‘This is not the end. It is not even the beginning of the end. But it is, perhaps, the end of the beginning.’ Winston Churchill
Daniel St Johnston from the Wellcome/CRC Institute at the University of Cambridge was one of the two recipients of the EMBO Gold Medal in 2000. This medal was awarded in recognition of his extensive contribution to developmental biology, in particular his discoveries on how the main body axes are polarized in Drosophila and how specific mRNAs are localized. His account of his past and present research is presented in the Medal Review in this issue of The EMBO Journal on pages 6169--6179.
When I heard that I had won the EMBO gold medal, I was immensely flattered to receive such a prestigious award, but some of my less kind friends commented that it could only be downhill from here on. After all, medals are usually given to distinguished and aged professors near the end of their productive years. However, I hope that my title does not reflect the state of my career, but instead introduces the questions that I have spent the last 10 years studying, namely what is the origin of anterior– posterior polarity in Drosophila, and how does this lead to the localization of the determinants that define the two ends of the embryo? In other words, what is the beginning of the ends? Before discussing our work on this topic in detail, I would like to start with the customary autobiographical account of how I ended up where I am, so that I can give credit to all of the exceptional people who have helped me along the way.
In seeking inspiration on how to write this section of my review, I turned to those written by previous winners of the EMBO medal, and discovered to my surprise that I have followed in the footsteps of three other former recipients, Hugh Pelham, Jim Smith and Richard Treisman (Pelham, 1989; Smith, 1993; Treisman, 1995). We were all under graduates at Christ’s College, Cambridge, and fell under the benign influence of the Director of Studies in Biology, Dr Douglas Barker. Indeed, Dr Barker saved both me and Jim from specializing in other science subjects. On arriving in Cambridge, I intended to study Chemistry, but became so excited by Dr Barker’s infectious enthusiasm for his subject during his supervisions that I rapidly switched allegiance and ended up specializing in Genetics. Dr Barker and his family even took me into their home when I was in danger of cracking up under the stress of finals, and it is largely thanks to him that I got a degree at all. It is a great pity that there are no prizes for excellent teachers who inspire their students to do research, because he would be a very worthy recipient.
Several further influences during my last year as an undergraduate convinced me that I wanted to work on Drosophila developmental biology. The outstanding Development course taught by John Gurdon, Peter Lawrence and others made me realize how many important and interesting questions there were in this field, and how little was known about them. At the same time, Mike Ashburner provided me with an introduction to Drosophila genetics, while stealing my cigarettes during lectures. Finally, my final year saw the publication of Eric Wieschaus’ and Christiane Nüsslein-Volhard’s Nobel Prize-winning Nature paper, which revealed the power of developmental genetics in all its glory (Nüsslein-Volhard and Wieschaus, 1980). My first foray into the laboratory during my undergraduate research project was not with Drosophila, however, but with Escherichia coli, and was such a disaster that I almost gave up there and then. I was supposed to be looking at intragenic recombination between two alleles of LacZ in various mutants, only to discover with 1 week to go, that the strains I had been given all carried the identical LacZ mutation. I do not know how I managed to write a 50 page report on a project that could not possibly have worked, but it did teach me the important lesson that one should always check one’s stocks before starting the experiment.
Since I felt that I needed more experience before embarking on a PhD, I followed Dr Barker’s advice and went to the Department of Cellular and Developmental Biology at Harvard, so that I could take advantage of a year of research rotations in different groups. I eventually chose to do my PhD with William Gelbart on the molecular characterization of decapentaplegic (dpp), despite his attempts to put me off by telling me that his tenure decision was imminent and that he might soon have to move to Alabama. This proved a very fortunate choice in many ways. First, Bill got tenure and we remained in Cambridge, Massachusetts. Secondly, Michael Hoffman, who was then the senior post doc in the lab, took me under his wing, and taught me how to do molecular biology properly. Thirdly, dpp was becoming really interesting: Forrest Spencer, Mike and Bill had shown that it was required for the patterning of all the imaginal discs, and Vivian Irish had just found that it was also needed very early in embryogenesis for the formation of the dorsal– ventral axis (Spencer et al., 1982; Irish and Gelbart, 1987). Mike had already spent nearly 2 years walking along the chromosome towards dpp, before generously handing the project to me, and my biggest piece of luck was that I only had to take a few steps to clone the entire gene, and could therefore spend the rest of my thesis characterizing its organization and function (St Johnston et al., 1990).
Everyone in the lab was convinced that dpp must a homeobox gene, because all important developmental genes in those days were, while Gary Struhl had argued that it was probably a metabolic enzyme (Struhl, 1982). When Rick Padgett and I sequenced the first cDNAs we were therefore surprised to discover that it encodes a secreted transforming growth factor-β (TGF-β)-like protein, and was the first example of a signalling molecule required for early pattern formation (Padgett et al., 1987). I also found that dpp is one of the earliest zygotic genes to be transcribed in the embryo, where it is expressed in the dorsal part of the blastoderm (St Johnston and Gelbart, 1987). This allows the formation of a dorsal to ventral gradient of secreted DPP that acts as a morphogen to pattern the dorsal half of the embryo (Ferguson and Anderson, 1992; Wharton et al., 1993). It later became clear that this function has been conserved thoughout animal evolution, because the DPP homologue, BMP-4, plays a very similar role in dorsal–ventral axis formation in vertebrates, although this axis is inverted with respect to Drosophila (Holley et al., 1995). One of the best things about doing a PhD with Bill was that he took his responsibilities as a teacher very seriously, and this more than compensated for his obsession with terrible jokes about sheep. All in all, I had a great time as a graduate student at Harvard (Figure 1). My only regret is that my mother still believes that I was working on a gene called decaparaplegic.
Fig. 1. My farewell party at Bill’s house, July 1988. From left to right: Bill Gelbart, Marnie Gelbart, Vern Twombly, me, Nick Brown, Larry Zwiebel and Christos Delidakis.
As one is supposed to change subject on becoming a post doc, I rotated through 90°, by switching from a zygotic dorsal–ventral gene to the maternal genes that polarize the anterior–posterior axis. Since the beginning of the 20th century, it had been known that the posterior end of many insect eggs contains a specialized region of cytoplasm called the pole plasm, which is characterized by the presence of electron-dense polar granules, and Illmensee and Mahowald (1974) had demonstrated that this cytoplasm contains localized determinants that direct the formation of the primordial germ cells, the pole cells. Furthermore, Frohnhöfer et al. (1986) had shown that the pole plasm also contains a determinant that specifies the abdomen of the embryo, whereas the anterior cytoplasm of the egg contains another localized determinant that patterns the head and thorax. Since I found these localized maternal determinants a fascinating subject, I decided to work with the doyenne of this field, Dr Christiane Nüsslein-Volhard (or Janni to those who know her), and obtained an EMBO long-term fellowship to join her group in the Max Planck Institut für Entwicklungsbiologie in Tübingen.
