Abstract
Abnormalities in l-glutamic acid (glutamate) and GABA signal transmission have been postulated to play a role in depression, but little is known about the underlying molecular determinants and neural mechanisms. Microarray analysis of specific areas of cerebral cortex from individuals who had suffered from major depressive disorder demonstrated significant down-regulation of SLC1A2 and SLC1A3, two key members of the glutamate/neutral amino acid transporter protein family, SLC1. Similarly, expression of l-glutamate-ammonia ligase, the enzyme that converts glutamate to nontoxic glutamine was significantly decreased. Together, these changes could elevate levels of extracellular glutamate considerably, which is potentially neurotoxic and can affect the efficiency of glutamate signaling. The astroglial distribution of the two glutamate transporters and l-glutamate-ammonia ligase strongly links glia to the pathophysiology of depression and challenges the conventional notion that depression is solely a neuronal disorder. The same cortical areas displayed concomitant up-regulation of several glutamate and GABAA receptor subunits, of which GABAAα1 and GABAAβ3 showed selectivity for individuals who had died by suicide, indicating their potential utility as biomarkers of suicidality. These findings point to previously undiscovered molecular underpinnings of the pathophysiology of major depression and offer potentially new pharmacological targets for treating depression.
Keywords: bipolar disorder, GABAA receptors, glutamate transporters, major depression, suicide
Clinical depression, the phenotypic hallmark of the two leading mood disorders [major depressive disorder (MDD) and bipolar affective disorder (BPD)], is the most common psychiatric illness. It affects ≈121 million people worldwide, with 10–20% of women and 5–12% of men estimated to experience a depressive episode in any 1-year period, and with evidence of suicidality in 15% of those affected (ref. 1 and www.who.int). Yet, the etiology of the depressive disorders is largely unknown, although a joint contribution of genetic, environmental, and social factors is widely acknowledged (2, 3).
l-glutamic acid (glutamate) and GABA are the principal excitatory and inhibitory neurotransmitters in the central nervous system, respectively (4), and evidence suggests that alterations in both of these neurotransmitter systems may contribute to the pathophysiology of depression (5–7). For example, elevated levels of both neurotransmitters have been observed in the cerebral cortex of subjects diagnosed with MDD (8). Pharmacological evidence implicates NMDA receptors in the neuropathology of MDD and in the action of antidepressants (9, 10). Knockout mice and positive modulators of GABAA and GABAB receptors suggest dysfunction of the GABA system in depression (6). Although this evidence begins to implicate these neurotransmitters in major depression, the molecular mechanisms that may cause or contribute to the dysregulation of these systems remain unknown.
In the present study, we asked whether multiple molecular actors involved in the biosynthesis and regulation of these transmitters may be altered in major depression. To this end, we used microarray analysis (11), which permitted the discovery of genes that are differentially expressed in the cerebral cortex of depressed patients relative to healthy controls (12). We then validated these findings by using a combination of tools, including the analysis of an additional brain region in an independent subject cohort, cross-validation with an alternate custom microarray platform, and follow-up with in situ hybridization histochemistry (ISHH) (13).
The results identify a subset of functionally related molecules and suggest a unifying mechanism that could affect glutamate-GABA neurotransmission in depression. We report here the dysregulation in MDD, of a specific subset of genes encoding the glial high-affinity glutamate transporters, SLC1A2 (EAAT2; GLT1) and SLC1A3 (EAAT1; GLAST); glutamate-ammonia ligase (glutamine synthetase; GS; GLUL; GSII; EC 6.3.1.2); and various subunits of glutamate receptors and GABAA receptors. We compare the expression patterns of these genes in MDD versus BPD. We also identify two GABAA receptor subunits showing altered expression specifically in suicidal subjects, regardless of the type of the mood disorder.
Methods
Human Subjects. The demographic data, diagnosis, and medical examination of the subjects, as well as the brain quality control criteria, have been described (14–16). The cohort on which this study is based included seven healthy controls (one female, six male), nine MDD patients (two female, seven male), and six BPD patients (one female, five male). Control subjects had no clinical history of neurological or psychiatric disease or of substance abuse, and the patients had no record of potentially confounding clinical conditions. The use of human subjects in the study was approved by the University of California, Irvine, Institutional Review Board, and informed written consent was obtained from immediate family members of the tissue donors.
Tissue Acquisition and RNA Preparation. Brains were processed and stored frozen at –80°C (17). The experimental procedures used have been published (14–16). Samples of anterior cingulate cortex (AnCg; area 24) and left dorsolateral prefrontal cortex (DLPFC; areas 9 and 46) were dissected from the frozen brains. Total RNA was extracted (18), labeled, and hybridized (19) to Affymetrix (Santa Clara, CA) GeneChips (HG_U95Av2, each containing 12,626 probe sets).
