Abstract
The core fucosylation (α1,6-fucosylation) of glycoproteins is widely distributed in mammalian tissues, and is altered under pathological conditions. To investigate physiological functions of the core fucose, we generated α1,6-fucosyltransferase (Fut8)-null mice and found that disruption of Fut8 induces severe growth retardation and death during postnatal development. Histopathological analysis revealed that Fut8-/- mice showed emphysema-like changes in the lung, verified by a physiological compliance analysis. Biochemical studies indicated that lungs from Fut8-/- mice exhibit a marked overexpression of matrix metalloproteinases (MMPs), such as MMP-12 and MMP-13, highly associated with lung-destructive phenotypes, and a down-regulation of extracellular matrix (ECM) proteins such as elastin, as well as retarded alveolar epithelia cell differentiation. These changes should be consistent with a deficiency in TGF-β1 signaling, a pleiotropic factor that controls ECM homeostasis by down-regulating MMP expression and inducing ECM protein components. In fact, Fut8-/- mice have a marked dysregulation of TGF-β1 receptor activation and signaling, as assessed by TGF-β1 binding assays and Smad2 phosphorylation analysis. We also show that these TGF-β1 receptor defects found in Fut8-/- cells can be rescued by reintroducing Fut8 into Fut8-/- cells. Furthermore, exogenous TGF-β1 potentially rescued emphysema-like phenotype and concomitantly reduced MMP expression in Fut8-/- lung. We propose that the lack of core fucosylation of TGF-β1 receptors is crucial for a developmental and progressive/destructive emphysema, suggesting that perturbation of this function could underlie certain cases of human emphysema.
Keywords: fucosylation, glycobiology, matrix metalloproteinase
The physiological importance of fucose modifications on proteins has been highlighted recently by the description of human congenital disorders of glycosylation (CDG). The disease CDG-IIc is due to lack of the GDP-fucose transporter activity (1, 2). α1,2-, α1,3-, α1,4-, and α1,6-fucosylations have been described; α1,6-fucose is found linked to the Asn-linked GlcNAc in the N-glycan core. All of these fucosylations are terminal capping reactions, and the fucose residues have no substituent. In contrast, O-fucosylation, in which fucose is attached directly to a serine or threonine residue in a particular protein context such as an EGF repeat, undergoes the elongation of the oligosaccharide into a tetrasaccharide (NeuAcα2-3/6Galβ1-4GlcNAcβ1-3Fuc-Ser/Thr), in which Fringe has been found to be the N-acetylglucosaminyltransferase acting on O-linked fucose of the Notch receptor (3). Because Notch receptors play key roles in numerous developmental events, several features of the phenotype in CDG-IIc could be explained by defects in Notch function. However, Sturla et al. (4, 5) reported that reduced fucosylation is mainly confined to terminal fucosylation of N-glycans, and that protein O-fucosylation levels such as those that occur in Notch are unaffected in CDG-IIc; therefore, we speculate that core fucosylation may be responsible for the phenotype of CDG-IIc.
GDP-L-Fuc:N-acetyl-β-d-glucosaminide α1,6-fucosyltransferase (Fut8, EC 2.4.1.152) catalyzes the transfer of a fucose residue from GDP-fucose to position 6 of the innermost GlcNAc residue of hybrid and complex types of N-linked oligosaccharides on glycoproteins (6). Core Fut8 is the only core FucT in mammals, but there are core α1,3-Fuc residues in plants, insects, and probably other species. The Fut8 gene is expressed in most rat organs with a relatively high level of expression in brain and small intestine (7). In good agreement with the Fut8 gene expression, α1,6-fucosylated glycoproteins are widely distributed in mammalian tissues (8). Furthermore, the expression of Fut8 and the extent of core fucosylation are altered under pathological conditions such as hepatocellular carcinoma and liver cirrhosis (8, 9).
The molecular cloning of the Fut8 gene (10) enabled us to manipulate it and to remodel the N-glycans in animal models. Overexpression of the Fut8 gene caused steatosis in the liver and kidney due to a decreased lysosomal acid lipase activity accompanied by its over-fucosylation (11). Recently, it was reported that the core fucose-deficient IgG1 (produced in a fucose-deficient Chinese hamster ovary cell line) showed improved binding to FcγRIIIA. As a consequence, antibody-dependent cellular cytotoxicity activity mediated by their interaction was enhanced (12, 13). These findings strongly suggested that core fucosylation of N-glycans modifies the function of the glycoproteins.
To define the physiological roles of a particular glycosylation, gene targeting technology to disrupt the relevant glycosyltransferase gene function is considered as the best approach currently available. In fact, accumulating evidence on gene targeting for glycosyltransferases has elucidated a variety of novel functions of carbohydrates and provided new insights into their roles in vivo (14). Here we report the generation of Fut8-null mice and describe critical roles of core fucosylation in vivo.
Materials and Methods
Gene Targeting. A part of the mouse Fut8 gene spanning 13.9 kb, which includes the exon containing the translation-initiation site, was isolated by screening a mouse 129SvJ λ genomic library (Stratagene), using a SacI-SacI fragment of porcine Fut8 cDNA (nt -39 to 373) (10) as a probe. A targeting vector was constructed by replacing the 184-bp SacI-HindIII fragment containing the translation-initiation site with a 4.9-kb SacI-SalI fragment of the plasmid pGT1.8IresBgeo (15) that contains an internal ribosome entry site (IRES)-LacZ-Neor-polyadenylation signal (pA) cassette, flanked with a 1.5-kb XhoI-NotI fragment of the plasmid pMC1DTpA (16), which encodes diphtheria toxin A chain (DT-A) for negative screening (see Fig. 4, which is published as supporting information on the PNAS web site). The targeting vector was transfected into D3 embryonic stem cells, and clones were selected with G418. Southern blot analysis of selected clones with 5′ (A) and 3′ (B) probes (Fig. 4) revealed that 1.2% (4 of 343) of the embryonic stem clones had undergone correct homologous recombination. Targeted cell clones were then injected into blastocysts from B6C3F1 mice, which are F1 mice resulting from the intercross of female C57BL/6 and male C3H mice. Germ-line transmission of the mutant allele was achieved from male chimeras derived from two independent embryonic stem cell clones.
