Abstract
The established strand-displacement model for mammalian mitochondrial DNA (mtDNA) replication has recently been questioned in light of new data using two-dimensional (2D) agarose gel electrophoresis. It has been proposed that a synchronous, strand-coupled mode of replication occurs in tissues, thereby casting doubt on the general validity of the “orthodox,” or strand-displacement model. We have examined mtDNA replicative intermediates from mouse liver using atomic force microscopy and 2D agarose gel electrophoresis in order to resolve this issue. The data provide evidence for only the orthodox, strand-displacement mode of replication and reveal the presence of additional, alternative origins of lagging light-strand mtDNA synthesis. The conditions used for 2D agarose gel analysis are favorable for branch migration of asymmetrically replicating nascent strands. These data reconcile the original displacement mode of replication with the data obtained from 2D gel analyses.
Keywords: Replication, mtDNA, branch migration, atomic force microscopy, 2D agarose gels, strand displacement
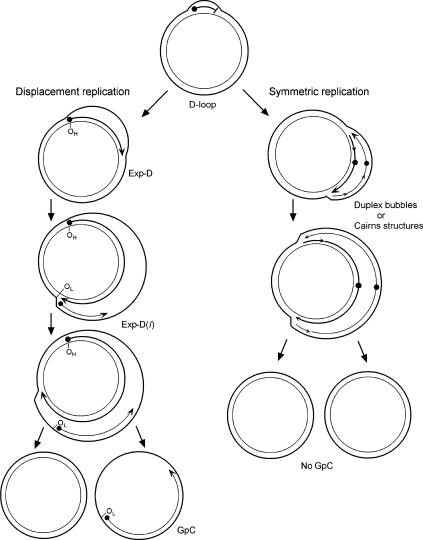
Replication of mammalian mtDNA has been thought to occur asymmetrically, using broadly separated strand-specific unidirectional origins of replication (Fig. 1). Replication of the leading strand initiates at the origin of heavy (H)-strand synthesis (OH) and proceeds unidirectionally, displacing the parental H-strand as single-stranded DNA (ssDNA). The enlargement of this displacement-loop (D-loop) proceeds for a distance of about two-thirds around the circular genome. The origin of replication for the lagging strand, termed the origin of light (L)-strand synthesis (OL), is then exposed, and synthesis of the nascent L-strand proceeds in the opposite direction on the single-stranded template of the expanded D-loop (Exp-D). A consequence of strand-asymmetric synthesis is that the daughter molecule with the new L-strand is delayed in completion with respect to the other daughter molecule, and segregates prior to completing replication. These molecules are termed gapped circles (GpC), since they contain a gap in newly synthesized L-strand DNA, leaving an H-strand template region that is still single-stranded. The asymmetric replication model is reviewed in detail elsewhere (Shadel and Clayton 1997).
Figure 1.
The asymmetric and strand-coupled models of mtDNA replication. Both models agree on the nature of the simple D-loop form of mtDNA. The displacement-model of replication is shown on the left and proceeds with single-stranded replication of the H-strand with further expansion and displacement of the D-loop. The intermediates are called expanded D-loops (Exp-D). This proceeds until the L-strand origin (OL) is exposed, with subsequent synthesis of the new L-strand in the opposite direction. Those intermediates are termed Exp-D(l). The asymmetry of strand synthesis leaves one segregated daughter molecule with an incompletely synthesized L-strand, called a gapped circle (GpC). The strand-coupled or synchronous model of replication is shown on the right. In this model, there is thought to be a zone of replication initiation within a broad area beyond the simple D-loop. Within this zone, both strands are synthesized bidirectionally as the double-stranded replication forks proceed through the length of the mtDNA. Exp-D, Exp-D(l), and GpC forms are excluded by this mode of replication.
Recently, several papers have appeared that re-examine this model of mammalian mtDNA replication (Holt et al. 2000; Yang et al. 2002; Bowmaker et al. 2003; Yasukawa et al. 2005). Restriction fragments of DNA that contain candidate replication origins and replication forks can be detected using two-dimensional (2D) agarose gel electrophoresis (Friedman and Brewer 1995). In this technique, DNA fragments are separated in the first dimension based on size and in the second dimension based on strand configuration. Under this analysis DNA fragments with classic synchronous leading- and lagging-strand replication forks display what is known as a y-arc. Linear fragments form a spot, and a distribution of double-stranded replicating fragments creates a y-arc that emanates from this linear spot. The arc ends in a position that is twice the size of the linear fragment, as would be expected as replication forks proceed through the end of the restriction fragment (see Fig. 6A, below). Using this technique, Holt, Jacobs, and coworkers (Holt et al. 2000) have revealed y-arcs within an area between the two origins described above. Replication forks of this type in this mtDNA region are inconsistent with the asymmetric strand-displacement model of replication. To account for the behavior of this y-arc under various experimental conditions, Holt, Jacobs, and collaborators (Young et al. 2002; Bowmaker et al. 2003) concluded that mtDNA replicates symmetrically, with leading- and lagging-strand synthesis progressing from multiple, bidirectional replication forks within a broad zone (see Fig. 1). They also propose that the strand-displacement model of replication is based on an artifact of preparation in which discontinuously synthesized “lagging” strands are preferentially degraded due to both a very high rate of ribo-substitution in that strand and an RNase H-like activity that is envisioned as a contaminant in those preparations. It was concluded that there is no asymmetric replication of mtDNA, despite overwhelming evidence to the contrary. These suppositions and other arguments have recently been presented, along with a more detailed examination of this current controversy (Bogenhagen and Clayton 2003a,b; Holt and Jacobs 2003).
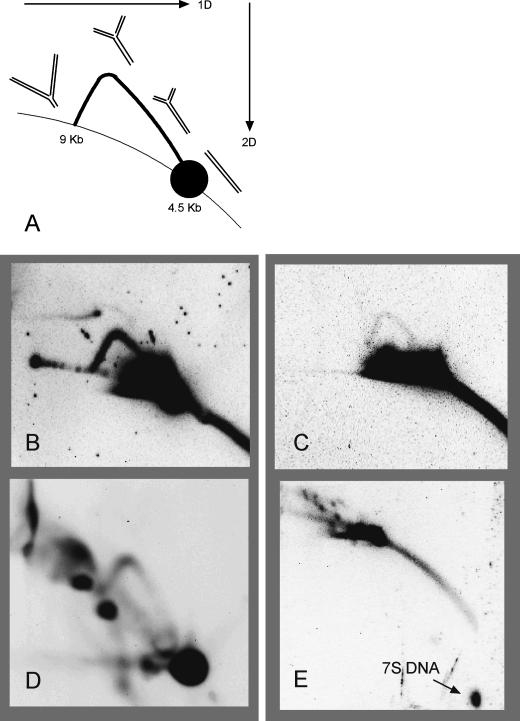
Figure 6.
