Abstract
IκB proteins associate with the transcription factor NF-κB via their ankyrin repeat domain. Bcl-3 is an unusual IκB protein because it is primarily nucleoplasmic and can lead to enhanced NF-κB-dependent transcription, unlike the prototypical IκB protein IκBα, which inhibits NF-κB activity by retaining it in the cytoplasm. Here we report the 1.9 Å crystal structure of the ankyrin repeat domain of human Bcl-3 and compare it with that of IκBα bound to NF-κB. The two structures are highly similar over the central ankyrin repeats but differ in the N-terminal repeat and at the C-terminus, where Bcl-3 contains a seventh repeat in place of the acidic PEST region of IκBα. Differences between the two structures suggest why Bcl-3 differs from IκBα in selectivity towards various NF-κB species, why Bcl-3 but not IκBα can associate with its NF-κB partner bound to DNA, and why two molecules of Bcl-3 but only one of IκBα can bind to its NF-κB partner. Comparison of the two structures thus provides an insight into the functional diversity of IκB proteins.
Keywords: Bcl-3/IκB proteins/NF-κB transcription factors
Introduction
Bcl-3 is a putative oncoprotein first discovered during studies of B-cell chronic lymphocytic leukemias (Ohno et al., 1990). Bcl-3 belongs to the IκB family of proteins, which modulate the DNA-binding activity and subcellular localization of the transcription factor NF-κB (reviewed in Ghosh et al., 1998). NF-κB regulates the expression of a wide variety of cellular and viral genes, including those involved in immune and stress responses, apoptosis and cellular proliferation. NF-κB is a homo- or heterodimer of proteins belonging to the Rel family, which includes p65 (RelA), RelB, c-Rel, p50 and p52, with the p50–p65 heterodimer constituting the principal NF-κB species. Rel proteins share a 300-residue Rel homology region (RHR) responsible for dimerization, DNA binding, nuclear localization and binding of IκB proteins. Crystal structures of the DNA-bound RHRs of several Rel proteins reveal that the RHR consists of two immunoglobin-like domains followed by a C-terminal basic type I nuclear localization signal (NLS) that is disordered (Ghosh et al., 1995; Müller et al., 1995; Cramer et al., 1997; F.E.Chen et al., 1998; Y.Q.Chen et al., 1998). The N-terminal immunoglobin-like domain (RHR-n) is primarily responsible for DNA binding specificity, whereas subunit dimerization is exclu sively mediated by the C-terminal domain (RHR-c). While p65, c-Rel and RelB contain a C-terminal trans-activation domain, p50 and p52 do not and thus behave as transcriptional repressors in their homodimeric forms (Baeuerle and Henkel, 1994).
The mammalian members of the IκB family include Bcl-3, the IκB proteins α, β, γ and ε, and the Rel protein precursors p100 and p105. These proteins are characterized by N- and C-terminal domains of variable length and sequence and by a conserved central ankyrin (ANK) repeat domain (ARD) that mediates the interaction with NF-κB. The ARD contains six or seven copies of the ANK repeat, a 33-residue sequence motif present in a large number of eukaryotic and prokaryotic proteins (reviewed in Michaely and Bennett, 1992; Bork, 1993). Crystal structures of several such proteins reveal that ANK repeats form an L-shaped structure composed of a β-hairpin and two antiparallel α-helices packing together to yield a left-handed superhelix (reviewed in Sedgwick and Smerdon, 1999).
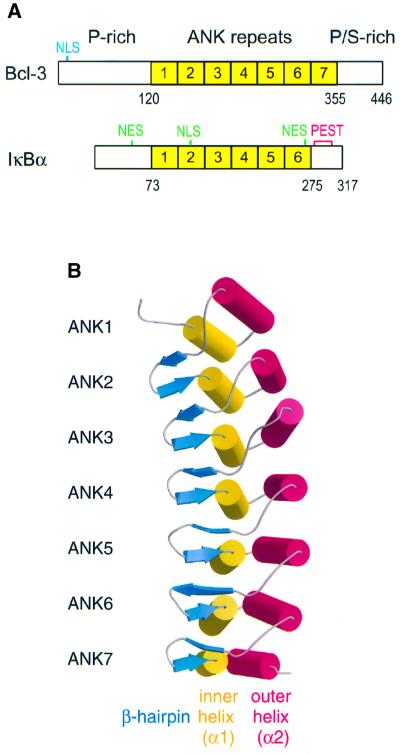
The various IκB proteins target different NF-κB species: IκBα and IκBβ bind preferentially to p50–p65 and p50–c-Rel heterodimers (Thompson et al., 1995), IκBε associates only with homo- or heterodimeric complexes containing p65 and c-Rel (Simeonidis et al., 1997; Whiteside et al., 1997), and Bcl-3 interacts specifically with p50 and p52 homodimers (Franzoso et al., 1992; Wulczyn et al., 1992). In contrast, p100 and p105, which are precursors of the Rel proteins p50 and p52, respectively, appear to bind efficiently to all mammalian Rel proteins. The best characterized IκB protein is IκBα, which contains an N-terminal ‘signal-receiving domain’ (SRD) that becomes phosphorylated in response to various extracellular stimuli, an ARD composed of six repeats and a C-terminal PEST domain involved in basal turn over (reviewed in Baeuerle, 1998; Ghosh et al., 1998; Figure 1A). IκBα retains NF-κB in the cytoplasm by masking the latter’s NLS. NF-κB-stimulating signals lead to the phosphorylation, polyubiquitylation and proteasome-mediated degradation of IκBα, permitting NF-κB to be translocated to the nucleus via its newly exposed NLS and to bind to target DNA sites. IκBα, which is rapidly resynthesized after degradation, can enter the nucleus and promote the dissociation of NF-κB from DNA.
Fig. 1. Structure of Bcl-3. (A) Domain organization of Bcl-3 and IκBα. The basic nuclear localization sequence (NLS) of Bcl-3 is indicated in blue, while the leucine-rich NLS and NES of IκBα are in green. The fragment of Bcl-3 that was crystallized spans residues 119–359 and encompasses the entire ARD. (B) Ribbon diagram of the Bcl-3 ARD. The molecule curves towards the α1 helices, which together with the β-hairpins forms the presumed binding surface for p50 and p52 homodimers. This figure and Figures 3B and C, 5 and 6 were made with Bobscript (Esnouf, 1999) and Raster3D (Merritt and Bacon, 1997).
The structure of a complex of partially deleted fragments of IκBα and of an NF-κB p50–p65 heterodimer reveals that the ARD of IκBα interacts extensively with the RHR moieties of NF-κB (Huxford et al., 1998; Jacobs and Harrison, 1998). ANK repeats 1 and 2 interact with and induce the p65 NLS, which is unstructured in solution, to adopt a helical conformation. ANK repeats 4–6 contact the p50–p65 dimerization interface located within the two RHR-c domains, while ANK6 and the C-terminal PEST residues interact with the p65 RHR-n domain, which becomes re-oriented with respect to its position when bound to DNA.
At present, the only reported structure of an IκB protein is that of IκBα bound to NF-κB. In order to better understand the NF-κB–IκB signaling system, we have undertaken structural studies of Bcl-3. Bcl-3 is unique among the IκB proteins in that it is primarily localized in the nucleus and is not degraded upon activation of NF-κB-stimulating pathways (Franzoso et al., 1993; Nolan et al., 1993; Zhang et al., 1994). Moreover, whereas IκBα and IκBβ belong to a subclass of IκB proteins containing six ANK repeats, Bcl-3 contains seven repeats and lacks an acidic PEST sequence, forming a distinct subclass together with p100, p105 and (probably) IκBε.
