Abstract
The recently published human genome with its relatively modest number of genes has highlighted the importance of post-transcriptional and post-translational modifications, such as alternative splicing or glycosylation, in generating the complexities of human biology. The human UDP-N-acetylglucosamine (UDPGlcNAc) pyrophosphorylases AGX1 and AGX2, which differ in sequence by an alternatively spliced 17 residue peptide, are key enzymes synthesizing UDPGlcNAc, an essential precursor for protein glycosylation. To better understand the catalytic mechanism of these enzymes and the role of the alternatively spliced segment, we have solved the crystal structures of AGX1 and AGX2 in complexes with UDPGlcNAc (at 1.9 and 2.4 Å resolution, respectively) and UDPGalNAc (at 2.2 and 2.3 Å resolution, respectively). Comparison with known structures classifies AGX1 and AGX2 as two new members of the SpsA-GnT I Core superfamily and, together with mutagenesis analysis, helps identify residues critical for catalysis. Most importantly, our combined structural and biochemical data provide evidence for a change in the oligomeric assembly accompanied by a significant modification of the active site architecture, a result suggesting that the two isoforms generated by alternative splicing may have distinct catalytic properties.
Keywords: alternative splicing/crystallography/human enzyme/oligomeric assembly/UDPGlcNAc pyrophosphorylase
Introduction
Carbohydrates, N- or O-linked to proteins via post-translational modifications, play diverse and crucial biological roles in eukaryotes. When present on cell surface glycoproteins, they are instrumental in cell–cell/matrix interactions, immune reactions and tumour development (Hakomori, 1991; Rudd et al., 2001). They are important for the transport, biological activity and clearance from the circulation of secreted glycoproteins and modify the structure of cytosolic and nuclear proteins with implications in transcription, translation, neuronal pathology and other biological processes (Hart et al., 1989; Dennis et al., 1999; Wells et al., 2001).
UDP-N-acetylglucosamine (UDPGlcNAc), the activated form of GlcNAc, is a key precursor of these N- and O-linked glycosylations (Hart et al., 1989; Herscovics and Orlean, 1993). It is also essential for the synthesis of chitin (a major component of the fungal cell wall) (Cabib et al., 1982) and of the glycosylphosphatidylinositol (GPI) linker which anchors a variety of cell surface proteins to the plasma membrane (Udenfriend and Kodukula, 1995). In bacteria, UDPGlcNAc represents an essential precursor for both peptidoglycan and lipopolysaccharide biosynthesis (Raetz, 1996).
UDPGlcNAc is synthesized in the cell cytoplasm by an enzyme which catalyses the following reversible reaction (in the presence of Mg2+/Mn2+):
UTP + GlcNAc1P ↔ PPi + UDPGlcNAc
This enzyme is named UDPGlcNAc pyrophosphorylase (UAP) or GlcNAc1P uridyltransferase, depending on the direction of the reaction considered. In eukaryotes, initial characterization of UAPs was undertaken on partially purified enzymes from yeast, Neurospora crassa, calf liver and sheep brain (Strominger and Smith, 1959; Pattabiraman and Bachhawat, 1961; Kawai et al., 1976; Yamamoto et al., 1976, 1979). Recent kinetic data have also been reported on purified enzymes from the pig liver cytosol (Szumilo et al., 1996) and the human pathogenic parasite Giardia lamblia (Bulik et al., 1998, 2000). The genes coding for the yeast and human UAPs were identified only recently (Mio et al., 1998). The human UAPs correspond to AGX1 and AGX2, two isoforms resulting from the alternative splicing of a single gene and differing in sequence by the presence, in AGX2, of an extra 17 amino acid peptide (Diekman and Goldberg, 1994). The functional characterization of these isoforms revealed that AGX1 was 2- to 3-fold more active with GalNAc1P as a substrate while AGX2 was 8-fold more active with GlcNAc1P, a result suggesting a role for the 17 amino acid peptide in modifying the enzyme substrate specificity (Wang-Gillam et al., 1998).
The significant sequence conservation observed between eukaryotic UAPs and UDPglucose pyrophosphorylases (UDPGlc Ppase) suggests that these proteins share a common fold (Figure 1). The recently solved crystal structures of Escherichia coli and Streptococcus pneumoniae GlmU (EcGlmU and SpGlmU), the bacterial UAP, revealed a GlmU Ppase domain reminiscent of the typical Rossmann fold (Brown et al., 1999; Kostrewa et al., 2001; Olsen and Roderick, 2001; Sulzenbacher et al., 2001). A similar domain was also reported in the bacterial dTTP-d-glucose Ppase RmlA (Blankenfeldt et al., 2000) and in a distantly related nucleotidyltransferase, MobA (Lake et al., 2000; Stevenson et al., 2000). This structural homology extends to glycosyltransferases such as SpsA (Charnock and Davies, 1999) and GnT I (Unligil et al., 2000), thereby defining a new family of nucleotide-binding proteins named the SpsA-GnT I core (SGC) superfamily (Unligil et al., 2000). Except for the signature motif LX2GXGTX6PK, shown by mutagenesis experiments to be essential for the enzymatic activity of Ppases (Mio et al., 1998; Wang-Gillam et al., 2000), no other sequence similarity could be detected between the bacterial enzyme and its eukaryotic counterparts (Figure 1). Extensive mutagenesis experiments performed on the eukaryotic enzymes led to the identification of putative nucleotide-sugar-binding and catalytic residues (Mio et al., 1998; Wang-Gillam et al., 2000). However, in the absence of structural template, details of the catalytic machinery, the topology of the active site and the role of the extra 17 amino acid peptide of AGX2 in catalysis remain unclear.
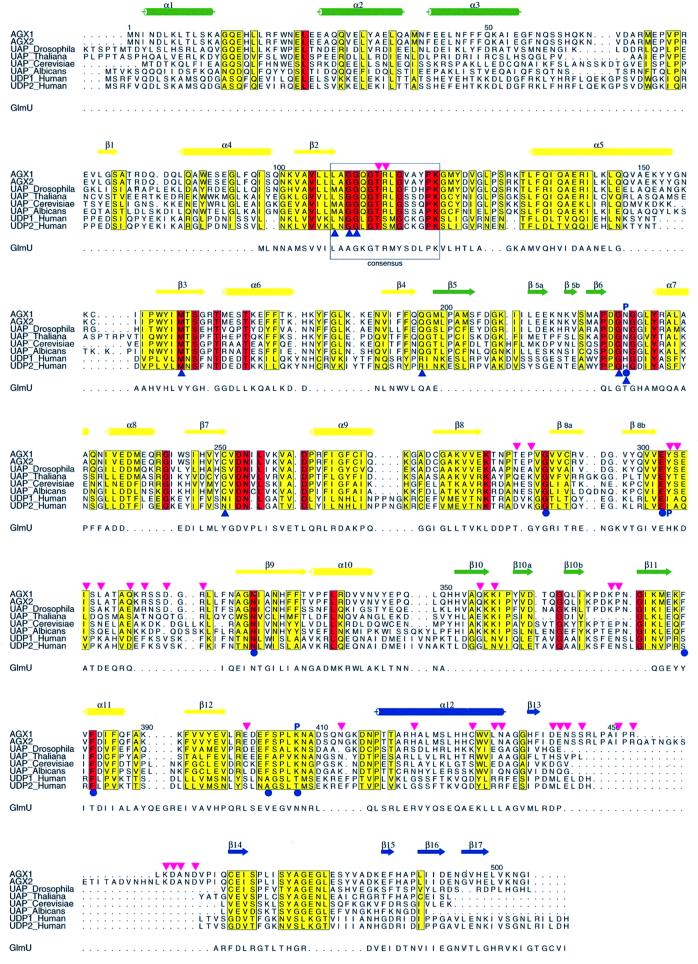
Fig. 1. Sequence alignment of pyrophosphorylases. The sequence alignment of AGX1 and AGX2 with other eukaryotic UAPs and two human UDPGlc Ppases (UDP1 and UDP2) is presented as well as that with GlmU, based on a structural comparison. Conserved and homologous residues between the eukaryotic enzymes are highlighted with a red and yellow background, respectively. The AGX secondary structure elements are shown above the sequences, with those forming the central core in yellow, and those part of the N- and C-terminal domains in green and blue, respectively. AGX residues involved in nucleotide-sugar binding are indicated by blue circles, blue triangles and a letter P for those contacting the nucleotide, the sugar and the phosphate moiety, respectively. The Ppase consensus sequence motif is boxed. Pink triangles identify residues buried at the dimer interface.