Oligosaccharide Structural Analyses of the Mouse Lungs. N-linked oligosaccharides of lung were liberated by hydrazinolysis at 100°C for 10 h and then re-N-acetylated. The reducing ends of the oligosaccharides were labeled with 2-aminopyridine (PA) as described in ref. 17. PA-oligosaccharides were subjected to HPLC analysis and further to liquid chromatography-electrospray ionization-MS analysis, which was performed on an HCT ion trap mass spectrometer (Bruker Daltonics, Bremen, Germany) equipped with an electrospray source working in positive ion mode.
Measurement of Lung Compliance and Ventilation. Lung compliance was measured by drawing static air pressure-volume relations in urethane-anesthetized (1.5 g/kg) mice tracheotomized with polyethylene tubing (O.D. = 0.8 mm). Total lung capacity was defined as the lung volume of full inflation as judged by visual inspection of the lung that fully occupied the chest cavity. Any obvious, continual decrease in the pressure was evidence of a leaking lung, which was then discarded. The lung volumes at each measured point were expressed as a percentage of the total lung capacity. Ventilation was measured by whole-body plethysmography in unanesthetized freely moving mice as reported in ref. 18.
Establishment of Embryonic Fibroblasts. For preparation of embryonic fibroblasts, a whole mouse embryo at 18.5 days postcoitus was dissected, and the head and all internal organs were removed. The carcasses were minced, incubated in PBS (-) containing 0.05% trypsin, 0.53 mM EDTA, and 40 μg/ml DNase at 37°C for 30 min with stirring three times, and then cells were plated on a 100-mm dish in DMEM supplemented with 10% FCS and incubated at 37°C in humidified air containing 5% CO2. To obtain immortal cells, Zeocine-resistant vector (pcDNA3.1) containing the SV40 gene was introduced to these primary embryonic fibroblasts. Transfectants were screened in the presence of 400 μg/ml Zeocine, and SW (wild-type cells immortalized with SV40 gene) and SK (knockout cells immortalized with SV40 gene) immortal cells were established from Fut8+/+ and Fut8-/- primary fibroblasts, respectively.
125I-TGF-β1-Binding Assays. The cells (1.5 × 105 per well) were cultured on 24-well plates, washed twice with 500 μl of PBS containing 0.1% BSA, and incubated with 200 μl of PBS containing different amounts of 125I-TGF-β1 in a concentration range of 0.1-1.0 ng and 10 ng of unlabeled TGF-β1. Nonspecific binding was determined by adding 100 ng of unlabeled TGF-β1. After incubation for 2 h at 4°C with shaking, the cells were washed and solubilized in 500 μl of 1 M NaOH. The radioactivity of the cell lysates was counted with a γ-counter.
Receptor Cross-Linking. The cells (5 × 105) were seeded onto a 60-mm culture dish and incubated for 48 h. After being washed with cool PBS containing 0.1% BSA, the cells were incubated for 2 h at 4°C with 250 pM 125I-TGF-β1 and then incubated with 1 mM cross-linking reagent BS3 for 30 min at room temperature. After washing, the cells were lysed, the protein concentrations of cell lysates were determined, and the same amounts of protein were loaded onto 10% SDS-polyacrylamide gel for analysis. The gels were exposed and quantified with the BAS-2500 bio-imaging analyzer (Fuji).
Immunohistochemical Analysis. To detect the P-Smad2 (Ser-465/467), matrix metalloproteinase (MMP)-12, or SP-C, whole lung tissues from animals after age indicated were fixed in 0.1 M PBS containing 4% paraformaldehyde and embedded in paraffin. For immunohistochemical analysis, the dewaxed sections were pretreated with avidin-biotin blocking and hydroxygen blocking for 10 min at 37°C, and then incubated with rabbit anti-P-Smad2 antibody (Cell Signaling Technology, Beverly, MA), anti-MMP-12 antibody, or anti-SP-C antibody (Santa Cruz Biotechnology) for 16 h at 4°C. Localization of the first antibody was visualized by an avidin-biotin coupling (ABC) immunoperoxidase technique, using a commercial kit (Vector Laboratories) according to the manufacturer's instructions.
Therapeutic Administration of Exogenous TGF-β1 to Fut8-/- Mice. We performed i.p. injection of recombinant TGF-β1 in a dose of 50 or 100 ng/g of mouse body weight to postnatal 18-day Fut8-/- mice. After 20 times of injection every 2 days, lung sections were subjected to hematoxylin/eosin staining or immunohistochemical study.