Bcl-3 has been reported to possess various activities, which partly depend on its concentration, phosphorylation state and interaction with other proteins (Wulczyn et al., 1992; Nolan et al., 1993; Bundy and McKeithan, 1997; Dechend et al., 1999). Many of these activities lead to increased transcription from the κB promoter, in contrast to IκBα, which appears to behave exclusively as an inhibitor. Thus, Bcl-3 can cause DNA-bound p50 homodimers to dissociate from the κB site, permitting these inhibiting NF-κB species to be replaced by trans-activating species containing p65, RelB or c-Rel (Franzoso et al., 1992, 1993; Inoue et al., 1993). Bcl-3 can also form a ternary complex with DNA-bound p50 or p52 homodimers and activate transcription directly, an activity that requires both N- and C-terminal domains of Bcl-3 (Bours et al., 1993; Fujita et al., 1993; Pan and McEver, 1995; Hirano et al., 1998). Furthermore, expression of Bcl-3 can cause p50 to be released from cytosolic p105–p50 complexes and to translocate to the nucleus in a trans-activating complex containing Bcl-3 (Watanabe et al., 1997; Heissmeyer et al., 1999).
Here we report the crystal structure of the ARD of human Bcl-3 at 1.9 Å resolution. Although the central ANK repeats are highly similar in structure to those of the IκBα ARD, there are several important differences at the N- and C-termini. Comparison of the two structures provides an insight into why Bcl-3 binds specifically to p50 or p52 homodimers whereas IκBα prefers p65–p50 heterodimers, why Bcl-3 but not IκBα can form a complex with its DNA-bound NF-κB partner, and why Bcl-3 can bind its NF-κB partner symmetrically under certain conditions (two Bcl-3 molecules per homodimer) (Bundy and McKeithan, 1997), whereas IκBα can only bind asymmetrically. The structure of the Bcl-3 ARD should also serve as a better model for the other seven-repeat IκB members of its subclass.
Results and discussion
Structure determination
Human Bcl-3 is a 446-residue protein composed of a central ARD flanked by N- and C-terminal domains that are rich in proline and serine residues (Figure 1A). Reasoning that these flanking domains would be poorly ordered and hamper crystallization, we set out to crystallize only the ARD. Accordingly, a 241-residue fragment spanning residues 119–359 was expressed in Escherichia coli and purified to near homogeneity. Although the protein aggregated at the high concentrations normally used during crystallization trials, it remained relatively monodisperse at low concentrations (∼3 mg/ml), yielding small, well-diffracting crystals. Two related monoclinic crystal forms were obtained, each with one molecule per asymmetric unit. The structure was solved by molecular replacement using the ARD of human IκBα and refined at 1.9 Å resolution in both crystal forms (Table I; Materials and methods). The structure is essentially identical in the two forms, apart from a few side chain conformations that reflect differences in crystal packing interactions. All residues of the expressed fragment are well ordered, except for a small number of N- and C-terminal residues (119–124 and 353–359), for which no density is visible in either crystal form. The final model includes residues 125–352 and either 195 (form I) or 198 (form II) water molecules.
Table I. Data collection and refinement statistics.
| Data set | Form I | Form II |
|---|---|---|
| Space group | P21 | P21 |
| Cell parameters | a = 31.7 Å b = 51.2 Å | a = 31.4 Å b = 50.9 Å |
| c = 64.7 Å β = 102.0° | c = 59.5 Å β = 102.0° | |
| Beamline | ID14-EH1 | ID14-EH1 |
| Resolution (Å) | 40.0–1.8 | 40.0–1.9 |
| Completeness (%)a | 89.8 (77.8) | 98.6 (94.4) |
| Total observations | 46 088 | 49 074 |
| Unique reflections | 17 021 | 14 381 |
| Redundancya | 2.71 (1.73) | 3.41 (2.68) |
| Rsym (%)a,b | 7.2 (69.7) | 9.7 (26.0) |
| I/σIa | 10.5 (1.65) | 15.1 (3.15) |
| Refinement | ||
| resolution (Å) | 20.0–1.86 | 20.0–1.9 |
| total number of protein atoms | 1729 | 1716 |
| number of water molecules | 195 | 198 |
| R.m.s.d. from ideal geometry | ||
| bond lengths (Å) | 0.007 | 0.007 |
| bond angles (°) | 0.94 | 1.00 |
| R-factor (%) (reflections)c | 19.9 (15 500) | 17.7 (13 489) |
| Rfree (%) (reflections)d | 22.9 (828) | 21.7 (709) |
aData for the highest resolution shell are given in parentheses.
bRsym = Σ|I – <I>|/ΣI, where I is the intensity of the individual reflections and <I> is the mean intensity over symmetrically equivalent reflections.
cR = Σ||Fobs| – |Fcalc||/Σ|Fobs|, where Fobs and Fcalc are observed and calculated structure factor amplitudes, respectively.
dRfree is the same calculation including only the randomly chosen 5% of reflections not used for refinement.
Structure of the Bcl-3 ARD
The seven ANK repeats of the Bcl-3 ARD form an elongated structure with approximate dimensions of 75 × 25 × 25 Å (Figure 1B). As in IκBα and other ANK repeat proteins (for review see Sedgwick and Smerdon, 1999), each repeat consists of an initial β-hairpin followed by a helix–turn–helix motif nearly perpendicular to it. The overall shape of the ARD has been compared with that of a ‘cupped hand’ (Jacobs and Harrison, 1998) such that the β-hairpins form the fingers, the α1 ‘inner’ helices form the palm and the α2 ‘outer’ helices form the back of the hand. The ANK repeats stack together with a slight left-handed twist, such that a complete turn of the superhelix would require a total of 32 repeats, with a 245 Å pitch and a 35 Å radius.
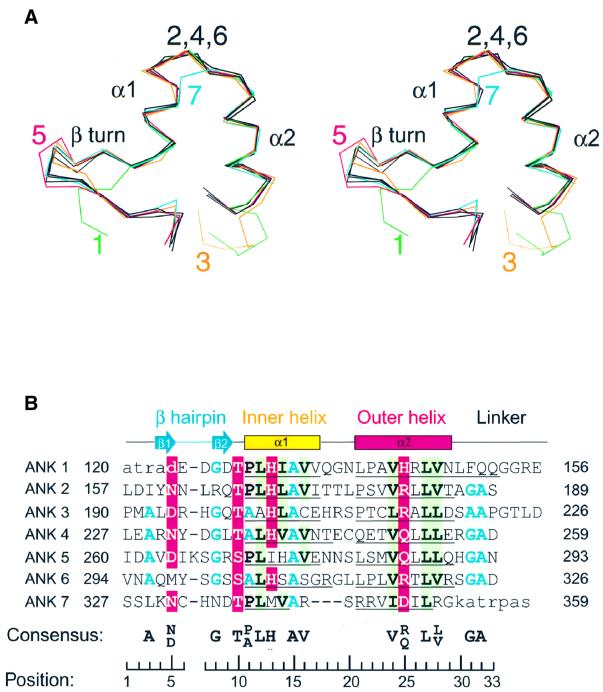
The seven ANK repeats are fairly uniform in backbone conformation [mean root mean square deviation (r.m.s.d.)Cα = 1.7 Å for all pairwise comparisons], as expected from their degree of sequence similarity (mean pairwise sequence identity = 27%) (Figure 2). The structures of ANK repeats 2, 4 and 6 are the most similar (r.m.s.d.Cα = 0.49–0.78 Å), reflecting their higher sequence identity (30–39%). ANK1 lacks the initial β-hairpin and contains a four-residue insertion after helix α2, as does ANK3, causing the ARD to bend slightly towards the interhelical turns of these repeats. Compared with the other repeats, ANK5 contains an additional residue, Lys266, inserted within the β-hairpin, creating a slightly wider ‘fingertip’ (Figure 2). A similar insertion, Cys215, occurs in the corresponding β-hairpin of IκBα. ANK7 deviates most from the other repeats in sequence (15–27% identity) but adopts a similar overall conformation, except that a three-residue deletion between the two helices results in an unusually short α1 helix (Figure 2).