Microarray Data Analysis. The built-in and spiked controls, 3′/5′ ratios of housekeeping genes, scaling factor, and raw noise (Q) levels documented in the report (RPT) file of each GeneChip were used for determining technical variability and the quality of array hybridizations. To define variations of interest in the brains of depressed subjects over controls, relative gene expression was estimated by using the fitPLM function of the robust multiarray average (rma) software package (http://bioconductor.org). The log-transformed PM (perfect match) values were directly compared array-to-array, after a global background adjustment and across-array quantile normalization to remove between-chip bias (observed variation) (20, 21). By using the output probe level summaries, t scores (case signal minus the control signal divided by the square root of the sum of the square of the error terms) were calculated for all 12,626 genes on the array (HG_U95Av2) by using Microsoft excel spreadsheet; and, redundant probes were eliminated by using a publicly available custom CDF file, (UG-4) (http://brainarray.mhri.med.umich.edu/brainarray) (14). Probe sets showing a P value (derived from the t score) of ≥0.05 were considered significant in differential expression; and from among them, those with a fold-change threshold of ≤1.175 were considered up-regulated, and those with a threshold of ≥0.85 down-regulated. The thresholds of 1.175 and 0.85 were chosen in keeping with the compressed nature of rma output, where FOLD CHANGE values always remain below a maximum of 2.0. Conversely, even a 15% (fold) change in rma output can be significant, unlike that in mas 5.0 (Affymetrix) output. Therefore, a combined filter of P value and FOLD CHANGE with higher thresholds applied to rma output can prove too stringent and result in type1 error (loss of true positives), especially in the case of rare transcripts that show relatively weak signals.
Transcript Profiling Using the Sentrix BeadChip Arrays. As an independent validation of altered expression of genes identified by Affymetrix GeneChip arrays, we arrayed 7 of the 13 selected candidate genes on Sentrix Custom BeadChip arrays (Illumina, San Diego), processed and analyzed according to the manufacturer's recommendations. The 7 genes tested were SLC1A2, GluR5, GluR-KA2, mGluR3, GABAAα5, GABAAδ, and GABAB1.
In Situ Hybridization. Representatives of candidate genes selected by microarray analysis were independently validated by using ISHH (13). Small blocks of tissue were excised from DLPFC and AnCg of the right hemispheres from the same patients and controls used for microarray analysis, and were matched by age, gender, and postmortem interval (PMI), wherever possible. The subject number, n, for each category is indicated in the figure legends. The frozen tissue blocks were allowed to rise to 4°C and then placed in chilled 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) overnight. The blocks were then infused with 30% sucrose in 0.1 M phosphate buffer, refrozen in dry ice, and stored at –85°C until sectioning. Serial sections (50 μm thick) were cut on a sliding microtome and collected in 4% paraformaldehyde in 0.1 M phosphate buffer (pH 7.4) in which they remained for ≤7 days, pending further processing.
Free-floating sections were prepared for in situ hybridization by washing in 0.1 M phosphate buffer containing 0.1 M glycine, followed by two washes in phosphate buffer and two washes in 2× saline sodium citrate (SSC; 1× SSC is 0.15 M sodium chloride and 0.015 M sodium citrate, pH 7.0). The pretreated sections were hybridized overnight at 60°C in a humid chamber by incubating with labeled probe in the hybridization solution, containing 50% formamide, 10% dextran sulfate, 0.7% ficoll, 0.7% polyvinyl pyrrolidone, 0.5 mg/ml yeast tRNA, 0.33 mg/ml denatured herring sperm DNA, 20 mM DTT, and 5.0 × 105 cpm/ml of the [α-33P]UTP-labeled antisense (or sense) riboprobe. The following linearized cDNA templates were prepared and used in transcription, in accordance with National Institutes of Health guidelines for recombinant DNA research: SLC1A2: Hs cDNA, 150 bp {5′>855–1005<3′}, (NM_004171; gi: 40254477); SLC1A3: Hs cDNA, 223 bp {5′>236–455<3′}, (BC054475; gi: 32450688); GS: Hs cDNA, 211 bp {5′>1025–1236<3′} (BC018992; gi: 17512037); mGLUR3: Hs cDNA, 322 bp {5′>2903–3225<3′} (BC041407; gi: 27552882); GluR1: Hs cDNA, 380 bp {5′>809–1188<3′} (NM_000827; gi: 6552333); GA BAAα5: Hs cDNA, 290 bp {5′>1430–1720<3′} (NM_000810; gi: 6031207).