Results and Discussion
Fut8 Gene Is a Unique Fucosyltransferase Responsible for the Core Fucosylation of N-Glycans. A targeted disruption of Fut8 was generated through homologous recombination in embryonic stem cells. The targeting vector was constructed by replacing exon 2 of Fut8, which contains the translation initiation site, with an IRES-LacZ-Neo-pA cassette (Fig. 4A). Genotypes of pups from intercrosses between heterozygous mice were determined by Southern blotting (Fig. 4B). The phenotype described here was identical in two lines. Northern blot analysis of lung and brain RNA revealed that expression of full-length Fut8 mRNA was abolished in homozygous mutant mice, whereas Fut8 transcripts were detected as a single 3.5-kb band in wild-type mice (Fig. 4C). Consistent with this finding, Fut8 activity could not be detected in Fut8-/- tissues including brain or lung, even with six times longer incubation than that used for Fut8+/+ specimens (Fig. 4D and data not shown). Furthermore, the analysis of N-glycan structures showed that the elution profile of Fut8-/- lung lacked the peaks of oligosaccharides with core fucose eluting at 30-35 min, as described in refs. 19 and 20 (Fig. 4E). These oligosaccharides were also confirmed by mass spectrometric analysis (Fig. 5, which is published as supporting information on the PNAS web site). This finding agrees with previous reports indicating that there are no additional genes homologous to Fut8 in mammals and lower organisms (21, 22). Thus, the Fut8 gene is the only one responsible for the core fucosylation of N-glycans in mouse tissues.
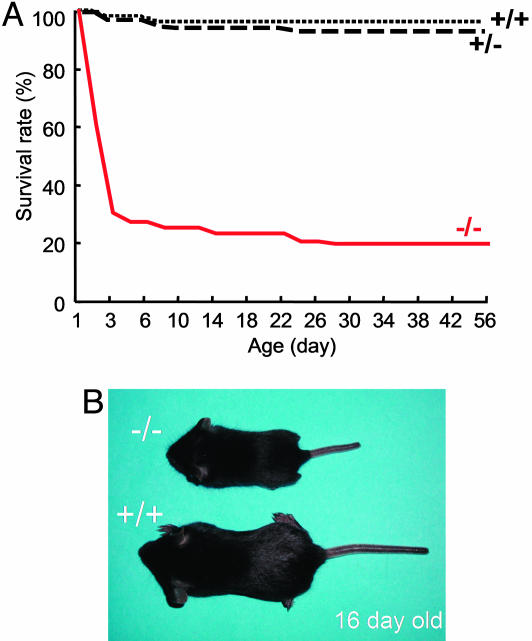
Core Fucosylation Is Essential for Mice Survival and Growth. Fut8-/- mice were born apparently healthy with almost the expected Mendelian inheritance: Of 277 pups, there were 59 (21.3%) Fut8-/-, 147 (53.1%) Fut8+/-, and 71 (25.6%) Fut8+/+ mice. At embryonic day 19, frequencies of Fut8-/-, Fut8+/-, and Fut8+/+ mice were 26, 52, and 22 of 100 embryos, and there were no apparent anomalies in Fut8-/- mice. In contrast to Fut8-/- mice, embryonic lethality was observed in mutant mice deficient in the FX gene, which encodes an enzyme in the de novo pathway for GDP-fucose synthesis and is responsible for all cellular fucosylation, e.g., α1,2; α1,3; α1,6; etc. (23). The appearance of Fut8-/- mice could not be distinguished from Fut8+/- and Fut8+/+ mice within 3 days of age, but ≈70% of them died during this period (Fig. 1A). Most of the survivors manifested severe growth retardation (Fig. 1B). This pattern observed in the Fut8-/- mice is quite different from other N-linked medial- and trans-Golgi glycosyltransferases, such as GnT-III-, GnT-V-, or ST6Gal1-null mice (14), suggesting that core fucose has a unique role in the regulation of proliferation and differentiation after birth.
Fig. 1.
Semilethality and growth retardation in Fut8-/- mice. (A) Survival ratio of Fut8-/- (-/-, solid line), Fut8+/- (+/-, broken line), and Fut8+/+ (+/+, dotted line) mice after birth. (B) A 16-day-old Fut8-/- pup (-/-) with a Fut8+/+ littermate (+/+).
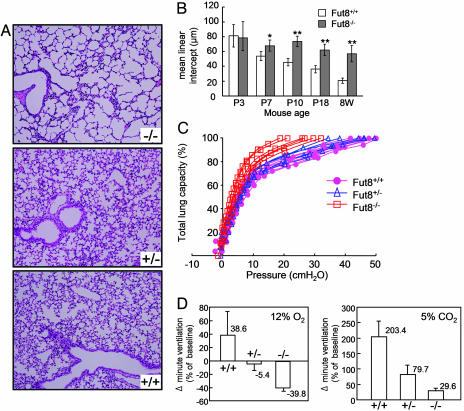
Progressive Emphysema-Like Changes in Fut8-/- Lungs. Histological analyses by hematoxylin/eosin staining of 3-, 7-, 10-, and 18-day-old and 8-week-old Fut8-/- mice showed a symmetrical reduction in the size of most organs, which otherwise appeared devoid of pathological signs. The lungs of Fut8-/- mice, however, apparently displayed generalized air-space enlargement and dilated alveolar ducts, compared with those of Fut8+/+ and Fut8+/- mice (Fig. 2A). The mean linear intercept was calculated at the ages indicated. From postnatal day 7, diameters of the pulmonary alveoli of Fut8-/- mice were increased significantly, compared with those in Fut8+/+ mice (Fig. 2B). To evaluate the functional relevance of this morphological abnormality, lung compliance was evaluated by a static air deflation curve (Fig. 2C). The Fut8-/- lungs had larger total lung capacities and increased lung compliance compared with Fut8+/- and Fut8+/+ lungs. These results indicate that the altered architecture of the enlarged Fut8-/- mouse airspace contributes to the increased compliance. We also measured pressure-volume relationships in body-weight-matched young wild types (≈7 days old) to cancel the difference in total lung capacity. Again, static lung compliance in Fut8-/- mice was greater than that in wild-type mice (data not shown). When Fut8-/- mice breathed room air under resting conditions, respiratory minute volume and rate, as determined by body plethysmography, were higher than in wild-type mice. Ventilatory responses to systemic hypoxia (12% O2) or hypercapnia (5% CO2/21% O2), or increases in the respiratory minute volume, were significantly attenuated in Fut8-/- mice, compared with Fut8+/+ and Fut8+/- mice (Fig. 2D). These findings, although not specific markers of emphysema, suggest that Fut8 is involved in the physiological control of ventilation. However, we do not believe that the main reason that Fut8-/- mice die is lung disorder. Interestingly, besides TGF-β1 receptor as described below, we also found that loss of core fucosylation resulted in modest down-regulation of several other receptors-mediated signaling, such as EGF receptor and integrins (unpublished data), which are responsible for cell growth and differentiation. Therefore, we would like to take the hypothesis that the growth retardation and early death of Fut8-/- mice can be attributed to dysregulation of many receptors-mediated signaling.