To address these questions, we have overexpressed and purified full-length AGX1 and AGX2, characterized them in solution and solved their crystal structures in complexes with UDPGlcNAc and UDPGalNAc. These four structures reveal an AGX fold composed of three domains, with a large central domain similar to that found in members of the SGC superfamily, and provide a template for related eukaryotic Ppases. AGX1 and AGX2 bind UDPGlcNAc and UDPGalNAc similarly, a result consistent with our enzyme kinetic data that did not show the change in substrate specificity reported by others (Wang-Gillam et al., 1998). Moreover, our structural and biochemical data indicate that AGX1 and AGX2 differ in their oligomeric arrangement in solution. This difference, which results in a modified active site architecture, suggests that the alternative splicing event could influence the activity of the enzyme. Finally, structural comparison with related members of the SGC superfamily, together with mutagenesis data, also provide insights into the catalytic mechanism of the enzyme.
Results and discussion
Structure determination
The crystal structure of full-length AGX1 was determined by the multiwavelength anomalous dispersion (MAD) phasing method using selenomethionine-substituted protein. The model could be readily traced from residue 1 to 500, with the exception of a disordered surface loop encompassing residues Glu54–Arg68. The structure was refined against a native protein data set at 1.9 Å resolution to an R-factor of 18.2% and Rfree of 22% and has an excellent stereochemistry. Although no UDPGlcNAc was added to the purification or the crystallization buffers, a clear unbiased density corresponding to UDPGlcNAc was observed in the active site, suggesting that the sugar nucleotide was captured by the enzyme in the cytoplasm of the E.coli expression cells.
Full-length native AGX2 crystallized in similar conditions and identical space group as AGX1 (see Materials and methods). However, crystals of AGX2 only grew in the presence of UDPGlcNAc, UDPGalNAc or GlcNAc1P, suggesting that AGX2, unlike AGX1, did not capture the UDPGlcNAc present in the E.coli cytoplasm. The structure of AGX2 in complex with UDPGlcNAc was obtained by molecular replacement using AGX1 as a template and refined to 2.4 Å resolution. The structures of UDPGalNAc-complexed AGX1 and AGX2 are also reported. Data collection and refinement statistics are presented in Table I.
Table I. Structural statistics.
| Data collectiona |
f′′ max |
f′ min |
Remote |
High resolution |
||||
| SeAGX1–GlcNAc1P |
SeAGX1–GlcNAc1P |
SeAGX1–GlcNAc1P |
SeAGX1–GlcNAc1P |
AGX1–UDPGlcNAc |
AGX1–UDPGalNAc |
AGX2–UDPGlcNAc |
AGX2–UDPGalNAc |
|
| beamline | BM14 | BM14 | BM14 | ID14-3 | ID14-2 | ID14-2 | ID14-3 | ID14-4 |
| |
||||||||
| λ (Å) | 0.9785 | 0.9787 | 0.8856 | 0.9310 | 0.9330 | 0.9330 | 0.9310 | 0.9464 |
| resolution (Å) | 2.9 | 2.9 | 2.9 | 2.6 | 1.9 | 2.1 | 2.4 | 2.3 |
| Rsymb (%) | 10.4 | 9.6 | 12.6 | 8.3 (28.2) | 5.6 (30.5) | 6.6 (37.3) | 6.2 (27.3) | 6.2 (25.8) |
| Ranomc (%) | 9 | 8 | 9.3 | – | – | – | – | – |
| I/σ (I) | 6.6 | 7.6 | 5.7 | 4 (1.8) | 10.6 (2.2) | 7.5 (1.8) | 6.2 (2.6) | 8.2 (1.8) |
| completeness (%) | 98.1 | 96.2 | 93.9 | 97 (95.1) | 99.6 (97.4) | 97.6 (98.2) | 99.8 (99.8) | |
| anomalous completeness | 95.9 | 91.8 | 88.9 | – | – | – | – | – |
| |
||||||||
| Phasing statistics | ||||||||
| phasing powerd (centric/acentric) | 0.17/0.22 | 0.40/0.57 | 0.00/0.00 | |||||
| mean figure of merit (centric/acentric/all) | 0.226/0.257/0.255 | |||||||
| |
||||||||
| Refinement statistics | ||||||||
| AGX1–UDPGlcNAc |
AGX1–UDPGalNAc |
AGX2–UDPGlcNAc |
AGX2–UDPGalNAc |
|
| resolution range (Å) | 25–1.9 | 25–2.2 | 20–2.4 | 20–2.3 |
| Rfactor/Rfree (%) | 18.2/22 | 18.9/23.3 | 19.3/25.1 | 20.1/23.9 |
| total reflectionse | 90 377 (2706) | 56 389 (1684) | 43 565 (2180) | 49 655 (1986) |
| total atoms (protein/ligands/solvent) | 7817/78/844 | 7786/78/615 | 7674/78/310 | 7673/78/310 |
| Mean B-factors (Å2) | ||||
| main chain/side chain | 22.2/26.2 | 35.4/38.1 | 37.2/39.2 | 33.2/36.1 |
| solvent/ligand | 32.6/18.2 | 39.3/30.7 | 36.3/32.9 | 32.2/27.3 |
| r.m.s.d. bonds (Å) | 0.017 | 0.011 | 0.015 | 0.012 |
| r.m.s.d. angles/dihedrals/impropers (°) | 1.7/24.3/1.06 | 1.5/24.0/1.57 | 2.0/24.8/1.2 | 1.5/24.0/1.58 |
aNumbers in parentheses refer to values for the highest resolution shell.
bRsym = Σ|I – <I>|/Σ|I|, where I is intensity and <I> is the average I for all equivalent reflections.
cRanom = Σ|<I+> – <I–>|/Σ(|<I+> + <I–>|), where I+ and I– are the intensities of Bijvoet positive and negative reflections, respectively.
dPhasing power = <|FH(calc)|>/<E>, where E is the residual lack of closure error.
eValues in parentheses refer to the numbers of reflections used in the random test set.
Overall structure
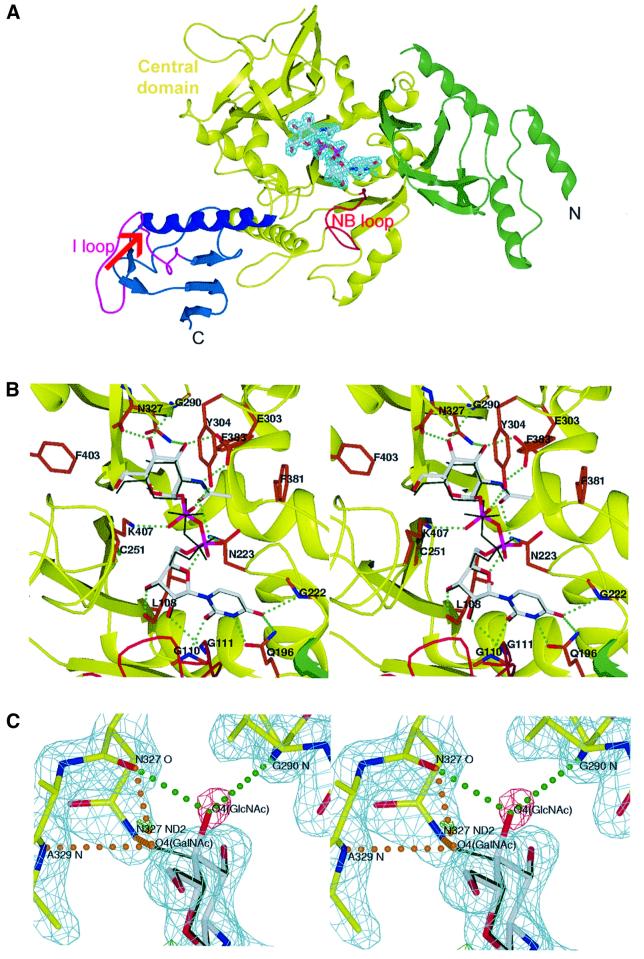
AGX1 and AGX2 share a common fold composed of a large central core flanked by two smaller extra domains, located at each end of the polypeptide chain and defined as the N- and C-terminal domains (Figure 2A). The central main domain displays an α/β structure resembling the Rossmann fold (Rossmann et al., 1975). It consists of a central mixed eight-stranded β-sheet with strand order 1-12-8-9-7-2-3-4, surrounded by eight helices (α4–α11) and topped at one end by a small two-stranded β-sheet (β8a, β8b). The small C-terminal extra domain comprises the last 68 residues and consists of a β-sheet structure connected to the central domain via a long 18 residue α-helix (α12). In contrast to the C-terminal domain, the N-terminal domain is formed by discontinuous segments in the amino acid sequence (Figure 1). It encompasses the first 55 residues and two additional segments (residues 198–220 and 352–379), which protrude from the central core. These segments form a four-stranded antiparallel β-sheet flanked by three α-helices on one side and topped by two short β-fingers. The fold of this N-terminal domain was screened against the DALI database of known structures (Holm and Sander, 1995) and no homologous structure could be found; hence this domain is likely to define a new fold.