After hybridization, the sections were washed twice in 4× SSC at 60°C, digested with 20 μg/ml of ribonuclease A (pH 8.0) for 30 min at 45°C, and washed with SSC of descending concentrations, down to 0.5×. They were then mounted on gelatin-coated slides, dried, and exposed to Amersham Pharmacia β-max film for 15 days. After the film was developed, the sections were lipid-extracted in chloroform, dipped in Kodak NTB2 emulsion (diluted 1:1 with water), exposed for 8 days at 4°C, developed in Kodak D-19, fixed, and counterstained with cresyl violet.
Film autoradiograms of hybridized sections were quantified by using a microcomputer imaging device (MCID/M5; Imaging Research, St. Catharine's, Ontario, Canada). Mean gray levels, recorded by the system from each sample and reported as integrated optical density (IOD) values, were converted to radioactivity units (nCi/g) (1 Ci = 37 GBq), based on a set of 14C radioactive standards (Amersham Pharmacia) exposed on each film. A minimum of five measurements were taken within each cortical layer in at least three sections, from each brain region. The background signal, measured as that over the white matter signal was subtracted from each individual measurement in all layers of each cortical region. For each cortical region and layer, the mean values of target mRNA levels were calculated, and the mean and standard error for each cohort were calculated from the 10 individual means. Statistical significance of the differences between the mean values of the diseased and controls was determined by repeated measures analysis of variance (RMANOVA), by using cohorts as groups and cortical layers as variates. In addition, Student's two-tailed paired t test was used to independently determine the significance of differences observed between cohorts for each cortical layer.
Results
Discovery of Altered Expression of GABA and Glutamate-Related Genes in MDD Using GeneChip Arrays. In MDD, AnCg (249) and DLPFC (252) exhibited similar numbers of genes with expression changes compared with controls (Table 1). Both cortical areas showed significant down-regulation of the glial highaffinity glutamate transporters SLC1A2 and SLC1A3. In addition, both cortical areas showed significant down-regulation of GS. Concomitantly, the glutamate receptor genes AMPA1 and GluR-KA2 were up-regulated in AnCg; and AMPA3, GluR5, and GluR-KA2 were up-regulated in DLPFC (Table 2). GABAAβ3, GABAAδ, and GABAAγ2 were also up-regulated in DLPFC (Table 2).
Table 1. A summary of gene expression changes observed in the AnCg and DLPFC of brains from patients with MDD and BPD.
| Number of genes showing expression change
|
|||||||
|---|---|---|---|---|---|---|---|
| AnCg
|
DLPFC
|
||||||
| Disease | Genes | Total | Glutamate | GABA | Total | Glutamate | GABA |
| MDD | Total | 249 | 4 | 0 | 252 | 5 | 3 |
| Up-regulated | 115 | 0 | 0 | 77 | 2 | 3 | |
| Down-regulated | 134 | 4 | 0 | 175 | 3 | 0 | |
| BPD | Total | 359 | 4 | 1 | 238 | 1 | 3 |
| Up-regulated | 285 | 3 | 1 | 214 | 1 | 3 | |
| Down-regulated | 74 | 1 | 0 | 24 | 0 | 0 | |
Table 2. List of candidate genes showing expression changes in MDD and BPD.
| Fold change
|
|||||
|---|---|---|---|---|---|
| Gene name | Symbol | Alias | Cytoband | AnCg | DLPFC |
| Major depressive disorder | |||||
| Glial high-affinity glutamate transporter, Na+-dependent | SLC1A2 | GLT-1; EAAT2 | 11p13 | 0.80 | 0.71 |
| Glial high-affinity glutamate transporter, Na+-dependent | SLC1A3 | GLAST; EAAT1 | 5p13 | 0.85 | 0.65 |
| Glutamine synthetase | GLUL | GS | 1q31 | 0.73 | 0.78 |
| Glutamate receptor, ionotropic, AMPA 1 | GRIA1 | AMPA1; IGluR1 | 5q31.1 | 1.3 | |
| Glutamate receptor, ionotropic, AMPA 3 | GRIA3 | AMPA3; IGluR3 | Xq25 | 1.18 | |
| Glutamate receptor, ionotropic, kainate 1 | GRIK1 | GluR5; EAA3 | 21q22.11 | 1.21 | |
| Glutamate receptor, ionotropic, kainate 5 | GRIK5 | GluR-KA2; EAA2 | 19q13.2 | 1.14 | 1.21 |
| GABAA receptor, beta 3 | GABARB3 | GABAARβ3 | 15q11.2 | 1.21 | |
| GABAA receptor, delta | GABRD | GABAARδ | 1p36.3 | 1.19 | |
| GABAA receptor, gamma 2 | GABARG2 | GABAARγ2 | 5q3.1 | 1.22 | |
| Bipolar affective disorder | |||||
| Glutamate receptor, ionotropic | GRIA1 | AMPA1; IGluR1 | 5q31.1 | 1.21 | |
| Glutamate receptor, ionotropic | GRIA3 | AMPA3; IGluR3 | Xq25 | 1.21 | |
| Glutamate receptor, ionotropic, kainate 1 | GRIK1 | GluR5; EAA3 | 21q22.11 | 0.78 | |
| Glutamate receptor, metabotropic 3 | GRM3 | mGluR3 | 7q21.1 | 1.24 | 1.26 |
| GABAA receptor, alpha 5 | GABRA5 | GABAARα5 | 15q11.2 | 1.24 | 1.20 |
| GABAB receptor 1 | GABBR1 | GABABR1 | 6p21.31 | 1.22 | |
Each record indicates the gene name, symbol, common name, cytoband, and fold changes (FC) of expression in the AnCg and/or DLPFC. Expression values, meeting a significance threshold of P ≤ 0.05 and a fold change of ≥1.175 were considered increases, and those with a fold change of ≤0.85 were considered decreases.