Janni’s lab was a very exciting place to be in the late 1980s, because many of the genes responsible for the production and localization of these determinants had been identified in large-scale genetic screens for maternal-effect mutations that disrupt the patterning of the embryo, and it was therefore becoming possible to investigate the molecular basis of axis formation for the first time (Schüpbach and Wieschaus, 1986; Nüsslein-Volhard et al., 1987). Hans Georg Frohnhöfer identified bicoid as the anterior determinant, and Thomas Berleth, Wolfgang Driever and others subsequently showed that bicoid mRNA is localized to the anterior of the egg, where it is translated to produce a morphogen gradient of a homeodomain protein that patterns the head and thorax (Frohnhöfer and Nüsslein-Volhard, 1986; Berleth et al., 1988; Driever and Nüsslein-Volhard, 1988a,b). Several years later, Ruth Lehmann’s group identified nanos as encoding the posterior determinant, and showed that nanos mRNA is enriched in the pole plasm (Lehmann and Nüsslein-Volhard, 1991; Wang and Lehmann, 1991). Furthermore, the analysis of the maternal-effect mutations revealed that there were in fact two other signals that pattern the embryo that do not involve localized cytoplasmic determinants. The entire dorsal–ventral axis is set up by a single extracellular cue that is controlled by the expression of pipe in the ventral cells of the follicular epithelium that surrounds the developing oocyte (Stein et al., 1991; Sen et al., 1998). After fertilization, this localized cue leads to the production of a signal that induces the formation in the embryo of a nuclear localization gradient of Dorsal protein, which then acts as a morphogen by regulating the transcription of zygotic target genes, such as dpp (Roth et al., 1989; Rushlow et al., 1989). The terminal system functions in a similar way to define the two extremities of the embryo. In this case, the inductive signal depends on the expression of torsolike in the terminal follicle cells that surround the anterior and posterior poles of the oocyte, while the receptor in the egg is the receptor tyrosine kinase, Torso (Klingler et al., 1988; Casanova and Struhl, 1989; Sprenger et al., 1989; Stevens et al., 1990).
Just before I arrived, Janni had a bet with Herbert Jäckle about when Gerd Jürgens was going to submit a paper, in which the forfeit doubled with every week that the manuscript was not finished. As a consequence, she won over a thousand miniatures of Calvados, and every time there was a pretext to celebrate, she would hand these ‘Herbertschens’ around the lab. We celebrated all important results, and the submission, acceptance, page proofs, publication and first reprint request of every paper. Given the great burst of discovery that was taking place in the lab, this was an almost weekly event. Although my own contribution was relatively minor, it was a lot of fun to watch my friends uncover most of the basic details of how the anterior–posterior (AP) and dorsal–ventral (DV) axes of the embryo are defined, and this classic work provides the foundation for much my subsequent research on axis formation (Figure 2; reviewed in St Johnston and Nüsslein-Volhard, 1992).
Fig. 2. A diagram of a stage 10a Drosophila egg chamber showing the localized signals that polarize the AP and DV axes of the embryo. bicoid mRNA (blue), oskar mRNA (red), gurken mRNA (green); pipe expression (dark green); and torsolike expression (magenta).
My project was to clone and characterize the posterior group gene staufen, which Trudi Schüpbach had shown to be the only gene in this class that is required for both anterior and posterior patterning (Schüpbach and Wieschaus, 1986). The embryos produced by staufen mutant females have a reduced head that often fails to involute correctly, and develop no abdomen or pole cells, because the pole plasm fails to form at the posterior of the egg. However, the name derives from the ‘grandchildless’ phenotype produced by weaker alleles, in which the embryos have enough pole plasm to form an abdomen, but not pole cells, so that the progeny develop into sterile adults. Like the other posterior group genes, vasa, valois and tudor, staufen is named after a royal family that died out through a lack of offspring, in this case the Hohen Staufen, the most famous of whom was the Holy Roman Emperor, Friedrich Barbarossa. My historical research suggests, however, that it was mainly infanticide rather than sterility that led to the demise of the family.
My first goal was to investigate the cause of the head defects in staufen mutants, and the obvious place to start was by looking at the localization of the anterior determinant, bicoid mRNA. I found that the mRNA is no longer tightly localized to the anterior of the egg, and instead forms a gradient that extends towards the middle, which explains why staufen mutants lack the anterior structures that require the highest concentrations of Bicoid protein. This work got me interested in how bicoid mRNA reaches the anterior pole in the first place, and I went on to show that exuperantia, swallow and staufen mutants disrupt localization at successively later stages of oogenesis, suggesting that there are multiple steps in the localization pathway (St Johnston et al., 1989).
At the same time, I was cloning staufen, with a lot of help from Denise Montell, who sent me a P element insertion that proved to be in the middle of the gene. Upon raising an antibody against the protein, I discovered that Staufen accumulates at the posterior of the oocyte at stage 9 of oogenesis, and is one of the first components of the polar granules to be localized (St Johnston et al., 1991). The significance of this result became much clearer when both Paul Macdonald’s group and Anne Ephrussi and Ruth Lehmann cloned the posterior group gene oskar, and showed that its mRNA localizes to the posterior at the same time as Staufen protein, and that this localization is abolished in staufen mutants (Ephrussi et al., 1991; Kim-Ha et al., 1991). Anne and Ruth also elegantly demonstrated that localized oskar mRNA is the key determinant for pole plasm formation, by localizing oskar RNA to the anterior of the oocyte by fusing it to the localization element in the bicoid 3′-untranslated region (3′-UTR) (Ephrussi and Lehmann, 1992). This ectopic oskar mRNA directs the assembly of fully functional pole plasm that induces the formation of both an anterior abdomen and pole cells. Furthermore, this does not require staufen, indicating that it is involved specifically in the localization of endogenous oskar mRNA to the posterior.
After 3 years in Tübingen, I obtained a Wellcome Trust Senior Research Fellowship to move to the new Wellcome/CRC Institute in Cambridge, and I started my own group there, sharing a lab with my old friend Nick Brown. Nick and I had lived in the same house at Harvard, and had collaborated extensively during our PhDs. We continued this close co-operation in our new domain, which was an enormous help during the difficult transition from post doc to group leader. At this point, it was already clear that the AP axis in Drosophila is specified by the localization of bicoid and oskar mRNAs to opposite poles of the oocyte, and this raised the two related questions that have been the focus of my research ever since. How does Staufen function in the localization of these mRNAs to the two ends of the same large cell, and how is the oocyte polarized in the first place to define the destination of these mRNAs?
The role of Staufen in mRNA localization
mRNA localization is a common mechanism for targeting proteins to the regions of a cell where they are required, and appears to occur in most, if not all, polarized cell types (Palacios and St Johnston, 2001). Most examples of mRNA localization in higher eukaryotes are microtubule dependent, and several lines of evidence show that this is also the case for bicoid and oskar mRNAs. The localization of both transcripts is disrupted by microtubule-depolymerizing drugs, such as colchicine, and the oocyte microtubule cytoskeleton appears to be polarized along the AP axis at the time that the mRNAs first localize, with the minus ends at the anterior and the plus ends extending towards the posterior pole (Pokrywka and Stephenson, 1991; Theurkauf et al., 1992; Clark et al., 1994, 1997). Apart from the role of the cytoskeleton, very little is known about the trans-acting factors involved in mRNA localization, or how they couple specific transcripts to the machinery that moves them. Since Staufen was the first example of such a trans-acting factor, I set out to exploit the advantages provided by the large size of the Drosophila oocyte and the power of Drosophila genetics to address the role it plays in the localization of bicoid and oskar mRNAs.