Fig. 2. The nucleotide sugar-complexed structures of AGX1. (A) Ribbon representation of the structure of UDPGlcNAc-complexed AGX1, colour-coded as in Figure 1. UDPGlcNAc is shown in its final 2Fo – Fc electron density map (in cyan, contoured at 2σ) with white carbon, blue nitrogen, red oxygen and purple phosphorus atoms. The loop (NB loop) found in all Ppases is highlighted in red. The α12–β14 loop (I loop), which contains the site of insertion (indicated by a red arrow) of the AGX2 17 amino acid peptide, is shown in purple. (B) Stereo view of the active site of the AGX1–UDPGlcNAc complex structure colour-coded as in (A) with residues involved in nucleotide-sugar binding displayed with orange carbon atoms. The superimposed UDPGalNAc molecule from the AGX1–UDPGalNAc complex structure is shown in thin dark green sticks. Green dotted lines indicate hydrogen bonds. (C) Stereo view of the final 2Fo – Fc electron density map (in cyan, contoured at 2σ) of AGX1 in complex with UDPGalNAc (shown as in B). The positive and negative peaks in the Fo – Fc electron density map (–3.5σ in green and 3.5σ in red) clearly indicate the partial substitution of UDPGlcNAc (shown as in A) by UDPGalNAc in the crystal. Hydrogen bonds found between protein residues and the sugar-O4 in the equatorial (UDPGlcNAc) or axial (UDPGalNAc) conformations are indicated by green and orange dotted lines, respectively.
Nucleotide-sugar-binding site
Although AGX1 and AGX2 have been reported to have different substrate specificities (Wang-Gillam et al., 1998), the mode of nucleotide-sugar binding is similar in the four crystal structures of UDPGlcNAc- and UDPGalNAc-complexed AGX1 and AGX2. The nucleotide-sugar binds at the bottom of a deep pocket located at the centre of the main core domain (Figure 2B). The nucleotide moiety establishes a majority of contacts with the first half of the central domain (residues 68–260) and, in particular, with the loop containing the Ppase consensus motif (consequently named the NB loop for nucleotide-binding loop) and a second loop comprising residues Asp221–Leu226, a result in agreement with previous mutagenesis data (Mio et al., 1998; Wang-Gillam et al., 1998). The sugar ring is stabilized mainly via hydrogen bonds to residues from the second half of the central domain (residues 261–417) (Table II).
Table II. The UDPGlc(Gal)NAc-binding site; the protein atoms that are hydrogen-bonded to the nucleotide-sugar in the structure of AGX1-UDPGlc(Gal)NAc are listeda.
| Interacting atoms | Distance (Å) | Interacting atoms | Distance (Å) | ||
|---|---|---|---|---|---|
| Uracil N3 | Q196 Oε1 | 2.6 | Glc(Gal)NAcO7 | N223 Nδ2 | 2.8 |
| Uracil O2 | G110 N | 2.9 | Glc(Gal)NAc N2 | E303 Oε2 | 2.8 |
| Uracil O4 | G222 N | 3.0 | Glc(Gal)NAc O3 | E303 Oε1 | 2.7 |
| Ribose O2′ | G111 N | 3.2 | Glc(Gal)NAc O3 | N327 Nδ2 | 2.7 |
| Ribose O2′ | H2O 296 | 2.8 | GlcNAc O4 | G290 N | 3.0 |
| Ribose O3′ | L108 N | 2.7 | GlcNAc O4 | N327 O | 2.6 |
| Ribose O4′ | N223 Nδ2 | 3.1 | GalNAc O4 | N327 Nδ2 | 3.0 |
| α-Phosphate O1α | H2O 80 | 2.7 | GalNAc O4 | N327 O | 2.6 |
| α-Phosphate O2α | K455 Nζb | 2.7 | Glc(Gal)NAc O6 | K407 Nζ | 3.0 |
| α-Phosphate O2α | H2O 900 | 3.0 | Glc(Gal)NAc O6 | H2O 44 | 3.0 |
| β-Phosphate O1β | R453 Nh2b | 3.6 | Glc(Gal)NAc O6 | H2O 226 | 2.8 |
| β-Phosphate O1β | K455 Nζb | 2.8 | H2O 226 | P288 O | 3.1 |
| β-Phosphate O1β | Y304 OH | 2.8 | H2O 80 | N223 N | 2.8 |
| β-Phosphate O2β | H2O 899 | 2.9 | H2O 80 | E379 Oε1 | 3.2 |
| β-Phosphate O2β | K407 Nζ | 2.6 | H2O 12 | N327 Nδ2 | 2.9 |
| Glc(Gal)NAc O7 | H2O 12 | 2.9 | H2O 44 | D253 Oδ1 | 2.7 |
aThis list (distance values and solvent molecule numbers) refers to the binding of the nucleotide-sugar to subunit A of the dimer in structures 1jv1 and 1jv3 (italic).
bThis residue belongs to the I loop of the second subunit in the dimer.
The protein accommodates either the GlcNAc or GalNAc moieties without any significant structural rearrangement. Indeed, the hydroxyl group at the sugar C-4 can bind either in an equatorial conformation, i.e. hydrogen-bonded to Gly290 and Asn327 backbone atoms, as found in the structures of the UDPGlcNAc–AGX1/2 complexes, or in the axial conformation, i.e. inserted between the Asn327 side chain and the Ala329 backbone amide, as found in the UDPGalNAc–AGX1/2 complex structures (Figure 2C). The sugar N-acetyl arm establishes numerous hydrogen bonds with Glu303, His331 and Asn223, and hydrophobic interactions with Phe381 and Phe383 (Table II). These extensive contacts suggest a specificity of the enzyme for acetylated hexosamine, a feature consistent with the substrate specificity of the pig and Saccharomyces cerevisiae UAPs (Szumilo et al., 1996; Mio et al., 1998). In contrast to the sugar or nucleotide moieties, the phosphate groups are less well stabilized and present static disorder (Figure 2B) (B-factors of 27.8 and 35.4 Å2 for the Pα atoms of the two UDPGlcNAc molecules bound to each AGX1 molecule in the dimer).
The structural similarities between the nucleotide-sugar-complexed AGX1 and AGX2 are in agreement with our kinetic analysis of the human UAPs, which shows no significant change of specificity between the two enzymes. Indeed, both AGX1 and AGX2 have UDPGlcNAc and UDPGalNAc Ppases activities with UDPGlcNAc as the preferred substrate (Table III). Although the substrate specificity could be characterized in the reverse reaction only, these enzymes are unlikely to behave differently in the forward direction. These results are in contradiction to the difference in substrate specificity reported by others (Wang-Gillam et al., 1998) and thus re-address the biological implication of the AGX gene alternative splicing event.
