Abstract
Inactivation of the Caenorhabditis elegans gene clk-1, which is required for ubiquinone biosynthesis, increases lifespan by an insulin signaling-independent mechanism. We find that homozygous inactivation of mclk1, the mouse ortholog of clk-1, yields ES cells that are protected from oxidative stress and damage to DNA. Moreover, in the livers of old mclk1+/- mice, hepatocytes that have lost mclk1 expression by loss of heterozygosity undergo clonal expansion, suggesting that their resistance to stress allows them to outcompete cells that still express the gene. mclk1+/- mice, whose growth and fertility are normal, also display a substantial increase in lifespan in each of three different genetic backgrounds. These observations indicate that the distinct mechanism by which clk-1/mclk1 affects lifespan is evolutionarily conserved from nematodes to mammals and is not tied to a particular anatomy or physiology.
Keywords: clk-1, mclk1, aging, loss of heterozygosity, reactive oxygen species, ubiquinone
The power of using the genetic approach to elucidate the mechanisms of aging has been underscored by the possibility of identifying long-lived mutants in invertebrate animal models of aging. Indeed, when a loss-of-function mutation in a gene prolongs lifespan, one has to conclude that the normal function of that gene limits lifespan in the organism under study. In the nematode Caenorhabditis elegans, this approach has been used to identify a number of mechanisms that affect aging; that is, (1) the insulin signaling pathway (Kenyon et al. 1993; Kimura et al. 1997); (2) the clk-1-dependent mechanism (Wong et al. 1995; Lakowski and Hekimi 1996; Ewbank et al. 1997); (3) caloric restriction (Lakowski and Hekimi 1998); (4) a mitochondrial mechanism that alters resistance to oxidative stress and does not affect animal size (Feng et al. 2001; Hekimi and Guarente 2003); (5) a mitochondrial mechanism that acts during development and appears distinct from mechanism 4 in terms of its effects on oxidative stress (Dillin et al. 2002; Lee et al. 2002; Hekimi and Guarente 2003); (6) a pathway linked to germ cell multiplication that might be distinct from the insulin pathway (Hsin and Kenyon 1999) although it involves some of the same molecular players, such as daf-2 and daf-16; (7) a mechanism that has links to telomere length (Benard et al. 2001; Joeng et al. 2004); and (8) the TOR pathway (Vellai et al. 2003; Jia et al. 2004).
In spite of the extensive study of these pathways in invertebrates, in particular C. elegans and Drosophila, and with the exception of caloric restriction, which was discovered in rodents, there is promising but limited evidence as to whether the effects of these pathways on longevity is evolutionarily conserved (Kenyon 2001, 2005). In this regard, the best studied pathway is the insulin signaling pathway. One study of mice heterozygous for a knockout of the insulin-like growth factor I receptor (a homolog of daf-2) found an increase in the lifespan of these animals (Holzenberger et al. 2003), and an adipose tissue-specific knockout of the insulin receptor itself is similarly effective (Bluher et al. 2003). On the other hand, although overexpressing catalase in the mitochondria increases mouse lifespan (Schriner et al. 2005), another study of mice heterozygous for a knockout that disrupts the function of the manganese superoxide dismutase (sod2) and results in high oxidative stress failed to reveal an effect on lifespan (Van Remmen et al. 2003), in spite of the wealth of evidence supporting the oxidative stress theory of aging.
The gene clk-1, which affects aging and numerous other physiological rates and rhythms in the nematode C. elegans (Wong et al. 1995), encodes an enzyme that is necessary for the biosynthesis of ubiquinone (coenzyme Q; UQ) (Marbois and Clarke 1996; Ewbank et al. 1997; Miyadera et al. 2001), an essential cofactor in numerous redox reactions, including mitochondrial respiration, as well as a membrane antioxidant, and an oxygen sensor (Georgellis et al. 2001). clk-1 mutants accumulate the biosynthetic intermediate demethoxyubiquinone (DMQ) instead of ubiquinone, but also contain ubiquinone of dietary origin, which is necessary for their survival (Jonassen et al. 2001; Hihi et al. 2002). clk-1 mutants have low levels of reactive oxygen species (ROS) (Shibata et al. 2003; Kayser et al. 2004), and, as a result, low levels of oxidative damage to lipoproteins and decreased activation of oncogenic ras signaling (Shibata et al. 2003).
A complete knockout of mclk1, the murine homolog of clk-1, leads to embryonic lethality as well as to a complete absence of ubiquinone in embryos and in mclk1-/- embryonic stem (ES) cells (Levavasseur et al. 2001). It also severely affects the activity of mitochondrial complex II, but not complex I and complex III. The lethality appears to be due to a developmental defect of the placenta (X. Liu and S. Hekimi, unpubl.). Heterozygous animals, however, are completely viable and newborns have normal levels of ubiquinone, suggesting that mclk1 is fully recessive for ubiquinone biosynthesis.
Here we investigate the phenotype of mclk1-/- ES cells and the effect of the loss of one copy of mclk1 in mclk1+/- mice. We find that the loss of mclk1 results in decreased ROS levels, decreased ROS sensitivity, and decreased ROS damage. We also find that mclk1+/- animals have an increased lifespan and lose mclk1 expression in a large subset of liver cells by a mechanism of loss of heterozygosity (LOH), likely because mclk1-/- cells have a growth advantage. We conclude that the longevity-promoting effect of reducing clk-1/mclk1 activity that was initially observed in C. elegans is conserved in mice, supporting the idea that some molecular mechanisms of aging are shared throughout the animal kingdom.
Results
Phenotypic analysis of mclk1-/- ES cells
We have previously derived mclk1-/- ES cells by cultivating mclk1-/- blastocysts derived from mclk1+/- mothers. A mclk1+/+ line from the same mothers was also derived and serves as control. In addition to the absence of ubiquinone and the mitochondrial respiration defect observed previously, we have now characterized a number of additional phenotypes of mclk1-/- ES cells, including (1) slow cell multiplication, (2) reduced tendency to differentiate in the presence of low levels of leukemia inhibitory factor (LIF), (3) low levels of basal and induced ROS measured by dye-dependent fluorescence, and (4) resistance to apoptosis induced by the ROS-generating compound menadione (Table 1).
Table 1.