Candidate Gene Discovery in BPD and Suicide Using GeneChip Arrays. In BPD, AnCg exhibited more genes with expression changes (359) than DLPFC (238) (Table 1). Both cortical areas in BPD also showed significant changes in expression of the glutamate receptor subunits AMPA1, AMPA3, GluR5, and mGluR3; of the GABAA receptor subunit GABAAα5; and of the GABAB receptor subunit GABAB1 (Table 2). Regardless of the type of mood disorder, however, two other GABAA receptor subunit mRNAs, GABAAα1 and GABAAβ3, were consistently upregulated in AnCg in suicide victims afflicted with MDD (seven suicidal, six nonsuicidal) or BPD (six suicidal, three nonsuicidal), compared with nonsuicidal controls (n = 22).
Validation of Candidate Genes Using BeadChip Arrays. The upregulation of GABAAα5 in both cortical areas and GABABR1 in DLPFC was further confirmed independently, by using focused arrays (Sentrix BeadChip; Illumina, San Diego). DLPFC from BPD brains showed up-regulation of both GABAAα5 and GABAB1, whereas AnCg showed up-regulation of just the former, and MDD brains showed no significant changes.
Validation of Candidate Genes Using ISHH. To validate microarray findings we used ISHH and analyzed four genes (SLC1A2, SLIC1A3, GS, and GABAAα5) in MDD, three genes (GluR1, mGluR3, and GABAAα5) in BPD, and all six genes in control samples. In MDD, the changes of GS, SLC1A2, and SLC1A3 expression showed a downward trend (Fig. 1), whereas GABAAα5 expression was significantly higher in both cortical regions (Fig. 2 c and d). Robust ISHH validation, consistent with microarray findings of SLC1 transcripts, may require probing enriched populations of a specific cell type, e.g., astroglia. However, the decreased expression of SLC1A3 and GS was additionally replicated in the amygdala from an additional independent cohort of MDD patients [(n = 5 (1 female, 4 male); control subjects n = 6 (1 female, 5 male)].
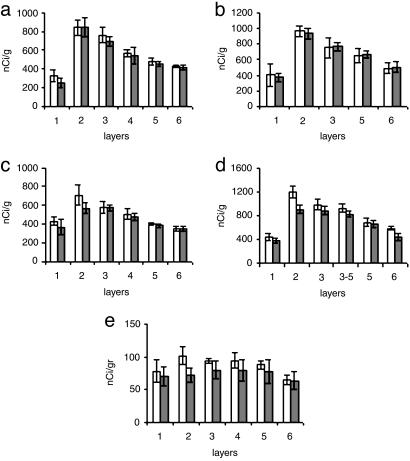
Fig. 1.
Histograms from the analysis of film autoradiograms, showing levels of expression of GS, SLC1A2, and SLC1A3 mRNAs in the DLPFC and AnCg in control subjects and in patients with MDD. Shown are histograms for GS (a and b), SLC1A2 (c and d), SLC1A3 (e), DLPFC (a, c, and e), and AnCg (b and d). Open columns, control subjects (n = 6); gray columns, patients with MDD (n = 8).
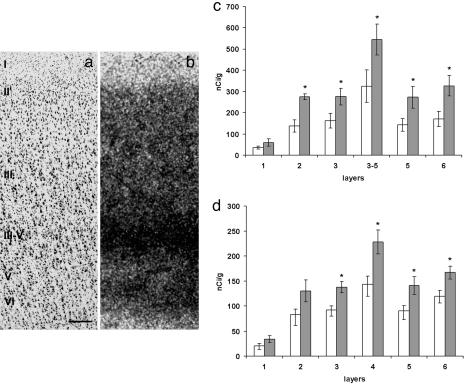
Fig. 2.
gabaAα5 expression in the human AnCg and DLPFC. (a and b) Paired photomicrographs from AnCg. (a) Nissl staining of a perpendicular section from a control subject showing the AnCg cytoarchitecture. (b) Film autoradiogram from an adjacent section hybridized with GABAAα5 probe. Shown are histograms showing the distribution patterns of GABAAα5 mRNA in different layers of AnCg (c) and DLPFC (d) in control subjects (n = 5; white columns) and in bipolar patients (n = 5; gray columns). (Scale bar in a, 250 mm.)