Table III. Enzymatic activities of wild-type and mutant AGX1 and AGX2.
| Wild-type enzymesa | Substrate | AGX1 |
AGX2 |
||
|---|---|---|---|---|---|
| Km (µM) | kcat/Km (µM−1 s−1) | Km (µM) | kcat/Km (µM−1 s−1) | ||
| Forward | UTP | 53 | 1.2 | 49 | 1.4 |
| GlcNAc1P | 5.3 | 12.2 | 6 | 11.2 | |
| Reverse | UDPGlcNAc | 32 | 2.0 | 32 | 2.2 |
| UDPGalNAc | 500 | 0.18 | 530 | 0.2 | |
| Mutagenesis data | AGX1 |
AGX2 |
|||
| |
|
wild-type |
R115A |
wild-type |
R115A |
| Specific activityb (µmol/min/mg) | 69 | 1.5 | 68 | 0.9 | |
aSee Materials and methods for conditions of the experiments.
bThe activity was measured at fixed GlcNAc1P and UTP concentrations of 250 µM and 1 mM, respectively.
The 17 amino acid insertion modifies the oligomeric assembly
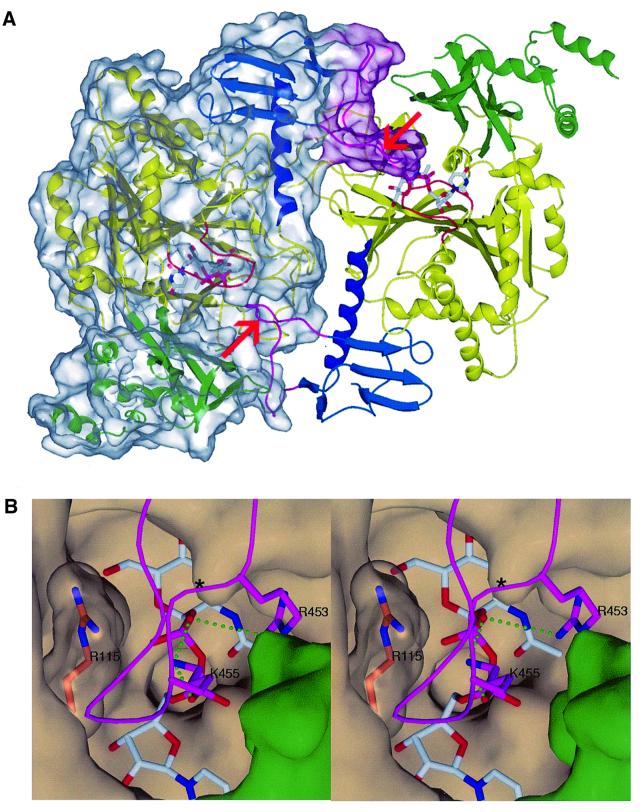
The AGX1 structure reveals a dimeric arrangement of molecules in the crystal: the two molecules are positioned very close to each other, burying a total protein surface area of ∼2920 Å2, a value consistent with a biological dimeric assembly (Figure 3A) (Janin, 1995). Interestingly, in the AGX1 C-terminal domain, the loop in which the extra 17 residues insert in AGX2 (named the I loop for insertion loop) largely contributes to the dimer formation. In the dimer, the I loop of one subunit inserts into the active site pocket of the second subunit, burying 875 Å2 of protein surface area. This loop is stabilized by extensive intersubunit contacts and also provides additional nucleotide-sugar-binding residues, such as Lys455 and Arg453 (Figure 3B). A similar dimer is observed in the AGX2 structures, but the I loop is ill defined in the electron density maps and the extra 17 amino acid segment could not be traced. Since this loop has not been cleaved (as verified by mass spectrometry), it is disordered in the crystal structures of AGX2.
Fig. 3. The AGX1 dimer. (A) Ribbon representation of the AGX1 dimer. Each subunit is coloured as in Figure 2A. One subunit is shown with a transparent molecular surface. The red arrows indicate the site of insertion of the 17 residue peptide in AGX2 in each subunit. (B) Stereo view of the AGX1 UDPGlcNAc-binding pocket of one subunit occluded by the I loop of the other subunit. I loop residues Lys455 and Arg453 (purple carbon atoms) are hydrogen-bonded to UDPGlcNAc phosphate groups. Arg115, which lies at the dimer interface, is shown under a transparent surface with orange carbon atoms. An asterisk indicates the site of insertion of the 17 residue peptide in AGX2. The protein surface corresponding to the central core and the N-terminal domains is coloured in beige and green, respectively.
Gel filtration experiments performed in ionic strength conditions close to the average physiological level (i.e. 150 mM NaCl) show that, in the 0.5–10 µM concentration range, the majority of AGX1 is dimeric whereas AGX2 is exclusively monomeric (see Materials and methods). However, diluting the protein to a concentration of 1 nM or increasing the NaCl concentration to 500 mM led to the dissociation of the AGX1 dimer. These data are in agreement with the large number of electrostatic interactions found at the crystalline dimer interface, thereby suggesting that the AGX1 dimer in solution corresponds to the crystalline dimer. In contrast, the crystallographic dimer of AGX2 was not observed in solution, a result which indicates that the additional bulky and flexible 17 residue peptide hampers dimer formation in solution.
These results provide evidence for a role for the 17 amino acid insertion in dictating the oligomeric assembly of the enzyme in solution. Most importantly, our structural data show that the dimerization process significantly modifies the structural environment of the active site: whilst wide open in monomeric AGX2, this site is occluded at the AGX1 dimer interface. Moreover, the presence of the alternatively spliced loop (I loop) at the dimer interface of AGX1, in the vicinity of the active site with Lys455 and Arg453 salt-bridged to UDPGlcNAc, contrasts with the position of the equivalent residues which, in monomeric AGX2, are situated ∼32 Å away from the active site. These structural differences support a functional role for the I loop and, overall, an influence of the dimerization process on the human UAP enzymatic activity. Such a biological implication of the alternative splicing in modulating the activity of protein isoforms via a modification of the dimerization properties has already been described; it prevails particularly in gene transcription control by alternatively spliced transcriptional factor isoforms (Lopez, 1995). Whether the AGX1 dimer formation in solution is physiologically relevant and whether it is competent for catalysis remain to be investigated. Indeed, structural rearrangements are required at the dimer interface to allow trafficking of substrates and products. AGX1 dimer dissociation occurs in the nanomolar concentration range, as deduced from our gel filtration experiments. Thus, in the experimental conditions of the kinetic assay, both AGX1 and AGX2 are monomeric, a result that explains why the kinetics of the two isoforms are very similar, despite the observed structural differences.
Structural comparison
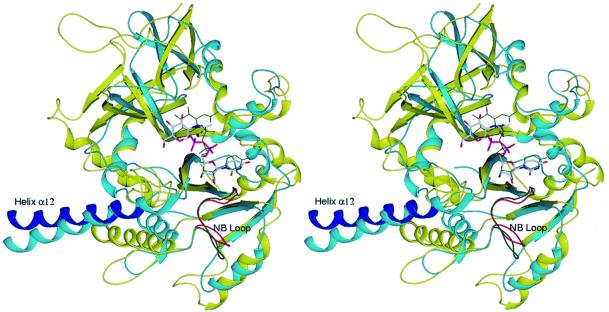
Comparison of the AGX fold with known structures revealed high homologies with other Ppases/nucleotidyltransferases such as GlmU (Brown et al., 1999; Kostrewa et al., 2001; Olsen and Roderick, 2001; Sulzenbacher et al., 2001), the Glc1P thymidylytransferase (RmlA) (Blankenfeldt et al., 2000), the CMP-acylneuraminate (CMP-NeuAc) synthetase (Mosimann et al., 2000) and the distantly related MobA, an enzyme synthesizing molybdopterin guanine dinucleotide (Lake et al., 2000; Stevenson et al., 2000). These homologies mainly concern the Rossmann fold-like domains of AGX1/2, RmlA, CMP-NeuAc synthetase, MobA and GlmU, but interesting similarities are also observed beyond that domain (Figure 4). In particular, the long helix at the start of the C-terminal domain of AGX1/2 is also found in RmlA (helix α9), CMP-NeuAc synthetase (helix αI), MobA (helix α7, although significantly shorter) and GlmU, in which it corresponds to the long helical arm (helix α9) connecting the LβH structure to the Ppase domain. The remarkable conservation of these structural elements between these four enzymes, which otherwise show no significant sequence identity, not only suggests a common ancestor but also highlights the importance of these elements for the Ppase/nucleotidyltransferase activity.