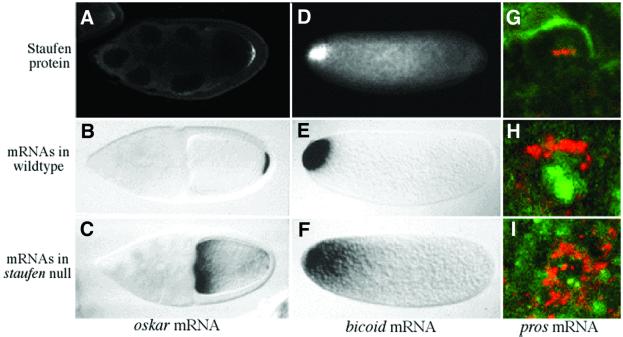
The discovery that Staufen co-localizes with oskar mRNA to the posterior of the oocyte suggested that it might also co-localize with bicoid mRNA, and this turned out to be the case. Staufen does not localize with bicoid mRNA during the stages of oogenesis when it mediates the posterior localization of oskar mRNA, but co-localizes with the RNA at the anterior once the egg has been laid (Figure 3). Furthermore, this localization is bicoid mRNA dependent, since no Staufen is seen at the anterior in exu or swallow mutants, which block bicoid localization at an earlier stage, while much more protein is recruited to the anterior of the eggs laid by females that carry several extra copies of the bicoid gene. Thus, the localization of Staufen to the anterior requires bicoid mRNA, and vice versa, strongly suggesting that the two associate to form the substrate for localization.
Fig. 3. The role of Staufen in the localization of bicoid, oskar and prospero mRNAs. (A–C) Staufen is required for the posterior localization of oskar mRNA. Staufen protein (A) localizes to the posterior of the oocyte at stage 9, with oskar mRNA (B). (C) In a staufen null mutant, oskar mRNA remains at the anterior of the oocyte. (D and E) Staufen anchors bicoid mRNA at the anterior of the egg. Staufen (D) and bicoid mRNA (E) co-localize at the anterior of the egg in wild type, whereas bicoid mRNA diffuses to form a gradient in a staufen null (F). (G–I) Staufen mediates the basal localization of prospero mRNA during the asymmetric divisions of the neuroblasts. Staufen (G) localizes to the basal side of the cell at metaphase with prospero mRNA (H). (I) In staufen mutants, prospero mRNA is delocalized.
To test this more directly, Dominque Ferrandon and I transcribed the bcd localization signal in vitro, and injected this RNA into early embryos (Ferrandon et al., 1994). Injected bcd 3′-UTR recruits the free cytoplasmic Staufen protein into large particles, which then localize to the poles of the mitotic spindles in a microtubule-dependent manner. This activity is highly specific to the bicoid 3′-UTR, since no other regions of the RNA, nor any of the other transcripts we tested, recruit Staufen into particles that localize. The bicoid localization signals have been mapped to a 640 nucleotide region of its 3′-UTR, which is predicted to form an extensive secondary structure with several large double-stranded stems (Macdonald and Struhl, 1988; Macdonald, 1990). The injection assay provided a very convenient way to map the sequences within this region that are required for its interaction with Staufen. By injecting a series of linker-scanning mutations, Dominique and I showed that particle formation depends on three non-contiguous regions of the 3′-UTR, each of which corresponds to a large stem–loop in the proposed secondary structure of the RNA.
Although these in vivo experiments suggested that Staufen is an RNA-binding protein, its amino acid sequence contained none of the known RNA-binding domains. The protein did contain five copies of a repeated motif, however, and database searches with the consensus derived from these repeats identified similar sequences in a number of other proteins that interact with double-stranded RNA (dsRNA), such as the mammalian dsRNA- dependent protein kinase (PKR) and E.coli RNase III. I therefore expressed one of the Staufen repeats in bacteria to examine its ability to bind to RNA on northwestern blots, and found that it binds specifically to dsRNA. At the same time, Michael Jantsch in Joe Gall’s laboratory was reaching an identical conclusion from his work on a Xenopus protein, Xlrbpa, and we decided to publish a joint paper on the identification of the dsRNA-binding domain (dsRBD) (St Johnston et al., 1992).
We were then very fortunate to be able to collaborate with Mark Bycroft and his student, Mark Proctor, who determined the structure of the domain by NMR (Bycroft et al., 1995). This information was not particularly revealing to a developmental biologist like myself, but Mark showed it to Alexei Murzin, who is a great resource for Cambridge structural biology because he has every known protein structure stored in his head. Even though no amount of database searching could reveal it, Alexei spotted that the domain has the same protein fold as the N-terminal region of prokaryotic ribosomal protein S5. Once we knew what to look for, it became clear that the amino acids that form the structural core of the domain are conserved.
The structure of the dsRBD did not explain why it binds specifically to dsRNA, but it did tell us which amino acids lie on the surface of the domain, and which might therefore interact directly with RNA. Stefan Grünert in my lab introduced mutations into many of these amino acids to map the RNA-binding face of the domain, and also showed that it binds optimally to 12 bp or more of dsRNA. With this information in hand, Andres Ramos and Gabriel Varani succeeded in solving the structure of the domain in a complex with dsRNA (Ramos et al., 2000). This reveals that the dsRBD binds to the bases in the minor groove of dsRNA, which is much shallower and wider than that in DNA, and to the phosphate backbone across the major groove, which is much narrower. Thus, the domain recognizes the unique structure of dsRNA, and does not require specific bases for its interaction. This raises the question of how the dsRBDs in Staufen mediate its specific association with oskar and bicoid mRNAs in vivo, if the single domains do not recognize specific sequences. We still have no answer to this question, but one possibility is that Staufen recognizes the tertiary structure of the RNAs through the correspondence between positions of the dsRBDs in the native protein and that of double-stranded regions in the RNAs.