Phenotype of mclk1-/- ES cells
|
mclk1+/+
|
mclk1–/–
|
|||||||
|---|---|---|---|---|---|---|---|---|
| ES cell phenotypea | Without treatment | Sodium pyruvate | UQ9 | Sodium pyruvate and UQ9 | Without treatment | Sodium pyruvate | UQ9 | Sodium pyruvate and UQ9 |
| Resistance to menadione [% viability]b | 22.7 ± 5.4 (100) | 23 ± 7.5 (101) | 24 ± 2.9 (105) | 23.9 ± 6.9 (105) | 68.4 ± 5.4 (304) | 68.9 ± 6.5 (304) | 91.3 ± 3.4 (403) | 92.7 ± 4.2 (403) |
| Oxygen consumption [10–3 μl O2/sec/μg of protein] | 9.3 ± 0.6 (100) | 9.1 ± 0.4 (98) | 8.96 ± 0.8 (96) | 8.96 ± 0.6 (97) | 4.9 ± 0.5 (53) | 7.9 ± 0.4 (85) | 6.8 ± 0.7 (72) | 8.8 ± 0.7 (95) |
| Cell multiplication [×105 cells]c | 25.2 ± 5.2 (100) | 25.0 ± 4.8 (99) | 25.3 ± 5.5 (100) | 25.7 ± 5.5 (102) | 9.4 ± 1.5 (39) | 18.9 ± 2.8 (75) | 17.5 ± 0.4 (69) | 26.8 ± 5.7 (106) |
| LIF requirement [% undifferentiated colonies, n = 300]d | 43.3 ± 10.6 (100) | 39.3 ± 12.7 (90) | 39.0 ± 18.5 (90) | 44.7 ± 11.0 (102) | 293.3 ± 11.5 (676) | 181.3 ± 3.2 (418) | 193.7 ± 40.8 (446) | 133.3 ± 0.1 (306) |
| Basal ROS levels [fluorescence unit × 100/μg protein] | 4.0 ± 0.5 (100) | 4.1 ± 0.3 (100) | 3.8 ± 0.2 (94) | 3.6 ± 0.2 (90) | 2.4 ± 0.3 (59) | 2.3 ± 0.2 (57) | 2.4 ± 0.3 (59) | 2.3 ± 0.3 (57) |
| Induced ROS levelse [fluorescence unit × 100/μg protein] | 11.5 ± 0.8 (100) | 11.8 ± 1.2 (102) | 11.5 ± 0.5 (100) | 11.7 ± 1.1 (101) | 8.0 ± 0.7 (69) | 8.1 ± 1.1 (70) | 6.7 ± 1.2 (57) | 6.7 ± 1.1 (57) |
| DNA damagef [% cells with tails] | 45.3 ± 8.4 (100) | n.d. | n.d. | n.d. | 28.7 ± 3.2 (63) | n.d. | n.d. | n.d. |
| Lipid peroxidation MDA equivalentsg [pmol/μg protein) | 18.1 ± 0.6 (100) | n.d. | n.d. | n.d. | 10.7 ± 1.5 (59) | n.d. | n.d. | n.d. |
| Mitochondrial complex II activity without exogenous Q1 [nmol/min/mg protein]h | 15.1 ± 1.6 (100) | n.d. | n.d. | n.d. | 4.7 ± 0.2 (31) | n.d. | 9.1 ± 0.5 (60) | n.d. |
| Mitochondrial complex II activity with exogenous Q1 [nmol/min/mg protein]h | 61 ± 6.0 (100) | n.d. | n.d. | n.d. | 23.1 ± 3.5 (38) | n.d. | 39.0 ± 3.5 (64) | n.d. |
The number in brackets in each cell represents the phenotype as a percent of the wild-type phenotype (mclk1+/+ ES cells)
Cell viability is expressed as the proportion of cells after 24 h of growth in menadione (6 μM), with the number on day 1 being 1 × 105
Cell multiplication is expressed as the number of cells on day 5 of growth, with the number on day 1 being 1 × 105
LIF requirement is expressed as the percentage of colonies on day 3 of growth consisting of undifferentiated cells only when the cells are grown in medium with 0.008 M LIF
Oxidative stress was induced by treatment with 0.1 mM FeSO4 and 0.2 mM sodium ascorbate
DNA damage was measured by a “comet” assay (see also Fig. 1). Cells were counted as damaged when a “comet tail” could be seen, but the size of the tail was not taken into account; the experiment was repeated three times and the means and standard errors are given
The measure of lipid peroxidation is by the TBARS assay and is given in malondialdehyde (MDA) equivalents
Exogenous Q1 is added in vitro to increase the activities measured. Significantly, complex II activity is higher when the mclk1–/– cells have grown in Q9-containing medium, even in the presence of an excess of Q1
To further explore these phenotypes we treated mclk1-/- and control cells with ubiquinone or sodium pyruvate, or both. Sodium pyruvate promotes the growth rate of cells with mitochondrial impairment by facilitating the regeneration of cytosolic NAD+ (King and Attardi 1989). Treatment with exogenous UQ at 0.16 μM did not reconstitute normal intracellular levels of UQ, which remained at least four times lower than in wild-type cells (1.12 × 10-5 vs 5.73 × 10-5 nmol/mg protein); yet, exogenous UQ was able to reach deep intracellular sites, like the mitochondrial matrix, as indicated by its effect on restoring the function of mitochondrial complex II (Table 1). Treatment with either UQ or sodium pyruvate ameliorated the low respiration, slow growth, and LIF resistance phenotypes, and treatment with both compounds almost completely rescued these phenotypes. However, resistance to menadione and ROS levels were unaffected (Table 1). This suggests that the low ROS levels and concomitant resistance to menadione-induced oxidative stress are not the result of low respiration or slow growth rate.
To reinforce our conclusion that resistance to menadione is not secondary to other phenotypes, we tested the effects of one treatment (serum starvation) as well as of a number of compounds (etoposide, anisomysin, staurosporine, all-trans retinoic acid, and sodium azide) that induce cell death, but not specifically by raising ROS levels (Supplementary Fig. 1). mclk1-/- cells were neither resistant nor hypersensitive to sodium azide and staurosporine, but these cells were resistant to etoposide, anisomycin, all-trans retinoic acid, and serum withdrawal. However, upon treatment with sodium pyruvate, which partially rescues growth rate (Table 1), the resistance of the mclk1-/- cells became indistinguishable from that of the mclk1+/+ cells (Supplementary Fig. 1). Thus, except for the resistance to menadione, the resistance to cell death-inducing agents appears to be a secondary effect of other mclk1-/- phenotypes, presumably the slow growth rate.
Reduced damage in mclk1-/- ES cells
ROS are toxic molecules that damage proteins, lipids, and nucleic acids. We wondered therefore whether the mclk1 phenotype resulted in a decrease of oxidative damage. Oxidative damage to lipids was examined by the thiobarbituric acid-reactive substances (TBARS) assay (Janero 1990), and found to be significantly lower in mclk1-/- cells than in control cells (Table 1).
To examine DNA damage, we used the comet assay (Collins 2004), an in situ method that minimizes artefacts due to extraction procedures, and in which damaged DNA is visualized as a smear of DNA coming out of lysed nuclei under electrophoresis. DNA damage in mclk1-/- cells was much less pronounced than that in the isogenic wild-type cells that were cultured in parallel (Fig. 1A,B). Furthermore, the differences seen in Figure 1B are likely to be an underestimate, as the numbers given do not take into account the sizes of the smears, which were systematically larger in the wild-type cells (Fig. 1A).
Figure 1.