Fig. 4. Structural alignment of the central domain of AGX1 and the Ppase domain of GlmU. Ribbon representations of the superimposed structures of the UDPGlcNAc-complexed forms of AGX1 and EcGlmU, which aligned with an r.m.s.d. of 1.8 Å over 180 Cα. AGX1 is coloured as in Figure 2A whilst GlmU is shown in cyan with bound UDPGlcNAc in green thin sticks and the NB loop highlighted in green.
More distant but still significant structural homologies have also been detected between AGX1/2 and glycosyltransferases from the SGC superfamily (Unligil and Rini, 2000; Unligil et al., 2000). Structural alignments of AGX1 with SpsA (Charnock and Davies, 1999) and GnT I (Unligil et al., 2000) (r.m.s.d. of 2.3 Å over 90 Cα atoms) show that the central domain of AGX1/2 possesses most of the secondary structural elements characterizing the SGC domain (i.e. central β-sheet plus α-helices 5, 6, 9, 10 and 11), thereby defining AGX1 and AGX2 as new members of the SGC superfamily. These results, together with the sequence conservation observed between AGX1/2 and UDPGlc Ppases (Figure 1), suggest a wide representation of the SGC domain not only among glycosyltransferases (Unligil and Rini, 2000; Unligil et al., 2000) but also among nucleotide-sugar Ppases and, overall, possibly XDP-sugar-binding enzymes.
The mode of binding of the nucleotide entity in AGX1/2 is similar to that observed in other members of the SGC superfamily. On the other hand, the mode of sugar binding differs markedly between AGX1/2 and glycosyltransferases from this superfamily, but is comparable with that of GlmU; comparisons were not possible with MobA and CMP-NeuAc synthetase (their modes of sugar substrate binding have not been determined) and with RmlA (the coordinates of the RmlA–dTTP-d-glucose complex structure are not available). Superimposition of the UDPGlcNAc-complexed structures of AGX1 and EcGlmU (Brown et al., 1999) shows that whilst the nucleotide moieties are almost perfectly aligned, the positions of the sugar moieties are shifted by ∼2.5 Å (Figure 4). Yet, most of the hydrophobic and hydrogen bonds between the protein and the amino sugar are conserved: of the 16 AGX1/2 and 14 EcGlmU residues contacting UDPGlcNAc, 12, of which seven are strictly conserved, lie in a similar position. In addition, the sequence alignment of AGX1 and eukaryotic UDPGlc Ppases shows that the majority of the nucleotide-sugar-binding residues are conserved (Figure 1). This conservation, however, does not extend to the region encompassing the sugar N-acetyl arm in the nucleotide-sugar-complexed AGX1/2 structures. In particular, residues AGX Ala329 and Asn223 are replaced in the human UDPGlc Ppases by more bulky amino acids (tryptophan and histidine, respectively), a substitution which might hinder binding of the N-acetyl group and account for the substrate specificity of UDPGlc Ppases. In summary, the strong conservation of substrate-binding residues highlighted by this sequence and structural comparison supports the use of the AGX structure as a valuable template for the studies of UDPGlc Ppases and is likely to reflect common catalytic features.
Finally, AGX1/2 diverges from GlmU, RmlA, CMP-NeuAc synthetase, MobA and other members of the SGC superfamily by the presence of its two extra domains and particularly its N-terminal domain, which is unique to the eukaryotic enzymes (Figure 1). This N-terminal domain sits on top of one side of the active site pocket, partly reducing its accessibility (Figure 3B). This position suggests a possible role for this domain in regulating the enzymatic activity, a hypothesis in agreement with a previous report showing that modification at the N-terminal end of the human liver UDPGlc Ppase significantly modifies the kinetic behaviour of the enzyme in vitro (Duggleby et al., 1996).
Implication for catalysis
A single-step mechanism involving the stereochemical inversion at the phosphate α has been proposed for the UDPGlc Ppases (deduced from NMR spectra of thio-substituted nucleotides) (Sheu and Frey, 1978), and for other Ppases such as GlmU, RmlA and ADPGlucose Ppases (based on structural and/or kinetic data) (Paule and Preiss, 1971; Blankenfeldt et al., 2000; Kostrewa et al., 2001). Such a mechanism involves the formation of a P–O bond between a non-esterified oxygen atom of the GlcNAc1P phosphate group and the α-phosphate of UTP, with subsequent release of the UTP β- and γ-phosphate groups as pyrophosphate (Paule and Preiss, 1971; Blankenfeldt et al., 2000; Kostrewa et al., 2001). Given their close structural homology to GlmU and the sequence conservation of most residues involved in substrate binding (Figure 1), AGX1 and AGX2 are likely to use a similar mechanism.
Facilitating the correct positioning of the two negatively charged substrates and stabilizing the presumed pentacoordinate reaction intermediate require the presence of counterbalancing positively charged groups. These include divalent cations, shown to be crucial for the activity of several XDP-sugar Ppases (Fukui et al., 1993). Mg2+ is the best cofactor for the UAP in most organisms studied to date, although the pig liver enzyme is more active with Mn2+ (Szumilo et al., 1996). Co-crystallization experiments were undertaken to localize the position of the cation in the active site but were unsuccessful. Nevertheless, the mode of cation binding has been described for the UDPGlcNAc-complexed EcGlmU and SpGlmU (Kostrewa et al., 2001; Olsen and Roderick, 2001; Sulzenbacher et al., 2001), and for the structurally related glycosyltransferases SpsA (Charnock and Davies, 1999) and GnT I (Unligil et al., 2000), and presents some remarkably well-conserved features allowing us to model the binding of the metal ion in the AGX active site. Octahedral coordination of Mg2+ probably involves the two phosphate groups of UDPGlcNAc, the side chain of the sequence-conserved Asp253 and water molecules.
The active site of AGX1/2 is lined with positively charged residues which could provide additional counterbalancing charges to the substrate phosphate groups during catalysis. In particular, the conserved Lys122, Lys407 and Arg115 are possible candidates. Lys122 is highly conserved in UAPs, GlmU and UDPGlc Ppase, as well as in related nucleotidyltransferases (RmlA, MobA and CMP-NeuAc). A crucial role for the lysine residue in charge compensation of the reaction transition state is supported by the S.cerevisiae UAP (ScUAP) and EcGlmU mutagenesis data as well as recent structural data (Mio et al., 1998; Brown et al., 1999; Blankenfeldt et al., 2000). As for Arg115, an essential role possibly in UTP binding of EcGlmU Arg18 and of ScUAP Arg116 (equivalent to AGX Arg115) is supported by mutagenesis (Mio et al., 1998; Brown et al., 1999) and structural data (Blankenfeldt et al., 2000). Surprisingly, the R115A mutation did not significantly affect the enzymatic activity of AGX1 (Wang-Gillam et al., 2000). Could this unexpected difference result from the dimeric arrangement of AGX1? Arg115 lies at the AGX1 dimer interface where it is liganded stably to residues from both dimer subunits (Figure 3B). A role in substrate binding would thus require Arg115 to move, a positional change which appears unlikely but cannot be excluded since rearrangements at the active site must occur to allow the substrates to enter and the products to exit.
To elucidate the paradox and further investigate the role of Arg115, we replaced it by an alanine residue in AGX1 and in AGX2. Both AGX1 and AGX2 R115A mutant activities were drastically affected (Table III). Moreover, the gel filtration analysis showed that the AGX1 R115A mutant does not associate as a dimer in solution. These results indicate a crucial role for Arg115 for the dimerization of AGX1 as well as for the activity of the monomeric form of the enzyme. The structural and sequence conservation of this arginine suggests a role in UTP binding as proposed for GlmU (unpublished results) or RmlA (Blankenfeldt et al., 2000).
The loss of activity of the AGX1 R115A mutant is in contradiction to previously reported results obtained from a GST–AGX1 fusion protein (Wang-Gillam et al., 2000). Considering that GST is a dimeric enzyme, could the dimeric assembly of the AGX1 R115A mutant be preserved and the activity rescued by residues from the second subunit? Indeed, additional positively charged residues, such as Lys455 and Arg453 of the second subunit, are positioned in the vicinity of the active site of the first subunit via dimerization and could participate in the catalysis of AGX1 (Figure 3B). The role of these residues remains to be investigated, a study which would address simultaneously the possibility of catalytic differences between the two isoforms generated by alternative splicing.