Fig. 2. Alignment of the seven ANK repeats in Bcl-3. (A) Stereo view of superimposed Cα atom traces of the ANK repeats. ANK repeats 2, 4 and 6 (black traces) are the most similar, while larger deviations occur in ANK repeats 1, 3, 5 and 7 (green, orange, red and cyan, respectively). (B) Structure-based sequence alignment of the seven ANK motifs. Highly conserved polar, hydrophobic and small residues are highlighted on a red, green and gray background, respectively. The consensus numbering of residue positions shown at the bottom is that of Jacobs and Harrison (1998). Lower case residues at the N- and C-termini are disordered in the crystal structure. The precise limits of the α1 and α2 helices are shown by underlining.
A structure-based alignment of the seven ANK motifs (Figure 2B) shows that half of the residue positions are highly conserved. Residues Ala(15) in helix α1 and Leu(27) in helix α2 are invariant across all repeats, and together with five other conserved hydrophobic residues (at positions 12, 14, 16, 24 and 28 on the two helices) are involved in packing interactions within and between repeats, giving rise to a continuous hydrophobic core running the length of the molecule. As noted for IκBα (Huxford et al., 1998; Jacobs and Harrison, 1998), helix α1 contains hydrophobic residues (Ala or Val) smaller than those in helix α2 (mostly Leu), causing the stack of ANK repeats to curve slightly towards the α1 helix. Residue positions 10–13 comprise the signature TPLH motif characteristic of ANK repeats; these form a tight turn initiating helix α1, stabilized by hydrogen bonds involving residues Thr(10) and His(13). A conserved asparagine or aspartic acid residue at position 5 stabilizes the β-hairpin by hydrogen bonding with backbone nitrogen atoms at positions 7 and 9. Conserved glycines at positions 8 and 31 adopt backbone dihedral angles in the αL region of the Ramachandran plot, allowing a sharp turn in chain direction within the β-hairpin and at the end of helix α2, respectively. A small alanine residue is preferred at position 3, where it packs against side chain atoms from the β-hairpin of the preceding repeat, and at position 32, where it is buried against conserved leucine residues at positions 12 and 27 of the same repeat. A conserved hydrophilic residue at position 25 in helix α2 is exposed to the solvent on the convex surface of the ARD.
Whereas ANK repeats 2–6 each possess two hydrophobic faces mediating interactions with the preceding and successive repeats, ANK repeats 1 and 7 have a unique solvent-exposed face and thus deviate from the consensus sequence to avoid exposing hydrophobic residues. On the bottom surface of ANK7, two arginines (Arg342 and Arg351) replace the consensus valine or leucine residues at positions 16 and 28, while a third (Arg344) at position 21 replaces the leucine or proline residue usually present in the other repeats. However, a small patch of hydrophobic residues (formed by Met339, Ile347 and Leu338) remains exposed on this surface, which may explain why the protein aggregates at high concentrations. On the top surface of ANK1, alanine and arginine residues are present at positions 23 and 26 on helix α2, which are usually hydrophobic residues in the other repeats, while the four-residue extension at the end of helix α2 contributes three polar residues (Gln151, Gln152 and Arg155) to this surface.
Comparison of Bcl-3 and IκBα tertiary structures
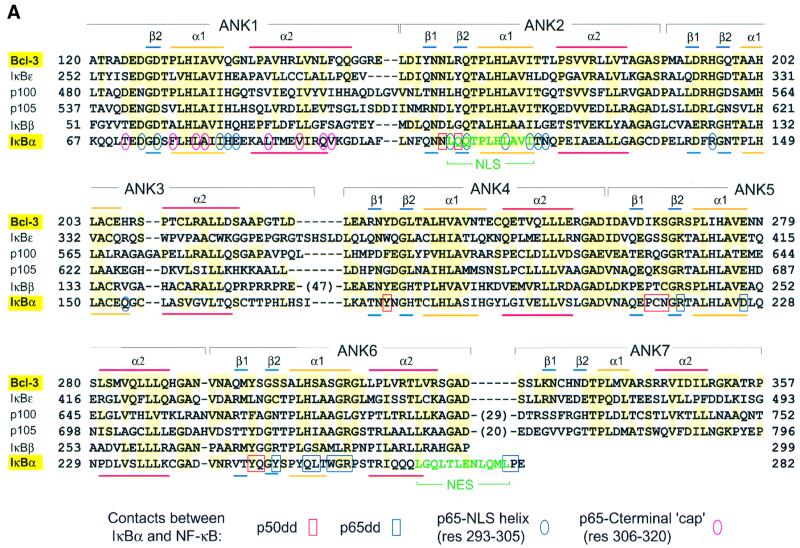
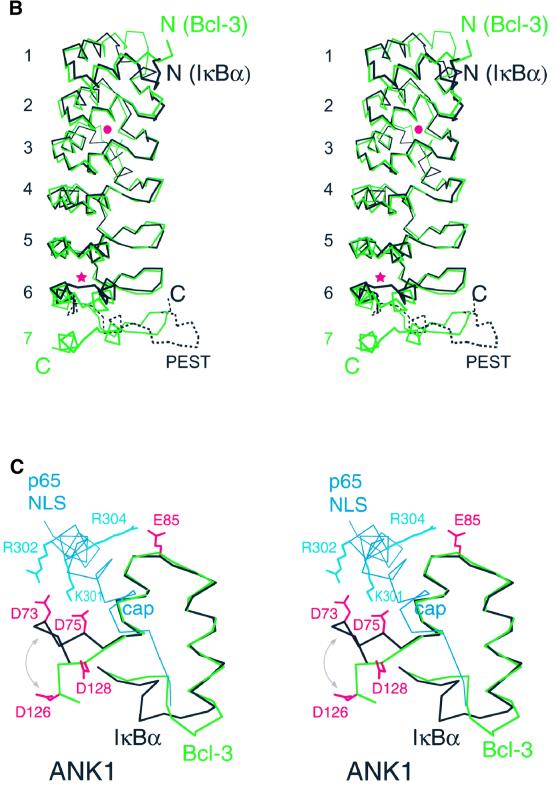
The ARDs of Bcl-3 and IκBα share 35% sequence identity over the first six ANK repeats (Figure 3A) and, as expec ted, have very similar overall structures (r.m.s.d.192Cα = 1.5 Å; Figure 3B). This indicates that a large conformational change is not required for IκB proteins to bind their NF-κB partners, as the structures compared are of IκBα in its bound state and Bcl-3 in its unbound state. Minor differences occur in residues joining ANK repeats 3 and 4, where the two proteins contain sequence insertions of differing length. This region contains a short helical segment in IκBα but has no regular secondary structure in Bcl-3 (Figure 3B, red dot).
Fig. 3. Comparison of the Bcl-3 and IκBα structures. (A) Sequence alignment of Bcl-3 homologs. Secondary structure elements are indicated above the Bcl-3 and below the IκBα sequences. Large insertions in the IκBβ, p100 and p105 sequences are indicated by numbers in parentheses. Residues are highlighted in yellow if identical or nearly identical to those in Bcl-3 [residues considered nearly identical are: (D,E), (R,K), (F,Y), (S,C) and (I,L,V)]. IκBα residues that interact with the p50 or p65 dimerization domains are boxed in red and blue, respectively. Those in contact with the NLS-containing helix of p65 are circled in blue and those in contact with the C-terminal p65 helix (involved in the hydrophobic capping interaction) are circled in red. Circled residues are contacts observed in the structure by Jacobs and Harrison (1998); contacts involving IκBα residues 280 and 281 are observed in that by Huxford et al. (1998). All other contacts are common to both structures. (B) Superimposition of the Bcl-3 and IκBα Cα backbones. Bcl-3 and IκBα are in green and black, respectively. The view is that of Figure 1B rotated by ∼180° about the vertical axis. The IκBα trace was generated by superimposing the two known IκBα structures, as one has a greater number of ordered residues within N-terminal ANK repeats (Jacobs and Harrison, 1998; solid line) while the other has a more complete PEST region (Huxford et al., 1998; broken line). Differences between Bcl-3 and IκBα within residues joining ANK repeats 3 and 4 and within the interhelical turn of ANK6 are marked by a dot and an asterisk, respectively. (C) Differences between Bcl-3 and IκBα within ANK1. Proteins are colored as in (B). The conformational difference of the N-terminal residues is indicated by a double-headed arrow. Residue Glu85 in IκBα is replaced by Gly138 in Bcl-3. Shown in blue are residues at the C-terminus of p65 that form two helices upon binding to IκBα: the first (residues 293–305) contains the basic NLS and the second (residues 305–320) is involved in a hydrophobic ‘capping’ interaction with ANK1.