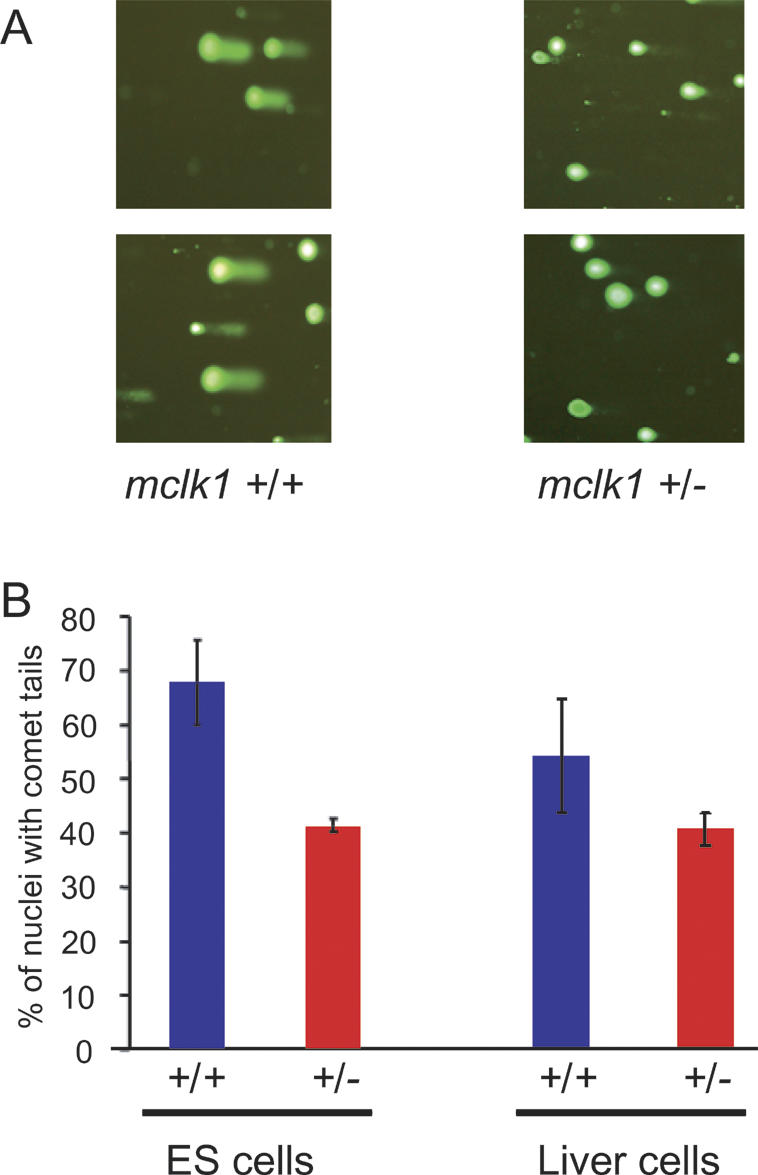
Reduction in the level of DNA damage in mclk1-/- ES cells and in mclk1+/- mice. (A) DNA damage measured by the comet assay. Staining is for DNA and the presence of a tail associated with a nucleus signals the presence of fast-migrating damaged DNA. Two independent fields of view are shown for each genotype. Much fewer mclk1-/- ES cells show nuclei with tails, and the tails are smaller. (B) The number of nuclei with tails, without consideration of the size of the tails, was determined for mclk1-/- and mclk1+/+ ES cells (three samples of 100 cells for each genotype) as well as for liver cells of mclk1+/- and mclk1+/+ mice (n = 7 mice for each genotype; three samples of 100 cells for each mouse). Error bars represent the standard deviation of the means.
Long lifespan of mclk1+/- mice
Given our observations of low levels of oxidative stress and DNA damage in mclk1-/- cells, and the fact that reducing clk-1 activity prolongs the lifespan of nematodes (Wong et al. 1995; Lakowski and Hekimi 1996), it was of interest to test the effect of reducing the activity of mclk1 on the lifespan of mice. Although 2-d-old mclk1+/- heterozygous mice display a reduced level of the mCLK1 protein (Levavasseur et al. 2001), they are fully viable. These mice are born at the expected frequency of two out of three of the live progeny of heterozygous parents (168 [65%] mclk1+/- and 89 [35%] mclk1+/+ pups from 43 litters). The growth rates and the adult weights of +/- and +/+ females are similar, but adult male +/- might be somewhat heavier than +/+ (Supplementary Fig. 2). The fertility of females is not different from that of control animals by various measures (Table 2).
Table 2.
Fertility of mclk1+/- females
| mclk1+/+ | mclk1+/- | |
|---|---|---|
| Female fertile age (weeks)a | 5.8 ± 0.5 (n = 5) | 5.7 ± 0.6 (n = 6) |
| Number of embryosa,b | 8.5 ± 2.9 (n = 8) | 8.0 ± 3.1 (n = 4) |
| Number of live newbornsc | C57BL/6J: 8.7 ± 2.0 (n = 13)d | C57BL/6J: 9.3 ± 1.8 (n = 10) |
| 129Sv/J: 7.5 ± 2.7 (n = 8) | 129Sv/J: 5.8 ± 1.7 (n = 4) | |
| Balb/c: 8.3 ± 2.7 (n = 12) | Balb/c: 7.3 ± 2.1 (n = 4) | |
| Estrus cycle length at 6 mo (days)a | 5.0 ± 0.8 (n = 5) | 5.3 ± 0.9 (n = 4) |
The genetic background of the mice was 129Sv/J
The number of embryos was determined by dissecting the embryos from the uteri of pregnant females and genotyping them at E10.5
The males used were mclk1+/- for the mclk1+/+ females, and mclk1+/+ for the mclk1+/- to obtain an equal genotype distribution for the pups
The sample size is the number of litters of live newborns examined
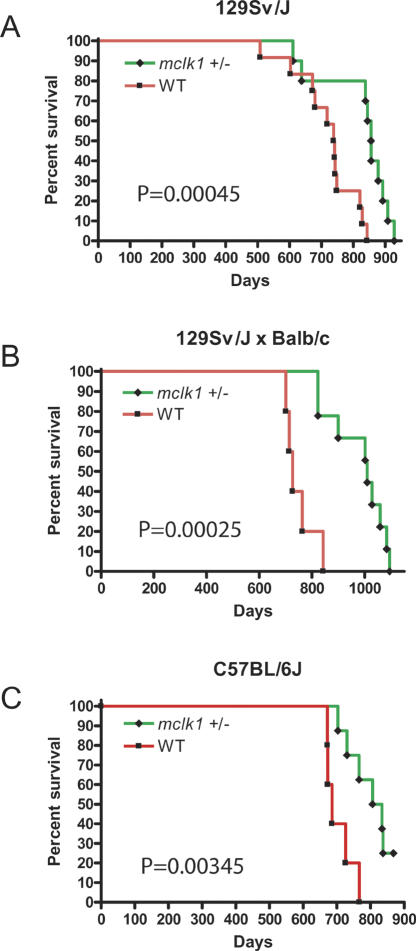
To date, we have examined the effect of mclk1 on lifespan in three genetic backgrounds; the study in the 129Sv/J background and in F1 animals from a 129Sv/J cross with Balb/c have been completed, and the study in the C57BL/6J background is ongoing (Fig. 2). We analyzed survival by the Mantel-Haenszel test and found significantly greater survival of the mclk1+/- in all three studies, p = 0.00045, p = 0.00025, and p = 0.00345, respectively (Fig. 2). In the 129Sv/J background, the maximum lifespan was 928 d for the mclk1+/- mice (n = 10) and 843 d for the wild-type animals (n = 12), and mclk1+/- mice lived on average 15% longer than their wild-type littermates (824.8 ± 103.8 vs. 720.2 ± 96.1 d). In the 129Sv/J × Balb/c background, the maximum lifespan of mclk1+/- animals (n = 9) was 1092 d and only 843 d for their wild-type siblings (n = 5). The mclk1+/- mice lived, on average, 31% longer than their wild-type littermates (980.4 ± 105.9 vs. 749.8 ± 57.2 d). Although the study in the C57BL/6J background is not complete, the available data shows a median survival of 686 d for mclk1+/+ (n = 5) and of 820 d for mclk1+/- (n = 8), a difference that is already significant at p = 0.00345.
Figure 2.
Increased lifespan of mclk1+/- mice. Kaplan-Meier survival curves are shown with p values calculated by the Mantel-Haenszel log rank test. Green indicates the mclk1+/- mice and red the wild-type mice in all three panels. (A) Lifespan extension in the 129Sv/J genetic background. mclk1+/- mice (n = 10) lived, on average, 15% longer than their wild-type (n = 12) littermates (824.8 ± 103.8 vs. 720.2 ± 96.1 d; p = 0.00045). All animals were female. (B) Lifespan extension in the 129Sv/J × Balb/c background. mclk1+/- mice (n = 9) live, on average, 31% longer than their wild-type littermates (n = 5) (980.4 ± 105.9 vs. 749.8 ± 57.2 d; p = 0.00025). (C) Lifespan extension in the C57BL/6J background. There are both males and females in the C57BL/6J study, and both sexes behave similarly. Although the study in the C57BL/6J background is not finished, the available data show a median survival of 686 d for mclk1+/+ (n = 5) and of 821.5 d for mclk1+/- (n = 8), a difference that is already significant at p = 0.00345. Currently, the median lifespan of the males is 726 d for mclk1+/+ (n = 3) and 837 d for mclk1+/- (n = 5) (p = 0.026).
