Abstract
Fungal conventional kinesins are unusually fast microtubule motor proteins. To compare the functional organization of fungal and animal conventional kinesins, a set of C-terminal deletion mutants of the Neurospora crassa conventional kinesin, NcKin, was investigated for its biochemical and biophysical properties. While the shortest, monomeric construct comprising the catalytic core and the neck-linker (NcKin343) displays very high steady-state ATPase (kcat = 260/s), constructs including both the full neck and adjacent hinge domains (NcKin400, NcKin433 and NcKin480) show wild-type behaviour: they are dimeric, show fast gliding and slower ATP turnover rates (kcat = 60–84/s), and are chemically processive. Unexpectedly, a construct (NcKin378, corresponding to Drosophila KHC381) that includes just the entire coiled-coil neck is a monomer. Its ATPase activity is slow (kcat = 27/s), and chemical processivity is abolished. Together with a structural analysis of synthetic neck peptides, our data demonstrate that the NcKin neck domain behaves differently from that of animal conventional kinesins and may be tuned to drive fast, processive motility.
Keywords: ATPase kinetics/domain analysis/fungal kinesin/processivity/synthetic peptide
Introduction
Conventional kinesins from fungi are microtubule-dependent motor proteins that presumably play a role in the intracellular transport of secretory vesicles. In comparison with their relatives from animals, they move ∼3-fold faster (Kirchner et al., 1999; Steinberg, 2000). Despite this distinctive feature, fungal kinesins move processively in a stepwise fashion, like their animal counterparts (Crevel et al., 1999). The analysis of their core motor domains reveals a high degree of sequence conservation with conventional kinesins from human, rat and fly. Consequently, fungal kinesins can be considered as natural mutants of their animal counterparts, which makes them interesting models for studying motor function. The conventional kinesin NcKin from N.crassa was the first fungal kinesin described and therefore the proto type of this group of kinesins (Steinberg and Schliwa, 1995). Today, similar kinesins from other ascomycetes, basidiomycetes and zygomycetes are known (Lehmler et al., 1997; Grummt et al., 1998a; Wu et al., 1998; Requena et al., 2001).
Like animal conventional kinesins, their fungal relatives consist of several domains (Vale and Fletterick, 1997; Woehlke and Schliwa, 2000). The conserved core motor domain, which is responsible for the catalytic activity, is located at the N-terminus, and is linked to the so-called neck domain via a short neck-linker that seems to be important for the generation of motility (Rice et al., 1999). Therefore, the shortest construct in this study (NcKin343) comprises the motor core and the entire neck-linker, the so-called ‘head’. In animal kinesins, the neck forms a two-stranded α-helical coiled-coil, causes dimerization of the motor molecule and joins the motor domain to the flexible, extended hinge region (Seeberger et al., 2000; Woehlke and Schliwa, 2000). The domains further C-terminal, the stalk and tail domains, seem to provide the cargo-binding site and a mechanical linker to the functional motor domain, and have been shown not to alter the motile behaviour in vitro (Grummt et al., 1998b; Seiler et al., 2000).
The functional organization of fungal conventional kinesins has been inferred from studies on the Syncephalastrum racemosum kinesin (Grummt et al., 1998b). These studies suggest that regions outside the core motor domain play important roles in determining the velocity and processivity of the motor. Thus, deletions of the neck and hinge domains, either alone or in combination, cause a dramatic decrease in the gliding velocity accompanied by an increase in the ATP turnover. Interestingly, replacement of the fungal hinge by the corresponding domain from the Drosophila melanogaster kinesin heavy chain, DmKHC, restores the gliding velocity but still leaves one significant parameter altered: the chemical processivity.
The ability to move processively is a feature of kinesin motors and refers to the fact that a single motor molecule can remain attached to its substrate filament during several mechanical steps. This behaviour can be observed in single-molecule microscopic assays, but is also reflected in kinetic properties (Hackney, 1995). In an ATPase assay, a processive motor appears to possess a lower half-maximal activation constant for microtubules (K0.5MT) than expected from its binding properties because the re-binding process necessary to initiate a new catalytic cycle is circumvented. Formally, the ratio kcat/K0.5MT = kbiATPase describes a second-order rate constant that characterizes kinesin–microtubule binding (Fersht, 1984). In the simplest Michaelis–Menten case, the ratio kcat/KM reflects a bimolecular rate constant for the association of substrate and enzyme. For processive enzymes such as conventional kinesins, however, the simple Michaelis– Menten assumptions do not apply. The kbiATPase value is connected to the processivity of the enzyme, with high kbiATPase values indicating high processivity. Following this kinetic line of evidence for the S.racemosum kinesin, we suspect that the neck and hinge domains play important roles in conferring processivity to the molecular motor (cf. Romberg et al., 1998).
To test this hypothesis and to map the structural determinants of the fast velocity of fungal kinesins, we use here a series of C-terminally truncated NcKins, and characterize them in a way comparable with the well-known Drosophila kinesin heavy chain. The ends of the constructs used here follow closely the domain borders neck-linker–neck, neck–hinge and hinge–coil 1. The results indicate that the core motor domain alone is a slow, non-processive motor despite its extremely high ATPase activity. The entire neck and at least part of the hinge domain are required to link efficiently the catalytic events in the core motor domain to motility. These findings suggest that the fast velocity originates in the core motor domain and is transmitted by highly optimized fungal-specific neck and hinge domains. Sequence features well conserved among all fungal kinesins and unusual properties of peptides derived from the neck support this hypothesis. Moreover, as demonstrated by truncated mutants and synthetic peptides, dimerization through the neck domain appears to be less favoured than in the animal case, but, in conjunction with the hinge domain, the motor displays fast, processive wild-type behaviour.
Results
To address the question of whether fungal conventional kinesins possess the same functional domain organization as their animal relatives, we expressed a series of C-terminally truncated versions of the N.crassa conventional kinesin, NcKin (Steinberg and Schliwa, 1995). The ends of these constructs were chosen to (i) fit domain boundaries as closely as possible (Kozielski et al., 1997) and (ii) be comparable with published Drosophila kinesin mutants (Jiang et al., 1997). Figure 1 shows a sequence alignment of fungal and animal kinesins indicating the positions at which the mutants of this study terminate. As only two-headed conventional kinesins are known to possess processive motility (Hancock and Howard, 1998), the dimerization characterisics of NcKin were investigated.
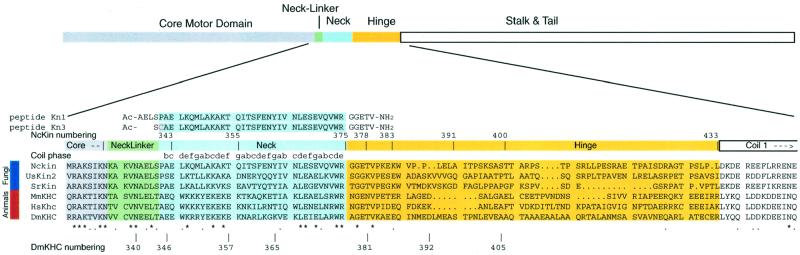
Fig. 1. Domain structure of kinesin. The location of domains relevant for this study is indicated in the top part of the figure. Below, the sequence alignment of three fungal (upper sequences) and three animal conventional kinesins highlights the differences of fungal and animal kinesins. In the core motor domain and the neck-linker, the homology, as indicated by asterisks for amino acid identity and dots for similarity, of all representatives is obvious. The central parts of the neck domains clearly differ between fungal and animal kinesins. In the hinge, all representatives diverge significantly. The position of the truncations used for this study is indicated above the alignment. For comparison, published Drosophila kinesin truncations are marked below (Jiang et al., 1997). The positions of the peptides Kn1 and Kn3 are indicated above the alignment. Domain borders were derived from the dimeric rat crystal structure (Kozielski et al., 1997) and coiled-coil prediction programs for the hinge and coil 1 domains (Lupas, 1996). The phase of the neck coiled-coil is indicated above the sequence alignment. Kinesins from: NcKin, N.crassa (SwissProt accession No. L47106); UmKin2, Ustilago maydis (U92845); SrKin, S.racemosum (AJ225894); MmKin, Mus musculus (X61435); HsKHC, Homo sapiens ubiquitous kinesin (X65873); DmKHC, D.melanogaster (M24441).