Fig. 2.
Emphysematous change and impaired ventilatory response in Fut8-/- lung. (A) Representative histological sections (hematoxylin/eosin staining) of the lung of 18-day-old Fut8+/+ (+/+), Fut8+/- (+/-), and Fut8-/- (-/-) pups show enlarged alveoli indicative of emphysema in Fut8-/- mice. (B) Lung morphometry at different stages. The mean linear intercepts of pulmonary alveoli were calculated at postnatal 3-, 7-, 10-, 18-day-old and 8-week-old mice. The diameters of the pulmonary alveoli were shown as the mean ± SD from three independent experiments. Statistical analysis was performed by using Student's t test. *, P < 0.01; **, P < 0.001 (versus the matched age of Fut8+/+ mice). P, postnatal. (C) Lung compliance test. Static pressure-volume curves were plotted by using 19- to 20-day-old Fut8-/- (brown), Fut8+/+ (pink), and Fut8+/- (blue) pups. (D) Comparison of changes in respiratory minute volume in response to hypoxic or hypercapnic air in conscious 19- to 20-day-old littermate Fut8+/+ (+/+), Fut8+/- (+/-), and Fut8-/- (-/-) mice.
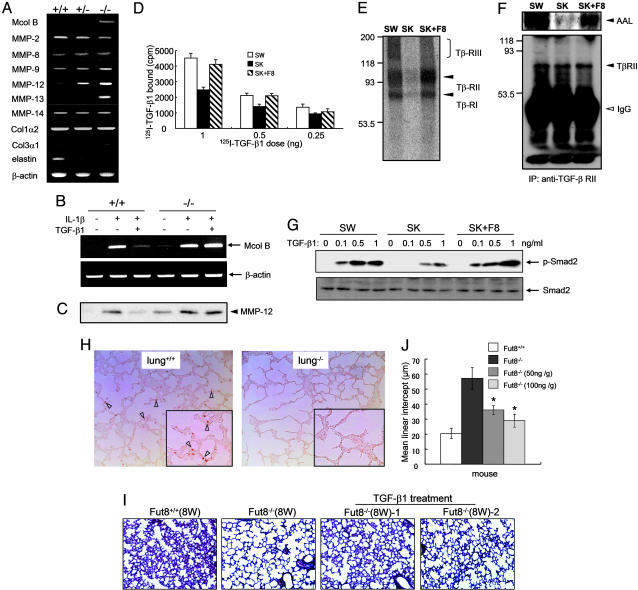
Enhanced Expression Levels of MMPs in Fut8-/- Lungs and Fut8-/- Cells. Pulmonary emphysema is believed to result from decreased structural integrity of connective tissues due to a defect in their formation or to an abnormal proteolysis. Elastin and fibrillar collagen are major components of the extracellular matrix (ECM), which sustains the normal lung architecture. On the other hand, MMPs are a group of zinc- and calcium-dependent proteinases that have an important role in the normal turnover of ECM components. Abnormal production of MMPs is implicated in the induction of emphysema. Thus, transgenic mice expressing human MMP-1 develop emphysema through destruction of collagen fibrils (24). Furthermore, MMP-2, MMP-9, and MMP-12 lead to emphysema by degradation of elastin fibers (25, 26). Therefore, expression levels of collagens, elastin, and a number of MMPs of putative relevance in lung pathology, including McolB (a mouse orthologue of human MMP-1) (27) and MMP-2, -8, -9, -12, -13, and -14, were examined in the lungs of Fut8-mutant mice. RT-PCR analysis showed that there were no significant changes in the expression levels of collagens, MMP-2, -8, and -14 in lung tissues from Fut8-/-, Fut8+/-, and Fut8+/+ mice (Fig. 3A). However, expression levels of McolB, MMP-12, and MMP-13 were greatly enhanced (Fig. 3A); in addition, a slight increase in MMP-9 levels was detected in lungs from Fut8-/- by using real-time RT-PCR (Fig. 6A and Table 2, which are published as supporting information on the PNAS web site). Conversely, elastin expression was down-regulated in lungs from Fut8-deficient mice. On the other hand, fragmentation and a significantly reduced number of elastic fibers were observed by elastin staining in Fut8-/- mice (Fig. 6B), supporting the view that the degrading phenotype, i.e., emphysematous changes, occurs in Fut8-/- lung. Actually, emphysema-like changes were coincident with increased expression levels of MMP-12 from postnatal day 7 of Fut8-/- mice (Fig. 6C). Furthermore, only in the bronchoalveolar lavage fluid of Fut8-/- mice did macrophages look vacuolated, which, along with MMP-12 expression, indicates activation (Fig. 6I). In addition, an enhanced expression of CD68, a marker of macrophage, was clearly observed in Fut8-/- lung (data not shown). Recently, Morris et al. (28) reported that the loss of the epithelial integrin αVβ6, which causes a local deficiency in active TGF-β1, results in the increased expression of MMP-12 and leads to a slowly progressive, age-related emphysema. Likewise, McolB is a murine orthologue of human MMP-1, an enzyme that has been associated repeatedly with lung pathology, including emphysema (24), whereas MMP-13 or collagenase-3 is a very potent enzyme with wide substrate specificity, which is also associated with pulmonary diseases. It will also be interesting to examine whether core fucosylation affects MMP proteolytic function, because some MMPs such as MMP-9 contain N-glycans with core fucose. Taken together, these results suggest that overexpression of a set of MMPs might be causally linked to the development of emphysema in Fut8-/- mice.