The 129Sv/J animals tested were all females. The F1 and the C57BL/6J animals tested were both male and female. In the F1 study, only three of the heterozygotes and one wild-type animal were male. The average lifespans of the tested F1 females only (1019 ± 98 d for mclk1+/- vs. 762 ± 54 d for the wild type) were not meaningfully different from the average lifespans that include the males. There are both males and females in the C57BL/6J study. and both sexes behave similarly. Currently, the median lifespan of the males is 726 d for mclk1+/+ (n = 3) and 837 d for mclk1+/- (n = 5) (p = 0.0231). Overall, it appears that the lifespan of males and females are similarly affected.
A number of facts indicate that the lifespan increases we observe are robust: (1) The total number of mice in the three studies is significant (22 wild type and 27 heterozygotes); (2) all three studies show an increased lifespan for the mclk1+/- animals; (3) the observed differences in each of the three studies are statistically significant; (4) the three experiments give similar results in spite of the differences of genetic background; (5) the three experiments were carried out independently, over different time periods; (6) the control wild-type animals tested were the siblings of the heterozygotes; and (7) although the female data is more extensive, males and females show similar effects. In spite of these considerations, we are currently conducting a study with larger sample sizes to confirm these results.
Low DNA damage in mclk1+/- mice
We tested the possibility that mclk1+/- mice were experiencing lower levels of ROS damage to DNA by using the comet assay to compare the livers of mclk1+/- to those of mclk1+/+ animals (age range, 14–18 mo; n = 7 for each genotype). The mclk1+/- mice experience significantly (p < 0.05) lower levels of DNA damage by this measure (Fig. 1B).
Loss of mclk1 expression in the liver of aged mice
The magnitude of the effect on lifespan of the mclk1+/- heterozygous condition was surprising to us because, in a previous study, we did not observe a reduced level of ubiquinone in young heterozygotes (Levavasseur et al. 2001). If the phenotypic effects of reduced clk-1/mclk1 activity observed in worms and in ES cells are entirely mediated by a reduction of the level of ubiquinone, then we should not expect to observe an effect on the lifespan of the heterozygous mice. We wondered whether increased lifespan could be due to a phenomenon of LOH, and whether old heterozygous mice contained populations of mclk1-/- cells. Our findings with mclk1-/- ES cells suggested that homozygous somatic cells produced by spontaneous LOH might experience reduced oxidative stress, which could confer a growth or survival advantage resulting in expanded mclk1-/- clones. We chose to examine the liver for such a phenomenon because of the large regenerative potential of hepatocytes and other hepatic cell types, which can produce large clones in regenerating livers.
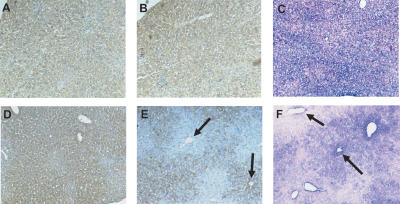
We discovered the presence of large groups of cells that do not express mclk1 in the livers of every old mclk1+/- animal examined (age range: 25–36 mo; n = 6), but not in the livers of either old mclk1+/+ (age range: 25–27 mo; n = 3), or young mclk1+/- or mclk1+/+ animals (age range: 4–6 mo; n = 3 for each genotype) (Fig. 3). Cells lacked mclk1 at the protein level, as detected with a mCLK1-specific antiserum (Fig. 3E; Jiang et al. 2001), as well as at the RNA level, as demonstrated by in situ hybridization (Fig. 3F).
Figure 3.
Groups of cells lacking mCLK1 expression can be observed in the livers of mclk1+/- mice with extended longevity. Immunohistochemical analyses with anti-mCLK1 antibody revealed that groups of cells lacked mCLK1 expression in the livers of old mclk1+/- mice only. Uniform staining is seen in young (5-mo-old) mclk1+/+ (A) and mclk1+/- (D) mice. However, while there is uniform staining in 25-mo-old mclk1+/+ mice (B), the staining is patchy in similar mclk1+/- mice (E). (E) Large groups of cells without staining surround the central veins (upper arrow) and appear to expand throughout the whole classical hepatic lobule, which is the region drained by a central vein. (E) Other central veins appear surrounded by mCLK1-positive cells only (e.g., lower arrow). RISH with antisense DIG-labeled probe for mclk1 similarly showed uniformly positive cells in 25-mo-old mclk1+/+ mice (C), but in similar mclk1+/- mice (F) there were groups of cells that either lacked (e.g., left arrow) or expressed (e.g., right arrow) the signal for the mclk1 transcripts (blue). The nuclei (pink) were counterstained by nuclear fast red.
It is of note that although we were originally looking for evidence of loss of mclk1 expression to explain the increased longevity of the mclk1+/- mice, no data beyond the sheer existence of clones supports or refutes a causal link between the two phenomena (see Discussion).
LOH at the mclk1 locus
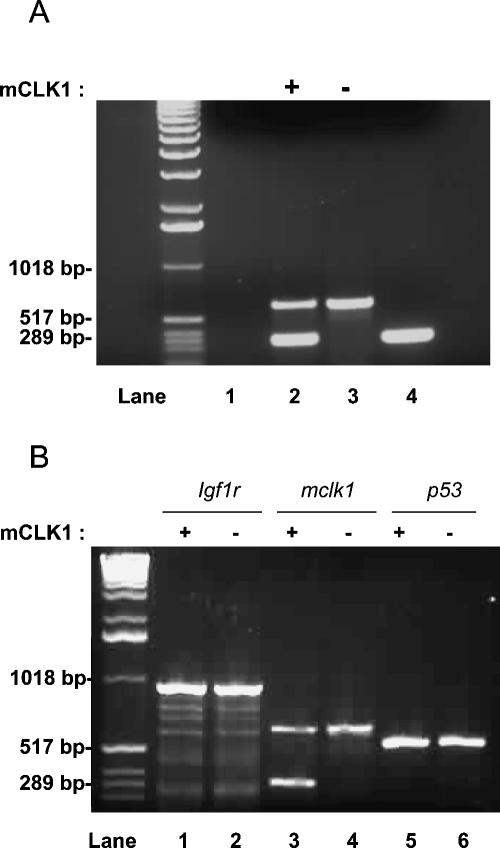
A classic mechanism to account for the total loss of expression of a gene in a subset of cells of a heterozygous (+/-) animal is LOH, the spontaneous mutational deletion of the wild-type allele (Thiagalingam et al. 2002). To investigate whether this is the mechanism behind the appearance of mclk1-/- clones, we used laser-capture microdissection (LCM) (De Preter et al. 2003). Cells from liver sections in which mclk1-expressing and nonexpressing areas were identified by immunocytochemistry, were obtained by LCM. DNA extracted from the captured cells was submitted to whole-genome amplification by the multiple-strand displacement amplification (MDA) technique (Dean et al. 2002). The amplified DNA was then analyzed by PCR for the presence of mclk1-, igf1r-, and p53-specific sequences. mclk1 and igf1r are both on chromosome 7 and p53 is on chromosome 11. Cells from eight mCLK1-expressing, and eight mCLK1-negative clones were examined in this way. In six of eight cases, we found that mCLK1-negative cells contained only sequences from the allele disrupted by targeted recombination (Fig. 4). In two of eight cases, for reasons that are not yet clear, we could not observe any mclk1-specific amplification from the mCLK1-negative cells. However, in all cases, we could amplify mclk1-specific products from mCLK1-expressing clones. Furthermore, we could always amplify igf1r- and p53-specific sequences from mCLK1-negative and mCLK1-expressing captured cells (Fig. 4B). In conclusion, the loss of expression of mCLK1 appears to be linked to the specific loss of the wild-type allele of mclk1.