Once Staufen has bound to either bicoid or oskar mRNA, it must do something to facilitate their localization, and it therefore seemed likely that it somehow acts to link each transcript to the microtubule-dependent machinery that localizes it. This led us to hypothesize that Staufen interacts with other proteins as well as RNA, and David Micklem set out to identify domains that might mediate such interactions by searching for conserved regions of the protein (Micklem et al., 2000). He identified clear Staufen homologues in worms, other insect species, mice and humans, but found that the only regions that had been maintained during evolution are the five dsRBDs. However, Stefan Grünert and Jan Adams found that two of these domains, dsRBD2 and 5, do not interact with dsRNA in vitro. Knowing the structure of the dsRBD, it was pretty obvious why these domains cannot bind dsRNA: domain 2 contains a conserved insertion that separates the two regions of the domain that contact the major and minor grooves, whereas dsRBD5 contains all of the residues that form the structural core of the domain, but lacks the RNA-binding amino acids on the surface. Thus, these domains must have been conserved for some reason other than their ability to bind dsRNA. Since this made these dsRBDs good candidates for domains that link Staufen–RNA complexes to other trans-acting factors, David constructed a staufen transgene in which the insert in domain 2 had been removed (StauΔloop2), and another that lacked dsRBD5 (StauΔdsRBD5), and tested their ability to rescue oskar mRNA localization in a staufen null mutant. The StauΔloop2 protein still associates with oskar mRNA, but the resulting complexes are not transported to the posterior of the oocyte. In contrast, the StauΔdsRBD5 protein mediates wild-type localization of oskar mRNA, but the mRNA is not translated once it is localized at the posterior. Thus, the insert in domain 2 is required for the microtubule-dependent localization of oskar mRNA, whereas dsRBD5 is essential for the derepression of oskar mRNA translation, once localized. Finally, we found that both of these domains are required for the anterior anchoring of bicoid mRNA, but that other regions of the protein are important for bicoid translation.
Although all our work had focused on the role of Staufen in the localization of bicoid and oskar mRNAs, it is also required for the localization of prospero mRNA in embryonic neuroblasts (Li et al., 1997; Broadus et al., 1998). These cells are the stem cells for the nervous system, and divide asymmetrically to produce a neuroblast and ganglion mother cell (GMC). During these divisions, prospero mRNA and Staufen protein localize to the basal side of the cell, and segregate into the GMC, where Prospero protein acts as a determinant for GMC fate. Unlike bicoid and oskar mRNAs, the localization of prospero mRNA does not require microtubules and depends instead on the actin cytoskeleton (Broadus and Doe, 1997). Thus, Staufen can mediate both actin- and microtubule-dependent mRNA localization.
To investigate how Staufen can couple transcripts to two different cytoskeletal systems, Jan Adams and I collaborated with Andrea Brand’s group on the floor above us, and found that StauΔloop2 localizes normally in the neuroblasts, whereas StauΔdsRBD5 does not. This implicated Staufen dsRBD5 in the actin-dependent localization of prospero mRNA, and Jan therefore performed a yeast two-hybrid screen with this domain and identified a single interacting protein, which is required for Staufen and prospero mRNA localization, and co-localizes with them throughout the neuroblast cell cycle (Schuldt et al., 1998). This turned out to be Miranda, which had been identified independently shortly before as a protein that binds to Prospero protein, and mediates its localization to the basal side of the neuroblast (Shen et al., 1997; Ikeshima-Kataoka et al., 1998). Miranda therefore localizes prospero mRNA by binding to Staufen, and Prospero protein through a direct interaction. This made Miranda the first example of an essential trans-acting factor that recognizes an RNA-binding protein associated with a localized RNA, but it is still unclear how Miranda itself is localized.
Our analysis of Staufen has revealed that it plays an essential role in the localization of three different mRNAs, and that distinct domains of the protein are required for actin- and microtubule-based transport. However, we still know very little about how Staufen–RNA complexes are linked to the motors that are presumed to transport them. It has been shown recently that the localization of oskar mRNA requires the plus end-directed microtubule motor protein, kinesin 1, whereas indirect evidence suggests that bicoid mRNA localization may involve the minus end-directed motor, dynein (Brendza et al., 2000; Schnorrer et al., 2000). Thus, one of the major challenges for the future will be to determine whether these motors actually transport oskar and bicoid mRNAs along microtubules and, if so, how they are linked to their cargoes. This is not a trivial problem, for several reasons. Unlike other intracellular transport processes, mRNA localization does not involve movement between membrane-bound compartments that can easily be purified, and the amounts of a specific mRNA that are localized are usually very small. Furthermore, many transcripts, including bicoid mRNA, seem to be assembled into large particles before they are localized. We will therefore need to understand how the mRNAs are packaged into transport-competent complexes, which probably contain many proteins, only a few of which have been identified. Finally, several lines of evidence indicate that the nuclear history of a transcript can determine its fate in the cytoplasm, and this also seems to be the case for oskar mRNA. We have characterized two other proteins, Mago nashi and Barentsz, that are essential for oskar mRNA localization and which co-localize with the mRNA at the posterior pole (Micklem et al., 1997; van Eeden et al., 2001). Mago nashi is predominantly nuclear, however, while Barentsz associates with the nuclear envelope, suggesting that they may be loaded onto the mRNA sequentially as it exits the nucleus.
All of these issues indicate how much we still have to learn about mRNA localization in Drosophila, but this may provide a useful model for understanding mRNA localization in other systems. For example, it has long been speculated that the translation of localized mRNAs at specific post-synaptic sites in the dendrites of neurons plays an important role in activity-dependent synaptic plasticity. Recent work from Michael Kiebler’s group suggests that mammalian Staufen homologues play a similar role to the Drosophila protein in the microtubule-dependent transport of some of these mRNAs (Kiebler et al., 1999; Köhrmann et al., 1999). Thus, it will be interesting to see whether what we learn from Drosophila will prove relevant to mRNA localization and translational control in the human brain.
The origin of anterior–posterior polarity
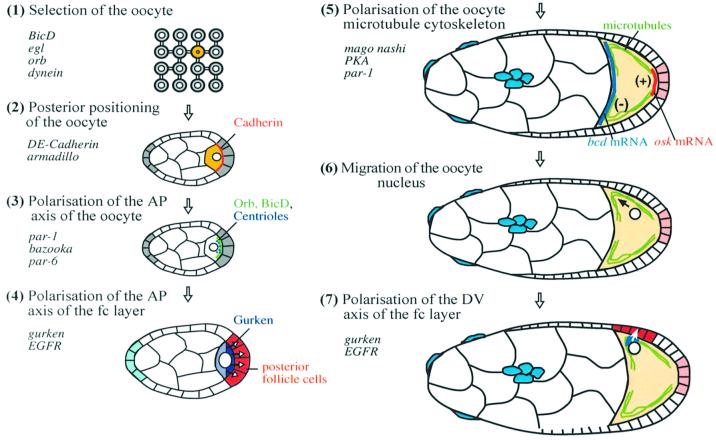
Since the localization of bicoid and oskar mRNAs is microtubule dependent, the organization of the microtubule cytoskeleton in the oocyte defines the destination of these transcripts, and hence the AP axis of the embryo. When I started my lab in Cambridge, it was totally unclear where this original AP polarity comes from, and I was very keen to investigate this question. I was extremely fortunate to recruit an outstanding first post doc, Acaimo González-Reyes and, during his five and a half years in the lab, we succeeded in unravelling most of the key steps in this process (Figure 4).
Fig. 4. A summary of the steps that lead to the polarization of the AP and DV axes.