Oligomerization studies
To map the regions responsible for dimerizing NcKin, the sizes of the deletion mutants were determined (Table I). According to the gel filtration and sucrose density gradient experiments, the short motor versions NcKin343, 378 and 383 are monomeric; constructs longer than 390 residues are dimeric. By analogy with animal kinesins, the neck domain was expected to dimerize the heads. Hence, the failure of NcKin378 and 383 to dimerize came as a surprise (Figure 1). The NcKin378 protein ends at a residue homologous to DmKHC381, which clearly is dimeric (Figure 1; Jiang et al., 1997). It is unlikely that the C-termini were degraded since we also obtained this result with the C-terminally cysteine-tagged versions and were still able to label the tags.
Table I. Oligomerization characteristics of NcKin.
| Construct | rStokes (nm) | Density (Sw,20) | Derived mass (kDa) | Predicted mass (kDa) | Oligomerization state |
|---|---|---|---|---|---|
| NcKin343 | 2.8 ± 0.1 n = 2 | 3.3 ± 0.2 n = 4 | 38 | 37.4 | monomer |
| NcKin378 | 2.9 ± 0.2 n = 2 | 3.5 ± 0.2 n = 3 | 42 | 41.9 | monomer |
| NcKin383 | 3,6 ± 0.1 n = 2 | 3.7 ± 0.0 n = 2 | 55 | 42.5 | monomer |
| NcKin433-Δneck | 4.3 ± 0.1 n = 2 | 3.3 ± 0.0 n = 2 | 58 | 44.2 | monomer |
| NcKin391 | 4,8 ± 0.1 n = 2 | 4.6 ± 0.2 n = 2 | 90 | 43.4 | dimer |
| NcKin400 | 4.2 ± 0.1 n = 2 | 4.1 ± 0.2 n = 3 | 71 | 43.8 | dimer |
| NcKin433 | 4.7 ± 0.1 n = 3 | 4.3 ± 0.1 n = 2 | 83 | 47.2 | dimer |
| NcKin480 | 5.0 ± 0.1 n = 1 | 4.2 ± 0.2 n = 6 | 87 | 52.8 | dimer |
| NcKin928 | 5.6 ± 0.1 n = 5 | 9.3 ± 0.3 n = 4 | 215 | 105.0 | dimer |
As dimerization is a quantitative process, we estimated the dimerization constant, Kd. In the sizing experiments, at least 8 µM kinesin was used, yet no signal for a dimeric form of NcKin378 could be detected. We therefore conclude that the Kd for dimerizing NcKin378 lies significantly above 8 µM. To test whether ionic conditions prevented dimerization of NcKin378, the gel filtration was performed in the presence of 75 and 200 mM KCl. However, the elution volume relative to the standard proteins was unchanged, demonstrating that NcKin378 remains monomeric under low salt conditions. It was not possible to test even lower ionic strengths because of interactions between the Sephadex matrix and the sample. The slightly longer NcKin383, in contrast, showed a second peak in the gel filtration in some preparations. This peak corresponded to the the dimer and contained <10% of the total protein, as determined by ATPase rates. Accordingly, the dissociation constant Kd is expected to range above 300 µM under these experimental condi tions (38 µM NcKin, 200 mM KCl). In comparison, DmKHC381, which resembles NcKin378, dimerizes below 1 µM, as a construct comprising amino acids 1–413 (Rosenfeld et al., 1996; Jiang et al., 1997). From these studies, we conclude that the NcKin neck domain has a much lower capability to dimerize the motor heads compared with animal kinesins.
The NcKin391 construct is only eight amino acids longer than NcKin383 and clearly forms stable dimers under all salt conditions tested, and all longer NcKin versions also form dimers. To test whether the hinge domain alone is able to dimerize, a mutant of NcKin433 lacking the neck was investigated. This Δneck construct is monomeric, indicating that a combination of neck and hinge sequence motifs acts cooperatively to dimerize NcKin in solution.
Peptide studies
To elucidate the structural basis of the abberant dimerization behaviour, synthetic peptides derived from the neck domain were investigated. The similarity of animal and fungal kinesin neck domains is low in its N-teminal part but becomes obvious in the C-terminal 12 amino acids (Figure 1). Algorithms predict a low coiled-coil-forming propensity for the N-terminal part of the animal kinesin neck due to unfavourable, bulky or charged residues in the coiled-coil positions a and d. In contrast, peptide studies showed that the neck does form a stable two-stranded α-helical coiled-coil, which was later confirmed by the dimeric rat kinesin crystal structure (Kozielski et al., 1997; Morii et al., 1997; Tripet et al., 1997). For the fungal neck, the prediction indicates coiled-coil formation (Lupas, 1996), but experimental evidence has been lacking so far.
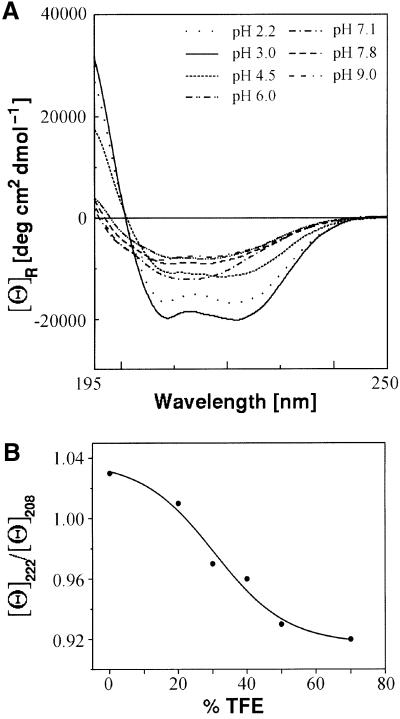
We therefore synthesized a NcKin neck domain peptide comprising amino acids 338–379 (peptide Kn1, Figure 1), which exceeds the expected coiled-coil by five amino acid residues at both the N- and C-termini. The solubility and conformational state of this peptide were found to be strongly pH dependent (Figure 3A). Only under acidic conditions (pH 3), the circular dichroism (CD) spectrum resembled those usually reported for two-stranded α-helical coiled-coils ([Θ]222/[Θ]208 = 1.03). This observation suggests an important role for the ionized glutamic acid residues for the onset of this type of fold. Moreover, at pH 3, the molar ellipticity [Θ]222 decreases sigmoidally from –6000 to –19 000°/cm2/dmol, with peptide concentrations rising from 10–6 to 10–3 M, indicating formation of intermolecular associations (data not shown). In agreement with this interpretation, ESI-MS spectra clearly reveal masses that can be assigned to peptide dimers (Supplementary figure 1, available at The EMBO Journal Online; Li et al., 1993; Przybilski and Glocker, 1996).