Fig. 3.
Enhancement of some MMPs expression through down-regulation of TGF-β-receptor-mediated signaling in Fut8-/- lung and embryonic fibroblasts. (A) RT-PCR analysis of emphysema-relating genes (see Table 1, which is published as supporting information on the PNAS web site). Total RNAs from 18-day-old Fut8+/+ (+/+), Fut8+/- (+/-), and Fut8-/- (-/-) lungs were used as template. β-Actin RNA is shown as a loading control. (B) Effects of IL-1β and TGF-β1 on McolB expression. These fibroblasts were preincubated with or without TGF-β1(1ng/ml) for 3 h and then further incubated with or without IL-1β (2 ng/ml) for 24 h. Total RNA was isolated and used as template. (C) Effects of IL-1β and TGF-β on protein expression of MMP-12 secreted into culture media. These fibroblasts were incubated with or without IL-1β and TGF-β1 in the absence of FCS. After incubation for 24 h, the conditioned media were concentrated and subjected to electrophoresis for Western blot. (D) Binding of 125I-TGF-β1 to its receptors on the cell surface. These cells from Fut8+/+ (+/+) and Fut8-/- (-/-) primary fibroblasts immortalized with SV40 large T, SW, and SK, respectively, or SK restored with Fut8 (SK+F8), were incubated with different amounts of radiolabeled TGF-β1 as indicated and 10 ng of cold TGF-β1 for 2 h on ice. Cell lysate radioactivity was measured. (E) 125I-TGF-β1 was bound and cross-linked to its receptors on cell surface. The cultured cells were incubated with 250 pM 125I-TGF-β1 for 2 h at 4°C, and then cross-linked with reagent BS3.(F) Analysis of fucosylation levels on TGF-β receptor II. TGF-β1 receptor II was immunoprecipitated from whole-cell lysates and then subjected to electrophoresis on 8% SDS/PAGE. After electroblotting, blots were probed by ALL lectin (Upper) and anti-TGF-β receptor II (Lower). (G) Effects of phosphorylated Smad2 levels on TGF-β1 stimulation. Serum-starved cells were treated with or without TGF-β1 at the indicated concentrations for 5 min and solubilized in lysis buffer as described in Materials and Methods. The cell lysates were detected by immunoblotting of anti-phospho-Smad2 antibody (Upper) and anti-Smad2 antibody (Lower). (H) The lung sections of 4-day-old mice were pretreated with hydroxygen blocking for 10 min at 37°C and then incubated with rabbit anti-human P-Smad2 antibody for 16 h at 4°C. The arrowheads indicate positive staining in Fut8+/+ lung. (I) Therapeutic administration of recombinant TGF-β1 rescues emphysema-like changes in Fut8-/- mice. The surviving postnatal 18-day-old Fut8-/- mice were treated with or without recombinant TGF-β1 (50 or 100 ng/g of mouse body weight) for 20 times of injection every 2 days, and then the lung sections were subjected to hematoxylin/eosin staining. (J) Quantitative analyses of the pulmonary alveolar sizes were performed by mean linear intercept as described above. The diameters of the pulmonary alveoli were shown as the mean ± SD from three independent experiments. Statistical analysis was performed by using Student's t test. *, P < 0.01 (Fut8-/- mice treated with TGF-β versus the matched age of mice without treatment).
To gain insight into the mechanism of induction of these MMPs in Fut8-/- mice, we established embryonic fibroblasts from Fut8+/+ and Fut8-/- mice. Consistent with the data from the lungs shown in Fig. 3A, under resting conditions, McolB mRNA transcripts (Fig. 3B) and MMP-12 (Fig. 3C) and MMP-13 (data not shown) proteins secreted into the media were barely detected in wild-type embryonic fibroblasts, whereas they were clearly detected in Fut8-/- cells. To examine whether these changes in MMP expression levels were also manifested in the respective MMP proteolytic activities, gelatin and casein zymographies were performed by using the conditioned media from these embryonic cells. A differential band of ≈45 kDa, likely corresponding to the gelatinolytic activity of MMP-12, was observed in the gelatin zymogram of Fut8-/- cells. Likewise, a band of ≈55 kDa was detected in the casein zymogram of Fut8 -/- cells (Fig. 6 D and E). In fact, the increased expression levels of both MMP-12 and MMP-13 secreted into media in the Fut8-/- cells were also confirmed by a Western blot (Fig. 6F). Consistent with these results, the enhancement of MMP-12 expression was also observed in lung tissues of Fut8 -/- mice (Fig. 6G). These results demonstrate that the increased RNA and protein expression levels of some MMPs are accompanied by an increase in the proteolytic potential of cells from Fut8-deficient mice.