The Drosophila ovary is composed of ∼16 ovarioles, each of which is a long tube that contains a series of progressively more mature egg chambers (Spradling, 1993). The germline stem cells reside at the anterior tip of the ovariole in a region called the germarium, and these cells divide asymmetrically to produce a new stem cell and a cystoblast, which then goes through four rounds of mitosis with incomplete cytokinesis to form a cyst of 16 germ cells that are connected to each other by ring canals. One of these 16 cells is selected to become the oocyte and moves to the posterior of the cyst, while the remaining cells become polyploid nurse cells. At the same time, somatic follicle cells migrate to form a monolayered epithelium around each cyst, to give rise to the complete egg chamber. During the next stages of oogenesis (stages 1–6), the arrangement and relative size of the three cell types in the egg chamber remain fairly constant, but the oocyte begins to increase rapidly in volume at stage 7, through the transport of material through the ring canals from the nurse cells and the uptake of yolk from the follicle cells, and the majority of the follicle cells also start to migrate to surround the oocyte as it grows. During this stage, the original microtubule-organizing centre (MTOC) at the posterior of the oocyte disappears, and a new diffuse MTOC forms at the anterior, and organizes the polarized microtubule network that directs the localization of bicoid and oskar mRNAs (Theurkauf et al., 1992; Clark et al., 1994, 1997).
The first clue to how this microtubule reorganization occurs came from the work of Hannele Ruohola-Baker (Ruohola et al., 1991), who showed that a conditional mutant in the Notch receptor results in the development of a symmetric AP axis, in which bicoid mRNA localizes to both poles of the oocyte, and oskar mRNA to the centre. Furthermore, a marker for the plus ends of the microtubules, kinesin-βgal, localizes to the middle of these oocytes with oskar mRNA rather than at the posterior pole (Clark et al., 1994). Clonal analysis revealed that Notch is not required in the germline cells of the egg chamber, indicating that it must function in the somatic follicle cells. This led to the proposal that the follicle cells that surround the posterior of the oocyte signal to polarize its microtubule cytoskeleton, and that the AP axis therefore arises from the patterning of the follicle cell epithelium.
When Acaimo and I thought about this model, it struck us that there is an obvious asymmetry in the egg chamber long before any AP patterning is visible in the follicle cell layer, which is the positioning of the oocyte posterior to the nurse cells. We therefore set out to test the relationship between this early asymmetry in the arrangement of the germline cells and the patterning of the follicle cells by analysing the phenotype of mutants in spn-C, which produce some egg chambers in which the oocyte lies in the middle of the cyst with nurse cells on either side (González-Reyes and St Johnston, 1994). In these cases, the follicle cells at the posterior of the egg chamber differentiate as anterior follicle cells, and the oocyte develops symmetrically with bicoid mRNA at both ends and oskar mRNA in the middle. This phenotype is germline dependent, indicating that the correct positioning of the oocyte is required for the AP patterning of the follicle cells, and not vice versa. The AP axis is therefore specified early in oogenesis when the oocyte migrates to the posterior of the cyst. This polarity must then be transmitted to the follicle cell layer, suggesting that the oocyte signals to the adjacent follicle cells to induce them to adopt a posterior fate. Finally, these posterior follicle cells signal back to the oocyte later in oogenesis to define the AP axis of the embryo.
Our next goal was to determine the nature of the signal from the oocyte to the posterior follicle cells, and an important clue came from the work in Trudi Schüpbach’s lab on the role of gurken in DV axis formation (Schüpbach, 1987; Neuman-Silberberg and Schüpbach, 1993). At about stage 7 of oogenesis, gurken mRNA becomes localized above the oocyte nucleus at the dorsal anterior corner of the oocyte, and is translated there to produce a TGF-α-like protein that signals to the adjacent follicle cells to induce them to adopt a dorsal fate, thereby defining the DV axis of the follicle cell layer and the embryo. gurken mutants therefore produce completely ventralized eggs. However, Trudi also reported that these eggs sometimes develop a micropyle at their posterior end. Because of our work on spn-C, we knew that the micropyle is produced by two specific populations of anterior follicle cells. This suggested that Gurken might be required to induce posterior follicle cell fate, and this turned out to be the case (González-Reyes et al., 1995; Roth et al., 1995). In the strongest gurken mutant combinations, all of the posterior cells adopt the default anterior fate, even though the oocyte is positioned correctly at the posterior. As a consequence, the oocyte develops a symmetric AP axis in which bicoid mRNA localizes to both poles of the oocyte, and oskar mRNA localizes to the centre. Furthermore, gurken is required in the germline for the induction of posterior follicle cell fate, whereas its receptor, the Drosophila epidermal growth factor receptor (EGFR), is required in the follicle cells themselves (González-Reyes et al., 1995; González-Reyes and St Johnston, 1998b). Thus, Gurken signals from the oocyte to activate the EGFR in the adjacent follicle cells, thereby inducing them to become posterior.
The discovery that Gurken–EGFR signalling polarizes both the AP and DV axes raised the question of how the same signalling molecule can signal in two different directions from the same cell. During early oogenesis, gurken mRNA accumulates on one side of the nucleus at the posterior of the oocyte, where it can act as a localized source for Gurken to signal to the posterior follicle cells. The nucleus then migrates from the posterior pole to one corner of the anterior margin of the oocyte at stage 7. gurken mRNA also relocalizes to form a dorsal/anterior cap above the nucleus, where Gurken signals for a second time to induce dorsal follicle cell fate. Thus the direction of each Gurken signal depends on the localization of gurken mRNA, which is controlled by the position of the oocyte nucleus.
It had been known for a long time that the anterior movement of the nucleus requires microtubules, and that the organization of the microtubule cytoskeleton depends on the polarizing signal from the posterior follicle cells (Koch and Spitzer, 1983; Ruohola et al., 1991). We therefore reasoned that nuclear migration should require the induction of the posterior follicle cells by the first Gurken signal, and this turned out to be the case. In a proportion of gurken or EGFR mutants, the nucleus remains at the posterior of the oocyte, along with gurken mRNA. Although the AP and DV axes previously had been thought to be completely independent, this result shows that the positioning of the second Gurken signal depends on the first. The AP axis is therefore the primary axis in Drosophila, because it must be polarized for the DV axis to form in the right place.
The formation of two perpendicular axes must involve some mechanism that fixes the orientation of one axis relative to the other. In Drosophila, this function is fulfilled by the migration of the oocyte nucleus, which positions the DV axis at right angles to the AP axis. This is demonstrated most convincingly by the phenotype of mago nashi mutants. Although we still know almost nothing about the polarizing signal from the posterior follicle cells or how it is transduced by the oocyte, David Micklem and Ramanuj Dasgupta demonstrated that mago nashi is required in the oocyte for the response to this signal (Micklem et al., 1997). Gurken signalling to the posterior follicle cells is unaffected in mago nashi mutants, but the microtubule cytoskeleton does not rearrange in response to the signal back from these cells, and the nucleus therefore often fails to migrate. As a consequence, gurken mRNA remains at the posterior of the oocyte, and the second Gurken signal is sent in the same direction as the first. These eggs therefore develop parallel AP and DV axes, with a dorsalized posterior and a ventralized anterior.