Neutral–neutral 2D agarose gel analysis of mouse mtDNA. (A) Diagram of a y-arc. The orientations of the first and second dimensions are shown with arrows. The formation of the y-arc is described in the text. In B and C, the 4.5-kb DraI fragment was probed with a portion of the ATPase 6 gene (nucleotides 8032–8497) and was treated with S1 nuclease. DNA in D was digested with BglI and probed with a fragment of the ND4 gene (nucleotides 10577–11193). DNA in E was probed for the 7S DNA strand, corresponding to the D-loop region (nucleotides 16027–15510). DNA in D and E was not treated with S1 nuclease. (B) 300 ng liver mtDNA. (C) 500 ng LA9 mtDNA. (D) 500 ng liver mtDNA. (E) 150 ng liver mtDNA.
Here we present new experimental results and interpret them as evidence that the conclusions above are incorrect. To this end, we purified mtDNA from mouse liver tissue using procedures that yield the y-arc patterns displayed by the 2D gel technique. We then characterized these molecules using atomic force microscopy (AFM) and find evidence that strand-displacement replication is the dominant or even sole mode of mtDNA replication in mouse liver. In addition, AFM mapping indicates that there are alternative L-strand initiation sites. We propose that these alternate L-strand initiation sites can explain the 2D agarose gel patterns in a way that is consistent with a strand-displacement mode of replication. Finally, we demonstrate that branch migration of replicating DNA strands during preparation for 2D gel analysis may also contribute to the erroneous conclusion that strand-synchronous replication is the principal mode of replication in mammalian mitochondria.
Results
Direct visualization of mtDNA by atomic force microscopy
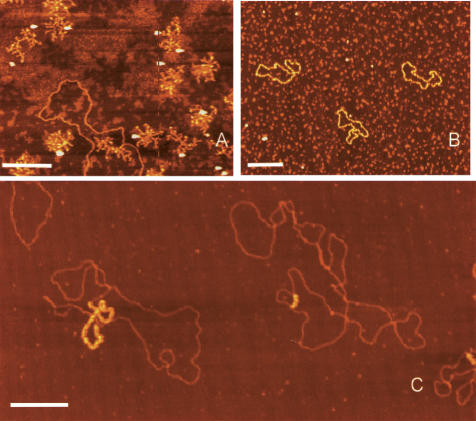
In AFM images, ssDNA appears thinner than double-stranded DNA (dsDNA) and, under the ionic strength used for imaging, forms a condensed structure that prevents determination of the contour length. We therefore developed a procedure to bind Escherichia coli ssDNA-binding protein (SSB) to the mtDNA just prior to immobilization for AFM (Fig. 2). M13 phage DNA is a single-stranded circular molecule of 6407 nucleotides and appears as a collapsed self-annealed molecule under standard AFM imaging conditions (Fig. 2A). Upon incubation with SSB, the M13 molecule is resolved (Fig. 2B). Similarly, ssDNA regions of mtDNA are resolved and are highlighted after binding with SSB (Fig. 2C). The SSB-coated ssDNA appears thicker than dsDNA in AFM images due to wrapping of the DNA around multimers of SSB protein. This allowed easy recognition of ssDNA regions of mtDNA even in large scan sizes with lower resolution.
Figure 2.
AFM images of M13 phage and mtDNA. M13 phage genomes before (A) and after (B) binding to SSB. (C) Two mtDNA molecules with single-stranded regions coated with SSB. Bars, 0.25 μm.
Table 1 summarizes the data collected using AFM. In order to quantify the relative number of replicative intermediates in this sample, we counted and classified each molecule within nonoverlapping 4-μm2 regions of the mica grid. From a total of 615 molecules thus observed, 78 had extended regions of ssDNA, as defined by SSB binding, which represents 12.7% of the total molecules. These are considered to be in various stages of replication. Another 66 molecules (10.7%) were identified that contained only a short D-loop, presumably at the classically defined control region (Kasamatsu et al. 1971). Molecules with Exp-D were the most abundant replicative intermediates in our preparations (21), followed closely by GpC (20). Those intermediates containing both daughter molecules in various stages of replication were less abundant (8). The remaining molecules with extended ssDNA fell into areas with multiple overlapping circles, which prevented a clear classification. We also observed five mtDNA molecules that were twice the normal length. Finally, we were able to measure unambiguously the contour lengths of 192 molecules. Our average length measurement of 5 μm for mouse mtDNA agrees with earlier electron microscopic data (Borst and Kroon 1969). In following these contours, and in deliberate searching, we found no molecules that appeared to have opposing double-stranded replication forks or Cairns structures, which are predicted by the strand-coupled model of replication. These would have been revealed either by following the contour path for the forks, or by finding molecules that were longer than expected for a simple circle. Supplementary Figure S1 illustrates the results of a control experiment demonstrating that our AFM imaging technique is capable of revealing double-stranded Cairns structures, even though they were not seen in our mtDNA preparations. Therefore, by direct visualization, we find no molecules that support the symmetric replication model of mouse mtDNA as proposed by Holt, Jacobs, and coworkers (Holt et al. 2000, Yang et al. 2002; Bowmaker et al. 2003; Yasukawa et al. 2005).
Table 1.
Summary of mtDNA molecules observed using atomic force microscopy
| Total molecules screened | 615 |
| Molecules with extended ssDNA | 78 (12.7%) |
| Molecules with short D-loops | 66 (10.7%) |
| Expanded D-loop molecules | 21 |
| Molecules with partial synthesis of both daughters | 8 |
| Gapped circular (β daughter) molecules | 20 |
| Dimer molecules | 5 |
| Contours measured | 192 |
| Mean length (micrometers) | 5.04 ± 0.18 |
The top panel represents molecules scored from random sampling. The center panel also includes additional molecules that were not part of the random sample. The lower panel represents the contour length measurements made on simple circular and D-loop-containing molecules.
(ssDNA) Single-stranded DNA.