Fig. 3. Conformational state of the synthetic neck peptide Kn1. (A) The CD spectrum of the peptide Kn1 at varying pH values shows that an acidic environment below pH 4.5 induces an α-helical conformation, indicating the importance of protonated glutamic acid residues. (B) The ellipticity ratio [Θ]222/[Θ]208 of 1.03 in 50 mM phosphate buffer at pH 3, 20°C, indicates a coiled-coil formation, which is disrupted by increasing amounts of TFE, as seen by the sigmoidal decrease of [Θ]222/[Θ]208 to 0.96.
Evidence that this interaction is due to coiled-coil formation was obtained from 2,2,2-trifluoroethanol (TFE) titration experiments. TFE is known to stabilize secondary structures, but to disrupt higher order structures such as coiled-coils (Soennichsen et al., 1992; Bodkin and Goodfellow, 1996). The ratio [Θ]222/[Θ]208 was found to change in a sigmoidal manner from 1.03 in phosphate buffer, pH 3, to 0.96 in the presence of 70% TFE, indicating a cooperative transition from a two-stranded α-helical coiled-coil to a single-stranded α-helix (Figure 3B). To stabilize the dimeric form of the neck peptide, a second, slightly shorter peptide was used, which was cross-linked via a disulfide bridge at the a position of the first heptad repeat by replacing the endogenous proline with cysteine (Kn3, Figure 1). Taking the ellipticity ratio [Θ]222/[Θ]208 as an index for coiled-coil formation, the dimeric peptide Kn3 was found to fold into this type of conformation at neutral pH (50 mM phosphate buffer pH 7). The slightly lower ellipticity ratio of ∼1 compared with Kn1 (1.03) could result from the constraints introduced by the disulfide bridge.
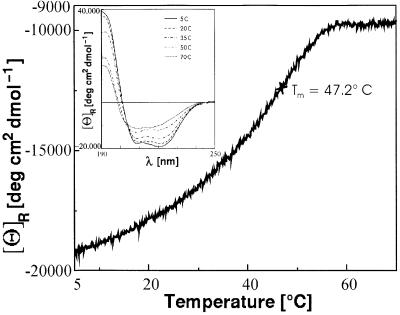
To determine the stability of the NcKin neck coiled- coil, thermal denaturation of the longer, wild-type-sequence peptide Kn1 was performed. Thermal unfolding of the peptide proceeds cooperatively with a Tm of 47.2°C, and an isodichroic point at 203 nm, which is indicative of a two-state transition from a folded to an unfolded state (Figure 4). During the recooling process, however, formation of β-structure takes place, thus preventing the extraction of thermodynamic parameters. Due to the low solubility of the Kn1 peptide, related NMR conformational analyses could not be carried out. Similarily, attempts to purify a bacterially expressed fragment comprising the NcKin neck and hinge domains also failed due to solubility problems.
Fig. 4. Thermal stability of the wild-type neck peptide Kn1 conformation. Thermal denaturation of the peptide Kn1 leads to an irreversible disintegration of the helical conformation. The isodichroic point in the CD spectra (inset) indicates a two-state transition from a folded to an unfolded conformation.
These results suggest that the neck domain is required but not sufficient for NcKin dimerization. Its unfavourable protonation state at physiological pH prevents folding into a two-stranded α-helical coiled-coil. Additional stabilizing motifs present in hinge residues 384–400 apparenty facilitate helix and/or coiled-coil formation by an unknown mechanism.
To characterize the impact of dimerization and C-terminal deletions on motility, ATPase and processivity, functional assays were performed (Table II).
Table II. Gliding assays and steady-state ATPase.
| State | Construct | Gliding velocity (µm/s) | Basal ATPase, k0 (/s) | Turnover, kcat (/s) | K0.5MTa (µM tubulin) | No. of independent ATPase assays | kbiATPase = kcat/K0.5MT (/µM/s) | kbiADP (/µM/s) | kbi ratiob |
|---|---|---|---|---|---|---|---|---|---|
| Monomeric | NcKin343 | 0.65 ± 0.06 | 0.052 ± 0.012 | 260 ± 74 | 0.40 ± 0.23 | 4 | 650 | 15.8 ± 0.3 | 41 |
| Monomeric | NcKin378 | 1.05 ± 0.13 | 0.015 ± 0.002 | 27 ± 10 | 0.86 ± 0.74 | 4 | 31 | 5.0 ± 0.3 | 6 |
| Monomeric | NcKin383 | 0.83 ± 0.18 | 0.016 ± 0.004 | 24 ± 4 | 0.47 ± 0.03 | 2 | 51 | 5.9 ± 0.5 | 9 |
| Dimeric | NcKin391 | 1.16 ± 0.20 | 0.039 ± 0.006 | 72 ± 1 | 0.04 ± 0.01 | 2 | 2000 | 5.3 ± 0.5 | 377 |
| Dimeric | NcKin400 | 2.61 ± 0.17 | 0.026 ± 0.011 | 84 ± 17 | 0.05 ± 0.03 | 3 | 1680 | 4.7 ± 0.1 | 357 |
| Dimeric | NcKin433 | 2.14 ± 0.22 | 0.047 ± 0.005 | 61 ± 8 | 0.02 ± 0.03 | 4 | 3050 | 4.2 ± 0.5 | 726 |
| Dimeric | NcKin480 | 2.15 ± 0.38 | 0.042 ± 0.016 | 60 ± 2 | 0.01 ± 0.01 | 4 | 6000 | 13.9 ± 2.5 | 432 |
| Dimeric | NcKin928 | 2.60 ± 0.30 | 0.010 ± 0.007 | 57 ± 8 | 0.05 ± 0.04 | 4 | 1140 | c | c |
aHalf-maximal microtubule activation constant.
bBiochemical processivity.
cLabelling with mantADP not possible.
Motility assays
Motility assays were performed on streptavidin-coated coverslips with biotin–maleimide-labelled motors, derived from the cysteine-tagged protein versions. Unlabelled motor proteins did not adhere to the glass surface by themselves. Since the ATPase activities of truncated Neurospora kinesins with a C-terminal cysteine tag were indistinguishable from those of the corresponding untagged versions, the tag is unlikely to alter the motile behaviour. Using gliding assays under multiple motor conditions (motor concentration 0.1–0.5 mg/ml), all investigated constructs were shown to be motile (Table II). Low dilutions of the motors (1:4) already caused a lack of microtubule binding and thus prevented observation of single-molecule gliding events. Within the standard deviation, the NcKin400, 433 and 480 motors were as fast as wild-type full-length NcKin. The monomeric NcKin versions and NcKin391 were also motile, but significantly slower. NcKin391 may be slower than the other dimers because of an unfavourable linkage to the glass surface.
Steady-state ATPase measurements
All constructs were also active in basal and microtubule-dependent ATPase assays, with activation factors of ∼1300–5700 (Table II). Whereas the dimeric NcKin391, 400, 433 and 480 were essentially indistinguishable from the full-length wild-type motor in the microtubule-dependent ATPase assay with average turnovers of 60–84/s and extremes of 50–104/s, monomeric constructs exhibited distinctly different behaviour. The shortest construct in this study, NcKin343, which lacks the entire neck region, showed an extremely fast ATPase with a turnover of kcat = 260/s. The turnover was variable, ranging from 200 to 400/s, probably due to temperature effects. A slightly longer construct, NcKin355, behaved very similarly to the NcKin343 version and yielded kcat values ranging from 75 to 360/s (n = 5), with an average of 216/s.