Figure 4.
LOH at the mclk1 locus. LCM of groups of 20–30 cells were obtained from mCLK1-negative (-) or mCLK1-positive (+) regions of sections from livers of old mclk1+/- animals stained for the mCLK1 protein by immunocytochemistry. DNA isolated from these cells was then amplified by whole-genome MDA. Amplified DNA was used for PCR amplification with mclk1-specfic primers. This yields two products from mclk1 heterozygous DNA: one corresponding to the wild-type gene (300 bp) and a larger one corresponding to the disrupted allele (600 bp). (A) DNA specifically corresponding to the wild-type mclk1 allele is lost from cells that do not express mCLK1. (Lane 1) Negative control provided by LCM buffer, without any captured cells, but which subsequently underwent all procedures (DNA extraction, MDA, and PCR). (Lane 2) PCR from captured cells expressing mCLK1. (Lane 3) PCR from captured cells, not expressing mCLK1. (Lane 4) PCR production from DNA of a wild-type mouse tail (positive control). (B) Control for extracted DNA quality. Wild-type DNA from the igf1r locus (on chromosome 11) and p53 locus (on chromosome 7) can be unfailingly PCR amplified from both mCLK1-negative and mCLK1-positive cells. The same sample obtained by LCM and whole-genome MDA is being used for PCR in lanes 1, 3, and 5 from a mCLK1-positive group of cells, and in lanes 2, 4, and 6 from a mCLK1-negative group of cells.
Clonal expansion of mclk1-/- cells
Strikingly, the distribution of mclk1-/- cells was not random with respect to the main microanatomical compartment of the liver, the lobule, which is the region drained by a single vein of the microvasculature. In fact, the clones generally appeared to be of a similar size and frequently appeared to correspond to entire lobules (Fig. 3). Affected lobules were quite numerous, representing as much as 50% of the tissue in certain regions of the liver. The existence of large clones that have lost mclk1 expression suggests a mechanism in which loss of mclk1 expression confers a growth advantage to single hepatocytes, which over time allows their progeny to replace all hepatocytes in the lobule in which they originated.
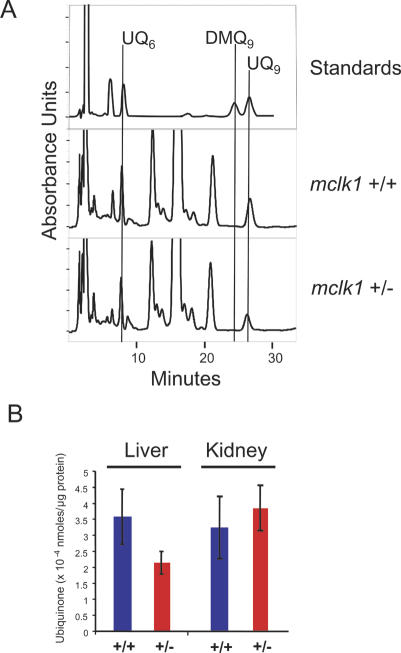
We also found that the livers, but not the kidneys of relatively old mclk1+/- animals (age range: 14–22 mo) contained less ubiquinone relative to protein than those of mclk1+/+ animals (Fig. 5B). As no difference in ubiquinone content is observed in the liver of young animals (Levavasseur et al. 2001), these findings are consistent with an age-dependent increase of liver cells that have lost mclk1 expression. Interestingly, we did not observe the presence of DMQ in these livers (Fig. 5A), nor did we detect DMQ in any other organ (data not shown). This suggests that, in contrast to what is observed in ES cells (Levavasseur et al. 2001), the UQ synthesis pathway is turned off in adult hepatocytes in the absence of the mCLK1 protein.
Figure 5.
Quinones in mclk1+/- mice. (A) Reverse-phase HPLC chromatograms show the elution of UQ6, DMQ9, and UQ9 standards, and the elution of quinones from representative livers of mclk1+/- and mclk1+/+ mice. UQ6 is added in the liver samples as an internal standard. No DMQ9 peak was detected in any of the liver samples from mclk1+/- animals (n = 7; age range: 14–22 mo). (B) Ubiquinone levels in livers and kidneys of mclk1+/- mice. In the livers, but not in the kidneys, ubiquinone levels were significantly decreased compared with that in wild-type littermates (n = 7 for each genotype; three measurements were taken for each liver; p = 0.0024). The error bars represent the 95% confidence interval (approximately two times the standard error of the mean).
Discussion
Aging and oxidative stress
We find that reducing the activity of mclk1 reduces ROS levels, oxidative stress, and oxidative damage in mouse cells, and prolongs the lifespan of whole animals. Such a correlation between lifespan and the level of oxidative stress and its consequences has frequently been observed and has led to the oxidative stress theory of aging. Decreased oxidative stress must at least be considered a marker for a physiological condition that favors increased lifespan. An increased resistance to some type of oxidative stress has been frequently found in association with increased lifespan in genetic models, including in the long-lived dwarf mice (Hauck et al. 2002), igf1r+/- mice (Holzenberger et al. 2003; Baba et al. 2005) and p66shc-/- mice (Migliaccio et al. 1999; Nemoto and Finkel 2002; Napoli et al. 2003). Our results with mclk1 strengthen the generality of the observation that resistance to oxidative stress accompanies increased lifespan. In addition, the fact that reduction of mclk1 activity has similar ROS-reducing and lifespan-lengthening effects in mice and in nematodes, and that the mclk1 gene product is an enzyme involved in the production of a major cellular redox cofactor also suggests a causal link between ROS reduction and increased longevity.
Increased fitness of mclk1-/- cells
We find that in the livers of every old mclk1+/- animal examined, entire hepatic lobules have lost mclk1 expression. Hepatic lobules appear to be either entirely positive or entirely negative for mclk1 expression. This suggests a model in which random mclk1 inactivation in a single cell of a lobule leads to clonal expansion within the microanatomical compartment of the lobule. It is reasonable to expect that the hepatocytes in which mclk1 is inactivated will have a number of properties in common with the mclk1-/- ES cells. Thus, the observed phenomenon of clonal expansion might be due to increased resistance of these cells to age-dependent oxidative stress and apoptosis. As the capacity of the liver to regenerate decreases with age (Fry et al. 1984), the mclk1-/- cells might be the only cells in these old livers that have sustained sufficiently little damage and are sufficiently resistant to stress to be capable of extensive propagation.
To date, we do not know when the mclk1-/- clones arise during the life of the animals, except that none are observed at 5 mo of age. Thus, it is possible that, rather than being the result of the response to acute age-related stresses, the clones arise gradually as the result of the normal process of cell turnover. Although mclk1-/- grow slowly in culture, the rate of cell division during normal cell turnover, which is a very slow process, is probably not limiting. Given the importance of ROS in the regulation of apopotosis, it is possible that the expected low level of ROS in mCLK1-negative hepatocytes makes spontaneously arising mclk1-/- and their descendants resistant to the physiological apoptosis that is part of normal cell turnover in tissues, which would allow them to expand slowly at the expense of cells that sustain higher levels of apoptosis.
LOH at the mclk1 locus
The phenomenon of LOH can be observed in animals heterozygous for a loss-of-function mutation in a tumor-suppressor gene (Knudson 1993; Devilee et al. 2001). The cells that spontaneously lose the second allele of the gene, for example, through the loss of an entire chromosome or a large section of a chromosome, escape normal growth controls and clonally expand into a tumor. LOH is but one of the consequences of the accumulation of somatic mutations that is one of the proposed mechanisms of aging (Hasty et al. 2003). However, our results suggest that, under special circumstances, these normally deleterious processes could act favorably by inactivating the remaining wild-type allele in animals heterozygous for a gene whose normal activity limits lifespan.