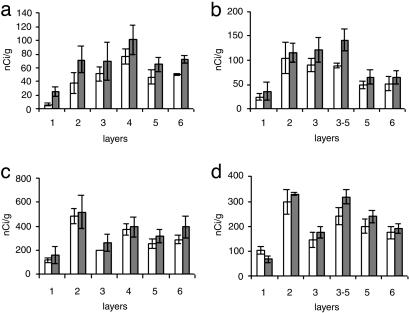
In controls as well as diseased samples, AnCg showed higher levels of GABAAα5 (Fig. 2 a–d), SLC1A2, and GS mRNAs than DLPFC (Fig. 1). By contrast, GluR1 mRNA was found at higher levels in DLPFC, whereas mGluR3 mRNA levels were comparable in both regions (Fig. 3). Overall, the expression of GABAAα5 (Fig. 2), GluR1 (Fig. 3), and mGluR3 mRNAs (Fig. 3) was highest in deep layer III and superficial layer V of AnCg, and in layer IV of DLPFC. The GS, SLC1A2, and SLC1A3 expression was highest in layer II (Fig. 1).
Fig. 3.
Histograms from the analysis of film autoradiograms, showing levels of expression of mGLUR3 and GLUR1 mRNAs in the DLPFC and AnCg of control subjects and in patients with BPD. Shown are histograms for mGluR3 (a and b), GluR1 (c and d), DLPFC (a and c), and AnCg (b and d). Open columns, control subjects (n = 6); gray columns, patients with BPD (n = 5).
In BPD, GABAAα5 (Fig. 2 c and d) was up-regulated in both cortical regions by ≤35% in layers II, III, V, and VI of AnCg and in layers III, IV, V, and VI of DLPFC (Fig. 2d). mGLUR3 and GLUR1 showed a definite upward trend (Fig. 3).
Discussion
The present study provides evidence for the dysregulation of genes related to the glutamatergic and GABAergic transmitter systems in the AnCg and DLPFC of individuals diagnosed with major depression. Consistent alterations in the expression of glutamate transporter genes suggest major involvement of neuroglia in the pathology of depression, whereas changes in expression of certain GABAA receptor subunit genes in suicide victims from both MDD and BPD suggest their value as potential biomarkers for predicting suicidal behavior in both affective disorders.
The glial high-affinity glutamate transporters SLC1A2 and SLC1A3 are members of SLC1, a seven-member family of amino acid transporters (22–24). SLC1A2 and SLC1A3 are primarily astroglial in distribution (25). Deficits in these molecules could impair reuptake of glutamate from the synaptic cleft by astroglia, prolonging synaptic activation by glutamate (26, 27). Accumulations of extracellular glutamate could not only perturb the ratio of excitatory-inhibitory neurotransmitter levels (7), but also potentially cause cytotoxic damage to neurons and glia (4, 28–30). Significantly reduced numbers of neuroglia in MDD have been reported (31).
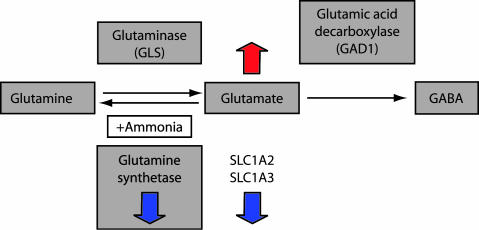
Glutamate is recycled in brain cells through several alternate mechanisms. Two mechanisms are important in this context. First, glutamate serves as the starting material for the biosynthesis of GABA (Fig. 4), catalyzed by glutamic acid decarboxylase (GAD), an enzyme consistently implicated in mood disorders as well as in schizophrenia (18, 32–34). Equally importantly, glutamate is also the starting material for the biosynthesis of glutamine catalyzed by the 42-kDa astroglial enzyme GS, which in the process neutralizes ammonia (35). The significantly lowered expression of GS observed in AnCg and DLPFC in MDD is consistent with a previous report (7) of reduced glutamate-glutamine cycling in the plasma and cerebrospinal fluid of depressed patients. Down-regulated GS can conceivably elevate residual glutamate concentrations and trigger feedback inhibition of glutamate transport/synthesis and/or allow a rise in extracellular glutamate with a potential for excitotoxic effects.
Fig. 4.