The kcat of NcKin378 was ∼10-fold slower than that of NcKin343, with a turnover of 27 ± 10/s. This low rate is remarkable because all longer constructs, including the full-length wild-type NcKin, exhibit a faster kcat of 60– 80/s. However, a slightly shorter (NcKin375) and a slightly longer construct (NcKin383) show similar turnovers (kcat = 24 ± 6 and 24 ± 4/s, respectively). To decide whether these constructs are intrinsically slow, or whether they contain a large number of inactive molecules, the C-terminal end of the neck domain of NcKin378 was digested with a mixture of carboxypeptidase A and B. SDS–PAGE analysis confirmed that the digest yielded one major kinesin fraction that appeared to be intermediate in size between NcKin378 and NcKin343, although the exact extent of the proteolytic degradation is not known. The ATPase activity after the treatment was increased to 50 ± 6/s (n = 2), corresponding to a 2-fold activation. Similar activation factors were obtained with two carboxypeptidase-treated NcKin375 preparations. The absolute turnover is not as high as in NcKin343 but is clearly above the NcKin378 level, indicating that the partial removal of the neck domain releases an inhibition of catalysis.
These data suggest that dimerc NcKin motors hydrolyse ATP at 60–84/s. The slower rate in NcKin378 and 383 is probably not due to a large fraction of inactive protein but to an inhibitory action of the neck. NcKin343 without the neck shows a very fast, unrepressed catalysis.
Chemical processivity
The steady-state microtubule-stimulated ATPase measurements also contain information on the processive behaviour of the enzyme. In particular, the ratio of the apparent second-order constant kbiATPase = kcat/K0.5MT from steady-state assays, and the measured binding rate, kbi, indicate the number of ATPase cycles induced by one productive diffusional encounter of kinesin and microtubule (Hackney, 1995). This ratio is called the kbi ratio (‘biochemical processivity’) in Table I. The binding rate kbi can be determined indirectly by following the ADP release because each productive initial encounter of kinesin with microtubules releases one ADP from the motor. Therefore, NcKin was incubated with a fluorescent ATP species, N-methylanthranoyl-ATP (mantATP), which yields a kinesin–mantADP complex because ATP is being hydrolysed quickly, even in the absence of microtubules. The fluorescence decrease upon mixing kinesin with varying, substoichiometric amounts of microtubules was then measured. The use of low microtubule concentrations ensures that the initial binding event is rate limiting, and not the subsequent catalysis which releases the second bound mantADP of a dimeric kinesin molecule. Hence, kinesin–microtubule binding causes a monoexponential fluorescence decrease, which reflects the rate of the binding reaction. Under microtubule-limiting conditions, this rate depends linearly on the microtubule concentration and reveals the second-order rate constant of kinesin– microtubule binding, kbiADP.
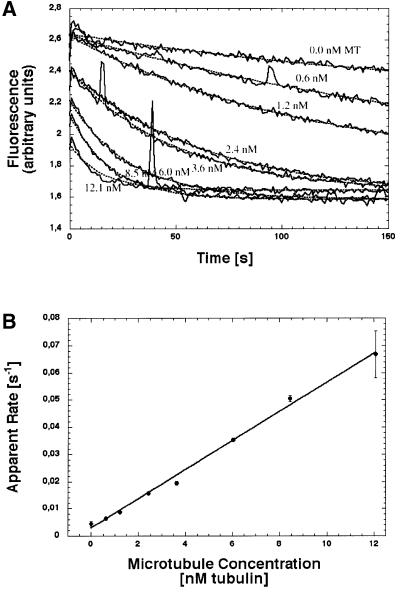
Table II lists the second-order binding rate constants of the NcKin constructs under investigation, as determined by mantADP release, kbiADP. The values were derived from time traces of mantADP release upon microtubule binding (Figure 2). The binding rates range from kbiADP = 4.2 to 15.8/µM/s. Most constructs show a kbiADP at ∼4.5/µM/s but the shortest and the longest ones deviate with values of kbiADP = 15.8 and 13.9/µM/s. These values are comparable with those found for DmKHC (0.6–7.6/µM/s; Jiang et al., 1997) or human kinesin (Vmax/KMMt = 40/s/13 µM = 3.1/µM/s; Ma and Taylor, 1995).
Fig. 2. Second-order rate constant of microtubule-activated mantADP release, kbiADP, from NcKin378cys. This example shows the apparent mantADP release rates upon mixing 100 nM mantADP–kinesin complex with variable amounts of microtubules (A). The time traces were fit to single exponentials (dotted lines). The resulting apparent rates depend linearly on the microtubule concentration, with the slope of the curve signifying kbiADP (B). Since the substoichiometric amount of microtubules limits the reaction, the kbiADP value reflects the rate of kinesin–microtubule binding. The full set of kbiADP values for all NcKin constructs is listed in Table II.
The comparison of apparent and measured microtubule binding rate constants (kbi ratio, Table I) indicates highly divergent chemical processivities for the investigated constructs. The NcKin378 and 383 mutants, comprising the core motor domain, neck-linker and the presumable neck helix, have the lowest values, suggesting that the construct dissociates from the filament after one or a few catalytic cycles. At the other extreme, the NcKin433 construct displays a ratio of 726 and may therefore stay on the microtubule for several hundreds of cycles. The shortest construct, NcKin343, possesses a kbi ratio of 41 and may stay bound at the microtubule due to the lack of a proper release mechanism (Jiang and Hackney, 1997). These results agree with experiments on truncated DmKHC constructs, with the exception of the monomeric, slow, non-processive NcKin378 and 383 variants whose homologue (DmKHC381) is chemically processive.
Discussion
This study addressed the question of the structural requirements for fast velocity and processive behaviour in conventional kinesins. In animal kinesins, the neck domain appears to confer processive motility to the motors, and its absence leads to mechanically and kinetically aberrant properties (Inoue et al., 1997; Jiang et al., 1997; Young et al., 1998). The findings presented here demonstrate that a fungal kinesin representative, NcKin, shares some features with animal kinesins, but also exhibits several unique and functionally significant properties. Thus, on the one hand, the motile and ATPase characteristics of NcKin clearly depend on the oligomerization state of the molecular motor, as in the animal case. Multiple motor gliding velocities for the shorter, monomeric versions NcKin343 and 378 are significantly slower than for the dimeric constructs, which are indistinguishable from the bacterially expressed full-length kinesin. Similar observations have been made for DmKHC (Young et al., 1998). As in animal kinesins, processivity can only be observed in dimeric variants. The chemical processivity described here agrees with biophysical studies on NcKin motility (Crevel et al., 1999), and a number of steps similar to processive ATP turnovers reported here have been observed in single-molecule motility assays on a dimeric NcKin483 kinesin (S.Lakämper and E.Meyhöfer, in preparation). Also in agreement with animal kinesins, short NcKin monomers (NcKin343) possess a faster ATPase than dimeric constructs (260 versus 60–80/s). For human kinesin, kcat values of 80 and 40/s have been reported for monomeric and dimeric forms, respectively (Ma and Taylor, 1997); for DmKHC, the values are 84 versus 20/s (Gilbert et al., 1995; Moyer et al., 1998) and 64 versus 44/s (Jiang et al., 1997).
On the other hand, there are also obvious differences between fungal and animal kinesins. For one, the fungal NcKin kinesin tail inhibition is less pronounced than in animals, and the dimeric truncated versions NcKin400, 433 and 480 have almost the same kcat values as the full-length protein. These findings are in agreement with the observation that the head–tail interaction in NcKin is so salt sensitive that under our assay conditions (ionic strength ∼50 mM), the interaction is weakened (Seiler et al., 2000).
