GSK3β sites within and just N-terminal to the SLSSL sequence interacting with β-catenin. An SLSSL sequence is conserved in the C-terminal region of the 20 amino acid β-catenin binding repeats of APC (Figures 1B and 4A, bottom panel), suggesting that this region of the 20 amino acid repeats can interact with the SLSSL binding region. This hypothesis is consistent with mutagenesis experiments (Graham et al., 2000), which show that the binding of β-catenin to an APC construct containing both 15 and 20mer repeats was affected by mutations in the SLSSL binding site of β-catenin (Phe253Asp/Phe293Asp, Ala295Trp/Ile296Trp) (Figure 4A) (Graham et al., 2000; Huber and Weis, 2001). As several hydrophobic residues from the SLSSL region of E-cadherin make contacts with β-catenin, binding at the SLSSL site need not be entirely dependent upon phosphorylation. However, phosphorylation of the GSK3β consensus sites in E-cadherin increases its affinity for β-catenin 1000-fold (Huber and Weis, 2001; A.H.Huber and W.I.Weis, in preparation). Likewise, GSK3β phosphorylation of the 20 amino acid repeat region of APC has been shown to increase its affinity for β-catenin (Rubinfeld et al., 1996).
Alignment of the SLSSL sequence of an APC 20mer (e.g. hAPC-1, residues 1278–82) with that from E-cadherin (residues 690–694) results in the alignment of a conserved 20mer serine (hAPC-1 Ser1272) with an upstream GSK3β site in E-cadherin (pSer686) (Figure 4A, bottom panel). This serine corresponds to the last residue of the 20mer core homology region. However, E-cadherin pSer686 binds to β-catenin >18 Å from the end of the core homology region-binding site (APC-rA Ser1028) (Figure 4A). Thus, it would be impossible for a single 20mer repeat to bind by both mechanisms simultaneously. As the structural, sequence and mutagenesis data provide evidence for each binding mechanism we suggest that the 20mers can adopt two distinct binding modes, and that phosphorylation may act as a switch between these modes.
What might be the role of two binding modes in β-catenin degradation? Although the role of APC in the degradation complex is not fully understood, it is likely to include sequestration of β-catenin from Lef/Tcf before degradation and presentation of β-catenin for phosphorylation and degradation. The two binding modes could simply increase the number of potential binding sites, and hence the effective affinity between APC and β-catenin. Alternatively, the two binding modes could have distinct roles; for example, the unphosphorylated 20mers, in conjunction with the 15mers, may mediate the initial binding and sequestering of β-catenin, whereas subsequent phosphorylation of the 20mers and a switch in their mode of binding to β-catenin could be important for efficient presentation of β-catenin to GSK3β. The existence and roles, if any, of these alternatives await further experimentation.
Implications for drug design
A drug that could disrupt the Tcf–β-catenin complex would be expected to halt the transcription of oncogenes by this complex in many types of cancers. However, any such compound must specifically disrupt the Tcf– β-catenin complex without disrupting the interactions of β-catenin with cadherins or APC. Disruption of the β-catenin–cadherin complex could lead to decreased cell adhesion and an increased likelihood of metastasis (Guilford et al., 1998; Perl et al., 1998); interfering with the β-catenin–APC interaction would result in more free β-catenin that is available for Tcf complex formation. The structure of the APC-rA–β-catenin complex suggests the C-terminal portion of the extended region, in the area of the APC-rA bulge, as a potential target for such therapeutics. Although the interactions of XTcf3 in this region are the same as for E-cadherin, the extensive interaction interface (6100 Å2 buried surface area) observed in the E-cadherin complex suggests that this complex may be refractory to localized interference. This is supported by the observation that single alanine mutations were unable to disrupt the β-catenin– E-cadherin complex (W.Birchmeier, personal communication). Alternatively, regions of Tcf–β-catenin interaction outside of the extended region may also present attractive targets. However, interactions N-terminal to the extended region have not been shown to be critical for Tcf binding, and the C-terminal SLSSL-binding region is likely to be important in 20mer binding (Graham et al., 2000; von Kries et al., 2000). Thus, detailed structural information about the interaction of the 20mers with β-catenin will be required for rational design of any such therapeutic.