Conclusion
Alternative splicing is a widespread and efficient device for generating protein diversity. The sequence differences between alternatively spliced gene products can concern an entire protein domain or only a few amino acids, and have varied biological consequences ranging from a clear functional switch to subtle modifications of protein function (for reviews see Smith et al., 1989; Graveley, 2001). However, for the vast majority of alternative splicing events, the functional and structural consequences on the gene product remain unknown. Here, we describe for the first time the three-dimensional structures of two human UAP isoforms differing by an alternatively spliced 17 residue segment. Our structural data support the different oligomeric assemblies of the two isoforms observed in solution, and show how this difference modifies the active site architecture. This suggests that subtle functional differences exist between AGX1 and AGX2, although no significant differences could be deduced from our kinetic analysis in solution. A definite conclusion on whether the alternative splicing of the human AGX gene has a major biological role or simply generates functional redundancy must await further studies. In particular, the release of other genome sequences will allow us to examine whether the alternative splicing event has been maintained through evolution and whether the alternatively spliced 17 residue segment and residues Lys455 and Arg453 are conserved. Preliminary studies on the pig liver UAPs suggest that this alternative splicing event also occurs in pigs (Szumilo et al., 1996). Considering the key role of UDPGlcNAc in development (Boehmelt et al., 2000) and the possible implication of protein O-GlcNAc modifications in signal transduction cascades (Wells et al., 2001), tight regulation of UDPGlcNAc synthesis might be of great benefit to the cell. Whether the AGX gene alternative splicing plays a role in such a regulation merits being assessed experimentally.
Materials and methods
Expression and purification of the native and selenomethionyl proteins
The nucleotide sequences for full-length human AGX1 from astrocytes and AGX2 from cartilage differ from those referenced in the literature by two single base differences changing Ser445 into Gly445 in the AGX1 and AGX2 amino acid sequences, and Gln454 into Ser454 in AGX2 only (inspection of the human Expressed Sequence Tags database indicates that these mutations most probably reflect gene polymorphism rather than a cloning artefact). The AGX1 and AGX2 genes were subcloned into the pRU277 expression vector and transformed into Xl1 blue E.coli cells. Protein expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 20 h at 25–28°C, in RFB MIL11/04 medium (Korz et al., 1995) supplemented with 60 g/l glycerol and 20 g/l casamino acids. The recombinant enzymes were purified by anion exchange, phenyl interaction and size exclusion chromatographies and dialysed against 10 mM Tris–HCl pH 7.6, 2.5 mM dithiothreitol (DTT). The protein solutions of AGX1 and AGX2 were concentrated to 6.6 and 4 mg/ml, respectively.
For production of selenomethionyl AGX1 (Se-AGX1), the pRU277-AGX1 plasmid was transferred to a Met A-β 180 E.coli strain (Barton, 1993); protein expression was performed as described above, except that selenomethionine was added to the medium instead of casamino acids. The purification procedure was identical to that used for the native enzyme.
Site-directed mutagenesis
Site-directed mutagenesis was performed on the pRU277-AGX1 and pRU277-AGX2 plasmids using a Quickchange Site-directed Mutagenesis Kit (Stratagene), according to the manufacturer’s instructions. Codons for AGX1 and AGX2 Arg115 were mutated to alanine using the appropriate oligonucleotides. All the mutations were verified by sequencing. The mutants were expressed as described above and purified via anion exchange and phenyl interaction chromatographies. The mutants activities were assayed in the forward direction as described below.
Enzymatic assays
Kinetic studies were performed at 30°C in 100 mM Tris–HCl pH 8.0, 5 mM MgCl2, 20 mM KCl. The activity was measured in the forward direction by coupling with 2.5 U/ml yeast inorganic pyrophosphatase (Sigma), 2.5 U/ml purine nucleoside phosphorylase and 0.2 mM 2-amino-6-mercapto-7-methyl-purine riboside, available from the EnzChek™ Phosphate Assay Kit (Molecular Probes). The formation of the final product was monitored at λ = 360 nm using a Spectramax microplate reader (Molecular Devices). For Km determinations, either the GlcNAc1P concentration varied from 2.5 to 250 µM at a fixed concentration of 1 mM UTP, or the UTP concentration varied from 25 µM to 2 mM at a constant concentration of 250 µM GlcNAc1P. The enzyme concentration was 50 ng/ml. The activity in the reverse direction was followed by HPLC (Waters 2690, 996 detector) on a PL SAX anion exchange column (8 µm, 1000 Å, 4.6 mm × 150 mm, Polymer Laboratories). Elution of UDPGlcNAc (or UDPGalNAc), UTP and UDP used a linear gradient from 17.5 to 100% buffer B (500 mM Na2H2PO4 pH 4) in buffer A (100 mM Na2H2PO4 pH 4) for 3 min (flow rate 1 ml/min), and was monitored by optical density measurements in the 200–400 nm range. For Km determinations, UDPGlcNAc and UDPGalNAc concentrations varied from 0 to 500 µM and 10 mM, respectively, at a fixed concentration of 500 µM PPi. The UDPGlcNAc and UDPGalNAc activities were measured with a final enzyme concentration of 25 and 50 ng/ml, respectively. The reaction was stopped by addition of 1 vol of 2% phosphoric acid.
Gel filtration chromatography
Gel filtration chromatographies of AGX1 and AGX2 were performed at 8°C by FPLC on a pre-packed Superdex S200 HR 10/30 column (Amersham Pharmacia Biotech), equilibrated with 10 mM Tris–HCl pH 7.6, 2.5 mM DTT and variable amounts of NaCl (50, 150 and 500 mM). Aldolase (158 kDa), ovalbumin (43 kDa) and chymotrypsinogen (25 kDa) were used as molecular weight standards. AGX2 always eluted as a single peak with an apparent mol. wt of 74 kDa; AGX1 eluted as one major peak (apparent mol. wt 120 kDa) at 50 and 150 mM NaCl and as two peaks (apparent mol. wts 74 and 120 kDa) at 500 mM NaCl. The presence of magnesium, temperature increase (25–30°C) or prior incubation of the protein with its substrates did not modify these elution patterns. In the conditions of the kinetic experiments (1 nM protein concentration in 100 mM Tris–HCl, 20 mM KCl, 5 mM MgCl2), AGX1 eluted as a single peak (apparent mol. wt 74 kDa) which was detected using the enzymatic assay described above. The AGX1 and AGX2 R115A mutants were analysed at 150 mM NaCl.
Crystallization
Crystals were grown using the vapour diffusion technique at 20°C in hanging drops over 34% PEG600, 0.2 M imidazole/malate pH 5.7. The drops were prepared by mixing equal volumes of the protein and the reservoir solutions. Thin plate-shaped AGX1 crystals appeared within a week; they belong to the monoclinic space group P21 with cell dimensions a = 86.4, b = 71.0, c = 95.7, β = 95.4 and two molecules per asymmetric unit. AGX2 crystallized in similar conditions with identical space group but slightly different cell parameters (a = 86.5, b = 71.3, c = 91.4, β = 92.8). In contrast to AGX1, AGX2 crystals only grew in the presence of 2 mM UDPGlcNAc, UDPGalNAc or GlcNAc1P. Growth of Se-AGX1 crystals required the presence of 2 mM UDPGlcNAc/UDPGalNAc or GlcNAc1P. Crystals of the AGX2– UDPGalNAc complex were obtained by incubating AGX2 overnight with 2 mM UDPGalNAc prior to the crystallization assays. In contrast, to obtain the AGX1–UDPGalNAc complex crystals, AGX1 was incubated with 50 mM UDPGalNAc in the presence of 500 mM NaCl in order to dissociate the AGX1 dimer. This protein solution was then dialysed extensively against 10 mM Tris–HCl pH 7.6, 2 mM UDPGalNAc, 2.5 mM DTT, to eliminate NaCl prior to the crystallization assays. For data collection, all crystals were frozen in liquid nitrogen without additional cryoprotectant.