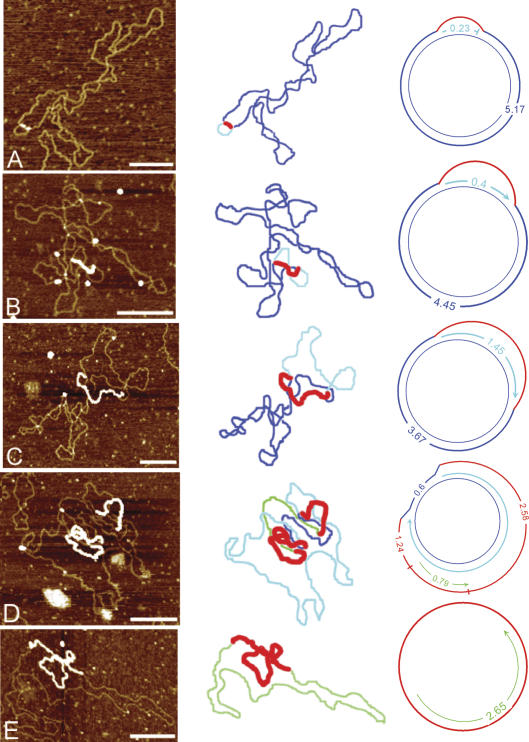
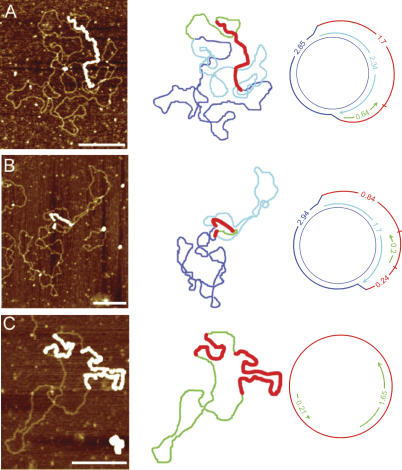
Figure 3 presents examples of the various forms of mouse liver mtDNA seen with AFM, their contour paths, and classification type. Figure 3A shows a molecule with a simple D-loop. This structure is formed by the synthesis of a short H-strand initiating at OH and terminating at the end of the noncoding control region. Figure 3B and C shows examples of Exp-D in which one strand is displaced as ssDNA while unidirectional replication proceeds on the other strand. Figure 3D shows partial synthesis of both daughter molecules attached to the parental molecule (Exp-D[l]). The strand type and lengths from the replication forks are consistent with the use of the two origins OH and OL, defined as shown. Figure 3E is an example of a daughter molecule that has segregated with a partially synthesized L-strand annealed to the parental ssDNA (a GpC). These molecules all represent intermediates consistent with the strand-displacement model of mtDNA replication.
Figure 3.
Replicative intermediates of mtDNA seen by AFM. The first column displays the AFM images, the second column outlines the contour, and the third column diagrams the intermediate and shows the strand contour lengths. Blue lines represent the parental strands, red lines represent the compacted ssDNA coated with SSB protein, aqua lines represent nascent H-strand synthesis, and green lines represent nascent L-strand synthesis. (A) Simple D-loop. (B) Short Exp-D. (C) Large Exp-D. (D) Exp-D with L-strand (Exp-D[l]). (E) GpC molecule with partial synthesis of the L-strand. Bars, 0.25 μm.
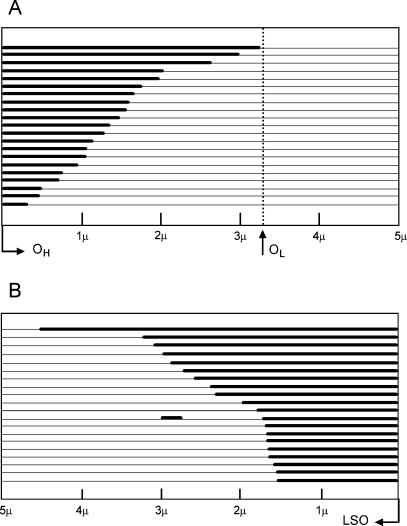
Figure 4A displays a summary diagram of the discernible lengths of the displaced regions of Exp-D. We assume here the location of the established leading-strand origin, OH, as described in the Materials and Methods. The length distribution of the Exp-D indicates that the single H-strand expansion boundary is restricted to the area between OH and OL. This result is also consistent with the displacement, or asymmetric, mode of mtDNA replication. Figure 4B is a summary diagram of the single-stranded lengths found in GpC molecules. The GpC molecules arise from the segregation of incompletely replicated daughter molecules with newly synthesized L-strands and single-stranded parental H-strands. These results may indicate that L-strand synthesis is either initiated at multiple locations and/or is frequently paused. Neither of these possibilities can be eliminated from this analysis. Most importantly, it is difficult to reconcile any of these GpC molecules with the strand-coupled model of replication, in which completion of replication is predicted to segregate two entirely replicated daughter molecules.
Figure 4.
Length distribution of newly synthesized nascent-strand intermediates. Individual mtDNA molecules are shown in linear form. Thick lines represent new complementary strand synthesis, and parental strands are shown as thin lines. (A) The lengths of the newly synthesized heavy strands measured from Exp-D molecules. All Exp-D in this analysis occur within the boundaries of OH and OL. (B) The lengths of nascent L-strands measured from GpC molecules. The position of OH cannot be determined from the GpC forms and is therefore not shown. The L-strand origin (LSO) is displayed to the far right.
Figure 5 shows three molecules that indicate that there are alternative L-strand origins in addition to the previously defined OL shown in Figure 3D. The molecule in Figure 5A has a clear double-stranded fork that defines one of the origins. There are two possible orientations. If we assume that this fork represents the H-strand origin, then the L-strand origin begins on the distal side of the double-stranded region. This interpretation would place the L-strand origin only 0.64 μm away from OH, at about mtDNA nucleotide 14,000, which is near the 5′ end of the ND6 gene. However, it is also possible that the double-stranded fork represents replication pausing at an L-strand origin. In that orientation, the total distance between the two forks defines the position of the L-strand origin. The length of the double-stranded region of the displaced loop is again 0.64 μm, and the adjacent single-stranded region is ∼1.7 μm. Therefore, the total length of the displaced H-strand is 2.34 μm. This interpretation would place the L-strand origin at approximately mtDNA nucleotide 8400, which is located within the 3′ region of the ATPase 6 gene. Figure 5A displays this latter interpretation. Length determinations of SSB-coated regions were less precise due to DNA wrapping (see Materials and Methods). Therefore, we rely more heavily on the dsDNA length measurements when corresponding displaced and replicated strand values are not identical.
Figure 5.
Replicating mtDNA with alternate L-strand origins. The first column displays the AFM images, the second column outlines the contour, and the third column diagrams the intermediate and shows the strand contour lengths. Blue lines represent the parental strands, red lines represent the compacted ssDNA coated with SSB protein, aqua lines represent the nascent H-strand synthesis, and green lines represent nascent L-strand synthesis. Although the third column shows the molecules in one particular orientation relative to OH, the opposite orientation is also possible, with OH located at the other fork. Bars, 0.25 μm.
The molecule in Figure 5B indicates the presence of yet another L-strand origin. In this case, the displaced H-strand has two ssDNA regions, separated by a short stretch of dsDNA. One of the forks at the boundary of the single-stranded region defines the position of OH. The L-strand origin is at one or the other of the two ends of the double-stranded region between the two single strands. If the fork adjacent to the short single-stranded region is OH, then the position of the L-strand origin would lie ∼0.44 μm away, which is estimated to be within the cytochrome b gene at approximately nucleotide 14,600. If the fork adjacent to the longer single-stranded region is the H-strand origin, then the L-strand origin is ∼1.46 μm away, which is calculated to be within the ND4 gene at approximately nucleotide 11,300. Again, Figure 5B displays the latter of the two possible interpretations.