Materials and methods
Expression and purification of proteins
The armadillo repeat region of β-catenin (residues 134–671 of murine β-catenin) and glutathione S-transferase (GST)–β-catenin were produced as described previously (Huber et al., 1997, 2001).
To produce APC-2,3, amino acids 1362–1539 of human APC were over-expressed with a C-terminal His6 tag in Escherichia coli BL21-CodonPlus-RIL cells (Stratagene), using the pET-29 vector (Novagen). Cells were grown in Luria–Bertani medium at 37°C to an OD of 0.6–0.8, induced with isopropyl-β-d-thiogalactopyranoside (IPTG) to a concentration of 0.2 mM, and grown for another 3–4 h at 37°C. Cells were spun down at 3500 g, and pellets were resuspended in a buffer containing 50 mM Tris pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.1% β-mercaptoethanol and 0.5 mM phenylmethylsulfonyl fluoride (PMSF). Before lysis, protease inhibitors were added to a cell suspension to final concentrations of 1 mM PMSF, 2.1 µg/ml aprotinin, 2.5 µg/ml pepstatin A, 1 µg/ml leupeptin and 0.5 µg/ml E64. Cells were lyzed using an SLM-AMINCO French Press at a maximum pressure of 1200 p.s.i., and lyzates were spun at 300 000 g for 30 min at 4°C to remove any insoluble material. Protein was batch-purified by incubation with 1.25 ml Ni-NTA–agarose (Qiagen) per 1 l cell culture. Beads were washed in a buffer containing 50 mM Tris pH 8.0, 300 mM NaCl, 20 mM imidazole, 0.05% Tween-20 and 0.1% β-mercaptoethanol plus the above-mentioned protease inhibitors, and eluted in the same buffer with the imidazole concentration increased to 250 mM. The protein was concentrated and exchanged into Mono Q buffer [50 mM Tris pH 8.5, 2 mM dithiothreitol (DTT), and the above-mentioned protease inhibitors] using an Amicon Centricon Plus 20 concentrator (PES membrane, 5000 MWCO), then purified using anion-exchange chromatography (Mono Q; Pharmacia), with elution by a NaCl gradient. The protein was then further purified using size-exclusion chromatography (Superdex 200 HR; Pharmacia) in 50 mM Tris pH 8.5, 150 mM NaCl, 2 mM DTT plus the above-mentioned protease inhibitors.
APC-fA (amino acids 1001–1069) was expressed as for APC-2,3, except with an N-terminal GST tag using the pGEX-2T vector (Pharmacia). Cells were resuspended in a buffer containing 1× phosphate-buffered saline (PBS), 1 M NaCl, 0.05% Tween-20, 2 mM EDTA, 0.1% β-mercaptoethanol and 0.4 mM PMSF, and lysed and spun as for APC-2,3. Protein was batch purified using glutathione agarose (5 ml beads/l of original culture), cleaved with thrombin to remove the GST tag (6 U thrombin/1 ml beads, 90 min at 22°C) and purified using the same chromatographic steps as for APC 2,3.
The APC-rA (residues 1021–1035 of human APC), APC-rAL (residues 1015–1040), Tcf-ext (residues 15–25 of XTcf3) and Cad-ext (residues 673–683 of murine E-cadherin) peptides were produced by Fmoc synthesis and purified to >95% purity by HPLC (Biopeptide Co.).
Crystallization, data collection and processing
Crystals resembling clusters of plates were obtained using the hanging drop vapor diffusion method at 20°C, with a well solution of 10% PEG 8000, 100 mM Na/K phosphate pH 6.2, 200 mM NaCl, 5 mM DTT and a protein solution of 75 µM β-catenin and 92 µM APC-rA. A single plate of ∼200 × 200 × <5 µm was dissected from a cluster and soaked in well solution plus 1.3 mM APC-rA for 1 week. Later examination of the crystal packing indicated that the peptide was almost certainly co-crystallized, as the crystal packing was too tight to allow the peptide to have entered by diffusion. Additionally, the fact that this crystal form has not been observed previously for the β-catenin arm-repeat domain suggests that this form is unique to this peptide complex.