Data collection
Data were collected at the ESRF on the BM14 and ID14 beamlines (Grenoble, France). A three-wavelength MAD experiment was performed at beamline BM14 on a GlcNAc1P-complexed Se-AGX1 crystal at 2.9 Å resolution. High resolution data sets were collected at 2.6 and 1.85 Å resolution on a Se-AGX1-GlcNAc1P complex crystal and a native AGX1 crystal, respectively. Diffraction data were also collected from crystals of AGX2 complexed with either UDPGlcNAc, UDPGalNAc or GlcNAc1P and of AGX1 co-crystallized with UDPGalNAc or GlcNAc1P. Data collection statistics are presented in Table I.
Structure solution and refinement
All data were processed and reduced using DENZO (Otwinowski and Minor, 1997) and the CCP4 program suite (CCP4, 1994). The positions of 20 out of the 22 Se atoms present in the asymmetric unit of the Se-AGX1 crystal were determined using the program Shake n’Bake (Miller et al., 1994) and refined with MLPHARE (CCP4, 1994) which was used subsequently to calculate the phases. The resulting experimental MAD electron density map was improved by solvent flattening, non-crystallographic symmetry (NCS) averaging and phase extension with DM (CCP4, 1994) using the 2.6 Å resolution data set collected from a Se-AGX1–GlcNAc1P crystal. The AGX1 structure was built from the resulting 2.6 Å map using TURBO-FRODO (Roussel and Cambillau, 1991) and refined against the 1.85 Å resolution native protein data set using CNS (Brünger et al., 1998), including bulk solvent and anisotropic B-factor corrections. No NCS restraints were applied. Although AGX1 was not crystallized in the presence of UDPGlcNAc, Fourier difference maps clearly revealed bound UDPGlcNAc in the two molecules of the asymmetric unit. High temperature factors and weak electron density are associated with residues in the α1–α2 loop of one molecule in the asymmetric unit, while, in both molecules, the region from Glu54 to Arg68 was ill defined and could not be modelled. The two molecules have an average r.m.s.d. of 0.5 Å for all Cα atoms. The structure of AGX1 co-crystallized with UDPGalNAc (see above) was solved from rigid-body refinement using the AGX1–UDPGlcNAc complex as a starting model, and refined at 2.2 Å resolution. Fourier difference maps show that the UDPGlcNAc present in the active site of AGX1 has only been partly replaced by UDPGalNAc, an observation consistent with the partial dissociation deduced from the gel filtration experiments.
The structure of AGX2 in complex with UDPGlcNAc was solved by the molecular replacement method with the program AMORE (Navaza, 1994) using the AGX1–UDPGlcNAc model, and refined to 2.4 Å resolution. This structure was then used as a starting model to solve the structure of the AGX2–UDPGalNAc complex, which was refined to 2.3 Å resolution. The NB loop was disordered in only one molecule of the asymmetric unit, whilst part of the α3–β1 and I loops could not be modelled in both molecules, which are aligned with an r.m.s.d. of 0.5 Å for all remaining Cα atoms. The crystal structures of the nucleotide-sugar-complexed AGX1 and AGX2 are very similar to each other (r.m.s.d. of 0.77 Å for all remaining Cα atoms). The only significant differences concern conformational positioning in surface loop regions in one subunit of the dimer (Gly113–Tyr120 and Asn408–Gln412) and a slight rigid-body motion (0.9 Å) of the N-terminal and core domains. The structures of GlcNAc1P-complexed AGX1 and AGX2 were found to be similar to the UDPGlcNAc-complexed protein structures.
The stereochemistry of the refined models was analysed by PROCHECK (Laskowski et al., 1993); no residue was found in the disallowed regions of the Ramachandran plot. The coordinates of the AGX1–UDPGlcNAc, AGX1–UDPGalNAc, AGX2–UDPGlcNAc and AGX2–UDPGalNAc complexes have been deposited in the Protein Data Bank (accession codes 1JV1, 1JV3, 1JVD and 1JVG). Figure 1 was generated with Alscript (Barton, 1993) and Figures 2–4 with SPOCK (Christopher, 1998) and Raster3D (Merritt and Bacon, 1997).
Acknowledgments
Acknowledgements
We thank Gerlind Sulzenbacher, Pascale Marchot and Bernard Henrissat for helpful discussions and critical reading of the manuscript; Marie-Thérèse Bocquel, Pascal Sempé, Kieron Brown and Christophe Bignon for support and assistance in the project; James Rini and Aiping Dong for providing us with the coordinates of GnT I and RmlA, respectively; and the ESRF staff, in particular Gordon Leonard and Andy Thompson (BM14), for precious technical support and assistance during X-ray data collection. This work was funded by a GIP-Aventis grant and the CNRS to Y.B. C.P. is holder of a CNRS PhD fellowship.
References
- Barton G.J. (1993) ALSCRIPT: a tool to format multiple sequence alignments. Prot. Eng., 6, 37–40. [DOI] [PubMed] [Google Scholar]
- Blankenfeldt W., Asuncion,M., Lam,J.S. and Naismith,J.H. (2000) The structural basis of the catalytic mechanism and regulation of glucose-1-phosphate thymidylyltransferase (RmlA). EMBO J., 19, 6652–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehmelt G. et al. (2000) Decreased UDPGlcNAc levels abrogate proliferation control in EMeg32-deficient cells. EMBO J., 19, 5092–5104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K., Pompeo,F., Dixon,S., Mengin-Lecreulx,D., Cambillau,C. and Bourne,Y. (1999) Crystal structure of the bifunctional N-acetylglucosamine 1-phosphate uridyltransferase from Escherichia coli: a paradigm for the related pyrophosphorylase superfamily. EMBO J., 18, 4096–4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bulik D.A., Lindmark,D.G. and Jarroll,E.L. (1998) Purification and characterization of UDP-N-acetylglucosamine pyrophosphorylase from encysting Giardia. Mol. Biochem. Parasitol., 95, 135–139. [DOI] [PubMed] [Google Scholar]
- Bulik D.A., van Ophem,P., Manning,J.M., Shen,Z., Newburg,D.S. and Jarroll,E.L. (2000) UDP-N-acetylglucosamine pyrophosphorylase, a key enzyme in encysting Giardia, is allosterically regulated. J. Biol. Chem., 275, 14722–14728. [DOI] [PubMed] [Google Scholar]
- Cabib E., Roberts,R. and Bowers,B. (1982) Synthesis of the yeast cell wall and its regulation. Annu. Rev. Biochem., 51, 763–793. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Charnock S.J. and Davies,G.J. (1999) Structure of the nucleotide-diphospho-sugar transferase, SpsA from Bacillus subtilis, in native and nucleotide-complexed forms. Biochemistry, 38, 6380–6385. [DOI] [PubMed] [Google Scholar]
- Christopher J.A. (1998) SPOCK: The Structural Properties Observation and Calculation Kit Program Manual. The Center for Macromolecular Design, Texas A&M University, College Station, TX.