The second dissimilarity is the high catalytic rate of the NcKin motor head. The extremely fast ATPase turnover of NcKin343 implies that the catalytic motor core contains determinants for the fast gliding velocity of NcKin. The microtubule-stimulated ATPase rate is >2.5-fold faster than that of the even shorter Drosophila kinesin DmKHC340 (kcat = 96/s), which presumably lacks a part of the functionally important neck-linker and, thus, is expected to display the higher ATPase rate (Jiang et al., 1997; Rice et al., 1999; Tomishige and Vale, 2000). The turnover required to drive motility in steps of 8 nm at 2.6 µm/s is 165/s per head, assuming one ATP per step. In part, the accelerated ATPase of NcKin compared with DmKHC may be due to a quicker rate of productive binding, as reflected in the apparent bimolecular rate constant, kbiADP. For NcKin, it ranges from 4.2 to 15.9/µM/s; DmKHC has values between 0.58 and 7.6/µM/s (Jiang et al., 1997). It will be interesting in the future to characterize further the striking correlation between the faster NcKin motor core ATPase (∼2.5-fold) and the faster gliding velocity of the NcKin motor (∼3-fold) compared with animal kinesins in pre-steady-state experiments.
Finally, the most striking difference between fungal and animal kinesins is the dimerization behaviour. Whereas animal kinesins form stable dimers when they contain the neck domain, the fungal NcKin requires additional motifs present in the hinge domain. In animal constructs that dimerize by virtue of their neck domains, dissociation constants below 1 µM have been reported (Rosenfeld et al., 1996; Jiang et al., 1997). On the basis of NcKin383, a dissociation constant at least 300-fold higher can be estimated. Differences between fungal and animal kinesin become most obvious when comparing NcKin378 and DmKHC381, which both include their respective neck domains and end at homologous residues (Figure 1). Whereas the truncation mutant DmKHC381 is dimeric, the NcKin378 construct is not. The neck of NcKin thus differs markedly from its animal counterparts.
What causes NcKin to dimerize? As in animal kinesins, the neck domain is able to form an α-helical coiled-coil, as seen in synthetic neck peptides under suitable conditions. Since the neck deletion construct NcKin433-Δneck contains the hinge domain but does not dimerize, the neck coiled-coil most probably causes dimerization. In the context of the protein, however, the neck alone is not sufficient and requires additional elements contributed by the hinge. The mechanism by which the hinge acts on the neck domain remains unclear, but may involve a pKa shift of glutamic acid residues, because α-helical coiled-coil formation requires an acidic environment. This pH dependence hints at the importance of protonation of one or more glutamic acid residues in g or e coil positions, as previously observed for coiled-coil model peptides (Kohn et al., 1998). In fact, NcKin contains Glu360 in a potential e position, which is not found in animal conventional kinesins (Figure 1). Peptides derived from the human kinesin neck were soluble at neutral pH and clearly assumed α-helical coiled-coil conformations (Morii et al., 1997; Tripet et al., 1997).
Dimerization of the NcKin neck in the context of the protein can be induced by the presence of a few key residues from the hinge domain. Thus the NcKin391 mutant was able to dimerize, whereas NcKin383 was not. Apparently, residues 384–391 favour the α-helical coiled-coil conformation of the neck domain. It is unlikely that these residues from the hinge domain contain a trigger sequence as described for other coiled-coils because, in those cases, trigger sequences are part of the α-helical coiled-coil and clearly contain coiled-coil heptad repeats (Burkhard et al., 2000). The hinge, however, has no predicted coiled-coil propensity at all, and contains many residues that are unfavourable for helical structures. Due to the low solubility of expressed hinge peptides, however, we were unable to test its conformational preferences.
Surprisingly, the presence of the neck domain is important for functional characteristics even in the monomeric NcKin378 and 383 versions that probably contain the neck in a non-helical conformation. These constructs displayed the slowest ATPase activities of all mutants. We cannot exclude rigorously that parts of the protein preparations are inactive since the basal activity was also slower by a factor of 2–3, compared with other constructs. However, the activation of NcKin378 by carboxypeptidases demonstates that the catalytic potential (kcat = 50/s) is at least as high as for fully functional dimeric constructs, suggesting that the ‘free’, undimerized neck domain of the monomer exerts an inhibitory effect on the catalytic core. As it is unknown how far the neck was digested, the inhibition may be even stronger than 1.9-fold [50/s (digested)/27/s (native)].
How does the neck affect ATPase and processivity? One possible hypothesis is that microtubule binding of NcKin378 induces dimerization and thus converts the molecule into a functional dimer (Crevel et al., 1999). However, when mantADP-charged NcKin378 is mixed with microtubules in the absence of ATP, 100% of the fluorescence signal is abolished, arguing for completely uncoupled heads. In dimeric constructs, only approximately half of the signal is lost (see Supplementary figure 2). The almost complete absence of chemical processivity of NcKin378 agrees with this observation and suggests that the microtubule-activated dimerization of NcKin378 may only be relevant at a high excess of kinesin over tubulin subunits.
Does the neck interact directly with the microtubule, and does this affect the ATPase and its processivity? Lysine residues from the neck have been suspected to interact with the acidic C-terminus of microtubules and thus to reduce the rate of detachment of the motor from the filament (Thorn et al., 2000; Wang and Sheetz, 2000). In fungal kinesins, three of these lysines are conserved (Figure 1). However, despite the presence of these residues, NcKin378 is not chemically processive.
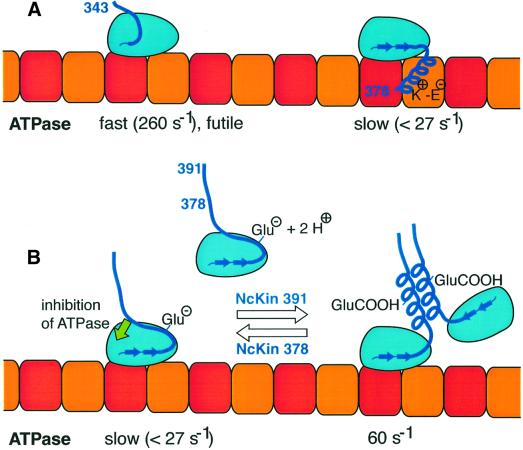
The neck–microtubule binding model might be extended by hypothesizing an equilibrium between a microtubule-bound form with disordered neck-linker and neck, which hydrolyses ATP quickly, and a slow microtubule-bound form that is tethered to the filament via the neck and an ordered neck-linker (Figure 5A). The conformation with a disordered neck-linker may be captured in the hydrolytically fast NcKin343 mutant, whereas filament binding of the neck domain in NcKin378 (and longer) induces ordering of the neck-linker and slow ATPase. Fully processive motility could only be established in dimeric constructs due to a strain-dependent release mechanism of the trailing head (Mandelkow and Johnson, 1998).
Fig. 5. Possible mechanisms of NcKin neck action. (A) A model in which the NcKin neck interacts with the microtubule by virtue of charged interactions (see Thorn et al., 2000; Wang and Sheetz, 2000). Binding of the neck by conserved lysines (K+) to the acidic, glutamate-rich C-terminus of tubulin (E–) might order the neck-linker, leading to a slow ATPase. The fast, futile ATP hydrolysis could occur with a disordered neck-linker, a state captured in NcKin343. (B) Alternatively, the slow ATPase can be understood by an inhibition of the ATPase by the neck, which would be absent in NcKin343. Dimerization is achieved by protonation of glutamates, resulting in wild-type ATP turnover. The coiled-coil neck conformation is facilitated by the presence of elements in the hinge domain.
Apart from being rather speculative, there is one argument against this model. Assuming an additional tether in NcKin378 in comparison with NcKin343, a longer dwell time τ at the microtubule would be expected. τ can be estimated by dividing the number of processive hydrolytic cycles (kbi ratio) by kcat, yielding τ = 41/260 s = 158 ms for NcKin343, and τ = 6/27 s = 222 ms for NcKin378. These values are quite similar, suggesting that either the K–E interactions between neck and filament are weak, or the release of the neck from the microtubule is tightly coupled to the catalytic cycle.