Data were measured from a single crystal at a wavelength of 0.97 Å at the Stanford Synchrotron Research Laboratory (SSRL) beamline 11-1. The crystal was adapted directly from the soak solution to a buffer consisting of well solution plus 25% glycerol, and frozen in a 100 K liquid nitrogen stream. A total of 180° of data were measured, using a 150 s exposure and 0.5° oscillation frames on an ADSC Quantum4 CCD detector. The crystal-to-detector distance was 225 mm. Radiation decay prevented the collection of data from a single crystal at higher redundancy, and scaling of data from multiple crystals failed due to non-isomorphism. Diffraction data were processed and scaled using DENZO and SCALEPACK (Otwinowski and Minor, 1997). The data were highly anisotropic, resulting in a relatively high Rmerge (Table I). There are two β-catenin–APC-rA complexes per asymmetric unit, with a solvent content of 44%.
Table I. Crystallographic statistics.
| Data collectiona | |
|---|---|
| Resolution (Å) | 30–3.1 (3.21–3.1) |
| Space group | P1 |
| Unit cell dimensions (Å) | |
| a | 51.09 |
| b | 69.26 |
| c | 95.10 |
| Unit cell dimensions (°) | |
| α | 68.70 |
| β | 87.04 |
| γ | 85.32 |
| Rmergeb | 8.2 (24.2) |
| Percent >3σ(I) | 73.0 (48.4) |
| Completeness (%) | 90.7 (87.8) |
| Average redundancy | 1.47 (1.39) |
| Refinement | |
| R values and temperature factors | |
| No. of reflections | |
| working set | 17 948 |
| test setc | 971 |
| Rcrystd | 0.234 |
| Rfreed | 0.273 |
| Average B (Å2) | |
| protein | 28.9 |
| solvent | 50.5 |
| Overall anisotropic B (Å2) | |
| B11 | 8.8 |
| B22 | –8.4 |
| B33 | –0.4 |
| B12 | –1.1 |
| B13 | –0.5 |
| B23 | –3.5 |
| Main chain-bond-related B r.m.s.d.e | 1.5 |
| Main chain-angle-related B r.m.s.d. | 2.0 |
| Side chain-bond-related B r.m.s.d. | 2.1 |
| Side chain-angle-related B r.m.s.d. | 3.0 |
| Model geometry | |
| Bond length r.m.s.d. from ideal (Å) | 0.005 |
| Bond angle r.m.s.d. from ideal (°) | 1.20 |
| Ramachandran plot | |
| percentage in most favored regions | 88.6 |
| percentage in additional allowed regions | 10.9 |
| percentage in generously allowed regions | 0.5 |
aValues in parentheses are for the highest resolution shell (3.21–3.10 Å).
bRmerge = 100 × Σh|I(h) – <I(h)>|/Σh<I(h)>
cThe ‘test set’ comprises a randomly selected subset of the data (5.1%) that was not included in the refinement of the model. The ‘working set’ contains the remaining reflections from the data set.
dR = Σh||Fobs(h)| – |Fcalc(h)||/Σh|Fobs(h)|. Rcryst and Rfree were calculated using the working and test reflection sets, respectively.
eRoot mean square deviation (r.m.s.d.).
Phasing, model building and refinement
All phasing and refinement was performed in CNS (Brünger et al., 1998). Phases were determined by molecular replacement, using β-catenin molecule A from the β-catenin–XTcf3 complex structure (Graham et al., 2000) (Protein Data bank ID code 1G3J) as the search model. After placement of both copies of β-catenin, rigid-body refinement was performed by breaking each copy of β-catenin into two halves (residues 134–390 and 391–664) to allow for the molecular flexibility previously observed in this protein (Huber et al., 1997). The R and Rfree values were 45.7 and 46.4%, respectively, after rigid-body refinement. The model was refined using a maximum likelihood target and all data between 30 and 3.1 Å. Rounds of simulated annealing (first two rounds) or minimization (all subsequent rounds) and group (first three rounds) or individual (all subsequent rounds) temperature factor refinement were interspersed with model building in O (Jones et al., 1991). Overall anisotropic temperature factor (Sheriff and Hendrickson, 1987) and bulk solvent corrections (Jiang and Brünger, 1994) were applied throughout the refinement. NCS restraints were applied to β-catenin (with each molecule again broken in two pieces) throughout refinement, and to the peptide only in the last round. The two copies of the complex were overlaid (using the lsq function in O; Jones et al., 1991) before using NCS restraints, in order to identify any regions of real difference in structure for which NCS restraints should not be applied (residue 1021 of APC-rA). Use of NCS restraints was validated at each round by comparison of Rfree values from minimization and B-factor refinement with and without NCS restraints, and at varying degrees of stringency. Refinement statistics are shown in Table I.