The most significant differences occur at the N- and C-termini. In ANK1, helix α2 in Bcl-3 is two residues longer than in IκBα, causing residues joining ANK1 with ANK2 to follow significantly different courses. More importantly, in IκBα the N-terminal residues 71–76 adopt a β-hairpin conformation similar to that observed in the canonical ANK motif structure, with residues Asp73 and Asp75 interacting with basic residues of the p65 NLS (Figure 3C). In contrast, the N-terminus of Bcl-3 follows a nearly perpendicular course, such that the corresponding Asp126 and Asp128 side chains point in the opposite direction. The absence of the N-terminal hairpin in Bcl-3 cannot be due to the choice of fragment used for crystallization, as the Bcl-3 construct (beginning at residue 119, equivalent to IκBα residue 66) contains three more residues at the N-terminus than the IκBα fragment (residues 69–288) which contains the N-terminal hairpin (Jacobs and Harrison, 1998). Moreover, there is ample room in the crystal for the N-terminal residues of Bcl-3 to form a β-hairpin. Instead, the observed conformation is likely to be due to crystal packing interactions, as Asp125 and Asp128 interact with Arg318 and Arg322 of a neighboring molecule. This suggests that a β-hairpin in ANK1 is relatively unstable or perhaps completely missing in the absence of a basic NLS peptide from a bound NF-κB species.
At the C-terminus, ANK repeat 7 of Bcl-3 and the PEST region of IκBα comprise approximately the same number of residues, occupying similar volumes below ANK repeat 6. However, the PEST backbone turns in the opposite direction to that taken by ANK7 residues, such that the C-termini of IκBα and Bcl-3 are on opposite faces of the ARD. Substantial deviations between the two protein backbones (up to a maximum of 5 Å) also occur within the helices and the interhelical turn of ANK repeat 6, where Bcl-3 helices α1 and α2 are slightly longer than their IκBα counterparts (Figure 3B, asterisk).
Comparison of Bcl-3 and IκBα molecular surfaces
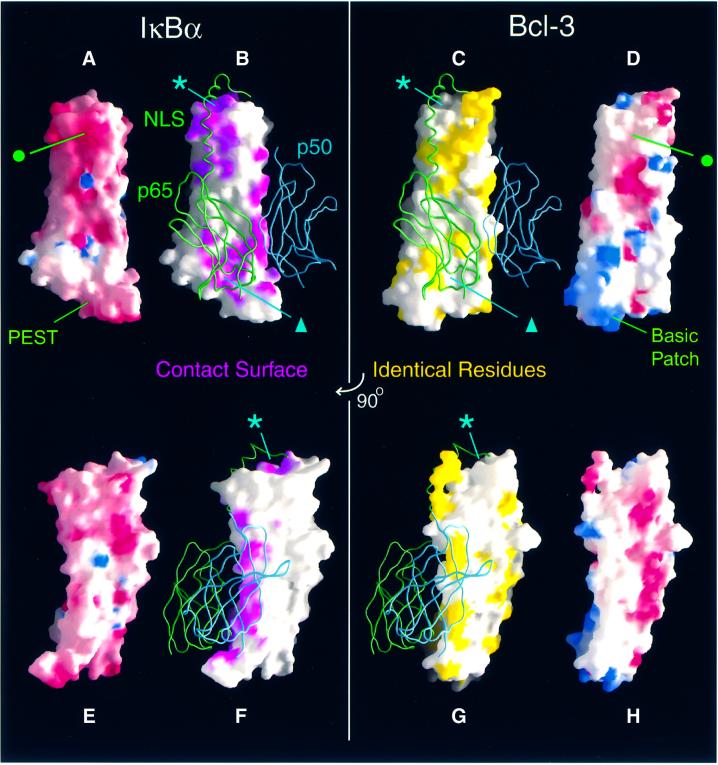
The electrostatic surface potentials of the Bcl-3 and IκBα structures are appreciably different (Figure 4). The ARD of Bcl-3 is considerably less acidic than that of IκBα: the theoretical pI value is 6.6 for Bcl-3 and 5.6 for IκBα (or 4.8 including the PEST sequence). This difference is partly due to the replacement of six glutamic acid residues in the first three ANK repeats of IκBα by uncharged or basic residues in Bcl-3. Consequently, the upper surface of IκBα is more negatively charged than that of Bcl-3 (Figure 4A and D, green dot). The difference in pI is also due to a greater number of basic residues in Bcl-3, which has a remarkable preference for arginine (17 residues) over lysine (only two). Seven arginine residues cluster in the outer helices and interhelical turns of ANK repeats 6 and 7, creating a large basic patch at the bottom of the ARD. In contrast, the corresponding surface of IκBα is highly acidic due to the presence of the PEST sequence (Figure 4, compare A with D and E with H).
Fig. 4. Comparison of Bcl-3 and IκBα molecular surfaces. The structures of IκBα (A, B, E and F) and Bcl-3 (C, D, G and H) are shown in equivalent orientations. The view of (A)–(D) is orthogonal to that of (E)–(H), which is approximately that of Figure 1B (i.e. with β-hairpins on the left and α2 helices on the right.) (A, D, E and H) Comparison of electrostatic surface potentials. Regions of negative and positive potential are shown in red and blue, respectively. The basic patch at the bottom of Bcl-3 is formed by arginine residues 311, 318, 322, 342, 344, 345 and 351. The corresponding surface of IκBα is formed by the acidic PEST region. (B, C, F and G) Conservation of the NF-κB contact surface. The C-terminal domains of p50 (blue) and p65 (green) are represented as ribbons bound to the surface of IκBα and, to facilitate comparison, to that of Bcl-3. In (B) and (F), regions of the IκBα surface within 4.5 Å of the p50 and p65 RHR-c domains are colored magenta. In (C) and (G), surface-exposed residues, which are identically conserved between Bcl-3 and IκBα, are shown in yellow. The asterisks and triangles indicate regions of the IκBα surface in contact with NF-κB that are composed of residues poorly conserved in Bcl-3. This figure was prepared using GRASP (Nicholls et al., 1991).
A large portion of the IκBα surface is involved in recognizing NF-κB (Figure 4B and F). ANK repeats 1 and 2 recognize the NLS-containing C-terminus of p65 (upper part of Figure 4B); the inner helices of ANK repeats 5 and 6 contact the dimerization domain of p65 (lower part of Figure 4B) and the β-hairpins of ANK repeats 4–6 interact with the p50 dimerization domain (Figure 4F). All three regions of the contact surface are relatively well conserved in Bcl-3 (Figure 4, compare B with C, and F with G), and over half of the IκBα residues involved in specific contacts with NF-κB are identical or nearly identical in Bcl-3 (Figure 3A). This suggests that many of the interactions observed in the IκBα–NF-κB complex are preserved in the Bcl-3–(p50)2 and Bcl-3–(p52)2 complexes. Indeed, because of the high degree of sequence and structural similarity between the p50 homodimer and the p50–p65 heterodimer (F.E.Chen et al., 1998), Bcl-3 is likely to bind to the homodimer with a nearly identical relative orientation to that of IκBα bound to the heterodimer. The strip of surface contacting p50 is particularly well conserved (Figure 4F and G), including Tyr181 and Tyr248 in the β-hairpins of ANK repeats 4 and 6 (Bcl-3 residues Tyr232 and Tyr299), which contact a cluster of hydrophobic residues in p50. This makes good sense, as the p50 subunit of the heterodimer recognized by IκBα is common to the homodimer recognized by Bcl-3.