We next asked how the response of the follicle cells to Gurken is controlled, so that they are induced to become posterior by the first signal and dorsal by the second. One can think of three possible mechanisms to explain this difference: (i) the two Gurken signals differ in some unknown way; (ii) the competence of the follicle cells is regulated temporally, so that they respond to an early signal by becoming posterior, and become dorsal if exposed to the signal later in oogenesis; and (iii) different populations of follicle cells have different competences to respond to Gurken. To address this issue, we examined the behaviour of the follicle cells in egg chambers in which the oocyte is mispositioned, and therefore sends the first signal in the wrong direction. If the oocyte lies at the anterior or the posterior of the cyst, the adjacent follicle cells express posterior markers, whereas the follicle cells in the middle cannot be induced to become posterior when the oocyte lies in the middle of the egg chamber. Thus, the follicle cell layer has already been subdivided into two populations before Gurken signalling occurs. By generating EGFR mutant follicle cell clones that cannot respond to Gurken, we showed that ∼200 ‘terminal’ cells at each pole of the egg chamber are competent to become posterior, and adopt the default anterior fate if they do not receive the signal. In contrast, the remaining ‘mainbody’ cells only respond later in oogenesis, and become dorsal rather than ventral. This means that the AP axis is already defined by the positioning of the terminal follicle cells. The Gurken signal then acts to polarize this axis by making the posterior terminal cells different from the anterior ones.
All of our work so far indicated that the initial asymmetry that polarizes the AP axis for the rest of development is provided by positioning of the oocyte at the posterior of the cyst, and we therefore set out to determine how it reaches this position. Our initial approach was to analyse the phenotypes of mutants in the spindle genes, since these often give rise to egg chambers with misplaced oocytes (González-Reyes et al., 1997). However, we found that spindle mutants delay the determination of the oocyte, and that if the oocyte has not yet been specified, it cannot move to the posterior of the cyst in region 2b of the germarium. Since the spindle mutants only affect oocyte positioning indirectly, we next screened for mutants that alter the position of the oocyte without affecting its determination, in order to identify factors that play a direct role in this process. We found that germline clones of mutants in the homophilic cell adhesion molecule DE-cadherin give rise to a high frequency of egg chambers in which the oocyte is not at the posterior, and observed an identical phenotype in mutants for Armadillo, which binds to the cytoplasmic tail of DE-cadherin to link it to the actin cytoskeleton (González-Reyes and St Johnston, 1998a). This indicated that the positioning of the oocyte involves cell adhesion, and prompted us to examine exactly how the oocyte reaches the posterior as the cyst moves from region 2b to region 3 of the germarium. The somatic follicle cells migrate to surround the cyst when it enters region 2b, and the cyst flattens to form a lens-shaped disc with the oocyte in its centre. As the cyst moves into region 3, the germ cells rearrange to form a sphere, with the oocyte at the posterior. During this transition, DE-cadherin and Armadillo are up-regulated in both the oocyte and the posterior follicle cells, and accumulate along the boundary between them. Thus, oocyte positioning appears to depend on DE-cadherin-mediated adhesion between the oocyte and the posterior follicle cells, which attaches the oocyte to the posterior as the cyst changes shape. If this is the case, DE-cadherin should also be required in the follicle cell layer, and we tested this prediction by generating DE-cadherin mutant follicle cell clones. As expected, these clones also disrupt the positioning of the oocyte, but only if they include the posterior cells. Furthermore, when the posterior follicle cells are mutant, the oocyte often moves so that it contacts the adjacent wild-type cells, suggesting that the increased adhesiveness of the oocyte causes it to out-compete the nurse cells for adhesion to the DE-cadherin-positive cells. This use of DE-cadherin in oocyte positioning is unusual in two respects. First, most examples of cadherin-based adhesion occur between cells of the same type, whereas the oocyte and the follicle cells derive from lineages that are separate from the blastoderm stage. Secondly, this heterotypic DE-cadherin-dependent adhesion is the first in vivo example of a morphogenetic movement that is mediated by quantitative differences in the levels of a single adhesion molecule.
Although we have traced the origin of AP polarity in Drosophila back to the very beginning of oogenesis, we still lack the final answer to how this axis is polarized, because it is unclear why the oocyte sticks to the posterior follicle cells and not the anterior ones. The anterior and posterior terminal follicle cells are believed to be equivalent until Gurken signals to specify posterior fate later in oogenesis, and both populations up-regulate DE-cadherin in the germarium. Indeed, the oocyte preferentially localizes to the anterior of the cyst if the posterior cells lack DE-cadherin, indicating that the anterior follicle cells are competent to recruit the oocyte to this pole (Godt and Tepass, 1998). Thus, something else must bias the adhesion of the oocyte to the posterior, and it is this unknown cue that generates the first AP asymmetry in development.
An important issue raised by our work is whether the mechanisms that generate the axes in Drosophila are related to those used by other organisms. One aspect that does appear to have been conserved is the localization of characteristic electron-dense particles containing the germline determinants to one pole of the primary axis of the egg (Houston and King, 2000). In Drosophila, localized oskar mRNA directs the formation of the polar granules at the posterior of the oocyte, while morphologically similar granules, called the P granules or the germinal granules, localize to the posterior pole in Caenorhabditis elegans and to the vegetal pole in Xenopus. Despite these similarities, studies in C.elegans suggest that the upstream events that polarize the egg are quite different. The localization of the P granules is actin dependent and occurs after fertilization, whereas oskar mRNA is localized by microtubules during oogenesis (Hill and Strome, 1990; Hird et al., 1996). Furthermore, genetic screens for mutants that disrupt AP axis formation in C.elegans have identified different sets of proteins from Drosophila, including PAR-3 and PAR-6, which form a complex at the anterior of the one-cell zygote, and PAR-1 and PAR-2 which localize to the posterior (Kemphues et al., 1988; Etemad-Moghadam et al., 1995; Guo and Kemphues, 1995; Boyd et al., 1996). When Joshua Shulman joined the lab, he wanted to test the relationship between these two polarity systems directly, by analysing the Drosophila homologue of PAR-1. He and Richard Benton found that PAR-1 localizes to the posterior of the oocyte with oskar mRNA and Staufen (Shulman et al., 2000). More importantly, par-1 mutants cause a novel polarity phenotype, in which bicoid mRNA still localizes to the anterior, but oskar mRNA is mislocalized to a dot in the centre of the oocyte. Further analysis revealed that this phenotype results from a defect in the polarization of the microtubule cytoskeleton, in which a marker for the plus ends localizes to the middle of the oocyte rather than to the posterior pole. Thus, PAR-1 is required for AP axis formation in both flies and worms and is one of the earliest markers for the posterior in each case. It therefore provides the first molecular link between the polarization of the AP axis in these two organisms, although it is still unclear how it functions in either system.