The final model contains 1026 amino acids of a possible 1106 (see Results and Discussion for details), 72 water molecules and one glycerol molecule. Coordinates and structure factors have been submitted to the PDB, under the accession code 1JPP.
Binding assays
For the APC-fA peptide competitive binding experiments (Figure 1C), 1 nmol of GST–β-catenin was combined with 50 µl of glutathione agarose, 5 nmol APC-fA and 5, 20, 80, 320 or 1280 nmol of peptide in a total volume (including beads) of 200 µl. Additional experiments at twice these protein concentrations yielded similar results, and are included in the data presented in Figure 1C. Protein concentrations were determined by absorbance at 280 nm, using extinction coefficients calculated from the protein sequence (Creighton, 1995). Proteins were incubated for 1 h at 4°C in the presence of 50 mM Tris pH 8.5, 150 mM NaCl, 0.05% Tween-20, 1 mM DTT and 1 mM PMSF. Beads were washed twice in 1 ml of the above buffer, resuspended in SDS loading dye and run on SDS–PAGE gels for analysis. The intensities of the APC-fA bands were quantified using the NIH Image program (developed at the US National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image/).
For the APC-fA/APC-2,3 competitive binding experiment (Figure 4B), 2 nmol of GST–β97 was combined with 50 µl of glutathione agarose, 2 nmol of APC 2,3, and 0, 0.66, 2, 6.6, 20 or 66 nmol of APC-fA in a total volume (including beads) of 250 µl. Reactions were incubated, beads were washed and run on SDS–PAGE gels as above. Proteins were then transferred to nitrocellulose membranes. Blots were blocked with 5% powdered milk in Tris-buffered saline containing 0.05% Tween-20, and APC detection was performed using a 1:1000 dilution of a rabbit polyclonal antibody to APC residues 1034–2130 (a gift from P.Polakis) in the same solution. Blots were developed using a 1:80 000 dilution of a horseradish peroxidase-linked anti-rabbit secondary antibody (Sigma) and the ECL system (Amersham).
Sequence analysis
For the MAST search for 15mer repeats, repeats A to C of human APC, A and B of Drosophila APC and A and B of Drosophila APC2 were aligned using BLOCK MAKER (Henikoff et al., 1995). The resulting block was used to search the non-redundant protein database using MAST (Bailey and Gribskov, 1998). hAPC-D, rAPC-D and mAPC-D scored as highly significant matches, with E-values (expected number of random sequences that would score that high, given the size of the database searched) of 3.2 × 10–11, 5.8 × 10–11 and 5.8 × 10–11, respectively. The sequences used to generate the block had E-values between 1.3 × 10–14 (eAPC-B) and 4.8 × 10–18 (eAPC-A) in the same search. hAPC-A had an E-value of 8.6 × 10–10 in a similar search using a block generated from the other six 15mers. A fourth sequence from Xenopus APC (residues 1883–1897) also scored as significant, although with a significantly lower E-value (4.8 × 10–5). However, several residues involved in contacts in the APC-rA structure were not conserved in this sequence.
Acknowledgments
Acknowledgements
We thank A.Huber, A.May and D.Daniels for helpful discussions and comments on the manuscript, A.May for assistance with data collection, and P.Polakis for the APC polyclonal antibody. This work is based on research conducted at the Stanford Synchrotron Radiation Laboratory (SSRL), which is funded by the Department of Energy (BES, BER) and the National Institutes of Health (NCRR, NIGMS). K.E.S. is a Howard Hughes Medical Institute Predoctoral Fellow. This work was supported by grant GM56169 to W.I.W. from the National Institutes of Health.