The molecule in Figure 5C is a GpC molecule with two regions of dsDNA. Since newly synthesized strands on GpC molecules initiate at L-strand origins, this molecule has used two L-strand origins. This case represents further evidence that the traditional OL is not the sole origin for L-strand synthesis. Without defined forks, it is impossible to map the location of these L-strand origins. However, it is clear that at least one of these two L-strand origins has not been previously recognized.
Confirmation of qualitative 2D gel analyses
Figure 6A is a diagram of the y-arc components in which the arc begins at the position of 4.5-kb linear DNA, in this case, and ends along the line of linear fragments at the 9-kb location. dsDNA with a distribution of fork positions and strand lengths forms the arc pattern between these boundaries. Figure 6B shows that the mouse liver mtDNA, from mitochondria isolated as described by Yang et al. (2002) and that we have characterized by AFM, displays a y-arc pattern. This result demonstrates that there are no substantial differences between our preparations of mtDNA and those of Holt and colleagues (Holt et al. 2000; Yang et al. 2002). More importantly, since no synchronously replicating Cairns-like molecules are seen with AFM, the 2D gel y-arc is probably formed by strand-asynchronous replicative intermediates. Alternatively, this y-arc could represent an extremely minor fraction of symmetric intermediates that escaped the AFM screen. Figure 6C shows that the y-arc is also formed using mtDNA from LA9 cells, using the method used historically for previous studies of mtDNA replication. This result indicates that the two methods of mtDNA preparation yield similar results, casting further doubt on the assertion that the LA9 cell purification method generates artifactual replication intermediates that have been degraded by RNase H (Yang et al. 2002). The LA9 y-arc also contradicts an earlier conclusion that cultured cell lines use a fundamentally different mode of replication than tissue mtDNA (Holt et al. 2000). Figure 6D displays further evidence that the integrity of our liver mtDNA preparation is equivalent to that of Holt and coworkers (Yang et al. 2002). Slow-moving y-arcs are thought by those authors to represent extensively ribo-substituted replicative intermediates that are not cleaved by the restriction enzyme BglI. This panel demonstrates that such molecules are present in our AFM preparation and, therefore, regardless of the extent of ribosubstitution, we find no evidence for degradation by an RNase H as they have proposed (Yang et al. 2002). In addition, we were unable to detect RNase H activity in our mitochondrial lysates (Supplementary Fig. S2). Figure 6E represents an analysis of DraI-cleaved mtDNA without S1-nuclease treatment that has been probed for the D-loop region mtDNA sequence. The nascent replication-paused strand of DNA, known as 7S DNA and shown as a single strand in the D-loop molecule in Figure 1 (top), has been released from the mtDNA due to branch migration and subsequent reannealing of the parental strands. This observation demonstrates that branch migration of asymmetrically replicating intermediates is favored under the conditions used for 2D gel analysis.
Discussion
The historical model of mammalian mtDNA replication
The strand-asynchronous displacement model for mammalian mtDNA replication was initially proposed based on the topology of single- and double-stranded replicative intermediates observed using electron microscopy (EM). An extensive series of detailed studies followed that were facilitated by the ability to isolate individually pure H- and L-strands of mtDNA and label them selectively due to the exclusive presence of the mitochondrial-specific form of thymidine kinase in certain mutated cell lines. This allowed mtDNA pulse and pulse-chase labeling experiments to be conducted that identify and characterize temporally the mtDNA molecules that are bona fide replicative intermediates (Berk and Clayton 1974). The advantages of mtDNA-specific labeling in cell culture systems are lacking in whole animal or tissue systems, which led to an experimental bias in cell sources and fewer studies using tissue sources. It therefore remained possible that an alternative mode of replication operates in tissues. However, EM studies from a variety of tissues did not reveal any substantial inconsistencies with the strand-displacement model of replication (Pikó and Matsumoto 1977; Rajamanickam et al. 1979; Pikó et al. 1984). The view that mtDNA replication in cell culture is fundamentally the same as in tissues has only recently been challenged by interpretation of data from 2D agarose gels (Holt et al. 2000; Yang et al. 2002; Bowmaker et al. 2003; Yasukawa et al. 2005).
Interpreting recent data on mammalian mtDNA replication
Our goal in this work was to clarify the mode of mtDNA replication in mouse liver tissue using approaches that were mindful of the previous interpretations of the 2D agarose gel data. We chose to use AFM in this analysis to avoid previous criticisms that the extensive sample processing, including the partial denaturation required for EM, might lead to artifacts in the images observed. Sample processing for AFM is limited to relatively mild buffer exchanges followed by immediate immobilization on the mica surface within minutes. Despite these differences, our results agree with earlier EM data. Using AFM, we searched for classic replication bubbles (or Cairns forms) that were either fully duplex or that contained short regions of template ssDNA for the lagging strand. Although not exclusive, this type of intermediate is a hallmark of strand-coupled replication and occurs in many organismal and viral DNA systems. We found no such intermediates, although we have demonstrated our ability to image these molecules. All of the replicative intermediates observed were uniquely consistent with the asynchronous, strand-displacement mode of replication. The extensively displaced single strands, the positions of the partially synthesized L-strands, and the prevalence of GpC with partial L-strand synthesis are all incompatible with strand-synchronous replication. Our results are also consistent with an earlier study in which native SSB–mtDNA protein complexes were purified from rat liver and visualized by EM, revealing D-loops, Exp-D, and GpC forms (Van Tuyle and Pavco 1985). Those preparations should have maintained the natural state of mtDNA replicative intermediates. While we cannot rule out the existence of any strand-coupled replicative intermediates, they would have to be present in our preparations at a level of <0.5% to be missed altogether.
Evidence for additional lagging L-strand origins
Interestingly, the AFM data provide new evidence for the existence of alternative L-strand origins. This is indicated by fork-to-fork measurements made within AFM images of Exp-D(l) molecules (Fig. 5). The distances between the displacement forks reveal that there must be other L-strand origins in addition to the orthodox OL. This observation requires a modification of a model for strand-displacement replication in which there is only a single L-strand origin. Prior evidence for multiple L-strand origins exists in the literature. Robberson et al. (1972) identified replication fork ends that mapped to locations on either side of the traditional OL. Koike and Wolstenholme (1974) found more dramatic evidence for the existence of such origins, and Pikó et al. (1984) also proposed the existence of additional L-strand origins.
However, the sequencing of the mtDNA genome showed that the traditional L-strand origin occurred within one of the rare noncoding regions of the genome. The only larger noncoding region was known to contain the H-strand origin. Further 5′-end mapping from cell culture systems supported the major OL site as the L-strand origin without the identification of any others (Bogenhagen et al. 1979; Tapper and Clayton 1981, 1982). The failure of these attempts to identify other ends likely indicates that they are individually less abundant than those at the major OL site in the cell lines used, and are probably broadly distributed. A comprehensive frequency and distribution of alternate OL usage in cultured cells and tissues remains to be determined.