Lack of Core Fucose in TGF-β Receptor Leads to Inhibiting Its Function. Recently, it became clear that modification by N-glycosylation can affect the biological functions of many glycoprotein receptors. As described above, we found that loss of core fucosylation resulted in down-regulation of several receptor-mediated signaling pathways, such as TGF-β1 receptor, EGF receptor, and integrins, which are responsible for cell growth and differentiation, and also emphysema. The TGF-β1 receptor-mediated signaling pathway is a key pathway for regulating expression of ECM proteins, including suppression of MMPs to produce a “synthetic” phenotype (29). When the embryonic fibroblasts were treated with IL-1β, which enhances MMP expression, McolB mRNA levels were elevated markedly in both Fut8+/+ cells and Fut8-/- cells (Fig. 3B). The enhancement of McolB expression stimulated by IL-1β was blocked by TGF-β1 treatment in Fut8+/+ cells but not in Fut8-/- cells (Fig. 3B), indicating that the deletion of Fut8 does not alter IL-1β-receptor-mediated function but diminishes TGF-β1-mediated signaling. The decreased response of TGF-β1 stimulation was also observed in protein expression levels of MMP-12 secreted into the media (Fig. 3C). To examine how core fucose affects TGF-β1-mediated signaling, we measured the binding activity of TGF-β1 for its surface receptor. The binding ability of 125I-TGF-β1 was reduced significantly in Fut8-/- cells compared with Fut8+/+ cells, which could be rescued by reintroducing Fut8 to Fut8-/- cells (Fig. 3D). Consistent with this, the amount of TGF-β1 bound and cross-linked to type I, II, and III receptors was suppressed dramatically in Fut8-/- cells; these features were recovered by reintroducing Fut8 to Fut8-/- cells (Fig. 3E). Actually, the levels of core fucosylation detected by AAL lectin in the TGF-β type II receptor, which is the primary binding subunit for TGF-β1 (30-32), were abolished in Fut8-/- cells, whereas they were recovered by restoring Fut8 (Fig. 3F). The TGF-β1 signaling via receptors to intracellular mediators of the Smad family was suppressed significantly in Fut8-/- cells (Fig. 3G). Smad2 is a direct substrate for the activated TGF-β type I receptor. In addition, Smad2 phosphorylation at C-terminal serine residues is required for its nuclear translocation (33). The down-regulation of Smad2 phosphorylation levels in Fut8-/- cells was rescued by reintroducing Fut8 (Fig. 3G). Consistently, immunohistochemical analysis of the phosphorylation levels of Smad2 in lung tissues revealed that P-Smad2 levels were greatly suppressed in Fut8-/- mice, compared with that in Fut8+/+ mice (Fig. 3H). Taken together, these results demonstrate that core fucosylation plays an important role in the regulation of TGF-β1 receptor function. Therefore, we assume that Fut8-/- lungs are committed to overexpressing MMPs, probably because they escape from the TGF-β1 suppressor mechanism, which operates in wild-type lungs, although other functions of core fucosylation of N-glycan-bearing glycoproteins might also be involved in the development of emphysema.
Exogenous TGF-β1 Treatment Rescued Emphysema-Like Changes in Fut8-/- Mice. We have performed rescue experiment with i.p. injection of exogenous TGF-β1 to postnatal-day-18 Fut8 knockout mice. Importantly, exogenous TGF-β1 resulted in a significant rescue of the emphysema-like phenotype (Fig. 3 I and J), stimulated the formation of elastin fiber (data not shown), and concomitantly reduced MMP-12 expression (Fig. 6H)in Fut8-/- lung. These data strongly support our hypothesis that the TGF-β1-mediated signaling pathway is down-regulated in Fut8-/- lungs. We do not exclude the possibility that aberrant regulation of other receptors may contribute partly to the emphysema-like changes.
In contrast to the mild and gradual formation of emphysema in integrin β6 knockout mice, which causes a local deficiency in active TGF-β1, the Fut8 deficiency as well as the induction of cytokines such as IL-13, TNF-α, and IFN-γ (25, 34, 35) results in the severe and rapidly progressive development of emphysema. Interestingly, the absence of β6 integrin leads mainly to MMP-12 overexpression in the lungs of mutant mice, whereas in Fut8-/- lungs, MMP-13 is also induced at even larger amounts than MMP12, as assessed by real-time PCR quantitative analysis (Fig. 6A). This fact, together with the wide substrate specificity of MMP-13, could contribute to the explanation of the differences in the severity of emphysema phenotypes between integrin β6- and Fut8-deficient mice. It has been reported that disruption of the latent TGF-β-binding protein 4 (LTBP-4), which regulates TGF-β targeting to ECM and TGF-β-mediated signaling, causes abnormal lung development (36). Using antibodies specific for surfactant protein C (SP-C), a marker of differentiated type-II alveolar epithelial cells, we found that expression levels of SP-C protein at each stage were slightly weaker in Fut8-/- lungs than in Fut8+/+ lungs (see Fig. 7, which is published as supporting information on the PNAS web site), suggesting that lung development was also disturbed by the loss of core fucosylation. The retarded alveolar epithelial cell differentiation may also contribute partly to emphysema-like changes of Fut8-/- lung from postnatal day 7 (Fig. 2B). Indeed, given the fact that alveolarization continues past day 7, at least part of the phenotype is related to abnormal lung development. Nevertheless, because it continues to increase after lung development, we conclude that both alveolar development and progressive (destructive) emphysema occur in the absence of Fut8.