Basis of the increased cellular fitness
It has been a question as to whether the increase in lifespan of clk-1 mutants in C. elegans was due to the presence of DMQ (Jonassen et al. 2002; Shibata et al. 2003). Here we find that the livers, but not the kidneys of relatively old mclk1+/- mice have lower levels of ubiquinone than those of mclk1+/+ animals (Fig. 5B). This is presumably due to the presence of mclk1-/- cells, as such difference in ubiquinone content between genotypes is not observed in younger animals (Levavasseur et al. 2001). However, there is no detectabe DMQ in these livers (Fig. 5A) or in any other organ of these animals (data not shown). This suggests that in old liver cells, in contrast to what was observed in ES cells (Levavasseur et al. 2001) and in worms (Miyadera et al. 2001), the entire pathway of ubiquinone biosynthesis is turned off when mCLK1 is absent, a phenomenon that is also observed in yeast (Marbois and Clarke 1996). Thus, the low level of DNA damage in old mclk1+/- livers, the apparent growth advantage of mclk1-/- cells, and the increased lifespan of these animals cannot be due to the presence of DMQ.
The above considerations suggest that, if the growth advantage and the decreased DNA damage are due to low oxidative stress, then the ubiquinone normally present in wild-type cells is, in fact, contributing to oxidative stress, as has been suggested for nematodes (Larsen and Clarke 2002). How could we explain the presence of deleterious amounts of ubiquinone in animal cells? The explanation likely hinges on the fact that ubiquinone has both pro-oxidant and antioxidant properties. For example, although ubiquinone is a membrane antioxidant, its function in the ubiquinone cycle of the respiratory chain promotes the formation of superoxide when it is in the ubisemiquinone state. Thus, the normal level of ubiquinone is likely a compromise between the need for antioxidant protection from acute stresses and its pro-oxidant role as cofactor. The amount of ubiquinone that is adequate to protect from acute oxidative stresses, such as can be brought about by heat stress, irradiation, or transient anoxia, might in fact participate in creating chronic oxidative stress.
Molecular basis of increased lifespan of mclk1+/- mice
The observations made above suggest two distinct possibilities for the increased lifespan of mclk1+/- animals. The first possibility is that the presence of clones of mclk1-/- cells could be sufficiently beneficial for the animal as a whole. This could be the case if there was a net loss of cells in some organs without the presence of mclk1+/- cells, or if these cells were somehow physiologically superior. However, all of the animals we examined were part of our aging study and were examined shortly before natural death, with most organs in a state of relative deterioration. Therefore, although we found clones only in the liver, the data for other organs such as the kidney and the gut was not of sufficient quality to be able to conclude firmly whether there were clones or not. Yet, even if mclk1-/- clones can develop only in the liver, due to the regeneration potential of hepatocytes, this might be sufficient to result in increased lifespan thanks to the important role of the liver in digestion, detoxification, and the regulation of circulating glucose levels.
The second possibility is that the presence of reduced amounts of mCLK1 protein in all of the cells of the mclk1+/- animals is the lifespan-lengthening factor. Studies in ES cells, in embryos, and in young animals (Levavasseur et al. 2001), as well as the results presented here with the kidneys of old mclk1+/- animals, suggest that mclk1 is recessive for ubiquinone biosynthesis. Yet it is possible that reduction of mclk1 expression might bring about an undetected minor reduction of ubiquinone levels, or a reduction in particular cell types, or during particular physiological conditions, that could be favorable for longevity by increasing resistance to damage at significant times and/or places.
Significance of evolutionary conservation
It is a broadly, if not universally accepted, view that aging is a consequence of the gradual accumulation of unrepaired molecular damage produced by endogenous processes or the environment. Thus, given that each species has its own physiology and environment, it is likely that there are species-specific processes that promote or protect from damage or damage accumulation. However, the evolutionary conservation of the longevity-promoting effect of clk-1/mclk1 mutations indicates that there are also processes that are shared between animals of such disparate morphologies, physiologies, and ecologies as worms and mice. This might have its basis in the universal conservation of the function of small molecular-weight effectors such as ubiquinone and ROS.
Materials and methods
Cell culture
ES cells were grown in high glucose Dulbecco's modified Eagle's medium (-pyruvate, -glutamine) supplemented with 20% fetal bovine serum, glutamine, β-mercaptoethanol, and LIF on feeder-free, gelatin-coated dishes at 37°C in an atmosphere of 5% CO2 and 95% air.
Growth rate and cell death assay
Cells were seeded in six-well dishes at 1 × 105/well in ES cell medium with or without supplements (UQ9: 0.16 μM, sodium pyvurate: 110 mg/L). These concentrations were established experimentally as the maximum concentrations at which no effect on the growth rate of wild-type cells is observed. At different time points, the cells were trypsinized and counted with a hemocytometer. The cells were also analyzed by the trypan blue exclusion method 24 h after treatment with menadione (6 μM).
LIF requirement assay
ES cells were plated at a density of 500 cells/cm2 in gelatin-coated six-well plates in ES cell medium containing various concentration of LIF. Three days after inoculation, the proportion of undifferentiated colonies was determined after scoring the morphology of 300 randomly chosen colonies.
ROS measurement
For the fluorimetric measurement of ROS, cells were incubated with 10 μM DCHF-DA (Molecular Probes) for 20 min at 37°C. ROS levels were measured fluorimetrically with excitation and emission wavelengths of 495 and 530 nm, respectively. Oxidative stress was induced by incubating the cells loaded with DCHF-DA with 0.1 mM FeSO4 and 0.2 mM sodium ascorbate.
DNA damage assay
DNA damage of cells was measured by using a single-cell gel electrophoresis assay (CometAssay, Trevigen) according to the manufacturer's instructions. The Comet tails were scored by examining the fixed and stained cells under a fluorescence microscope with ×10 Planoapo objective. One-hundred cells were scored per sample.
Lipid peroxidation assay
Lipid peroxidation was measured using a TBARS assay kit (ZeptoMetrix) according to the manufacturer's instructions. A standard curve was generated by using known amounts of malondialdehyde.
Mitochondrial activity
Mitochondrial complex II activity was measured as follows: mitochondria (containing 50 μg protein) were preincubated with reaction buffer (25 μM potassium phosphate, 5 μM MgCl2, 20 μM succinate) for 10 min at room temperature. The reaction was started by adding 2 μg/mL Antimycin A, 2 μg/mL Rotenone, 2 mM KCN, 50 μM 2,6-Dichlorophenolindophenol (Sigma) in the presence or absence of 65 μM Q1. The linear decrease in absorbance at 600 nm was recorded (ε = 21 mM-1 cm-1).
Animals
The mclk1 knockout mutant was described previously (Levavasseur et al. 2001) and was maintained in the heterozygous state in the 129Sv/J genetic background. By mating mclk1+/- males to 129Sv/J or Balb/c wild-type females, we generated isogenic mclk1+/- and mclk1+/+ littermates. mclk1+/- in the C57BL/6J background were obtained by back-crossing mclk1+/- animals in the 129Sv/J background to C57BL/6J animals six times, and then maintaining them by brother/sister matings. All of the animals were housed in a pathogen-free facility at McGill University and were given a standard rodent diet and water ad libitum. The mice were separated from their mother at 21 d of age and housed three to five per cage, with both genotypes present in each cage. Lifespan was determined by recording the age of spontaneous death, or when one of the following criteria was met: unresponsiveness to touch, slow respiration, coldness to touch, a hunched up position with matted fur, or sudden weight loss.
The onset of fertility was determined by mating mclk1+/- and mclk1+/+ female mice from 28 d onward with fertile wild-type males. We determined the estrus cycle by observing sexual behaviors, recording vaginal plugs as well as the resulting pregnancies and offsprings, and by examining vaginal smear histologically (daily for 2 wk at the age of 6 mo).