The qualitative 2D gel pattern of mammalian mtDNA is common
We also used 2D agarose gel analysis to characterize mtDNA from both mouse liver and the mouse LA9 cell line. These analyses lead to several important observations and conclusions. The first is that mouse liver mtDNA used for AFM analysis also yields y-arcs upon 2D gel analysis (Fig. 6B), indicating that we were successful in preparing mtDNA comparable to that used by Holt, Jacobs, and coworkers (Holt et al. 2000; Yang et al. 2002) in their 2D gel analyses. In addition to serving as a quality control, this result provided the first indication that the 2D gel patterns would have to be reconciled with the replicative forms observed by AFM.
The second observation is that LA9 mtDNA prepared by the procedure used historically in the Clayton laboratory also yields y-arcs upon 2D gel analysis (Fig. 6C). This finding contradicts the earlier conclusion that cell lines grown in culture normally replicate mtDNA via a different mode than in tissues (Holt et al. 2000). More importantly, it indicates that the numerous mtDNA replication studies in which LA9 and similar cell lines were used also had to be reconciled with the 2D gel pattern. Yang et al. (2002) have proposed that single-stranded replicative intermediates seen in these earlier studies are artifacts created during the isolation of mtDNA from cell lines. They have concluded that the nascent lagging strand is preferentially substituted with ribonucleotides relative to the leading strand. Upon isolation of mtDNA, the presence of an RNase H-like enzyme is inferred, which is posited to degrade the lagging strands and leave the parental strands intact. The postulated RNase H activity is thought by these authors to be eliminated during sucrose gradient purification of mitochondria. Our demonstration that the cell culture mtDNA preparation yields y-arcs is strong evidence against the notion of RNase H-mediated degradation of the lagging strand. This result would not be expected if one of the synchronously replicating strands were degraded.
There are further data that are inconsistent with the claim for an RNase H-induced artifact. First, the evidence for this enzyme contaminant is indirect, and L-strand degradation is contradicted by previous experiments in which newly synthesized L-strands were isolated intact (Robberson and Clayton 1972; Berk and Clayton 1974; Bogenhagen et al. 1979). Second, we have evaluated our procedures for artifact opportunities due to RNase H. We note that the use of AFM demanded a further purification since the bulk of DNA isolated directly from sucrose-gradient purified mitochondria is actually nuclear. It is remotely possible that by further purifying mtDNA in a CsCl–EtBr gradient we have introduced a step that would allow degradation of one strand as outlined above. However, the CsCl gradient step occurs after the sucrose gradient isolation of mitochondria that is argued to be the key step in eliminating RNase H. We have also assayed our mitochondrial lysates for RNase H and have found that the activity is below the level of detection, which is 0.01 U (Supplementary Fig. S2). Third, Figure 6D demonstrates that our liver mtDNA forms slow-moving y-arcs upon 2D gel analysis, showing that these preparations are equivalent to those of Holt and coworkers (Yang et al. 2002) and, by their criteria, free from RNase H contamination. Finally, the isolation of GpC molecules is strong evidence against a contaminating RNase H-like enzyme and is inconsistent with a strand-specific nuclease scheme. The double-stranded regions on these molecules are intact nascent L-strands and the complementary parental H-strands. These regions would have been at least partially degraded if they were highly ribosubstituted and exposed to an RNase H-like enzyme. These arguments and data leave little doubt that the single-strand-containing intermediates seen by AFM are the result of strand-displacement replication, not strand-specific degradation. It also indicates further that the 2D gel patterns from these mammalian mtDNA preparations cannot be interpreted in the simplest way, namely, that there are synchronous replication forks between OH and OL in mtDNA.
The 2D gel analysis is not necessarily inconsistent with strand-displacement replication
We hypothesize that the distribution of y-arcs and other 2D gel patterns might be explained by both alternative L-strand origins and branch migration of nascent strands in the strand-displacement mode of replication. Branch migration occurs when the parental strands reanneal and displace the nascent replicative strand. This process occurs rapidly and is promoted by low salt concentrations, nicking of closed circular DNA, and increased temperatures (Radding et al. 1977). It should be pointed out that the mtDNA preparation procedure employed by Holt, Jacobs, and colleagues (Yang et al. 2002; Bowmaker et al. 2003; Yasukawa et al. 2005) uses a final buffer solution with low salt, makes no attempt to isolate closed circular DNA, and generally employs a 50°C incubation. Thus, there are multiple conditions in those studies predicted to enhance branch migration. Figure 6E shows that branch migration of replication-stalled 7S DNA readily occurs during the preparation of the sample for 2D gel analysis. Since this mtDNA was purified as a closed circular mtDNA species, the release of 7S DNA occurred after isolation during the initial steps of the 2D gel procedure, either by the incubation in low-salt buffers or through the linearization required by the technique.
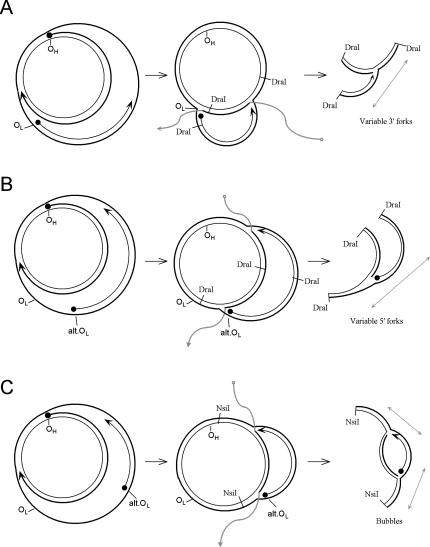
Figure 7 illustrates several ways in which branch migration of asymmetrically replicating mtDNA might lead to a set of molecules that distribute into the shape of the y-arc. In single-strand branch migration, the parental strands at either end could displace the leading strand of replication. The 5′ end of the leading strand would be quickly displaced until it meets the 3′ end of an L-strand. The AFM data can be interpreted to indicate that these 3′ ends are broadly distributed (Fig. 4B). Branch migration to these ends would create a distribution of “forks” that would be rendered clean by S1 nuclease treatment or by nicking at the branch point during the experiment. The parental strands in the opposite direction would similarly displace the 3′ end of the leading H-strand. This migration would slow or stop when meeting an alternative L-strand origin, creating a similar set of “forks.” “Bubbles” might also be formed by branch migration from both ends converging on a nascent L-strand. Indeed, this pattern has also been seen in the appropriate area with 2D gels (Bowmaker et al. 2003), casting doubt on the proposal of a bidirectional zone of initiation.
Figure 7.
Formation of y- and bubble-arc intermediates through alternate L-strand origins and branch migration of displacement replication strands. Branch-migrated arms of nascent strands are shown in gray. (A) Variable fork formation defined by the 3′ end of the L-strand. (B) Variable fork formation defined by the 5′ end of the L-strand. Variable formation of this end is dependent upon alternative L-strand initiation. (C) Variable fork formation at both ends to yield bubble-type intermediates.