Potential differences in recognition of the NF-κB C-terminus and implications for binding symmetry
In the IκBα–NF-κB structure, residues C-terminal to the dimerization domain of p65 form two helices. The first, consisting of p65 residues 293–305 and including the basic NLS, interacts with a negatively charged surface on IκBα formed by residues from ANK repeats 1, 2 and, to a lesser extent, 3 (Figures 3A and 4A). The corresponding surface in Bcl-3 is considerably less acidic (Figure 4A and D, green dot), suggesting that Bcl-3 may bind more weakly to a basic NLS. In particular, IκBα residue Glu85, which forms a salt bridge with Arg304 of the p65 NLS, is replaced by a glycine (Gly138) in Bcl-3; the two aspartate residues, which in IκBα interact with NLS residues Lys301 and Arg302, point away from the NLS site in the Bcl-3 crystal structure (although they may adopt a similar orientation in solution or upon the binding of p50 or p52; Figure 3C). The second C-terminal helix of p65 is formed by residues 306–320 and is involved in a hydro phobic ‘capping’ interaction with the top surface of the IκBα ARD, which is relatively poorly conserved in Bcl-3 (Figure 4, compare asterisks in B and C, and F and G). This surface is formed by the unique solvent-exposed face of ANK1, which is considerably less hydrophobic in Bcl-3 than in IκBα. Specifically, three bulky hydrophobic residues in IκBα (Phe77, Leu89 and Phe103) are replaced by smaller (Pro130, Ala142) or charged (Glu156) residues in Bcl-3 (Figure 3A).
Taken together, these differences suggest that basic NLS residues are bound less stably by Bcl-3 than by IκBα and that the hydrophobic ‘capping’ interaction observed with IκBα is absent from the Bcl-3 complex. This is reasonable given that Bcl-3 is mainly nucleoplasmic and that masking of the NLS is not its primary function. It is also consistent with early studies indicating that the p50 NLS does not contribute to binding (Wulczyn et al., 1992) and is not masked by Bcl-3 (Zhang et al., 1994). The IκBα-induced ordering of the p65 C-terminus in a helical conformation has been proposed to explain why only one molecule of IκBα binds to the NF-κB heterodimer, as the p65 C-terminus sterically prevents binding of a second IκBα molecule (related to the first by the pseudodyad symmetry of the heterodimer; Jacobs and Harrison, 1998). A different binding mode of the NLS by Bcl-3 would thus be consistent with the finding that under certain circumstances as many as two molecules of Bcl-3 can bind to a p50 or p52 homodimer (Wulczyn et al., 1992; Bundy and McKeithan, 1997).
Potential basis of selectivity towards NF-κB species
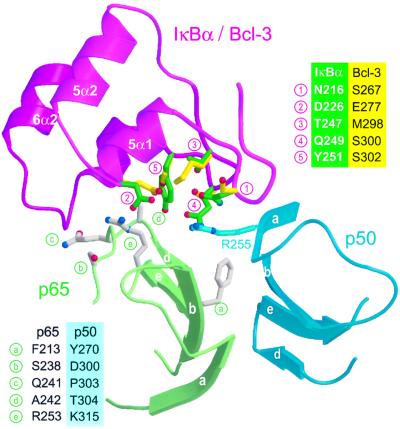
Bcl-3 binds better to p50 or p52 homodimers than to a p50–p65 heterodimer (Franzoso et al., 1992; Wulczyn et al., 1992), whereas the converse is true for IκBα. Careful inspection of the NF-κB binding surface of IκBα reveals a patch of residues involved in p65 recognition that are poorly conserved in Bcl-3 (Figure 4B and C, triangle). This patch consists of five residues located on the β-hairpins and inner helices of ANK repeats 5 and 6, close to the p50–p65 dimerization interface (Figure 5, positions 1–5). To see whether these residues could feasibly be involved in selectivity towards various NF-κB dimers, we superimposed the structures of Bcl-3 and a p50 homodimer onto that of the IκBα–NF-κB complex. Four surface-exposed residues of p65 lying in the vicinity of the non-conserved patch differ from their corresponding p50 residues (Figure 5, positions b–e). One of these, p65 residue Ala242 (position d), is in close contact with IκBα residue Tyr251 (5). In our superimposed models, the bulkier side chain of the corresponding p50 residue Thr304 clashes severely with IκBα residue Tyr251 but not with the smaller Bcl-3 residue Ser302. Both p50 and p52 have a threonine at position d, whereas p65, cRel and RelB all have an alanine; this position could therefore serve as an important recognition element for the discrimination of Rel subunits. IκBβ, which has a similar Rel dimer preference to IκBα, also has a large residue at position 5 (Arg275, corresponding to IκBα residue Tyr251), whereas IκBε, p100 and p105 have, like Bcl-3, small residues (Cys, Asn or Thr; Figure 3A).
Fig. 5. Potential site of selectivity by Bcl-3 and IκBα towards different NF-κB species. The view is that of the region marked by a triangle in Figure 4B and C, showing ANK repeats 5 and 6 of IκBα (magenta) and the β-sheets of p50 (blue) and p65 (green), which mediate heterodimer formation. The backbone structure of Bcl-3 (not shown) was locally aligned with that of IκBα repeats ANK 5 and 6, while that of a p50 homodimer (not shown) was superimposed onto that of the p50–p65 heterodimer. The Bcl-3 backbone is identical to that of IκBα in this region except for the interhelical turn of ANK6. Side chains are shown for IκBα residues (green), which are in close proximity to the p50–p65 heterodimer but are not identical to the corresponding Bcl-3 residues (yellow). Shown in gray are p65 side chains that are close to the IκBα surface but not conserved in p50. IκBα and Bcl-3 residues are identified in the legend at upper right; p65 residues and their p50 equivalents are shown in the legend at bottom left.
Next to position d are two other residues that differ between Rel subunits: p65 residues Ser238 and Gln241 (Figure 5, b and c) correspond to p50 residues Asp300 and Pro303, respectively. In p65, these residues interact with the interhelical turn of ANK repeat 6. The backbone conformation of this turn is significantly different between IκBα and Bcl-3 (Figure 3B, asterisk) and thus may also play an important role in selectivity. Also nearby is p65 residue Arg253 (position e), which forms a salt bridge with IκBα residue Asp226 (position 2). In the superimposed model of a Bcl-3–(p50)2 complex, this arginine is shortened to p50 residue Lys315 while the aspartate is lengthened to Bcl-3 residue Glu277, yielding compensatory substitutions that would preserve the salt bridge. Also close by on the ANK5 and ANK6 β-hairpins are IκBα residues Asn216 and Gln249 (positions 1 and 4), which are shortened to Bcl-3 residues Ser267 and Ser300. In IκBα, these residues recognize p50 residue Arg255, which in turn contacts p65 residue Phe213 (position a) in the dimerization interface. Phe213 is replaced by Tyr270 in p50, a residue critically important for stability of the p50 dimer (Sengchanthalangsy et al., 1999). Such substitutions in p50 and Bcl-3 might play a role in selectivity, as a bulkier p50 tyrosine could conceivably be sensed by the shorter Bcl-3 serines via an altered conformation of Arg255. Although we can not determine the role of specific residues without mutational analysis, the nature of the intermolecular contacts and the number of amino acid substitutions observed in this region suggest that ANK repeats 5 and 6 are likely to play an important role in the selectivity of IκBα and Bcl-3 towards different NF-κB dimers. The presence of the seventh repeat in Bcl-3 may also play an important role in selectivity. Indeed, in our hypothetical Bcl-3–(p50)2 model, residues Cys332, His333 and Asp335 in the β-hairpin of ANK7 point towards the unique p50 subunit (green subunit in Figure 5), while residue Arg342 in the interhelical turn of ANK repeat 7 is next to position c (Pro303 in p50 versus Gln241 in p65).