Schematic drawing showing the metabolic steps, substrates, and enzymes involved in glutamate recycling. The down-regulation of the glial high-affinity glutamate transporters and glutamine synthetase are shown by blue arrows pointing down, and potentially accumulating glutamate levels are shown by orange arrow pointing up.
We also detected up-regulation of certain ionotropic and metabotropic glutamate receptor subunits in depressed patient brains. Up-regulated AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid)-kainate receptors can allow excessive Ca2+ ions to enter through NMDA receptor-gated ion channels (36), which could in turn react with reactive oxygen species (ROS) and trigger apoptosis (37). This potential increase in vulnerability to apoptosis may be amplified by the dysregulation in growth factors we have observed in the same brains, particularly the FGF system (14). Indeed, it is conceivable that the dysregulation in growth factors plays a causative role in increased apoptosis and in the altered expression of glial-based glutamate transporters (38). In turn, because metabotropic glutamate receptors are coupled to G protein receptors and act on various second messenger systems, the effects could be even more far reaching through modulation of numerous downstream signaling mechanisms.
From our results, it seems that the coordination between different subunits of the GABAA complex may be disturbed in the DLPFC of MDD patients, as indicated by up-regulation of GABAAβ3, and GABAAδ and GABAAγ2, which control the function of the GABAA complex (5). This result is not surprising in view of the tight coupling of the biosynthesis of GABA with glutamate recycling (Fig. 4), but it remains to be deciphered whether this GABA up-regulation is primary or is secondary to the illness, and represents a compensatory response to the GABA deficit or glutamate overexcitability. Nevertheless, it is consistent with previous implications of changes in GABAAα5, GABAAβ3, and GABAAγ3 in MDD (6, 7, 39). Further, the genes encoding these receptor subunits map to 15q11-q13, within a span of 100 kb (40, 41), suggesting potential control at the genetic level.
Changes involving altered GABA/glutamate signaling are not unique to depression but also can be found in schizophrenia (32–35, 42) and in several neurological diseases (24). Our findings point to alterations in glutamate recycling, glutamate receptors, GABA receptors, and glial cells and open up new possibilities for treating depression and possibly other diseases sharing a common pathophysiology. Although further research is warranted to decipher whether the reduced elements of glutamatergic signaling systemrelated gene expression are a reflection of altered gene regulation leading to pathologic metabolism or more simply a reflection of diminished populations of glutamatergic neurons, they could provide pharmaceutical targets, specific to the biological defect, for drugs that modulate glutamate transporters or antagonize specific glutamate and GABAA subunits (6, 9, 43, 44).
Acknowledgments
We thank X. Fan, P. Nguyen, M. Yusufzai, S. Burke, M. Hoversten, K. Lopez, M. Atz, and K. Overman for technical assistance; C. R. Neal (University of Hawaii, Honolulu) and J. D. Stead (Carlton University, Ottawa, Canada) for cRNA probes; M. O. Lopez-Figueroa for discussions and critiquing the manuscript; T. Speed, F. Meng, B. Bolstad, J. Brettenbaugh, M. Dai, and Y. Wang for bioinformatics advice; E. Jakupovic, G. Chang, and A. Melcher for network support; K. Burke, J. Berndt, and the Orange County Coroner's Office for procuring brains; D. M. Walsh, P. Cartagena, and R. Stein for clinical characterization; F. W. Lovell for neuropathological evaluation of postmortem brains; W. W. Tourtellotte for facilities for processing and storing tissues; and the families of the patients involved. Funding was provided by the National Institute of Mental Health (NIMH) (Silvio O. Conte Center for Mood Disorders Research Grant 2P50MH060398-06 and NIMH Grants P01 MH42251, MH54844, and MH42251), the Pritzker Neuropsychiatric Disorders Research Fund L.L.C., the W. M. Keck Foundation, the Della Martin Foundation, and the William Lion Penzner foundation.
Author contributions: P.V.C., M.M., S.J.E., J.Z.L., R.M.M., W.E.B.J., H.A., S.J.W., and E.G.J. designed research; P.V.C., M.M., S.J.E., H.T., J.Z.L., M.P.V., and E.G.J. performed research; P.V.C., M.M., S.J.E., H.T., J.Z.L., M.P.V., and E.G.J. analyzed data; and P.V.C., M.M., W.E.B.J., H.A., and E.G.J. wrote the paper.
Conflict of interest statement: The authors are members of the Pritzker Neuropsychiatric Disorders Research Consortium, supported by the Pritzker Neuropsychiatric Disorders Research Fund L.L.C. The findings reported here are subjects of pending patent applications filed under a shared intellectual property agreement between the academic and philanthropic entities of the consortium.