Statistical analysis of survival
Survival was graphed by the Kaplan-Meier method and analyzed by the Mantel-Haenszel test, which is a log rank test designed to test the difference between two survival curves. We present the one-tailed p value because we are testing the hypothesis that mclk1+/- animals live longer, not longer or shorter than mclk1+/+ animals. For the unfinished study (in the C57BL/6J background), only the median survival is presented, as the lifespan of the most long-lived animals is unknown.
Immunohistochemistry
Immunohistochemical analysis of mCLK1 expression was performed on formalin-fixed paraffin sections (4 μm) of livers from mice sacrified at 5 mo of age or at natural death. The anti-mCLK1 serum was described previously (Levavasseur et al. 2001). All antibodies were used at a 1:100 dilution. The avidin-biotin-peroxidase method was used for visualization with 3,3′-diaminobenzidine-tetrahydrochloride as substrate, producing a brown reaction product. For the negative control, primary antibody was replaced with nonimmunized rabbit serum.
In situ hybridization
A mclk1 cDNA was cloned into the RNA expression vector pSPT18 and labeled with DIG following standard procedures. The vector was linearized to allow in vitro run-off synthesis of both sense- and antisense-oriented RNA probes. Paraffin-embedded tissue sections were subjected to a nonradioactive RNA in situ hybridization (RISH) using the DIG-labeled antisense mclk1 probes. Hybridization with the corresponding sense probes served as control. Sections were incubated with Anti-Dioxigenin-AP and NBT/BCIP color developing solution to visualize the mclk1 transcript signal; 0.1% nuclear fast red was used for counterstaining.
LCM and DNA extraction, whole genome amplification and PCR
We used a PixCell IIe Laser Capture Microdissection System (Arcturus) to pick up 20–30 cells from mCLK1-negative or mCLK1-positive regions of liver sections from old mclk1+/- animals stained for the mCLK1 protein by immunocytochemistry. DNA was then isolated with a PicoPure DNA extraction kit (Arcturus).
To use the minute quantities of DNA that are available through LCM, we performed whole-genome MDA with the GenomiPhi DNA amplification kit (Amersham Bioscience). PCR with amplified DNA was carried out using the specific primers for mclk1, igf1r, and p53. PCR conditions and primer sequences are available upon request.
HPLC analysis
Quinones were extracted as described (Miyadera et al. 2001), with slight modifications. Briefly, the quinones extracted in ethanol and hexane were evaporated with a freezing Speed Vac dryer and kept frozen at -80°C. Shortly after reconstitution with mobile phase (70% methanol and 30% ethanol), the samples were loaded on a reverse phase column (Inertsil ODS-3 C8-3, Ph-3, GL Science) and elution was monitored by a UV light detector at 275 nm. The amount of quinones was determined by comparison to known standards using the 32 Karat software (Beckman Coulter Inc.).
Acknowledgments
We thank Sophie Debellefeuille and Ying Wang for expert technical assistance and Françoise Levavasseur for initiating aging studies in our laboratory. We thank Robyn Branicky for reading the manuscript and for invaluable help with data analysis. We thank Dr. Svetlana Sadekova and Margarita Soul for access to and help with laser capture equipment, and Lily Li for help with histology. N.J. carried out all of the experiments with ES cells. S.H. is Strathcona Professor of Zoology. E.A.S. is an international scholar of the Howard Hughes Medical Institute and a senior investigator of the CIHR. This study was funded in part by a research contract from Chronogen Inc.
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1352905.
References
- Baba T., Shimizu, T., Suzuki, Y., Ogawara, M., Isono, K., Koseki, H., Kurosawa, H., and Shirasawa, T. 2005. Estrogen, insulin, and dietary signals cooperatively regulate longevity signals to enhance resistance to oxidative stress in mice. J. Biol. Chem. 280: 16417-16426. [DOI] [PubMed] [Google Scholar]
- Benard C., McCright, B., Zhang, Y., Felkai, S., Lakowski, B., and Hekimi, S. 2001. The C. elegans maternal-effect gene clk-2 is essential for embryonic development, encodes a protein homologous to yeast Tel2p and affects telomere length. Development 128: 4045-4055. [DOI] [PubMed] [Google Scholar]
- Bluher M., Kahn, B.B., and Kahn, C.R. 2003. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science 299: 572-574. [DOI] [PubMed] [Google Scholar]
- Collins A.R. 2004. The comet assay for DNA damage and repair: Principles, applications, and limitations. Mol. Biotechnol. 26: 249-261. [DOI] [PubMed] [Google Scholar]
- De Preter, K., Vandesompele, J., Heimann, P., Kockx, M.M., Van Gele, M., Hoebeeck, J., De Smet, E., Demarche, M., Laureys, G., Van Roy, N., et al. 2003. Application of laser capture microdissection in genetic analysis of neuroblastoma and neuroblastoma precursor cells. Cancer Lett. 197: 53-61. [DOI] [PubMed] [Google Scholar]
- Dean F.B., Hosono, S., Fang, L., Wu, X., Faruqi, A.F., Bray-Ward, P., Sun, Z., Zong, Q., Du, Y., Du, J., et al. 2002. Comprehensive human genome amplification using multiple displacement amplification. Proc. Natl. Acad. Sci. 99: 5261-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devilee P., Cleton-Jansen, A.M., and Cornelisse, C.J. 2001. Ever since Knudson. Trends Genet. 17: 569-573. [DOI] [PubMed] [Google Scholar]
- Dillin A., Hsu, A., Arantes-Oliveira, N., Lehrer-Graiwer, J., Hsin, H., Fraser, A.G., Kamath, R.S., Ahringer, J., and Kenyon, C. 2002. Rates of behavior and aging specified by mitochondrial function during development. Science 298: 2398-2401. [DOI] [PubMed] [Google Scholar]
- Ewbank J.J., Barnes, T.M., Lakowski, B., Lussier, M., Bussey, H., and Hekimi, S. 1997. Structural and functional conservation of the Caenorhabditis elegans timing gene clk-1. Science 275: 980-983. [DOI] [PubMed] [Google Scholar]
- Feng J., Bussiere, F., and Hekimi, S. 2001. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell 1: 633-644. [DOI] [PubMed] [Google Scholar]
- Fry M., Silber, J., Loeb, L.A., and Martin, G.M. 1984. Delayed and reduced cell replication and diminishing levels of DNA polymerase-α in regenerating liver of aging mice. J. Cell. Physiol. 118: 225-232. [DOI] [PubMed] [Google Scholar]
- Georgellis D., Kwon, O., and Lin, E.C. 2001. Quinones as the redox signal for the arc two-component system of bacteria. Science 292: 2314-2316. [DOI] [PubMed] [Google Scholar]
- Hasty P., Campisi, J., Hoeijmakers, J., van Steeg, H., and Vijg, J. 2003. Aging and genome maintenance: Lessons from the mouse? Science 299: 1355-1359. [DOI] [PubMed] [Google Scholar]
- Hauck S.J., Aaron, J.M., Wright, C., Kopchick, J.J., and Bartke, A. 2002. Antioxidant enzymes, free-radical damage, and response to paraquat in liver and kidney of long-living growth hormone receptor/binding protein gene-disrupted mice. Horm. Metab. Res. 34: 481-486. [DOI] [PubMed] [Google Scholar]