Model of a Bcl3–(p50)2 complex bound to DNA
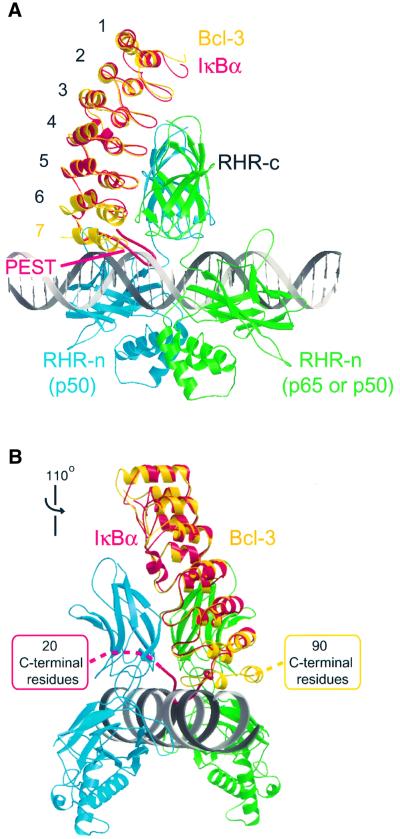
Unlike IκBα, which inhibits DNA-binding activity of NF-κB, Bcl-3 is able to form a stable complex with a DNA-bound p50 or p52 homodimer (Bours et al., 1993; Fujita et al., 1993; Bundy and McKeithan, 1997). We constructed a model of such a complex by superposing the structures of Bcl-3 and of a DNA-bound p50 homodimer onto the structure of the IκBα–NF-κB complex and compared it with a similarly constructed model of an (imaginary) IκBα–NF-κB–DNA complex (Figure 6). In the latter model, the acidic PEST region of IκBα is unfavorably located next to the negatively charged DNA phosphate groups, with several residues in a severe steric clash with the DNA (Figure 6A). In contrast, the corresponding surface of Bcl-3 is highly basic (Figure 4) and the more compact shape of the ANK7 repeat is better accommodated than the PEST region. Only small movements of ANK7 residues or the DNA backbone would be required to avoid any steric overlap. This is realistic given that the DNA is slightly bent in several NF-κB structures and could conceivably undergo additional minor adjustments in the presence of Bcl-3. Our model places the β-hairpin of ANK repeat 7 near the minor groove of the DNA, with several residues (Ser328, Lys330, His333, Asn334 and Thr336) as good candidates for making polar interactions with backbone atoms of both DNA strands.
Fig. 6. Hypothetical model of Bcl-3 bound to a DNA-bound p50 homodimer. The model was constructed based on the structure of the IκBα–NFκB complex, by structurally aligning Bcl-3 (yellow) onto IκBα (red) and a DNA-bound p50 homodimer (green and blue; Müller et al., 1995) onto the p50–p65 heterodimer (not shown, although it is nearly identical to the p50 homodimer, where the green monomer corresponds to the p65 subunit). A 30-base pair stretch of ideal B-form DNA was then superimposed onto the shorter duplex present in the p50–DNA crystal structure. (A) Side view showing that sufficient space is available next to the DNA to accommodate ANK repeat 7 of Bcl-3 but not the PEST region of IκBα. (B) View showing that the C-terminal domains of IκBα and Bcl-3 are positioned on opposite sides of the ARD. The view is that of (A) rotated by 110° about the vertical axis.
Figure 6 also shows that the C-terminal domain, which follows the ARD, occupies a different topological position in the two hypothetical models. Because the polypeptide chains of the PEST region and of ANK7 turn in opposite directions, the C-terminal domains of IκBα and Bcl-3 (which are missing from the crystal structures) are positioned on opposite sides of the ARD (Figure 6B). Thus, there is little room for the C-terminal domain of IκBα, constrained between the DNA, the p50 dimerization domain and the DNA-recognition (AB) loop of the RHR-n domain. In contrast, greater space is available for accommodating the C-terminal domain of Bcl-3. We emphasize that the model shown in Figure 6 is a hypothetical one. Understanding the precise nature of the interactions between Bcl-3, DNA and a p50 or p52 homodimer must await a crystal structure of the complex.
Bcl-3 is a highly phosphorylated protein and the level of phosphorylation can change dramatically depending on cell type and signal (Nolan et al., 1993; Caamano et al., 1996; Bundy and McKeithan, 1997). Phosphorylation of the C-terminal domain of Bcl-3 has been observed to hinder the formation of the three-species complex Bcl-3–(p52)2–DNA (Bundy and McKeithan, 1997). In our hypothetical model, the C-terminal domain emerges from the ARD close to the DNA. Phosphorylation might therefore destabilize the three-species complex by electrostatic repulsion with the DNA phosphate groups. Alternatively, the phosphorylated domain might interact with the basic patch of the Bcl-3 ARD (which is positioned adjacent to the DNA in our hypothetical model) and thus reduce the DNA-binding affinity of a preformed Bcl-3– (p52)2 complex by steric hindrance.
Subcellular distribution of Bcl-3 and IκBα
The different effects of Bcl-3 and IκBα on κB-dependent transcription depend in part on their different subcellular distributions. These in turn reflect the presence of different nuclear import and export sequences and their accessibility for recognition by nuclear transport factors. Bcl-3 appears to contain a classical NLS within its N-terminal domain (Zhang et al., 1994) and thus can be imported into the nucleus whether or not it is bound to p50 [or a p50 mutant lacking its own functional NLS (Zhang et al., 1994)], as this N-terminal NLS is remote from the presumed p50 interaction surface. In contrast, the import of IκBα is apparently mediated by an import sequence comprising residues within helix α1 of ANK repeat 2 (Sachdev et al., 1998, 2000; Figure 3A), which becomes partially occluded by the binding of p65, providing a plausible explanation for why IκBα is only imported in its unbound state. Conversely, two leucine-rich nuclear export sequences (NESs) have been identified in IκBα, located within helix α2 of ANK repeat 6 (Tam et al., 2000; Figure 3A) and within the N-terminal domain (Johnson et al., 1999; Sachdev et al., 2000). The N-terminal site is presumably remote from the NF-κB binding surface, while the site within ANK6 is only partly occluded in the structure of the IκBα–NF-κB complex, explaining why IκBα can be re-exported to the cytoplasm bound to NF-κB. In contrast, Bcl-3 lacks an obvious set of surface-exposed hydrophobic residues, which might constitute an NES (Rittinger et al., 1999). Those residues constituting the leucine-rich NES within ANK6 of IκBα are poorly conserved in Bcl-3 (Figure 3A) and adopt a substantially different conformation due to the presence of the additional ANK7 repeat.
Materials and methods
Protein expression, purification and crystallization
Construct Bcl-3ΔNΔC, which encodes amino acid residues 119–359 and spans the ANK repeat domain of human Bcl-3, was expressed from a pET11a vector in E.coli strain BL21(DE3) (Bours et al., 1993). Cells were grown at 37°C in 1 l of Luria–Bertani medium containing 100 μg/ml ampicillin until an OD600 of 0.6 was achieved. Protein expression was induced with 0.4 mM isopropyl-β-d-thiogalactopyranoside and cells were further grown overnight at 25°C before harvesting by centrifugation. Cells were resuspended in 30 ml of buffer A [20 mM HEPES pH 7.0, 300 mM NaCl, 5% glycerol, 10 mM dithiothreitol (DTT), 2 mM EDTA, protease inhibitors] and lysed by sonication. DNA was removed from the clarified lysate by precipitation with polyethylene imine at a final concentration of 0.6% (w/v). The protein was precipitated by adding 38% (w/v) ammonium sulfate, redissolved in 50 ml of buffer B (20 mM MES pH 6.5, 150 mM NaCl, 3 mM DTT) and loaded onto a 3 ml FAST-SP–Sepharose (Pharmacia) column pre-equilibrated in buffer B. After washing with 45 ml of buffer B, the protein was eluted using a 0.15–1.0 M gradient of NaCl. Fractions containing Bcl-3ΔNΔC were pooled, concentrated using a Centriprep-10 concentrator (Amicon) and loaded onto a Superdex-75 (Pharmacia) gel filtration column pre-equilibrated in 20 mM MES pH 6.5, 350 mM NaCl and 5 mM DTT. Peak fractions were pooled and brought to a final concentration of 3–10 mg/ml using a Centricon-10 concentrator (Amicon). Crystals were grown by the hanging drop vapor diffusion method from 5% PEG 6000, 100 mM MES pH 5.8 at 20°C. Crystals appeared as thin, triangular plates, which grew to their maximal size (longest edge 0.1 mm) within 3–4 weeks.