The most straightforward explanation for the observed effects is an inhibition of the catalytic domain by the presence of the neck domain (Figure 5B): NcKin378 and 383 are slower than NcKin343 because elements of the neck (which can be partially removed by C-terminal proteolysis) suppress the ATPase. The inhibition seems to be particularily effective in the absence of a coiled-coil conformation of the neck, and can be relieved by residues of the hinge domain. Protonation of glutamic acid residues in the neck and subsequent coil formation, induced by the presence of the hinge, may be involved in this process by an unknown mechanism. It is conceivable, therefore, that the neck may alternate between a coiled-coil and a molten conformation in the course of the ATP hydrolysis cycle, and this conformational change may be a prerequisite for the fast velocity of fungal kinesins. It will be interesting to test the implications of this model because point mutagenesis might identify single residues responsible for the inhibitory action of NcKin’s neck, and chimeric motor molecules may reveal whether the mechanism of action is a special adaptation of fungal kinesins, or a general feature of conventional kinesins.
Materials and methods
Cloning and expression
Truncated NcKin constructs were cloned using PCR with primers at the 5′ end of the NcKin gene and the desired stop codon. If necessary, a reactive cysteine tag was introduced (PSIVHRKCF*, Funatsu et al., 1997; NcKin480: KLGPSIVHRKCF*). The resulting fragments were cloned into pT7-based NcKin expression vectors (Henningsen and Schliwa, 1997), using the internal BamHI site in the NcKin gene, and a PstI site introduced after the stop codon. All plasmids were verified by sequence analysis. For the expression of NcKin protein, media flasks with 1 l of TPM (20 g/l tryptone, 15 g/l yeast extract, 2.5 g Na2HPO4, 1.0 g NaH2PO4, 10 mM glucose, 100 µg/ml ampicillin) were inoculated from a freshly transformed single colony of Escherichia coli BL21(DE3) (Novagene Corp.), pre-grown for 16–20 h at 22°C, and induced with 0.1 M isopropyl-β-d-thiogalactopyranoside (IPTG). The cells were incubated overnight at 22°C, harvested the next morning and stored at –70°C.
Purification was accomplished as described (Crevel et al., 1999; Supplementary figure 3). Briefly, 4 g of E.coli cells were resuspended in buffer A [20 mM Na-phosphate pH 7.4, 50 mM NaCl, 5 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol (DTT), 10 µM ATP, protease inhibitor mix (Roche Diagnostics)], sonified and centrifuged (35 min at 100 000 g). The supernatant was loaded on a 5 ml HiTrap SP Sepharose column (Amersham Pharmacia Biotech) and eluted in a manual NaCl step gradient. The peak fractions were pooled, frozen in liquid N2 with 10% glycerol and stored at –70°C.
C-terminal digestion of protein
To test whether NcKin is inhibited by its neck domain, NcKin378 and 375 preparations were digested using limited carboxypeptidase proteolysis (Ambler, 1972). Carboxypeptidases A and B (Sigma-Aldrich) were added to kinesin in 1:10 and 1:20 stoichiometries. The digest was incubated at 22°C and stopped by the addition of 5 mM EDTA after 1 or 2 h.
Oligomerization state
To characterize kinesin’s oligomerization state, sedimentation coefficients Sw,20 were measured by sucrose density centrifugation, and Stokes radii were determined by gel filtration. Sw,20 coefficients were determined by centrifugation of 5–10 µM protein solutions on 3–13% sucrose gradient cushions in buffer B (20 mM Na-phosphate pH 7.4, 50 or 200 mM NaCl, 10 µM ATP). The ionic strength was adjusted with NaCl as desired. The rotor (Beckman SW 50.1) was kept at 4°C and run at ∼20 000 g (37 000 r.p.m.) for 13 h. Aldolase, bovine serum albumin (BSA) and carboanhydrase (Sw,20 = 7.4, 4.3 and 3.2, respectively; Roche Diagnostics) were included at 0.1–0.2 mg/ml as internal standards. The gradients were fractionated and analysed on SDS–polyacrylamide gels. The positions of the peaks were used to determine the Sw,20 density values (NIH Image program, Kaleidagraph software).
The proteins’ Stokes radii were determined by gel filtration analysis. Protein solutions (8–35 µM) were loaded on a Superdex 200 column (Amersham Pharmacia Biotech) equilibrated with buffer B containing the desired concentrations of NaCl. The elution volumes of the samples were compared with standard proteins [ferritin, 5.9 nm radius; aldolase, 4.5 nm; BSA, 3.55 nm; carboanhydrase, 2.4 nm (or chymotrypsinogen, 2.24 nm); and cytochrome c, 1.64 nm; Roche Diagnostics and Sigma Chemical Corp.]. Stokes radii were calculated from a plot of elution volumes versus standard sizes (Andrews, 1970). The molecular weights of the kinesin constructs were calculated according to the equation of Cantor and Schimmel (1980):
Mr = (Sw,20 × nA × 6π × η × rStokes)/(1 – υ × ρ)
where nA is the Avogadro number, η is viscosity, υ is the specific volume of the protein = 0.725 cm3/g and ρ is the density of the medium.
Basal ATPase measurements
The ATPase rates in the absence of microtubules were measured using [γ-32P]ATP (Shimizu et al., 2000). NcKin was incubated in ATPase buffer 12A25 at 3 and 6 µM, along with a blank without kinesin. The reactions were started by the addition of 1 mM [γ-32P]ATP and stopped after 0, 5, 10, 20 and 30 min in 0.3 M perchloric acid in 1 mM NaH2PO4. The reactions were mixed with charcoal to absorb nucleotide and centrifuged to pellet the charcoal. The free [γ-32P]phosphate in the supernatant was quantified in a scintillation counter using Cerenkov radiation. The basal ATPase rates were calculated from linear fits of the time traces.
Steady-state microtubule-ATPase activity
Steady-state ATPase rates were determined using a coupled enzymatic assay (Huang and Hackney, 1994a; Grummt et al., 1998b). The assays were performed in a low ionic strength buffer (ATPase buffer: 12 mM, N-[2-acetamido]-2-aminoethanesulfonic acid (Aces)·KOH pH 6.8, 25 mM potassium acetate, 2 mM magnesium acetate, 0.5 mM EGTA). ATP was used at 1 mM; the reactions were started with kinesin. Control experiments omitting single components showed that the kinesin preparations did not contain measurable background activities that might interfere with the coupled assay. Microtubules from pig brain tubulin were polymerized freshly from pre-spun tubulin, stabilized with 20 µM paclitaxel and recentrifuged to remove excess nucleotide. The concentration of the resuspended microtubule solution was determined photometrically at 280 nm (Huang and Hackney, 1994b).
ADP release rates
ADP release rates were measured by mixing mantADP-charged kinesin with substoichiometric amounts of microtubules, and quantification of the fluorescence decrease. To load NcKin with mantADP, the kinesin mutants were incubated with a 4-fold molar excess of mantATP at 25°C for 15 min. The kinesin–mantADP complex was separated from excess nucleotide via Sephadex G25 spin columns. The concentration of the eluted protein was evaluated by a Bradford assay. This protocol was suitable for all truncated NcKin constructs but failed to label full-length NcKin. Kinesin–microtubule binding was monitored in an Aminco-Bowman spectrofluorometer with an excitation wavelength of 365 nm and emission at 445 nm. Kinesin (100–120 nM) was mixed with substoichiometric microtubule concentrations (0–10 nM). The resulting fluorescence decrease was monitored and fitted to single exponential curves from which the release rates were derived. The second mantADP, which is present in dimeric kinesin constructs, is being released upon addition of excess ATP at the rate kcat, which is much faster than the rates observed under our experimental conditions. Hence, the mantADP release rates are likely to reflect the rate of productive kinesin– microtubule binding. A plot of the release rates versus microtubule concentrations was used to calculate the bimolecular rate constant, kbiADP.