We note that branch migration has long been a concern in studies of DNA replication and recombination. Pikó et al. (1978) have reported that ∼20% of replicative mtDNA intermediates examined from multiple tissues showed some degree of branch migration. Indeed, the originators of the 2D agarose gel technique were aware of the potential dangers of branch migration and used inverted DNA repeats to prevent their occurrence (Bell and Byers 1983). Branch migration artifacts in 2D agarose gel analyses are likely to be more prominent in asymmetrically replicating DNA, in which a single nascent strand is more easily displaced than would be the case for two nascent strands at a more fully duplex replication fork. Infrequent or delayed lagging-strand initiation and synthesis could thus lead to branch migration and erroneous mapping of fork positions in other systems that share this characteristic with mtDNA.
Finally, it is probable that some of the y-arc-generating forks do not require branch migration at all to form those patterns. As shown in Figure 7, a stalled leading H-strand may actually coincide with the alternative initiation of an L-strand, perhaps through strand switching of the polymerase or priming by associated mitochondrial RNAs (Aloni and Attardi 1972; Carré and Attardi 1978; Mayhook et al. 1992). The molecule shown in Figure 5A may contain an example of such a fork. A distribution of these forks among separate molecules within this area is expected to form a y-arc upon 2D gel analysis. It is also important to point out that although we have evidence for multiple L-strand initiation sites, the AFM images indicate that this is not equivalent to frequent repeated initiations on any single molecule. If this were occurring, we would have seen evidence of traditional leading- and lagging-strand synthesis by AFM. Rather, the L-strand initiations appear to occur once or twice per molecule, in an asymmetric, strand-displacement fashion.
A coherent conclusion on the principal mode of mtDNA replication
All of the many investigations of the mode of vertebrate mtDNA replication from 1971 to date either directly support the strand-displacement model or provide no compelling experimental data to support any alternative. Indeed, the results obtained by 2D gel analyses of mtDNA, reported here by us and by others elsewhere, do not exclude the strand-displacement mechanism, nor do they alone constitute evidence for other possible modes of replication.
This conclusion does not preclude the possibility that further studies will continue to refine the original replication model. The existence of additional, alternative lagging L-strand origins opens the possibility that under some conditions they may initiate more frequently and thereby produce replicative intermediates whose formal appearance is more fully duplex. Mammalian alternative L-strand origins may reflect sequences or mechanisms more commonly used in avian species where the orthodox OL sequence is absent (Desjardins and Morais 1990). Indeed, the results presented here are likely to be relevant for the interpretation of recent 2D agarose gel data presented from chicken mtDNA (Reyes et al. 2005). Future comprehensive efforts to study various aspects of mammalian mtDNA replication should further inform its vital role in cellular processes.
Materials and methods
mtDNA preparation
Mouse liver mitochondria was isolated and homogenized as described by Yang et al. (2002) in order to maximize the recovery of the y-arc-forming replication intermediates visualized by 2D agarose gels. Briefly, mouse livers were isolated, minced, washed in HB (225 mM mannitol, 75 mM sucrose, 10 mM Tris-HCl at pH 7.6, 1 mM EDTA, 0.1% fatty acid-free BSA), and weighed. Ten volumes of buffer were then added and the liver tissue was Dounce-homogenized using 10 strokes with a tightfitting (type B) pestle. Centrifugation at 1000 × g for 5 min to pellet unbroken cells and nuclei was followed by centrifugation of the supernatant at 9000 × g for 10 min. The resulting mitochondrial fraction was resuspended, and both centrifugation steps were repeated. The mitochondria-enriched pellet was loaded onto a two-phase sucrose gradient with a lower phase of 1.5 M sucrose, 10 mM Tris-HCl (pH 7.6), 5 mM EDTA and an upper phase with 1.0 M sucrose and the same buffer. The gradient was centrifuged at 22,000 rpm in an SW28Ti rotor (Beck-man) for 45 min. Mitochondria were isolated from the interface, washed with three volumes of buffer, and pelleted at 15,000 × g for 15 min. Initial attempts at AFM using DNA from subsequent proteinase K/phenol/chloroform extraction were unsuccessful due to the predominance of contaminating nuclear DNA. In order to further purify the mtDNA, the mitochondrial fraction was lysed with SDS, and mtDNA was purified using a CsCl–EtBr gradient as described (Tapper et al. 1983). The lower band was isolated along with half the middle region upward toward the nuclear DNA band. The positions of the various mtDNA forms within this gradient have been fully characterized (Robberson and Clayton 1972; Robberson et al. 1972). This fraction is expected to contain clean circular molecules, D-loop molecules, Exp-D, GpC, and some of the doubly-initiated daughter forms (Exp-D[l]). Cairns-like structures are predicted to be in the lower band as demonstrated by the isolation of these circular forms from SV40 viral DNA preparations (Jaenisch et al. 1971; Sebring et al. 1971). Prior to AFM analysis, purified mtDNA was dialyzed twice against one liter of 100 mM NaCl, 50 mM Tris-HCl (pH 8.5), 10 mM EDTA (STE buffer) in Slide-a-lyzer 7K cassettes (Pierce) and quantified using the Roche Lightcycler as previously described (Brown and Clayton 2002). The STE buffer was exchanged for binding buffer (4 mM HEPES, 10 mM NaCl, 2 mM MgCl2) by passing the mtDNA through a Micro Bio-Spin 6 chromatography spin column (Bio-Rad) equilibrated with this buffer immediately prior to microscopy. Alternatively, mtDNA was stored in CsCl after the removal of EtBr through several extractions with CsCl-saturated isopropanol.
Mouse cell line LA9 mtDNA was prepared using the differential centrifugation and CsCl-gradient method previously described (Tapper et al. 1983).
AFM
E. coli SSB was purchased from Amersham Biosciences at a concentration of 4.75 μg/mL. Single-stranded M13 DNA was purchased from New England Biolabs. SSB was bound to mtDNA by incubation in binding buffer for 15 min at 37°C using a weight ratio of 5:1 (SSB:DNA). These conditions are similar to those previously used for EM (Griffith et al. 1984; Park et al. 1998). Immobilization of the DNA–protein complexes onto a mica surface was done as previously described (Rivetti et al. 1999). Briefly, a 20-μL volume of SSB/mtDNA was applied to freshly exposed mica and allowed to attach for 2 min. The mica surface was gently washed with distilled water for 45 sec and dried under a stream of nitrogen. Atomic force microscopy was performed with a Nanoscope III (Digital Instruments) in tapping mode, using tips from Nanosensors (pointprobes, type NCH-100). For random sampling data collection, an arbitrary site on the grid was selected. From this point, the probe offset was adjusted to scan four areas around that point, each being 4 μm2 in size. The position of the probe was then systematically moved along one axis to avoid resampling any area on the surface (Table 1). Additional replicating molecules were identified and characterized through deliberate searches for SSB/mtDNA.