Data collection and structure determination
Crystal forms I and II, which grew from identical crystallization conditions, resulted from the use of different cryoprotection protocols. Form I was obtained by transferring crystals to a cryoprotective solution [15% PEG 6000, 100 mM MES pH 6.0 and 25% (v/v) glycerol] immediately prior to flash cooling at 100 K in a cold nitrogen stream, whereas crystal form II was obtained upon stepwise transfer into solutions containing increasing amounts (5–25%) of glycerol over a 15 min period. Both crystal forms belong to space group P21 and have similar unit cell parameters, with one molecule per asymmetric unit (Table I). The stepwise transfer procedure caused partial dehydration of crystal form II, resulting in an ∼5 Å contraction of the crystallographic c axis and a decrease in solvent content from 38 to 32%.
Diffraction data were collected at ∼1.9 Å resolution from both crystal forms on a MarCCD detector at ESRF beamline ID14-1 (λ = 0.934 Å). Data were processed and reduced with DENZO and SCALEPACK (Otwinowski and Minor, 1997). Crystal form I was solved by molecular replacement using the program AMoRe (Navaza, 1994). Search models containing 4–6 ankyrin repeats were derived from residues 73–269 of human IκBα (PDB code 1ikn; Huxford et al., 1998), which share 35% sequence identity with residues 126–320 of Bcl-3. Cross rotation and translation searches were performed with a number of search models, resolution ranges and radii of integration. Considerable effort was required to find the correct solution among a large number of false ones, probably because close crystal packing of the asymmetrically shaped molecule contributed many intermolecular vectors to the Patterson function. Indeed, despite its higher data completeness, we were unable to solve the more tightly packed crystal form II using the IκBα structure. The correct solution appeared consistently within the top 30 solutions of all molecular replacement trials and exhibited good crystal packing. The best results (R-factor = 55.2% and correlation coefficient = 0.295 for 8–4 Å resolution data) were obtained using a search model that contained ANK repeats 1–5 plus half of repeat 6, in which residues differing from Bcl-3 were replaced by alanine.
Rigid body fitting of secondary structure elements with program CNS (Brünger, 1996) followed by simulated annealing torsion angle refinement using all reflections from 20–1.86 Å gave a preliminary model with an Rcryst of 42.9% and an Rfree of 46.5% (calculated using 5% of the data). A difference map revealed extra density where the seventh ankyrin repeat, absent from the search model, was expected. Programs ARP/wARP (Perrakis et al., 1999) and REFMAC (Murshudov et al., 1997) were then used to build automatically and refine a nearly complete model (Rcryst = 22.4%, Rfree = 30.2%) of the Bcl-3 ARD, which lacked only 10 N-terminal, 21 C-terminal and 15 internal residues. Most of these remaining residues could be built manually into |2Fo – Fc| and |Fo – Fc| electron density maps using the program TURBO (Roussel and Cambillau, 1991). The final model (Rcryst = 19.9%, Rfree = 22.9%) contains 195 water molecules and 228 amino acid residues, including five with double side chain conformations (Ser59, Leu63, Gln132, Arg150 and Ser188). The model accounts for the entire Bcl-3ΔNΔC protein except for residues 119–124 and 353–359 at the N- and C-termini, which are invisible in the electron density map and are presumably disordered.
The refined model of crystal form I was used to solve crystal form II by molecular replacement with AMoRe. The correct solution corresponded to the top peak in both the rotation and translation functions, with an Rcryst of 31.6% and a correlation coefficient of 73.5% (using 8–4 Å data) after rigid body fitting. Subsequent refinement against all 20–1.9 Å data using REFMAC (Murshudov et al., 1997) led to a final model with Rcryst = 17.7% and Rfree = 21.7%. The structure of Bcl-3ΔNΔC is essentially identical in the two crystal forms except for a few side chain conformations (r.m.s.d. is 0.83 Å for all atoms and 0.35 Å for main chain atoms). Both models show excellent stereochemistry as assessed by programs PROCHECK (Laskowski et al., 1993) and WHAT_CHECK (Hooft et al., 1996), with 91.5% of all non-glycine and non-proline residues in the most favored regions and 8.5% in the allowed regions of the Ramachandran plot. Atomic co-ordinates for crystal forms I and II have been deposited with the PDB under accession codes 1k1a and 1k1b, respectively.
Acknowledgments
Acknowledgements
We thank Hassan Belrhali for support during data collection at ESRF beamline ID14-1. M.S.-L. acknowledges support by an EMBO post-doctoral fellowship.
References
- Baeuerle P.A. (1998) Pro-inflammatory signaling: last pieces in the NF-κB puzzle? Curr. Biol., 8, R19–R22. [DOI] [PubMed] [Google Scholar]
- Baeuerle P.A. and Henkel,T. (1994) Function and activation of NF-κB in the immune system. Annu. Rev. Immunol., 12, 141–179. [DOI] [PubMed] [Google Scholar]
- Bork P. (1993) Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins, 17, 363–374. [DOI] [PubMed] [Google Scholar]
- Bours V., Franzoso,G., Azarenko,V., Park,S., Kanno,T., Brown,K. and Siebenlist,U. (1993) The oncoprotein Bcl-3 directly transactivates through κB motifs via association with DNA-binding p50B homodimers. Cell, 72, 729–739. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1996) Recent developments for crystallographic refine ment of macromolecules. Methods Mol. Biol., 56, 245–266. [DOI] [PubMed] [Google Scholar]
- Bundy D.L. and McKeithan,T.W. (1997) Diverse effects of BCL3 phosphorylation on its modulation of NF-κB p52 homodimer binding to DNA. J. Biol. Chem., 272, 33132–33139. [DOI] [PubMed] [Google Scholar]
- Caamano J.H., Perez,P., Lira,S.A. and Bravo R. (1996) Constitutive expression of Bc1-3 in thymocytes increases the DNA binding of NF-κB1 (p50) homodimers in vivo. Mol. Cell. Biol., 16, 1342–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.E., Huang,D.B., Chen,Y.Q. and Ghosh,G. (1998) Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature, 391, 410–413. [DOI] [PubMed] [Google Scholar]
- Chen Y.Q., Ghosh,S. and Ghosh,G. (1998) A novel DNA recognition mode by NFκB p65 homodimer. Nature Struct. Biol., 5, 65–73. [DOI] [PubMed] [Google Scholar]
- Cramer P., Larson,C.J., Verdine,G.L. and Müller,C.W. (1997) Structure of the human NF-κB p52 homodimer–DNA complex at 2.1 Å resolution. EMBO J., 16, 7078–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dechend R., Hirano,F., Lehmann,K., Heissmeyer,V., Ansieau,S., Wulczyn,F.G., Scheidereit,C. and Leutz,A. (1999) The Bcl-3 oncoprotein acts as a bridging factor between NF-κB/Rel and nuclear co-regulators. Oncogene, 18, 3316–3323. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Franzoso G., Bours,V., Park,S., Tomita-Yamaguchi,M., Kelly,K. and Siebenlist,U. (1992) The candidate oncoprotein Bcl-3 is an antagonist of p50/NF-κB-mediated inhibition. Nature, 359, 339–342. [DOI] [PubMed] [Google Scholar]