Abbreviations: AnCg, anterior cingulate cortex; BPD, bipolar affective disorder; DLPFC, dorsolateral prefrontal cortex; glutamate, l-glutamic acid; MDD, major depressive disorder; SSC, saline sodium citrate; GS, glutamine synthetase; ISHH, in situ hybridization histochemistry.
References
- 1.Nemeroff, C. B. (1998) Sci. Am. 278 (6), 42–49. [DOI] [PubMed] [Google Scholar]
- 2.Caspi, A., Sugden, K., Moffitt, T. E., Taylor, A., Craig, I. W., Harrington, H., McClay, J., Mill, J., Martin, J., Braithwaite, A. & Poulton, R. (2003) Science 301, 386–389. [DOI] [PubMed] [Google Scholar]
- 3.Lesch, K. P. (2004) J. Psychiatry Neurosci. 29, 174–184. [PMC free article] [PubMed] [Google Scholar]
- 4.Petroff, O. A. (2002) Neuroscientist 8, 562–573. [DOI] [PubMed] [Google Scholar]
- 5.Krystal, J. H., Snacora, G., Blumberg, H., Anand, A., Charney, D. S., Marek, G., Epperson, C. N., Goddard, A. & Mason, G. F. (2002) Mol. Psychiatry 7, S71–S80. [DOI] [PubMed] [Google Scholar]
- 6.Merali, Z., Du, L., Hrdina, P., Palkovits, M., Faludi, G., Poulter, M. O. & Anisman, H. (2004) J. Neurosci. 24, 1478–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cryan, J. F. & Kaupmann, K. (2005) Trends Pharmacol. Sci. 26, 36–43. [DOI] [PubMed] [Google Scholar]
- 8.Sanacora, G., Gueorguieva, R., Epperson, C. N., Wu, Y. T., Appel, M., Rothman, D. L., Krystal, J. H. & Mason, G. F. (2004) Arch. Gen. Psychiatry 61, 705–713. [DOI] [PubMed] [Google Scholar]
- 9.Skolnick, P., Layer, R. T., Popik, P., Nowak, G., Paul, I. A. & Trullas, R. (1996) Pharmacopsychiatry 29, 23–26. [DOI] [PubMed] [Google Scholar]
- 10.Berman, R. M., Cappiello, A., Anand, A., Oren, D. A., Heninger, G. R., Charney, D. S. & Krystal, J. H. (2000) Biol. Psychiatry 47, 351–354. [DOI] [PubMed] [Google Scholar]
- 11.Lockhart, D. J., Dong, H., Byrne, M. C., Follettie, M. T., Gallo, M. V., Chee, M. S., Mittman, M. Wang, C., Kobayashi, M., Horton, H. & Brown, E. L. (1996) Nat. Biotechnol. 14, 1675–1680. [DOI] [PubMed] [Google Scholar]
- 12.Bunney, W. E., Bunney, B. G., Vawter, M. P., Tomita, H., Li, J., Evans, S. J., Choudary, P. V., Myers, R. M., Jones, E. G., Watson, S. J. & Akil, H. (2003) Am. J. Psychiatry 160, 657–666. [DOI] [PubMed] [Google Scholar]
- 13.Molnar, M., Potkin, S. G., Bunney, W. E. & Jones, E. G. (2003) Biol. Psychiatry 53, 39–47. [DOI] [PubMed] [Google Scholar]
- 14.Evans, S. J., Choudary, P. V., Neal, C. R., Li, J. Z., Vawter, M. P., Tomita, H., Lopez, J. F., Thompson, R. C., Meng, F., Stead, J. D., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 15506–15511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tomita, H., Vawter, M. P., Walsh, D. M., Evans, S. J., Choudary, P. V., Li, J., Overman, K. M., Atz, M. E., Myers, R. M., Jones, E. G., et al. (2004) Biol. Psychiatry 55, 346–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li, J. Z., Vawter, M. P., Walsh, D. M., Tomita, H., Evans, S. J., Choudary, P. V., Lopez, J. F., Avelar, A., Shokoohi, V., Chung, T., et al. (2004) Hum. Mol. Genet. 13, 609–616. [DOI] [PubMed] [Google Scholar]
- 17.Jones, E. G., Hendry, S. H., Liu, X.-B., Hodgins, S., Potkin, S. G. & Tourtellotte, W. W. (1992) J. Neurosci. Methods 44, 133–144. [DOI] [PubMed] [Google Scholar]