Data analysis for AFM
Flattened images from the Nanoscope III were converted to files compatible with the analysis application, SigmaScan Pro 5 (SPSS, Inc.). Contour lengths of mtDNA strands were measured using the Trace Measurement mode. ssDNA regions bound by SSB appear short due to the wrapping of the DNA around tetramers and octamers of SSB (Lohman and Ferrari 1994). Using repeated measurements of SSB-coated M13 (see Fig. 2A,B) and clear GpC mtDNA molecules as length references, we found that the length of the SSB-bound ssDNA was shorter than linear dsDNA by a factor of ∼0.335. This value is similar to that found in electron microscopy studies (Park et al. 1998). Length determinations of SSB-bound ssDNA were much more variable than with measurements of dsDNA, perhaps due to obscured contour paths and a mixed mode of SSB binding. In defining the approximate locations of the L-strand origin, it is important to point out that multiple laboratories have repeatedly defined the unidirectional H-strand origin by a variety of approaches (Kasamatsu and Vinograd 1973; Crews et al. 1979; Tapper and Clayton 1982; Kang et al. 1997; Holt et al. 2000). This position of the H-strand origin is therefore used to map the L-strand origin, as done previously (Robberson et al. 1972).
Neutral–neutral 2D agarose gel electrophoresis
Mouse liver mtDNA was prepared as above, and 150–300 ng was digested with the restriction endonuclease DraI for 60 min as outlined by the manufacturer (New England Biolabs). This enzyme creates a relevant 4.5-kb (mtDNA #5274–#9815) fragment that lies between the classical H- and L-strand replication origins (Fig. 7). The restriction-digested mtDNA was ethanol precipitated and resuspended in S1-nuclease buffer. Where indicated, S1 nuclease was added (0.5 U) and the ssDNA was digested for 1 min at 37°C. The reaction was stopped by the addition of 0.02 M EDTA. For analysis of slow-moving y-arcs we used the enzyme BglI as described by Yang et al. (2002). The procedure of Friedman and Brewer (1995) was used for 2D agarose gel electrophoresis. Briefly, the DNA was electrophoresed in 0.4% agarose without EtBr in a 1× TBE gel at measured current of 0.7 V/cm for 20 h. The gel was then stained in TBE with 300 ng/mL EtBr for 20 min. The lane was then excised and embedded at a 90° angle at the top of a 1% agarose gel containing 300 ng/mL EtBr. Second-dimension electrophoresis was done at 4°C at 5V/cm for 4–5 h with buffer recirculation. The DNA was transferred to a nylon membrane, and Southern analysis was done using the nonisotopic DIG High Prime DNA detection kit (Roche). The DNA probe used to detect y-arcs in this region was synthesized by PCR using the primers 5′-CGCCTAATCAACAACCGTCT-3′ and 5′-TGGTAGCTGTTGGTGGGCTA-3′, spanning mtDNA positions 8032–8497, which lies within the ATPase 6 gene. This is identical to the probe used by Holt and colleagues (Holt et al. 2000). The probe used to detect 7S DNA that had been released by branch migration was also made by PCR using the primers 5′-CCTTAGGT GATTGGGTTTTGCGGAC-3′ and 5′-GGTTCAGGTCATAAAATAATCATCAAC-3′, spanning mtDNA positions 16027–15510. The probe used for the detection of the slow-moving y-arcs was made using the primers 5′-AACTGAACGCCTAAACGCAGGGA-3′ and 5′-CCATGATTATAGTACGGCTGTGGA-3′ spanning mtDNA positions 10577–11193 within the ND4 gene. Chemiluminescence image detection was done using a Lumi-Imager F1 (Roche).
Acknowledgments
We thank Drs. Story C. Landis and James F. Battey of the National Institute for Neurological Disorders and Stroke for generously hosting T.A.B., A.N.T., and D.A.C. as guests of that institute.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1352105.
Corresponding authors.
References
- Aloni Y. and Attardi, G. 1972. Expression of the mitochondrial genome in HeLa cells XI. Isolation and characterization of transcription complexes of mitochondrial DNA. J. Mol. Biol. 70: 363-373. [DOI] [PubMed] [Google Scholar]
- Bell L. and Byers, B. 1983. Separation of branched from linear DNA by two-dimensional gel electrophoresis. Anal. Biochem. 130: 527-535. [DOI] [PubMed] [Google Scholar]
- Berk A.J. and Clayton, D.A. 1974. Mechanism of mitochondrial DNA replication in mouse L-cells: Asynchronous replication of strands, segregation of circular daughter molecules, aspects of topology and turnover of an initiation sequence. J. Mol. Biol. 86: 801-824. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D.F. and Clayton, D.A. 2003a. The mitochondrial DNA replication bubble has not burst. Trends Biochem. Sci. 28: 357-360. [DOI] [PubMed] [Google Scholar]
- ____. 2003b. Concluding remarks: The mitochondrial DNA replication bubble has not burst. Trends Biochem. Sci. 28: 404-405. [DOI] [PubMed] [Google Scholar]
- Bogenhagen D., Gillum, A.M., Martens, P.A., and Clayton, D.A. 1979. Replication of mouse L-cell mitochondrial DNA. Cold Spring Harbor Symp. Quant. Biol. 43: 253-262. [DOI] [PubMed] [Google Scholar]
- Borst P. and Kroon, A.M. 1969. Mitochondrial DNA: Physico-chemical properties, replication, and genetic function. Int. Rev. Cytol. 26: 107-190. [DOI] [PubMed] [Google Scholar]
- Bowmaker M., Yang, M.Y., Yasukawa, T., Reyes, A., Jacobs, H.T., Huberman, J.A., and Holt, I.J. 2003. Mammalian mitochondrial DNA replicates bidirectionally from an initiation zone. J. Biol. Chem. 278: 50961-50969. [DOI] [PubMed] [Google Scholar]
- Brown T.A. and Clayton, D.A. 2002. Release of replication termination controls mitochondrial DNA copy number after depletion with 2′,3′-dideoxycytidine. Nucleic Acids Res. 30: 2004-2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carré D. and Attardi, G. 1978. Biochemical and electron microscopic characterization of DNA–RNA complexes from HeLa cell mitochondria. Biochemistry 17: 3263-3273. [DOI] [PubMed] [Google Scholar]
- Crews S., Ojala, D., Posakony, J., Nishiguchi, J., and Attardi, G. 1979. Nucleotide sequence of a region of human mitochondrial DNA containing the precisely identified origin of replication. Nature 277: 192-198. [DOI] [PubMed] [Google Scholar]
- Desjardins P. and Morais, R. 1990. Sequence and gene organization of the chicken mitochondrial genome. A novel gene order in higher eucaryotes. J. Mol. Biol. 212: 599-634 [DOI] [PubMed] [Google Scholar]