TGF-β activation also leads to emphysema formation. Marfan syndrome is a human autosomal dominant disorder of connective tissue caused by mutations in fibrillin-1. Fibrillin-1 usually functions to limit the activation of TGF-β, although the precise mechanism by which fibrillin-1 controls TGF-β activation is still unknown. Studies with mutant mice have revealed that fibrillin deficiency causes a pronounced TGF-β activation that triggers the developmental inhibition of alveolarization, induces apoptosis in the developing lung, and finally results in destructive emphysema (37). Nevertheless, very recent results have demonstrated that Marfan syndrome can also be caused by loss of TGF-β signaling function due to TGF-β receptor type II mutations in a group of patients lacking mutations in fibrillin (38). These findings emphasize the idea that the TGF-β signaling pathway plays an important role in lung integrity, and consequently, there is an absolute need to maintain the precise levels of all components of this complex pathway. Our finding that defects in core fucosylation profoundly dysregulate TGF-β activation and signaling in Fut8-/- mice adds a level of control to this pathway and opens the possibility that similar defects could be found in some cases of human emphysema.
Supplementary Material
Acknowledgments
We thank Dr. S. Koyota for excellent technical assistance on oligosaccharide structural analyses and Dr. J. Miyazaki for invaluable advice on this initial project. This work was partly supported by a Grant-in-Aid for Scientific Research, the Special Coordination Funds for Promoting Science and Technology, and the 21st Century COE Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
Author contributions: X.W., J.G., E.M., and N.T. designed research; X.W., S. Inoue, K.N., W.L., Y.M.-H., M.N., N.U., S. Ihara, S.H.L., A.K., and T.K. performed research; X.W., S. Inoue, and M.O. contributed new reagents/analytic tools; X.W., J.G., M.A., M.T., Y.I., Y.Y., Y.A., Y.T., J.F., K.S., S.D.S., C.L.-O., and K.H. analyzed data; and X.W. and J.G. wrote the paper.
Abbreviations: ECM, extracellular matrix; MMP, matrix metalloproteinase; PA, 2-aminopyridine.
See Commentary on page 15721.
References
- 1.Lubke, T., Marquardt, T., Etzioni, A., Hartmann, E., von Figura, K. & Korner, C. (2001) Nat. Genet. 28, 73-76. [DOI] [PubMed] [Google Scholar]
- 2.Luhn, K., Wild, M. K., Eckhardt, M., Gerardy-Schahn, R. & Vestweber, D. (2001) Nat. Genet. 28, 69-72. [DOI] [PubMed] [Google Scholar]
- 3.Moloney, D. J., Panin, V. M., Johnston, S. H., Chen, J., Shao, L., Wilson, R., Wang, Y., Stanley, P., Irvine, K. D., Haltiwanger, R. S. & Vogt, T. F. (2000) Nature 406, 369-375. [DOI] [PubMed] [Google Scholar]
- 4.Sturla, L., Rampal, R., Haltiwanger, R. S., Fruscione, F., Etzioni, A. & Tonetti, M. (2003) J. Biol. Chem. 278, 26727-26733. [DOI] [PubMed] [Google Scholar]
- 5.Sturla, L., Fruscione, F., Noda, K., Miyoshi, E., Taniguchi, N., Contini, P. & Tonetti, M. (2005) Glycobiology, in press. [DOI] [PubMed]
- 6.Wilson, J. R., Williams, D. & Schachter, H. (1976) Biochem. Biophys. Res. Commun. 72, 909-916. [DOI] [PubMed] [Google Scholar]
- 7.Miyoshi, E., Uozumi, N., Noda, K., Hayashi, N., Hori, M. & Taniguchi, N. (1997) Int. J. Cancer 72, 1117-1121. [DOI] [PubMed] [Google Scholar]
- 8.Miyoshi, E., Noda, K., Yamaguchi, Y., Inoue, S., Ikeda, Y., Wang, W., Ko, J. H., Uozumi, N., Li, W. & Taniguchi, N. (1999) Biochim. Biophys. Acta 6, 9-20. [DOI] [PubMed] [Google Scholar]
- 9.Noda, K., Miyoshi, E., Gu, J., Gao, C. X., Nakahara, S., Kitada, T., Honke, K., Suzuki, K., Yoshihara, H., Taniguchi, N., et al. (2003) Cancer Res. 63, 6282-6289. [PubMed] [Google Scholar]
- 10.Uozumi, N., Yanagidani, S., Miyoshi, E., Ihara, Y., Sakuma, T., Gao, C. X., Teshima, T., Fujii, S., Shiba, T. & Taniguchi, N. (1996) J. Biol. Chem. 271, 27810-27817. [DOI] [PubMed] [Google Scholar]
- 11.Wang, W., Li, W., Ikeda, Y., Miyagawa, J. I., Taniguchi, M., Miyoshi, E., Sheng, Y., Ekuni, A., Ko, J. H., Yamamoto, Y., et al. (2001) Glycobiology 11, 165-174. [DOI] [PubMed] [Google Scholar]
- 12.Shields, R. L., Lai, J., Keck, R., O'Connell, L. Y., Hong, K., Meng, Y. G., Weikert, S. H. & Presta, L. G. (2002) J. Biol. Chem. 277, 26733-26740. [DOI] [PubMed] [Google Scholar]