Labelling of reactive cysteine tags
NcKin kinesins containing a reactive cysteine tag were labelled with biotin–maleimide. The protein was incubated with a 6-fold molar excess of maleimide conjugate on ice for 60 min. The reaction was stopped with 10 mM DTT. Active kinesin was isolated by a microtubule binding and release step (Vale et al., 1985).
Gliding assays
Motility assays were performed with biotin-labelled kinesins in flow cells coated with streptavidin. After 10 min of incubation with 1 mg/ml streptavidin (Sigma Chemicals), the cells were washed with three chamber volumes of blocking buffer (1 mg/ml BSA and 0.8 mg/ml casein in BRB80: 80 mM PIPES–KOH pH 6.8, 1 mM MgCl2, 1 mM EGTA), and filled with biotin-labelled kinesin in blocking buffer. The assays were started after 10 min by floating the chambers with blocking buffer containing 6 mM MgCl2, 5 mM ATP, 5 mM phospho(enol)pyruvate, 200 mM KCl, 20 µM paclitaxel and microtubules. Motility was monitored in a Zeiss Axiophot using video-enhanced phase-contrast microscopy.
Peptide synthesis and purification
Two peptides derived from the NcKin neck region were used in this study, termed Kn1 and Kn3. The latter is a peptide dimer cross-linked by a disulfide bond between cysteine residues located in the first position of the heptad repeat of the presumed coiled-coil and was chosen to match the optimal location of disulfide bonds in supercoils according to previous studies (Figure 1; Kohn et al., 1998). Whereas peptide Kn1 was synthesized automatically by solid-phase 9-fluorenylmethoxycarbonyl (Fmoc)/tBu chemistry on TentaGel S-RAM resin on an Applied Biosystems model 431A peptide synthesizer, peptide Kn3 was synthesized manually with standard Boc/Bzl chemistry on a 4-methyl benzhydrylamine resin. For details, see Supplementary data.
Circular dichroism
The CD spectra were recorded on a Jasco J-715 spectropolarimeter at 20°C in quartz cuvettes (temperature controller PFD-350S). The concentrations of the filtered samples (0.45 µm) were determined by absorbance at 280 nm. The spectra (average of 10 scans) were normalized to their mean residue molar ellipticity [Θ]R (°/cm2/dmol). The spectra were recorded between 180 and 250 nm, with a scanning speed of 50 nm/min, a response of 1 s and a band width of 1.0 nm. Melting curves were measured by following the change of molar ellipticity at 222 nm versus temperature, with a temperature slope of 30°C/min, a response of 16 s and a band width of 1 nm. The melting temperature (Tm) was derived from the zero intercept of the second derivative of the respective melting curve.
Mass spectrometry
The mass spectra were taken on a PE SCIEX API 165 single quadrupole MS system. The spectrum for detecting the non-covalently bound dimers was collected in the range 700–2000 Da, with an infusion pump rate of 0.3 ml/h, an ion source high voltage of 4.5 kV, an orifice voltage of 15 V, a dwell time of 0.6 ms per scan and a step size of 0.2 Da with 10 scans summed.
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We thank Dr I.Crevel for sharing her experience, and Dr W.Heinzel (Polypeptide GmbH, Wolfenbuttel, Germany) for his help with the peptide synthesis. The authors’ work was supported by the Deutsche Forschungsgemeinschaft, the Volkswagen Foundation and the Fonds der Chemischen Industrie.
References
- Ambler R.P. (1972) Enzymatic hydrolysis with carboxypeptidases. Methods Enzymol., 25, 143–154. [DOI] [PubMed] [Google Scholar]
- Andrews P. (1970) Estimation of molecular size and molecular weights of biological components by gel filtration. Methods Biochem. Anal., 18, 1–53. [PubMed] [Google Scholar]
- Bodkin M.J. and Goodfellow,J.M. (1996) Hydrophobic solvation in aqueous trifluoroethanol solution. Biopolymers, 39, 43–50. [DOI] [PubMed] [Google Scholar]
- Burkhard P., Kammerer,R.A., Steinmetz,M.O., Bourenkov,G.P. and Aebi,U. (2000) The coiled-coil trigger site of the rod domain of cortexillin I unveils a distinct network of interhelical and intrahelical salt bridges. Structure Fold. Des., 8, 223–230. [DOI] [PubMed] [Google Scholar]
- Cantor C.R. and Schimmel,P.R. (1980) Techniques for the Study of Biological Structure and Function. Biophysical Chemistry. Vol. 2. W.H.Freeman, San Francisco, CA.
- Crevel I., Carter,N., Schliwa,M. and Cross,R. (1999) Coupled chemical and mechanical reaction steps in a processive Neurospora kinesin. EMBO J., 18, 5863–5872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. (1984) Enzyme Structure and Mechanism. W.H.Freeman and Co., New York, NY.
- Funatsu T., Harada,Y., Higuchi,H., Tokunaga,M., Saito,K., Ishii,Y., Vale,R.D. and Yanagida,T. (1997) Imaging and nano-manipulation of single biomolecules. Biophys. Chem., 68, 63–72. [DOI] [PubMed] [Google Scholar]
- Gilbert S.P., Webb,M.R., Brune,M. and Johnson,K.A. (1995) Pathway of processive ATP hydrolysis by kinesin. Nature, 373, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt M., Pistor,S., Lottspeich,F. and Schliwa,M. (1998a) Cloning and functional expression of a ‘fast’ fungal kinesin. FEBS Lett., 427, 79–84. [DOI] [PubMed] [Google Scholar]
- Grummt M., Woehlke,G., Henningsen,U., Fuchs,S., Schleicher,M. and Schliwa,M. (1998b) Importance of a flexible hinge near the motor domain in kinesin-driven motility. EMBO J., 17, 5536–5542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackney D.D. (1995) Highly processive microtubule-stimulated ATP hydrolysis by dimeric kinesin head domains. Nature, 377, 448–450. [DOI] [PubMed] [Google Scholar]
- Hancock W.O. and Howard,J. (1998) Processivity of the motor protein kinesin requires two heads. J. Cell Biol., 140, 1395–1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henningsen U. and Schliwa,M. (1997) Reversal in the direction of movement of a molecular motor. Nature, 389, 93–96. [DOI] [PubMed] [Google Scholar]
- Huang T. and Hackney,D. (1994a) Drosophila kinesin motor domain extending to amino acid position 392 is dimeric when expressed in Escherichia coli. J. Biol. Chem., 269, 16502–16507. [PubMed] [Google Scholar]
- Huang T.G. and Hackney,D.D. (1994b) Drosophila kinesin minimal motor domain expressed in Escherichia coli. Purification and kinetic characterization. J. Biol. Chem., 269, 16493–16501. [PubMed] [Google Scholar]
- Inoue Y., Toyoshima,Y.Y., Iwane,A.H., Morimoto,S., Higuchi,H. and Yanagida,T. (1997) Movements of truncated kinesin fragments with a short or an artificial flexible neck. Proc. Natl Acad. Sci. USA, 94, 7275–7280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W. and Hackney,D.D. (1997) Monomeric kinesin head domains hydrolyze multiple ATP molecules before release from a microtubule. J. Biol. Chem., 272, 5616–5621. [DOI] [PubMed] [Google Scholar]