References
- Aberle H., Bauer,A., Stappert,J., Kispert,A. and Kemler,R. (1997) β-catenin is a target for the ubiquitin-proteasome pathway. EMBO J., 16, 3797–3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey T.L. and Gribskov,M. (1998) Combining evidence using p-values: application to sequence homology searches. [Comment in: Bioinformatics (2000) 16, 488–489 UI: 20330595]. Bioinformatics, 14, 48–54. [DOI] [PubMed] [Google Scholar]
- Behrens J., von Kries,J.P., Kuhl,M., Bruhn,L., Wedlich,D., Grosschedl,R. and Birchmeier,W. (1996) Functional interaction of β-catenin with the transcription factor LEF-1. Nature, 382, 638–642. [DOI] [PubMed] [Google Scholar]
- Behrens J., Jerchow,B.A., Würtele,M., Grimm,J., Asbrand,C., Wirtz,R., Kühl,M., Wedlich,D. and Birchmeier,W. (1998) Functional interaction of an axin homolog, conductin, with β-catenin, APC and GSK3β. Science, 280, 596–599. [DOI] [PubMed] [Google Scholar]
- Brannon M., Gomperts,M., Sumoy,L., Moon,R.T. and Kimelman,D. (1997) A β-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev., 11, 2359–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. B, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Creighton T.E. (1995) Protein Structure: A Practical Approach. IRL Press, New York, NY.
- Fagotto F., Jho,E., Zeng,L., Kurth,T., Joos,T., Kaufmann,C. and Costantini,F. (1999) Domains of axin involved in protein–protein interactions, Wnt pathway inhibition and intracellular localization. J. Cell Biol., 145, 741–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan M.J., Grüning,W., Walz,G. and Sokol,S.Y. (1998) Wnt signaling and transcriptional control of Siamois in Xenopus embryos. Proc. Natl Acad. Sci. USA, 95, 5626–5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuchi T., Sakamoto,M., Tsuda,H., Maruyama,K., Nozawa,S. and Hirohashi,S. (1998) β-catenin mutation in carcinoma of the uterine endometrium. Cancer Res., 58, 3526–3528. [PubMed] [Google Scholar]
- Graham T.A., Weaver,C., Mao,F., Kimelman,D. and Xu,W. (2000) Crystal structure of a β-catenin/Tcf complex. Cell, 103, 885–896. [DOI] [PubMed] [Google Scholar]
- Guilford P. et al. (1998) E-cadherin germline mutations in familial gastric cancer. Nature, 392, 402–405. [DOI] [PubMed] [Google Scholar]
- Hart M.J., de los Santos,R., Albert,I.N., Rubinfeld,B. and Polakis,P. (1998) Downregulation of β-catenin by human axin and its association with the APC tumor suppressor, β-catenin and GSK3β. Curr. Biol., 8, 573–581. [DOI] [PubMed] [Google Scholar]
- He T.C., Sparks,A.B., Rago,C., Hermeking,H., Zawel,L., da Costa,L.T., Morin,P.J., Vogelstein,B. and Kinzler,K.W. (1998) Identification of c-MYC as a target of the APC pathway. Science, 281, 1509–1512. [DOI] [PubMed] [Google Scholar]
- Henderson B.R. (2000) Nuclear-cytoplasmic shuttling of APC regulates β-catenin subcellular localization and turnover. Nature Cell Biol., 2, 653–660. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff,J.G., Alford,W.J. and Pietrokovski,S. (1995) Automated construction and graphical presentation of protein blocks from unaligned sequences. Gene, 163, GC17–26. [DOI] [PubMed] [Google Scholar]
- Huber A.H. and Weis,W.I. (2001) The structure of the β-catenin/E-cadherin complex and the molecular basis of diverse ligand recognition by β-catenin. Cell, 105, 391–402. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Nelson,W.J. and Weis,W.I. (1997) Three-dimensional structure of the armadillo repeat region of β-catenin. Cell, 90, 871–882. [DOI] [PubMed] [Google Scholar]