- 18.Huntsmann, M. M., Tran, B. V., Potkin, S. G., Bunney, W. E., Jr., & Jones, E. G. (1998) Proc. Natl. Acad. Sci. USA 95, 15066–15071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans, S. J., Choudary, P. V., Vawter, M. P., Li, J., Meador-Woodruff, J. H., Lopez, J. F., Burke, S. M., Thompson, R. C., Myers, R. M., Jones, E. G., et al. (2003) Neurobiol. Dis. 14, 240–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gautier, L., Cope, L., Bolstad, B. M. & Irizarry, R. A. (2004) Bioinformatics 20, 307–315. [DOI] [PubMed] [Google Scholar]
- 21.Gentleman, R. C., Carey, V. J., Bates, D. M., Bolstad, B., Dettling, M, Dudoit, S., Ellis, B., Gautier, L., Ge, Y., Gentry, J., et al. (2004) Genome Biol. 5, R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gegelashvili, G., Robinson, M. B., Trotti, D. & Rauen, T. (2001) Prog. Brain Res. 132, 267–286. [DOI] [PubMed] [Google Scholar]
- 23.Kanai, Y. & Hediger, M. A. (2004) Pflügers Arch. 447, 469–479. [DOI] [PubMed] [Google Scholar]
- 24.Maragakis, N. J. & Rothstein, J. D. (2004) Neurobiol. Dis. 15, 461–473. [DOI] [PubMed] [Google Scholar]
- 25.Gegelashvilia, G., Dehnesb, Y., Danbolt, N. C. & Schousboea, A. (2000) Neurochem. Int. 37, 163–170. [DOI] [PubMed] [Google Scholar]
- 26.Auger, C. & Attwell, D. (2000) Neuron 28, 547–558. [DOI] [PubMed] [Google Scholar]
- 27.Danbolt, N. C. (2001) Prog. Neurobiol. 65, 1–105. [DOI] [PubMed] [Google Scholar]
- 28.Choi, D. W. (1988) Neuron 1, 623–634. [DOI] [PubMed] [Google Scholar]
- 29.Parsons, C. G., Danysz, W. & Quack, G. (1998) Drug News Perspect. 11, 523–569. [DOI] [PubMed] [Google Scholar]
- 30.Miller, G. (2005) Science 308, 778–781. [DOI] [PubMed] [Google Scholar]
- 31.Rajkowska, G. (2000) Biol. Psychiatry 48, 766–777. [DOI] [PubMed] [Google Scholar]
- 32.Akbarian, S., Huntsmann, M. M., Kim, J. J., Tafazzoli, A., Potkin, S. G., Bunney, W. E., Jr., & Jones, E. G. (1995) Cereb. Cortex 5, 550–560. [DOI] [PubMed] [Google Scholar]
- 33.Akbarian, S., Sucher, N. J., Bradley, D., Tafazzoli, A., Trinh, D., Hetrick, W. P., Potkin, S. G., Sandman, C. A., Bunney, W. E., Jr., & Jones, E. G. (1996) J. Neurosci. 16, 19–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lewis, D. A., Hashimoto, T. & Volk, D, W. (2005) Nat. Rev. Neurosci. 6, 312–324. [DOI] [PubMed] [Google Scholar]
- 35.Gunnersen, D. & Haley, B. (1992) Proc. Natl. Acad. Sci. USA 89, 11949–11953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novelli, A., Reilly, J. A., Lypsko, P. G. & Henneberry, R. C. (1988) Brain Res. 451, 205–212. [DOI] [PubMed] [Google Scholar]
- 37.Oki, T., Yamazaki, Y., Nomura, N., Furumai, T. & Igarashi, Y. (1999) J. Antibiot. 52, 449–454. [DOI] [PubMed] [Google Scholar]
- 38.Fernandez-Sanchez, M. T. & Novelli, A. (1993) FEBS Lett. 335, 124–131. [DOI] [PubMed] [Google Scholar]
- 39.Oruc, L., Verheyen, G. R., Furac, I., Ivezic, S., Jakovljevic, M., Raeymaekers, P. & Van Broeckhoven, C. (1997) Neuropsychobiology 36, 62–64. [DOI] [PubMed] [Google Scholar]
- 40.Sinnett, D., Wagstaff, J., Glatt, K., Woolf, E., Kirkness, E. J. & Lelande, M. (1993) Am. J. Hum. Genet. 52, 1216–1229. [PMC free article] [PubMed] [Google Scholar]
- 41.Russek, S. J. & Farb, D. H. (1994) Genomics 23, 528–533. [DOI] [PubMed] [Google Scholar]
- 42.Harrison, P. J. & Weinberger, D. R. (2005) Mol. Psychiatry 10, 40–68. [DOI] [PubMed] [Google Scholar]
- 43.Holden, C. (2003) Science 300, 1866–1868. [DOI] [PubMed] [Google Scholar]
- 44.Guidotti, A., Auta, J., Davis, J. M., Dong, E., Grayson, D. R., Veldic, M., Zhang, X. & Costa, E. (2005) Psychopharmacology 180, 191–205. [DOI] [PubMed] [Google Scholar]