- Hekimi S. and Guarente, L. 2003. Genetics and the specificity of the aging process. Science 299: 1351-1354. [DOI] [PubMed] [Google Scholar]
- Hihi A.K., Gao, Y., and Hekimi, S. 2002. Ubiquinone is necessary for Caenorhabditis elegans development at mitochondrial and non-mitochondrial sites. J. Biol. Chem. 277: 2202-2206. [DOI] [PubMed] [Google Scholar]
- Holzenberger M., Dupont, J., Ducos, B., Leneuve, P., Geloen, A., Even, P.C., Cervera, P., and Le Bouc, Y. 2003. IGF-1 receptor regulates lifespan and resistance to oxidative stress in mice. Nature 421: 182-187. [DOI] [PubMed] [Google Scholar]
- Hsin H. and Kenyon, C. 1999. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399: 362-366. [DOI] [PubMed] [Google Scholar]
- Janero D.R. 1990. Malondialdehyde and thiobarbituric acid-reactivity as diagnostic indices of lipid peroxidation and peroxidative tissue injury. Free Rad. Biol. Med. 9: 515-540. [DOI] [PubMed] [Google Scholar]
- Jia K., Chen, D., and Riddle, D.L. 2004. The TOR pathway interacts with the insulin signaling pathway to regulate C. elegans larval development, metabolism and life span. Development 131: 3897-3906. [DOI] [PubMed] [Google Scholar]
- Jiang N., Levavasseur, F., McCright, B., Shoubridge, E.A., and Hekimi, S. 2001. Mouse CLK-1 is imported into mitochondria by an unusual process that requires a leader sequence but no membrane potential. J. Biol. Chem. 276: 29218-29225. [DOI] [PubMed] [Google Scholar]
- Joeng K.S., Song, E.J., Lee, K.J., and Lee, J. 2004. Long lifespan in worms with long telomeric DNA. Nat. Genet. 36: 607-611. [DOI] [PubMed] [Google Scholar]
- Jonassen T., Larsen, P.L., and Clarke, C.F. 2001. A dietary source of coenzyme Q is essential for growth of long-lived Caenorhabditis elegans clk-1 mutants. Proc. Natl. Acad. Sci. 98: 421-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonassen T., Marbois, B.N., Faull, K.F., Clarke, C.F., and Larsen, P.L. 2002. Development and fertility in Caenorhabditis elegans clk-1 mutants depend upon transport of dietary coenzyme Q8 to mitochondria. J. Biol. Chem. 277: 45020-45027. [DOI] [PubMed] [Google Scholar]
- Kayser E.B., Sedensky, M.M., and Morgan, P.G. 2004. The effects of complex I function and oxidative damage on lifespan and anesthetic sensitivity in Caenorhabditis elegans. Mech. Age. Dev. 125: 455-464. [DOI] [PubMed] [Google Scholar]
- Kenyon C. 2001. A conserved regulatory system for aging. Cell 105: 165-168. [DOI] [PubMed] [Google Scholar]
- ____. 2005. The plasticity of aging: Insights from long-lived mutants. Cell 120: 449-460. [DOI] [PubMed] [Google Scholar]
- Kenyon C., Chang, J., Gensch, E., Rudner, A., and Tabtiang, R. 1993. A C. elegans mutant that lives twice as long as wild type. Nature 366: 461-464. [DOI] [PubMed] [Google Scholar]
- Kimura K.D., Tissenbaum, H.A., Liu, Y., and Ruvkun, G. 1997. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science 277: 942-946. [DOI] [PubMed] [Google Scholar]
- King M.P. and Attardi, G. 1989. Human cells lacking mtDNA: Repopulation with exogenous mitochondria by complementation. Science 246: 500-503. [DOI] [PubMed] [Google Scholar]
- Knudson A.G. 1993. Antioncogenes and human cancer. Proc. Natl. Acad. Sci. 90: 10914-10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowski B. and Hekimi, S. 1996. Determination of life-span in Caenorhabditis elegans by four clock genes. Science 272: 1010-1013. [DOI] [PubMed] [Google Scholar]
- ____. 1998. The genetics of caloric restriction in Caenorhabditis elegans. Proc. Natl. Acad. Sci. 95: 13091-13096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen P.L. and Clarke, C.F. 2002. Extension of life-span in Caenorhabditis elegans by a diet lacking coenzyme Q. Science 295: 120-123. [DOI] [PubMed] [Google Scholar]
- Lee S.S., Lee, R.Y., Fraser, A.G., Kamath, R.S., Ahringer, J., and Ruvkun, G. 2002. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet. 25: 25. [DOI] [PubMed] [Google Scholar]
- Levavasseur F., Miyadera, H., Sirois, J., Tremblay, M.L., Kita, K., Shoubridge, E., and Hekimi, S. 2001. Ubiquinone is necessary for mouse embryonic development but is not essential for mitochondrial respiration. J. Biol. Chem. 276: 46160-46164. [DOI] [PubMed] [Google Scholar]
- Marbois B.N. and Clarke, C.F. 1996. The COQ7 gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J. Biol. Chem. 271: 2995-3004. [DOI] [PubMed] [Google Scholar]
- Migliaccio E., Giorgio, M., Mele, S., Pelicci, G., Reboldi, P., Pandolfi, P.P., Lanfrancone, L., and Pelicci, P.G. 1999. The p66shc adaptor protein controls oxidative stress response and life span in mammals. Nature 402: 309-313. [DOI] [PubMed] [Google Scholar]
- Miyadera H., Amino, H., Hiraishi, A., Taka, H., Murayama, K., Miyoshi, H., Sakamoto, K., Ishii, N., Hekimi, S., and Kita, K. 2001. Altered quinone biosynthesis in the long-lived clk-1 mutants of Caenorhabditis elegans. J. Biol. Chem. 276: 7713-7716. [DOI] [PubMed] [Google Scholar]
- Napoli C., Martin-Padura, I., de Nigris, F., Giorgio, M., Mansueto, G., Somma, P., Condorelli, M., Sica, G., De Rosa, G., and Pelicci, P. 2003. Deletion of the p66Shc longevity gene reduces systemic and tissue oxidative stress, vascular cell apoptosis, and early atherogenesis in mice fed a high-fat diet. Proc. Natl. Acad. Sci. 100: 2112-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemoto S. and Finkel, T. 2002. Redox regulation of forkhead proteins through a p66shc-dependent signaling pathway. Science 295: 2450-2452. [DOI] [PubMed] [Google Scholar]
- Schriner S.E., Linford, N.J., Martin, G.M., Treuting, P., Ogburn, C.E., Emond, M., Coskun, P.E., Ladiges, W., Wolf, N., Van Remmen, H., et al. 2005. Extension of murine lifespan by overexpression of catalase targeted to mitochondria. Science 308: 1909-1911. [DOI] [PubMed] [Google Scholar]
- Shibata Y., Branicky, R., Landaverde, I.O., and Hekimi, S. 2003. Redox regulation of germline and vulval development in Caenorhabditis elegans. Science 302: 1779-1782. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S., Foy, R.L., Cheng, K.H., Lee, H.J., Thiagalingam, A., and Ponte, J.F. 2002. Loss of heterozygosity as a predictor to map tumor suppressor genes in cancer: Molecular basis of its occurrence. Curr. Opin. Oncol. 14: 65-72. [DOI] [PubMed] [Google Scholar]
- Van Remmen H., Ikeno, Y., Hamilton, M., Pahlavani, M., Wolf, N., Thorpe, S.R., Alderson, N.L., Baynes, J.W., Epstein, C.J., Huang, T.T., et al. 2003. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol. Genomics 16: 29-37. [DOI] [PubMed] [Google Scholar]
- Vellai T., Takacs-Vellai, K., Zhang, Y., Kovacs, A.L., Orosz, L., and Muller, F. 2003. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature 426: 620. [DOI] [PubMed] [Google Scholar]
- Wong A., Boutis, P., and Hekimi, S. 1995. Mutations in the clk-1 gene of Caenorhabditis elegans affect developmental and behavioral timing. Genetics 139: 1247-1259. [DOI] [PMC free article] [PubMed] [Google Scholar]