- Huber A.H., Stewart,D.B., Laurents,D.V., Nelson,W.J. and Weis,W.I. (2001) The cadherin cytoplasmic domain is unstructured in the absence of β-catenin. A possible mechanism for regulating cadherin turnover. J. Biol. Chem., 276, 12301–12309. [DOI] [PubMed] [Google Scholar]
- Huber O., Korn,R., McLaughlin,J., Ohsugi,M., Herrmann,B.G. and Kemler,R. (1996) Nuclear localization of β-catenin by interaction with transcription factor LEF-1. Mech. Dev., 59, 3–10. [DOI] [PubMed] [Google Scholar]
- Hülsken J., Birchmeier,W. and Behrens,J. (1994) E-cadherin and APC compete for the interaction with β-catenin and the cytoskeleton. J. Cell Biol., 127, 2061–2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda S., Kishida,S., Yamamoto,H., Murai,H., Koyama,S. and Kikuchi,A. (1998) Axin, a negative regulator of the Wnt signaling pathway, forms a complex with GSK-3β and β-catenin and promotes GSK–3β-dependent phosphorylation of β-catenin. EMBO J., 17, 1371–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J.-S. and Brünger,A.T. (1994) Protein hydration observed by X-ray diffraction. J. Mol. Biol., 243, 100–115. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Joslyn G. et al. (1991) Identification of deletion mutations and three new genes at the familial polyposis locus. Cell, 66, 601–613. [DOI] [PubMed] [Google Scholar]
- Kinzler K.W. and Vogelstein,B. (1996) Lessons from hereditary colorectal cancer. Cell, 87, 159–170. [DOI] [PubMed] [Google Scholar]
- Kinzler K.W. et al. (1991) Identification of FAP locus genes from chromosome 5q21. Science, 253, 661–665. [DOI] [PubMed] [Google Scholar]
- Knapp S. et al. (2001) Thermodynamics of the high-affinity interaction of TCF4 with β-catenin. J. Mol. Biol., 306, 1179–1189. [DOI] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT–A program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Merrit E.A. and Murphy,M.E.P. (1994) Raster3D version 2.0–a program for photorealistic molecular graphics. Acta Crystallogr. D, 50, 869–873. [DOI] [PubMed] [Google Scholar]
- Miyoshi Y. et al. (1992) Somatic mutations of the APC gene in colorectal tumors: mutation cluster region in the APC gene. Hum. Mol. Genet., 1, 229–233. [DOI] [PubMed] [Google Scholar]
- Molenaar M., van de Wetering,M., Oosterwegel,M., Peterson-Maduro,J., Godsave,S., Korinek,V., Roose,J., Destrée,O. and Clevers,H. (1996) XTcf-3 transcription factor mediates β-catenin-induced axis formation in Xenopus embryos. Cell, 86, 391–399. [DOI] [PubMed] [Google Scholar]
- Munemitsu S., Albert,I., Souza,B., Rubinfeld,B. and Polakis,P. (1995) Regulation of intracellular β-catenin levels by the adenomatous polyposis coli (APC) tumor-suppressor protein. Proc. Natl Acad. Sci. USA, 92, 3046–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Bharadwaj,R. and Honig,B. (1993) GRASP—graphical representation and analysis of surface properties. Biophys. J., 64, A116. [Google Scholar]
- Orford K., Crockett,C., Jensen,J.P., Weissman,A.M. and Byers,S.W. (1997) Serine phosphorylation-regulated ubiquitination and degradation of β-catenin. J. Biol. Chem., 272, 24735–24738. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Palacios J. and Gamallo,C. (1998) Mutations in the β-catenin gene (CTNNB1) in endometrioid ovarian carcinomas. Cancer Res., 58, 1344–1347. [PubMed] [Google Scholar]
- Peifer M., Berg,S. and Reynolds,A.B. (1994) A repeating amino acid motif shared by proteins with diverse cellular roles. Cell, 76, 789–791. [DOI] [PubMed] [Google Scholar]
- Perl A.K., Wilgenbus,P., Dahl,U., Semb,H. and Christofori,G. (1998) A causal role for E-cadherin in the transition from adenoma to carcinoma. Nature, 392, 190–193. [DOI] [PubMed] [Google Scholar]