During his characterization of par-1, Josh discovered that germline clones of a null mutation in the gene give rise to egg chambers with 16 nurse cells and no oocyte. Jean-René Huynh analysed this phenotype in more detail, and showed that the loss of PAR-1 blocks a novel step in oocyte determination (Huynh and St Johnston, 2000; Huynh et al., 2001a). In region 2 of the germarium, the centrioles and oocyte-specific proteins, such as BicD and Orb, still move from the nurse cells into the oocyte and accumulate at its anterior end. However, they fail to migrate from the anterior to the posterior of the oocyte in region 3, as they do in wild type, and the oocyte soon dedifferentiates to become a nurse cell. We next examined the oogenesis phenotype null mutations in the Drosophila homologues of par-3 (bazooka) and par-6. Germline clones of these mutants show an identical block to the AP movement of the centrioles and Orb in region 3, and also give rise to egg chambers with 16 nurse cells and no oocyte (Huynh et al., 2001b). Thus, this early AP polarity in the oocyte requires three of the same par genes as the polarization of the AP axis in C.elegans. This remarkable parallel suggests that these proteins act in a conserved pathway for generating the first AP asymmetry in the two organisms.
The end of the beginning
The last 10 years have seen the discovery of most of the important developmental events that lead to the polarization of the AP and DV axes in Drosophila, and many people might therefore think that the problem of axis formation in Drosophila has been finally solved. However, these results raise an equally large number of cell-biological questions. For example, we know almost nothing about how the posterior follicle cells signal to polarize the oocyte cytoskeleton, how the nucleus migrates to the anterior to set up the DV axis or how mRNA complexes are coupled to the motors that are presumed to transport them.
One reason why more is known about axis formation in Drosophila than any other organism is that screens for maternal-effect mutations have been very successful in identifying many of the genes that play key roles in this process. These screens could only recover mutations that are not lethal, however, and were therefore strongly biased towards the discovery of genes that are only needed for axis formation, such as gurken, bicoid and oskar. The next set of questions will require the identification of the more general factors that function in many cell types of the organism. Thanks largely to the work of Norbert Perrimon, it is now relatively straightforward to find these genes by performing genetic screens in germline clones (Chou and Perrimon, 1996). Indeed, Katia Litiere, Jean-René Huynh, Sophie Martin and Vincent Leclerc in my laboratory have already used this approach to identify many new mutants that disrupt the polarization of the oocyte, the organization of the microtubule cytoskeleton, and the localization of bicoid and oskar mRNAs. Thus, we can now ask some basic questions about the cell biology that underlies each step in axis formation, and I hope that this is the end of the beginning rather than the beginning of the end.
Acknowledgments
Acknowledgements
I would like to thank all of the people who have worked in my lab over the last 10 years, and did the experiments that won this medal. Many are already cited in this review, but I would particularly like to mention Hernán Lopéz-Schier, Isabel Palacios, Freek van Eeden, Uwe Irion, Hélène Doerflinger, Ruth McCaffrey, Lucie Whitehead, Rachel Smith and Vitaly Zimyanin, since I did not have space to cover their contributions. I am also extremely grateful to Douglas Barker, Bill Gelbart, Christiane Nüsslein-Volhard and John Gurdon for their unfailing support throughout my career, and to Mariann Bienz for nominating me for this medal. All of this work was generously supported by the Wellcome Trust in the form of a Wellcome Senior Fellowship and a Wellcome Principal Fellowship. Finally, I would like to thank my colleagues in the field of Drosophila oogenesis for making it such a friendly and exciting area in which to work.
References
- Berleth T., Burri,M., Thoma,G., Bopp,D., Richstein,S., Frigerio,G., Noll,M. and Nüsslein-Volhard,C. (1988) The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J., 7, 1749–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd L., Guo,S., Levitan,D., Stinchcomb,D.T. and Kemphues,K.J. (1996) PAR-2 is asymmetrically distributed and promotes association of P granules and PAR-1 with the cortex in C.elegans embryos. Development, 122, 3075–3084. [DOI] [PubMed] [Google Scholar]
- Brendza R.P., Serbus,L.R., Duffy,J.B. and Saxton,W.M. (2000) A function for kinesin I in the posterior transport of oskar mRNA and Staufen protein. Science, 289, 2120–2122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadus J. and Doe,C. (1997) Extrinsic cues, intrinsic cues and microfilaments regulate asymmetric protein localization in Drosophila neuroblasts. Curr. Biol., 7, 827–835. [DOI] [PubMed] [Google Scholar]
- Broadus J., Furstenberg,S. and Doe,C.Q. (1998) Staufen-dependent localization of prospero mRNA contributes to neuroblast daughter cell fate. Nature, 391, 792–795. [DOI] [PubMed] [Google Scholar]
- Bycroft M., Grünert,S., Murzin,A.G., Proctor,M. and St Johnston,D. (1995) NMR solution structure of a dsRNA binding domain from Drosophila Staufen protein reveals homology to the N terminal domain of ribosomal protein S5. EMBO J., 14, 3563–3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova J. and Struhl,G. (1989) Localized surface activity of torso, a receptor tyrosine kinase, specifies terminal body pattern in Drosophila. Genes Dev., 3, 2025–2038. [DOI] [PubMed] [Google Scholar]
- Chou T.-B. and Perrimon,N. (1996) The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics, 144, 1673–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark I., Giniger,E., Ruohola-Baker,H., Jan,L. and Jan,Y. (1994) Transient posterior localisation of a kinesin fusion protein reflects anteroposterior polarity of the Drosophila oocyte. Curr. Biol., 4, 289–300. [DOI] [PubMed] [Google Scholar]
- Clark I., Jan,L.Y. and Jan,Y.N. (1997) Reciprocal localization of Nod and kinesin fusion proteins indicates microtubule polarity in the Drosophila oocyte, epithelium, neuron and muscle. Development, 124, 461–470. [DOI] [PubMed] [Google Scholar]
- Driever W. and Nüsslein-Volhard,C. (1988a) A gradient of bicoid protein in Drosophila embryos. Cell, 54, 83–93. [DOI] [PubMed] [Google Scholar]
- Driever W. and Nüsslein-Volhard,C. (1988b) The bicoid protein determines position in the Drosophila embryo in a concentration-dependent manner. Cell, 54, 95–104. [DOI] [PubMed] [Google Scholar]