- Franzoso G., Bours,V., Azarenko,V., Park,S., Tomita-Yamaguchi,M., Kanno,T., Brown,K. and Siebenlist,U. (1993) The oncoprotein Bcl-3 can facilitate NF-κB-mediated transactivation by removing inhibiting p50 homodimers from select κB sites. EMBO J., 12, 3893–3901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita T., Nolan,G.P., Liou,H.C., Scott,M.L. and Baltimore,D. (1993) The candidate proto-oncogene bcl-3 encodes a transcriptional coactivator that activates through NF-κB p50 homodimers. Genes Dev., 7, 1354–1363. [DOI] [PubMed] [Google Scholar]
- Ghosh G., Van Duyne,G., Ghosh,S. and Sigler,P.B. (1995) The structure of NF-κB p50 homodimer bound to a κB site. Nature, 373, 303–310. [DOI] [PubMed] [Google Scholar]
- Ghosh S., May,M.J. and Kopp,E.B. (1998) NF-κB and Rel proteins: evolutionarily conserved mediators of immune responses. Annu. Rev. Immunol., 16, 225–260. [DOI] [PubMed] [Google Scholar]
- Heissmeyer V., Krappmann,D., Wulczyn,F.G. and Scheidereit,C. (1999) NF-κB p105 is a target of IκB kinases and controls signal induction of Bcl-3–p50 complexes. EMBO J., 18, 4766–4778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano F., Tanaka,H., Hirano,Y., Hiramoto,M., Handa,H., Makino,I. and Scheidereit,C. (1998) Functional interference of Sp1 and NF-κB through the same DNA binding site. Mol. Cell. Biol., 18, 1266–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooft R.W.W., Vriend,G., Sander,C. and Abola,E.E. (1996) Errors in protein structures. Nature, 381, 272. [DOI] [PubMed] [Google Scholar]
- Huxford T., Huang,D.B., Malek,S. and Ghosh,G. (1998) The crystal structure of the IκBα/NF-κB complex reveals mechanisms of NF-κB inactivation. Cell, 95, 759–770. [DOI] [PubMed] [Google Scholar]
- Inoue J., Takahara,T., Akizawa,T. and Hino,O. (1993) Bcl-3, a member of the IκB proteins, has distinct specificity towards the Rel family of proteins. Oncogene, 8, 2067–2073. [PubMed] [Google Scholar]
- Jacobs M.D. and Harrison,S.C. (1998) Structure of an IκBα/NF-κB complex. Cell, 95, 749–758. [DOI] [PubMed] [Google Scholar]
- Johnson C., Van Antwerp,D. and Hope,T.J. (1999) An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IκBα. EMBO J., 18, 6682–6693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski R.A., McArthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Michaely P. and Bennett,V. (1992) The ANK repeat: a ubiquitous motif involved in macromolecular recognition. Trends Cell Biol., 2, 127–129. [DOI] [PubMed] [Google Scholar]
- Müller C.W., Rey,F.A., Sodeoka,M., Verdine,G.L. and Harrison,S.C. (1995) Structure of the NF-κB p50 homodimer bound to DNA. Nature, 373, 311–317. [DOI] [PubMed] [Google Scholar]
- Murshudov G.N., Vagin,A.A. and Dodson,E.J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D, 53, 240–255. [DOI] [PubMed] [Google Scholar]
- Navaza J. (1994) AMoRe: an autamated package for molecular replacement. Acta Crystallogr. A, 50, 157–163. [Google Scholar]
- Nicholls A., Sharp,K. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Nolan G.P., Fujita,T., Bhatia,K., Huppi,C., Liou,H.C., Scott,M.L. and Baltimore,D. (1993) The bcl-3 proto-oncogene encodes a nuclear IκB-like molecule that preferentially interacts with NF-κB p50 and p52 in a phosphorylation-dependent manner. Mol. Cell. Biol., 13, 3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno H., Takimoto,G. and McKeithan,T.W. (1990) The candidate proto-oncogene bcl-3 is related to genes implicated in cell lineage determination and cell cycle control. Cell, 60, 991–997. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pan J. and McEver,R.P. (1995) Regulation of the human P-selectin promoter by Bcl-3 and specific homodimeric members of the NF-κB/Rel family. J. Biol. Chem., 270, 23077–23083. [DOI] [PubMed] [Google Scholar]
- Perrakis A., Morris,R. and Lamzin VS. (1999) Automated protein model building combined with iterative structure refinement. Nature Struct. Biol., 6, 458–463. [DOI] [PubMed] [Google Scholar]
- Rittinger K., Budman,J., Xu,J., Volinia,S., Cantley,L.C., Smerdon,S.J., Gamblin,S.J. and Yaffe,M.B. (1999) Structural analysis of 14-3-3 phosphopeptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell, 4, 153–166. [DOI] [PubMed] [Google Scholar]
- Roussel A., Inisan,A.G., Knoops-Mouthuy,E. and Cambillau,C. (1998) TURBO-FRODO, version Open GL 1. University of Marseille, Marseille, France.
- Sachdev S., Hoffmann,A. and Hannink,M. (1998) Nuclear localization of IκBα is mediated by the second ankyrin repeat: the IκBα ankyrin repeats define a novel class of cis-acting nuclear import sequences. Mol. Cell. Biol., 18, 2524–2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev S., Bagchi,S., Zhang,D.D., Mings,A.C. and Hannink,M. (2000) Nuclear import of IκBα is accomplished by a Ran-independent transport pathway. Mol. Cell. Biol., 20, 1571–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedgwick S.G. and Smerdon,S.J. (1999) The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem. Sci., 24, 311–316. [DOI] [PubMed] [Google Scholar]
- Sengchanthalangsy L.L., Datta,S., Huang,D.B., Anderson,E., Braswell,E.H. and Ghosh,G. (1999) Characterization of the dimer interface of transcription factor NFκB p50 homodimer. J. Mol. Biol., 289, 1029–1040. [DOI] [PubMed] [Google Scholar]
- Simeonidis S., Liang,S., Chen,G. and Thanos,D. (1997) Cloning and functional characterization of mouse IκBε. Proc. Natl Acad. Sci. USA, 94, 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W.F., Lee,L.H., Davis,L. and Sen,R. (2000) Cytoplasmic sequestration of rel proteins by IκBα requires CRM1-dependent nuclear export. Mol. Cell. Biol., 20, 2269–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J.E., Phillips,R.J., Erdjument-Bromage,H., Tempst,P. and Ghosh,S. (1995) IκBβ regulates the persistent response in a biphasic activation of NF-κB. Cell, 80, 573–582. [DOI] [PubMed] [Google Scholar]
- Watanabe N., Iwamura,T., Shinoda,T. and Fujita,T. (1997) Regulation of NFKB1 proteins by the candidate oncoprotein BCL-3: generation of NF-κB homodimers from the cytoplasmic pool of p50–p105 and nuclear translocation. EMBO J., 16, 3609–3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteside S.T., Epinat,J.C., Rice,N.R. and Israel,A. (1997) IκBε, a novel member of the IκB family, controls RelA and cRel NF-κB activity. EMBO J., 16, 1413–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulczyn F.G., Naumann,M. and Scheidereit,C. (1992) Candidate proto-oncogene bcl-3 encodes a subunit-specific inhibitor of transcription factor NF-κB. Nature, 358, 597–599. [DOI] [PubMed] [Google Scholar]
- Zhang Q., Didonato,J.A., Karin,M. and McKeithan,T.W. (1994) BCL3 encodes a nuclear protein which can alter the subcellular location of NF-κB proteins. Mol. Cell. Biol., 14, 3915–3926. [DOI] [PMC free article] [PubMed] [Google Scholar]