- Friedman K.L. and Brewer, B.J. 1995. Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Meth. Enzymol. 262: 613-627. [DOI] [PubMed] [Google Scholar]
- Griffith J.D., Harris, L.D., and Register III, J. 1984. Visualization of SSB–ssDNA complexes active in the assembly of stable RecA–DNA filaments. Cold Spring Harbor Symp. Quant. Biol. 49: 553-559. [DOI] [PubMed] [Google Scholar]
- Holt I.J. and Jacobs, H.T. 2003. Response: The mitochondrial DNA replication bubble has not burst. Trends Biochem. Sci. 28: 355-356. [DOI] [PubMed] [Google Scholar]
- Holt I.J., Lorimer, H.E., and Jacobs, H.T. 2000. Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell 100: 515-524. [DOI] [PubMed] [Google Scholar]
- Jaenisch R., Mayer, A., and Levine, A. 1971. Replicating SV40 molecules containing closed circular template DNA strands. Nature New Biol. 233: 72-75. [DOI] [PubMed] [Google Scholar]
- Kang D., Miyako, K., Kai, Y., Irie, T., and Takeshige, K. 1997. In vivo determination of replication origins of human mitochondrial DNA by ligation-mediated polymerase chain reaction. J. Biol. Chem. 272: 15275-15279. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H. and Vinograd, J. 1973. Unidirectionality of replication in mouse mitochondrial DNA. Nature New Biol. 241: 103-105. [DOI] [PubMed] [Google Scholar]
- Kasamatsu H., Robberson, D.L., and Vinograd, J. 1971. A novel closed-circular mitochondrial DNA with properties of a replicating intermediate. Proc. Natl. Acad. Sci. 68: 2252-2257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike K. and Wolstenholme, D.R. 1974. Evidence for discontinuous replication of circular mitochondrial DNA molecules from Novikoff rat ascites hepatoma cells. J. Cell Biol. 61: 14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohman T.M. and Ferrari, M.E. 1994. Escherichia coli single-stranded DNA-binding protein: Multiple DNA-binding modes and cooperativities. Annu. Rev. Biochem. 63: 527-570. [DOI] [PubMed] [Google Scholar]
- Mayhook A.G., Rinaldi, A.M., and Jacobs, H.T. 1992. Replication origins and pause sites in sea urchin mitochondrial DNA. Proc. Royal Soc. London—Series B: Biol. Sci. 248: 85-94. [DOI] [PubMed] [Google Scholar]
- Park K., Debyser, Z., Tabor, S., Richardson, C.C., and Griffith, J.D. 1998. Formation of a DNA loop at the replication fork generated by bacteriophage T7 replication proteins. J. Biol. Chem. 273: 5260-5270. [DOI] [PubMed] [Google Scholar]
- Pikó L. and Matsumoto, L. 1977. Complex forms and replicative intermediates of mitochondrial DNA in tissues from adult and senescent mice. Nucleic Acids Res. 4: 1301-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pikó L., Meyer, R., Eipe, J., and Costea, N. 1978. Structural and replicative forms of mitochondrial DNA from human leukocytes in relation to age. Mech. Ageing Dev. 3: 351-365. [DOI] [PubMed] [Google Scholar]
- Pikó L., Bulpitt, K.J., and Meyer, R. 1984. Structural and replicative forms of mitochondrial DNA in tissues from adult and senescent BALB/c mice and Fischer 344 rats. Mech. Ageing Dev. 26: 113-131. [DOI] [PubMed] [Google Scholar]
- Radding C.M., Beattie, K.L., Holloman, W.K., and Wiegand, R.C. 1977. Uptake of homologous single-stranded fragments by superhelical DNA: Branch migration. J. Mol. Biol. 116: 825-839. [DOI] [PubMed] [Google Scholar]
- Rajamanickam C., Merten, S., Kwiatkowska-Patzer, B., Chuang, C.H., Zak, R., and Rabinowitz, M. 1979. Changes in mitochondrial DNA in cardiac hypertrophy in the rat. Circ. Res. 45: 505-515. [DOI] [PubMed] [Google Scholar]
- Reyes A., Yang, M.Y., Bowmaker, M., and Holt, I.J. 2005. Bidirectional replication initiates at sites throughout the mitochondrial genome of birds. J. Biol. Chem. 280: 3242-3250. [DOI] [PubMed] [Google Scholar]
- Rivetti C., Guthold, M., and Bustamante, C. 1999. Wrapping of DNA around the E. coli RNA polymerase open promoter complex. EMBO J. 18: 4464-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberson D.L. and Clayton, D.A. 1972. Replication of mitochondrial DNA in mouse L cells and their thymidine kinase- derivatives: Displacement replication on a covalently closed circular template. Proc. Natl. Acad. Sci. 69: 3810-3814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberson D.L., Kasamatsu, H., and Vinograd, J. 1972. Replication of mitochondrial DNA. Circular replicative intermediates in mouse L cells. Proc. Natl. Acad. Sci. 69: 737-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebring E.D., Kelly Jr., T.J., Thoren, M.M., and Salzman, N.P. 1971. Structure of replicating simian virus 40 deoxyribonucleic acid molecules. J. Virol. 8: 478-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel G.S. and Clayton, D.A. 1997. Mitochondrial DNA maintenance in vertebrates. Annu. Rev. Biochem. 66: 409-435. [DOI] [PubMed] [Google Scholar]
- Tapper D.P. and Clayton, D.A. 1981. Mechanism of replication of human mitochondrial DNA. Localization of the 5′ ends of nascent daughter strands. J. Biol. Chem. 256: 5109-5115. [PubMed] [Google Scholar]
- ____. 1982. Precise nucleotide location of the 5′ ends of RNA-primed nascent light strands of mouse mitochondrial DNA. J. Mol. Biol. 162: 1-16. [DOI] [PubMed] [Google Scholar]
- Tapper D.P., Van Etten, R.A., and Clayton, D.A. 1983. Isolation of mammalian mitochondrial DNA and RNA and cloning of the mitochondrial genome. Meth. Enzymol. 97: 426-434. [DOI] [PubMed] [Google Scholar]
- Van Tuyle G.C. and Pavco, P.A. 1985. The rat liver mitochondrial DNA–protein complex: Displaced single strands of replicative intermediates are protein coated. J. Cell Biol. 100: 251-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M.Y., Bowmaker, M., Reyes, A., Vergani, L., Angeli, P., Gringeri, E., Jacobs, H. T., and Holt, I.J. 2002. Biased incorporation of ribonucleotides on the mitochondrial L-strand accounts for apparent strand-asymmetric DNA replication. Cell 111: 495-505. [DOI] [PubMed] [Google Scholar]
- Yasukawa T., Yang, M.Y., Jacobs, H.T., and Holt, I.J. 2005. A bidirectional origin of replication maps to the major noncoding region of human mitochondrial DNA. Mol. Cell 18: 651-662. [DOI] [PubMed] [Google Scholar]