- Riese J., Yu,X., Munnerlyn,A., Eresh,S., Hsu,S.C., Grosschedl,R. and Bienz,M. (1997) LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell, 88, 777–787. [DOI] [PubMed] [Google Scholar]
- Rosin-Arbesfeld R., Townsley,F. and Bienz,M. (2000) The APC tumour suppressor has a nuclear export function. Nature, 406, 1009–1012. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B., Souza,B., Albert,I., Müller,O., Chamberlain,S.H., Masiarz,F.R., Munemitsu,S. and Polakis,P. (1993) Association of the APC gene product with β-catenin. Science, 262, 1731–1734. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B., Souza,B., Albert,I., Munemitsu,S. and Polakis,P. (1995) The APC protein and E-cadherin form similar but independent complexes with α-catenin, β-catenin and plakoglobin. J. Biol. Chem., 270, 5549–5555. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B., Albert,I., Porfiri,E., Fiol,C., Munemitsu,S. and Polakis,P. (1996) Binding of GSK3β to the APC–β-catenin complex and regulation of complex assembly. Science, 272, 1023–1026. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B., Albert,I., Porfiri,E., Munemitsu,S. and Polakis,P. (1997a) Loss of β-catenin regulation by the APC tumor suppressor protein correlates with loss of structure due to common somatic mutations of the gene. Cancer Res., 57, 4624–4630. [PubMed] [Google Scholar]
- Rubinfeld B., Robbins,P., El-Gamil,M., Albert,I., Porfiri,E. and Polakis,P. (1997b) Stabilization of β-catenin by genetic defects in melanoma cell lines. Science, 275, 1790–1792. [DOI] [PubMed] [Google Scholar]
- Sheriff S. and Hendrickson,W.A. (1987) Description of overall anisotropy in diffraction from macromolecular crystals. Acta Crystallogr. A, 43, 118–121. [Google Scholar]
- Shtutman M., Zhurinsky,J., Simcha,I., Albanese,C., D’Amico,M., Pestell,R. and Ben-Ze′ev,A. (1999) The cyclin D1 gene is a target of the β-catenin/LEF-1 pathway. Proc. Natl Acad. Sci. USA, 96, 5522–5527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su L.K., Vogelstein,B. and Kinzler,K.W. (1993) Association of the APC tumor suppressor protein with catenins. Science, 262, 1734–1737. [DOI] [PubMed] [Google Scholar]
- Tetsu O. and McCormick,F. (1999) β-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature, 398, 422–426. [DOI] [PubMed] [Google Scholar]
- van de Wetering M. et al. (1997) Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell, 88, 789–799. [DOI] [PubMed] [Google Scholar]
- von Kries J.P., Winbeck,G., Asbrand,C., Schwarz-Romond,T., Sochnikova,N., Dell’Oro,A., Behrens,J. and Birchmeier,W. (2000) Hot spots in β-catenin for interactions with LEF-1, conductin and APC. Nature Struct. Biol., 7, 800–807. [DOI] [PubMed] [Google Scholar]
- Xu L., Corcoran,R.B., Welsh,J.W., Pennica,D. and Levine,A.J. (2000) WISP-1 is a Wnt-1- and β-catenin-responsive oncogene. Genes Dev., 14, 585–595. [PMC free article] [PubMed] [Google Scholar]
- Yap A., Brieher,W. and Gumbiner,B. (1997) Molecular and functional analysis of cadherin-based adherens junctions. Annu. Rev. Cell. Dev. Biol., 13, 119–146. [DOI] [PubMed] [Google Scholar]
- Yu X., Waltzer,L. and Bienz,M. (1999) A new Drosophila APC homologue associated with adhesive zones of epithelial cells. Nature Cell Biol., 1, 144–151. [DOI] [PubMed] [Google Scholar]
- Zurawel R.H., Chiappa,S.A., Allen,C. and Raffel,C. (1998) Sporadic medulloblastomas contain oncogenic β-catenin mutations. Cancer Res., 58, 896–899. [PubMed] [Google Scholar]