- 13.Shinkawa, T., Nakamura, K., Yamane, N., Shoji-Hosaka, E., Kanda, Y., Sakurada, M., Uchida, K., Anazawa, H., Satoh, M., Yamasaki, M., et al. (2003) J. Biol. Chem. 278, 3466-3473. [DOI] [PubMed] [Google Scholar]
- 14.Dennis, J. W., Granovsky, M. & Warren, C. E. (1999) BioEssays 21, 412-421. [DOI] [PubMed] [Google Scholar]
- 15.Mountford, P., Zevnik, B., Duwel, A., Nichols, J., Li, M., Dani, C., Robertson, M., Chambers, I. & Smith, A. (1994) Proc. Natl. Acad. Sci. USA 91, 4303-4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanagawa, Y., Kobayashi, T., Ohnishi, M., Tamura, S., Tsuzuki, T., Sanbo, M., Yagi, T., Tashiro, F. & Miyazaki, J. (1999) Transgenic Res. 8, 215-221. [DOI] [PubMed] [Google Scholar]
- 17.Koyota, S., Ikeda, Y., Miyagawa, S., Ihara, H., Koma, M., Honke, K., Shirakura, R. & Taniguchi, N. (2001) J. Biol. Chem. 276, 32867-32874. [DOI] [PubMed] [Google Scholar]
- 18.Nakamura, A., Fukuda, Y. & Kuwaki, T. (2003) J. Appl. Physiol. 94, 525-532. [DOI] [PubMed] [Google Scholar]
- 19.Lee, Y. C., Lee, B. I., Tomiya, N. & Takahashi, N. (1990) Anal. Biochem. 188, 259-266. [DOI] [PubMed] [Google Scholar]
- 20.Akama, T. O., Nakagawa, H., Sugihara, K., Narisawa, S., Ohyama, C., Nishimura, S., O'Brien, D. A., Moremen, K. W., Millan, J. L. & Fukuda, M. N. (2002) Science 295, 124-127. [DOI] [PubMed] [Google Scholar]
- 21.Oriol, R., Mollicone, R., Cailleau, A., Balanzino, L. & Breton, C. (1999) Glycobiology 9, 323-334. [DOI] [PubMed] [Google Scholar]
- 22.Roos, C., Kolmer, M., Mattila, P. & Renkonen, R. (2002) J. Biol. Chem. 277, 3168-3175. [DOI] [PubMed] [Google Scholar]
- 23.Smith, P. L., Myers, J. T., Rogers, C. E., Zhou, L., Petryniak, B., Becker, D. J., Homeister, J. W. & Lowe, J. B. (2002) J. Cell Biol. 158, 801-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Armiento, J., Dalal, S. S., Okada, Y., Berg, R. A. & Chada, K. (1992) Cell 71, 955-961. [DOI] [PubMed] [Google Scholar]
- 25.Zheng, T., Zhu, Z., Wang, Z., Homer, R. J., Ma, B., Riese, R. J., Jr., Chapman, H. A., Jr., Shapiro, S. D. & Elias, J. A. (2000) J. Clin. Invest. 106, 1081-1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Senior, R. M., Griffin, G. L., Fliszar, C. J., Shapiro, S. D., Goldberg, G. I. & Welgus, H. G. (1991) J. Biol. Chem. 266, 7870-7875. [PubMed] [Google Scholar]
- 27.Balbin, M., Fueyo, A., Knauper, V., Lopez, J. M., Alvarez, J., Sanchez, L. M., Quesada, V., Bordallo, J., Murphy, G. & Lopez-Otin, C. (2001) J. Biol. Chem. 276, 10253-10262. [DOI] [PubMed] [Google Scholar]
- 28.Morris, D. G., Huang, X., Kaminski, N., Wang, Y., Shapiro, S. D., Dolganov, G., Glick, A. & Sheppard, D. (2003) Nature 422, 169-173. [DOI] [PubMed] [Google Scholar]
- 29.Massague, J., Blain, S. W. & Lo, R. S. (2000) Cell 103, 295-309. [DOI] [PubMed] [Google Scholar]
- 30.Heldin, C. H., Miyazono, K. & ten Dijke, P. (1997) Nature 390, 465-471. [DOI] [PubMed] [Google Scholar]
- 31.Wrana, J. L., Attisano, L., Carcamo, J., Zentella, A., Doody, J., Laiho, M., Wang, X. F. & Massague, J. (1992) Cell 71, 1003-1014. [DOI] [PubMed] [Google Scholar]
- 32.Ebner, R., Chen, R. H., Lawler, S., Zioncheck, T. & Derynck, R. (1993) Science 262, 900-902. [DOI] [PubMed] [Google Scholar]
- 33.Macias-Silva, M., Abdollah, S., Hoodless, P. A., Pirone, R., Attisano, L. & Wrana, J. L. (1996) Cell 87, 1215-1224. [DOI] [PubMed] [Google Scholar]
- 34.Fujita, M., Shannon, J. M., Irvin, C. G., Fagan, K. A., Cool, C., Augustin, A. & Mason, R. J. (2001) Am. J. Physiol. Lung Cell Mol. Physiol. 280, L39-L49. [DOI] [PubMed] [Google Scholar]
- 35.Wang, Z., Zheng, T., Zhu, Z., Homer, R. J., Riese, R. J., Chapman, H. A., Jr., Shapiro, S. D. & Elias, J. A. (2000) J. Exp. Med. 192, 1587-1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sterner-Kock, A., Thorey, I. S., Koli, K., Wempe, F., Otte, J., Bangsow, T., Kuhlmeier, K., Kirchner, T., Jin, S., Keski-Oja, J. & von Melchner, H. (2002) Genes Dev. 16, 2264-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neptune, E. R., Frischmeyer, P. A., Arking, D. E., Myers, L., Bunton, T. E., Gayraud, B., Ramirez, F., Sakai, L. Y. & Dietz, H. C. (2003) Nat. Genet. 33, 407-411. [DOI] [PubMed] [Google Scholar]
- 38.Mizuguchi, T., Collod-Beroud, G., Akiyama, T., Abifadel, M., Harada, N., Morisaki, T., Allard, D., Varret, M., Claustres, M., Morisaki, H., et al. (2004) Nat. Genet. 36, 855-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.