- Dennis J.W., Granovsky,M. and Warren,C.E. (1999) Protein glycosylation in development and disease. BioEssays, 21, 412–421. [DOI] [PubMed] [Google Scholar]
- Diekman A.B. and Goldberg,E. (1994) Characterization of a human antigen with sera from infertile patients. Biol. Reprod., 50, 1087–1093. [DOI] [PubMed] [Google Scholar]
- Duggleby R.G., Chao,Y.C., Huang,J.G., Peng,H.L. and Chang,H.Y. (1996) Sequence differences between human muscle and liver cDNAs for UDPglucose pyrophosphorylase and kinetic properties of the recombinant enzymes expressed in Escherichia coli. Eur. J. Biochem., 235, 173–179. [DOI] [PubMed] [Google Scholar]
- Fukui T., Kazuta,Y., Katsube,T., Tagaya,M. and Tanizawa,K. (1993) Exploring the active site in UDP-glucose pyrophosphorylase by affinity labelling and site-directed mutagenesis. Biotechnol. Appl. Biochem., 18, 209–216. [PubMed] [Google Scholar]
- Graveley B.R. (2001) Alternative splicing: increasing diversity in the proteomic world. Trends Genet., 17, 100–107. [DOI] [PubMed] [Google Scholar]
- Hakomori S. (1991) Possible functions of tumor-associated carbohydrate antigens. Curr. Opin. Immunol., 3, 646–653. [DOI] [PubMed] [Google Scholar]
- Hart G.W., Haltiwanger,R.S., Holt,G.D. and Kelly,W.G. (1989) Nucleoplasmic and cytoplasmic glycoproteins. Ciba Found Symp., 145, 102–112. [DOI] [PubMed] [Google Scholar]
- Herscovics A. and Orlean,P. (1993) Glycoprotein biosynthesis in yeast. FASEB J., 7, 540–550. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1995) Dali: a network tool for protein structure comparison. Trends Biochem. Sci., 20, 478–480. [DOI] [PubMed] [Google Scholar]
- Janin J. (1995) Principles of protein–protein recognition from structure to thermodynamics. Biochimie, 77, 497–505. [DOI] [PubMed] [Google Scholar]
- Kawai H., Yamamoto,K., Moriguchi,M. and Tochikura,T. (1976) Purification of yeast UDP-N-acetylglucosamine pyrophosphorylase and its stabilization by dithiothreitol. J. Ferment. Technol., 54, 463–465. [Google Scholar]
- Korz D.J., Rinas,U., Hellmuth,K., Sanders,E.A. and Deckwer,W.D. (1995) Simple fed-batch technique for high cell density cultivation of Escherichia coli. J. Biotechnol., 39, 59–65. [DOI] [PubMed] [Google Scholar]
- Kostrewa D., D’Arcy,A., Takacs,B. and Kamber,M. (2001) Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 Å resolution and in complex with UDP-N-acetylglucosamine and Mg(2+) at 1.96 Å resolution. J. Mol. Biol., 305, 279–289. [DOI] [PubMed] [Google Scholar]
- Lake M.W., Temple,C.A., Rajagopalan,K.V. and Schindelin,H. (2000) The crystal structure of the Escherichia coli MobA protein provides insight into molybdopterin guanine dinucleotide biosynthesis. J. Biol. Chem., 275, 40211–40217. [DOI] [PubMed] [Google Scholar]
- Laskowski R., MacArthur,M., Moss,D. and Thornton,J. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 91–97. [Google Scholar]
- Lopez A.J. (1995) Developmental role of transcription factor isoforms generated by alternative splicing. Dev. Biol., 172, 396–411. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Miller R., Gallo,S.M., Khalak,H.G. and Weeks,C.M. (1994) SnB. J. Appl. Crystallogr., 27, 613–621. [Google Scholar]
- Mio T., Yabe,T., Arisawa,M. and Yamada-Okabe,H. (1998) The eukaryotic UDP-N-acetylglucosamine pyrophosphorylases. Gene cloning, protein expression and catalytic mechanism. J. Biol. Chem., 273, 14392–14397. [DOI] [PubMed] [Google Scholar]
- Mosimann S.C., Gilbert,M., Dombrowski,D., To,R., Wakarchuk,W.W. and Strynadka,N.C. (2000) Structure of a sialic acid activating synthetase, CMP acylneuraminate synthetase in the presence and absence of CDP. J. Biol. Chem., 11, 8190–8196. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an automated procedure for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Olsen L.R. and Roderick,S.L. (2001) Structure of the Escherichia coli GlmU pyrophosphorylase and acetyltransferase active sites. Biochemistry, 40, 1913–1921. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pattabiraman T.N. and Bachhawat,B.K. (1961) Purification of uridine diphosphoacetylglucosamine pyrophosphorylase from sheep brain. Biochim. Biophys. Acta, 50, 129–134. [DOI] [PubMed] [Google Scholar]
- Paule M.R. and Preiss,J. (1971) The kinetic mechanism of adenosine diphosphoglucose pyrophosphorylase from Rhodospirrillum rubrum. J. Biol. Chem., 246, 4602–4609. [Google Scholar]
- Raetz C.R.H. (1996) Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles. In Neidhardt,F.C., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd edn. ASM Press, Washington, DC, pp. 1035–1063.
- Rossmann M.G., Liljas,A., Branden,C.-I. and Bansazak,L.J. (1975) Evolutionary and structural relationships among deshydrogenases. In Boyer,P.D. (ed.), The Enzymes, Volume 11. Academic Press, New York, pp. 61–102.
- Roussel A. and Cambillau,C. (1991) The Turbo-Frodo graphics package. Silicon Graphics Geometry Partners Directory. Silicon Graphics Corp., Mountain View, CA, pp. 81.
- Rudd P.M., Elliott,T., Cresswell,P., Wilson,I.A. and Dwek,R.A. (2001) Glycosylation and the immune system. Science, 291, 2370–2376. [DOI] [PubMed] [Google Scholar]
- Sheu K.F. and Frey,P.A. (1978) UDP-glucose pyrophosphorylase. Stereochemical course of the reaction of glucose 1-phosphate with uridine-5′[1-thiotriphosphate]. J. Biol. Chem., 253, 3378–3380. [PubMed] [Google Scholar]
- Smith C.W., Patton,J.G. and Nadal-Ginard,B. (1989) Alternative splicing in the control of gene expression. Annu. Rev. Genet., 23, 527–577. [DOI] [PubMed] [Google Scholar]
- Stevenson C.E., Sargent,F., Buchanan,G., Palmer,T. and Lawson,D.M. (2000) Crystal structure of the molybdenum cofactor biosynthesis protein MobA from Escherichia coli at near-atomic resolution. Structure Fold. Des., 8, 1115–1125. [DOI] [PubMed] [Google Scholar]
- Strominger J.L. and Smith,M.S. (1959) Uridine diphosphoacetylglucosamine pyrophosphorylase. J. Biol. Chem., 234, 1822–1827. [PubMed] [Google Scholar]
- Sulzenbacher G., Gal,L., Peneff,C., Fassy,F. and Bourne,Y. (2001) Crystal structure of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase bound to acetyl-coenzyme A reveals a novel active site architecture. J. Biol. Chem., 276, 11844–11851. [DOI] [PubMed] [Google Scholar]
- Szumilo T., Zeng,Y., Pastuszak,I., Drake,R., Szumilo,H. and Elbein,A.D. (1996) Purification to homogeneity and properties of UDP-GlcNAc (GalNAc) pyrophosphorylase. J. Biol. Chem., 271, 13147–13154. [DOI] [PubMed] [Google Scholar]
- Udenfriend S. and Kodukula,K. (1995) How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu. Rev. Biochem., 64, 563–591. [DOI] [PubMed] [Google Scholar]
- Unligil U.M. and Rini,J.M. (2000) Glycosyltransferase structure and mechanism. Curr. Opin. Struct. Biol., 10, 510–517. [DOI] [PubMed] [Google Scholar]
- Unligil U.M., Zhou,S., Yuwaraj,S., Sarkar,M., Schachter,H. and Rini,J.M. (2000) X-ray crystal structure of rabbit N-acetylglucosaminyltransferase I: catalytic mechanism and a new protein superfamily. EMBO J., 19, 5269–5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang-Gillam A., Pastuszak,I. and Elbein,A.D. (1998) A 17-amino acid insert changes UDP-N-acetylhexosamine pyrophosphorylase specificity from UDP-GalNAc to UDP-GlcNAc. J. Biol. Chem., 273, 27055–27057. [DOI] [PubMed] [Google Scholar]
- Wang-Gillam A., Pastuszak,I., Stewart,M., Drake,R.R. and Elbein,A.D. (2000) Identification and modification of the uridine-binding site of the UDP-GalNAc (GlcNAc) pyrophosphorylase. J. Biol. Chem., 275, 1433–1438. [DOI] [PubMed] [Google Scholar]
- Wells L., Vosseller,K. and Hart,G.W. (2001) Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science, 291, 2376–2378. [DOI] [PubMed] [Google Scholar]
- Yamamoto K., Kawai,H., Moriguchi,M. and Tochikura,T. (1976) Purification and characterization of yeast UDP-N-acetylglucosamine pyrophosphorylase. Agric. Biol. Chem., 40, 2275–2281. [Google Scholar]
- Yamamoto K., Moriguchi,M., Kawai,H. and Tochikura,T. (1979) Purification and some properties of uridine diphosphate N-acetylglucosamine pyrophosphorylase from Neurospora crassa. Can. J. Microbiol., 25, 1381–1386. [DOI] [PubMed] [Google Scholar]