- Jiang W., Stock,M.F., Li,X. and Hackney,D.D. (1997) Influence of the kinesin neck domain on dimerization and ATPase kinetics. J. Biol. Chem., 272, 7626–7632. [DOI] [PubMed] [Google Scholar]
- Kirchner J., Woehlke,G. and Schliwa,M. (1999) Universal and unique features of kinesin motors: insights from a comparison of fungal and animal conventional kinesins. Biol. Chem., 380, 915–921. [DOI] [PubMed] [Google Scholar]
- Kohn W.D., Kay,C.M. and Hodges,R.S. (1998) Orientation, positional, additivity and oligomerization-state effects of interhelical ion pairs in α-helical coiled-coils. J. Mol. Biol., 283, 993–1012. [DOI] [PubMed] [Google Scholar]
- Kozielski F., Sack,S., Marx,A., Thormahlen,M., Schonbrunn,E., Biou,V., Thompson,A., Mandelkow,E.M. and Mandelkow,E. (1997) The crystal structure of dimeric kinesin and implications for microtubule-dependent motility. Cell, 91, 985–994. [DOI] [PubMed] [Google Scholar]
- Lehmler C., Steinberg,G., Snetselaar,K.M., Schliwa,M., Kahmann,R. and Bölker,M. (1997) Identification of a motor protein required for filamentous growth in Ustilago maydis. EMBO J., 16, 3464–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.-T., Hsieh,Y.-L., Henion,J.D., Senko,M.W., McLafferty,F.W. and Ganem,B. (1993) Mass spectrometric studies on noncovalent dimers of leucine zipper peptides. J. Am. Chem. Soc., 115, 8409–8413. [Google Scholar]
- Lupas A. (1996) Prediction and analysis of coiled-coil structures. Methods Enzymol., 266, 513–525. [DOI] [PubMed] [Google Scholar]
- Ma Y.Z. and Taylor,E.W. (1995) Mechanism of microtubule kinesin ATPase. Biochemistry, 34, 13242–13251. [DOI] [PubMed] [Google Scholar]
- Ma Y.Z. and Taylor,E.W. (1997) Interacting head mechanism of microtubule–kinesin ATPase. J. Biol. Chem., 272, 724–730. [DOI] [PubMed] [Google Scholar]
- Mandelkow E. and Johnson,K.A. (1998) The structural and mechanochemical cycle of kinesin. Trends Biochem. Sci., 23, 429–433. [DOI] [PubMed] [Google Scholar]
- Morii H., Takenawa,T., Arisaka,F. and Shimizu,T. (1997) Identification of kinesin neck region as a stable α-helical coiled coil and its thermodynamic characterization. Biochemistry, 36, 1933–1942. [DOI] [PubMed] [Google Scholar]
- Moyer M.L., Gilbert,S.P. and Johnson,K.A. (1998) Pathway of ATP hydrolysis by monomeric and dimeric kinesin. Biochemistry, 37, 800–813. [DOI] [PubMed] [Google Scholar]
- Przybilski M. and Glocker,O. (1996) Elektrospray-Massenspektrometrie von Biomakromolekülkomplexen mit nichtkovalenten Wechsel wirkungen—neue analytische Perspektiven fuer supramolekulare Chemie und molekulare Erkennungsprozesse. Angew. Chem., 108, 878–899. [Google Scholar]
- Requena N., Alberti-Segui,C., Winzenburg,E., Horn,C., Schliwa,M., Philipsen,P., Liese,R. and Fischer,R. (2001) Genetic evidence for a microtubule-destabilizing effect of conventional kinesin and analysis of its consequences for the control of nuclear distribution in Aspergillus nidulans. Mol. Microbiol., 42, 121–132. [DOI] [PubMed] [Google Scholar]
- Rice S. et al. (1999) A structural change in the kinesin motor protein that drives motility. Nature, 402, 778–784. [DOI] [PubMed] [Google Scholar]
- Romberg L., Pierce,D.W. and Vale,R.D. (1998) Role of the kinesin neck region in processive microtubule-based motility. J. Cell Biol., 140, 1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld S.S., Rener,B., Correia,J.J., Xing,J., Mayo,M.S. and Chung,H.C. (1996) Equilibrium studies of kinesin-nucleotide intermediates. J. Biol. Chem., 271, 9473–9482. [DOI] [PubMed] [Google Scholar]
- Seeberger C., Mandelkow,E. and Meyer,B. (2000) Conformational preferences of a synthetic 30mer peptide from the interface between the neck and stalk regions of kinesin. Biochemistry, 39, 12558–12567. [DOI] [PubMed] [Google Scholar]
- Seiler S., Kirchner,J., Horn,C., Kallipolitou,A., Woehlke,G. and Schliwa,M. (2000) Cargo binding and regulatory sites in the tail of fungal conventional kinesin. Nature Cell Biol., 2, 333–338. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Thorn,K.S., Ruby,A. and Vale,R.D. (2000) ATPase kinetic characterization and single molecule behavior of mutant human kinesin motors defective in microtubule-based motility. Biochemistry, 39, 5265–5273. [DOI] [PubMed] [Google Scholar]
- Soennichsen F.D., Van Eyck,J.E., Hodges,R.S. and Sykes,B.D. (1992) Effect of trifluoroethanol on protein secondary structure: an NMR and CD study using a synthetic actin peptide. Biochemistry, 31, 8790–8798. [DOI] [PubMed] [Google Scholar]
- Steinberg G. (2000) The cellular roles of molecular motors in fungi. Trends Microbiol., 8, 162–168. [DOI] [PubMed] [Google Scholar]
- Steinberg G. and Schliwa,M. (1995) The Neurospora organelle motor: a distant relative of conventional kinesin with unconventional properties. Mol. Biol. Cell, 6, 1605–1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorn K.S., Ubersax,J.A. and Vale,R.D. (2000) Engineering the processive run length of the kinesin motor. J. Cell Biol., 151, 1093–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M. and Vale,R.D. (2000) Controlling kinesin by reversible disulfide cross-linking. Identifying the motility-producing conformational change. J. Cell Biol., 151, 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet B., Vale,R.D. and Hodges,R.S. (1997) Demonstration of coiled-coil interactions within the kinesin neck region using synthetic peptides. Implications for motor activity. J. Biol. Chem., 272, 8946–8956. [DOI] [PubMed] [Google Scholar]
- Vale R.D. and Fletterick,R.J. (1997) The design plan of kinesin motors. Annu. Rev. Cell Dev. Biol., 13, 745–777. [DOI] [PubMed] [Google Scholar]
- Vale R.D., Reese,T.S. and Sheetz,M.P. (1985) Identification of a novel force-generating protein, kinesin, involved in microtubule-based motility. Cell, 42, 39–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z. and Sheetz,M.P. (2000) The C-terminus of tubulin increases cytoplasmic dynein and kinesin processivity. Biophys. J., 78, 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woehlke G. and Schliwa,M. (2000) Walking on two heads: the many talents of kinesin. Nature Rev. Mol. Cell Biol., 1, 50–58. [DOI] [PubMed] [Google Scholar]
- Wu Q., Sandrock,T.M., Turgeon,B.G., Yoder,O.C., Wirsel,S.G. and Aist,J.R. (1998) A fungal kinesin required for organelle motility, hyphal growth and morphogenesis. Mol. Biol. Cell, 9, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young E.C., Mahtani,H.K. and Gelles,J. (1998) One-headed kinesin derivatives move by a nonprocessive, low-duty ratio mechanism unlike that of two-headed kinesin. Biochemistry, 37, 3467–3479. [DOI] [PubMed] [Google Scholar]