- Ephrussi A. and Lehmann,R. (1992) Induction of germ cell formation by oskar. Nature, 358, 387–392. [DOI] [PubMed] [Google Scholar]
- Ephrussi A., Dickinson,L.K. and Lehmann,R. (1991) oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell, 66, 37–50. [DOI] [PubMed] [Google Scholar]
- Etemad-Moghadam B., Guo,S. and Kemphues,K.J. (1995) Asym metrically distributed PAR-3 protein contributes to cell polarity and spindle alignment in early C.elegans embryos. Cell, 83, 743–752. [DOI] [PubMed] [Google Scholar]
- Ferguson E.L. and Anderson,K.V. (1992) decapentaplegic acts as a morphogen to organize dorsal–ventral pattern in the Drosophila embryo. Cell, 71, 451–461. [DOI] [PubMed] [Google Scholar]
- Ferrandon D., Elphick,L., Nüsslein-Volhard,C. and St Johnston,D. (1994) Staufen protein associates with the 3′UTR of bicoid mRNA to form particles which move in a microtubule-dependent manner. Cell, 79, 1221–1232. [DOI] [PubMed] [Google Scholar]
- Frohnhöfer H.G. and Nüsslein-Volhard,C. (1986) Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature, 324, 120–125. [Google Scholar]
- Frohnhöfer H.G., Lehmann,R. and Nüsslein-Volhard,C. (1986) Manipulating the anteroposterior pattern of the Drosophila embryo. J. Embryol. Exp. Morphol., 97, 169–179. [PubMed] [Google Scholar]
- Godt D. and Tepass,U. (1998) Drosophila oocyte localisation is mediated by differential cadherin-based adhesion. Nature, 395, 387–391. [DOI] [PubMed] [Google Scholar]
- González-Reyes A. and St Johnston,D. (1994) Role of oocyte position in the establishment of anterior–posterior polarity in Drosophila. Science, 266, 639–642. [DOI] [PubMed] [Google Scholar]
- González-Reyes A. and St Johnston,D. (1998a) The Drosophila AP axis is polarised by the cadherin-mediated positioning of the oocyte. Development, 125, 3635–3644. [DOI] [PubMed] [Google Scholar]
- González-Reyes A. and St Johnston,D. (1998b) Patterning of the follicle cell layer along the anterior–posterior axis during Drosophila oogenesis. Development, 125, 2837–2846. [DOI] [PubMed] [Google Scholar]
- González-Reyes A., Elliott,H. and St Johnston,D. (1995) Polarization of both major body axes in Drosophila by gurken–torpedo signalling. Nature, 375, 654–658. [DOI] [PubMed] [Google Scholar]
- González-Reyes A., Elliot,H. and St Johnston,D. (1997) Oocyte determination and the origin of polarity in Drosophila: the role of the spindle genes. Development, 124, 4927–4937. [DOI] [PubMed] [Google Scholar]
- Guo S. and Kemphues,K.J. (1995) par-1, a gene required for establishing polarity in C.elegans embryos, encodes a putative Ser/Thr kinase that is asymmetrically distributed. Cell, 81, 611–620. [DOI] [PubMed] [Google Scholar]
- Hill D.P. and Strome,S. (1990) Brief cytochalasin-induced disruption of microfilaments during a critical interval in 1-cell C.elegans embryos alters the partitioning of developmental instructions to the 2-cell embryo. Development, 108, 159–172. [DOI] [PubMed] [Google Scholar]
- Hird S.N., Paulsen,J.E. and Strome,S. (1996) Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development, 122, 1303–1312. [DOI] [PubMed] [Google Scholar]
- Holley S.A., Jackson,P.D., Sasai,Y., Lu,B., De Robertis,E.M., Hoffmann,F.M. and Ferguson,E.L. (1995) A conserved system for dorsal–ventral patterning in insects and vertebrates involving sog and chordin. Nature, 376, 249–253. [DOI] [PubMed] [Google Scholar]
- Houston D.W. and King,M.L. (2000) Germ plasm and molecular determinants of germ cell fate. Curr. Top. Dev. Biol., 50, 155–181. [DOI] [PubMed] [Google Scholar]
- Huynh J. and St Johnston,D. (2000) The role of BicD, egl, orb and the microtubules in the restriction of meiosis to the Drosophila oocyte. Development, 127, 2785–2794. [DOI] [PubMed] [Google Scholar]
- Huynh J.R., Shulman,J.M., Benton,R. and St Johnston,D. (2001a) PAR-1 is required for the maintenance of oocyte fate in Drosophila. Development, 128, 1201–1209. [DOI] [PubMed] [Google Scholar]
- Huynh J.-R., Petronczki,M., Knoblich,J.A. and St Johnston,D. (2001b) Bazooka and PAR-6 are required with PAR-1 for the maintenance of oocyte fate in Drosophila. Curr. Biol., 11, 901–906. [DOI] [PubMed] [Google Scholar]
- Ikeshima-Kataoka H., Skeath,J.B., Nabeshima,Y.-i., Doe,C.Q. and Matsuzaki,F. (1998) Miranda directs Prospero to a daughter cell during Drosophila asymmetric divisions. Nature, 390, 625–629. [DOI] [PubMed] [Google Scholar]
- Illmensee K. and Mahowald,A.P. (1974) Transplantation of posterior pole plasm in Drosophila: induction of germ cells at the anterior pole of the egg. Proc. Natl Acad. Sci. USA, 71, 1016–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish V.F. and Gelbart,W.M. (1987) The decapentaplegic gene is required for dorsal–ventral patterning of the Drosophila embryo. Genes Dev., 1, 868–879. [DOI] [PubMed] [Google Scholar]
- Kemphues K.J., Priess,J.R., Morton,D.G. and Cheng,N.S. (1988) Identification of genes required for cytoplasmic localization in early C.elegans embryos. Cell, 52, 311–320. [DOI] [PubMed] [Google Scholar]
- Kiebler M.A., Hemraj,I., Verkade,P., Kohrmann,M., Fortes,P., Marion,R.M., Ortin,J. and Dotti,C.G. (1999) The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: implications for its involvement in mRNA transport. J. Neurosci., 19, 288–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim-Ha J., Smith,J.L. and Macdonald,P.M. (1991) oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell, 66, 23–35. [DOI] [PubMed] [Google Scholar]
- Klingler M., Erdelyi,M., Szabad,J. and Nüsslein-Volhard,C. (1988) Function of torso in determining the terminal anlagen of the Drosophila embryo. Nature, 335, 275–277. [DOI] [PubMed] [Google Scholar]
- Koch E. and Spitzer,R. (1983) Multiple effects of colchicine on oogenesis in Drosophila; induced sterility and switch of potencial oocyte to nurse-cell developmental pathway. Cell Tissue Res., 228, 21–32. [DOI] [PubMed] [Google Scholar]
- Köhrmann M., Luo,M., Kaether,C., DesGroseillers,L., Dotti,C.G. and Kiebler,M.A. (1999) Microtubule-dependent recruitment of Staufen– green fluorescent protein into large RNA-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell, 10, 2945–2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann R. and Nüsslein-Volhard,C. (1991) The maternal gene nanos has a central role in posterior pattern formation in the Drosophila embryo. Development, 112, 679–691. [DOI] [PubMed] [Google Scholar]
- Li P., Yang,X., Wasser,M., Cai,Y. and Chia,W. (1997) Inscuteable and Staufen mediate asymmetric localization and segregation of prospero RNA during Drosophila neuroblast cell divisions. Cell, 90, 437–447. [DOI] [PubMed] [Google Scholar]
- Macdonald P.M. (1990) bicoid mRNA localization signal: phylogenetic conservation of function and RNA secondary structure. Development, 110, 161–171. [DOI] [PubMed] [Google Scholar]
- Macdonald P.M. and Struhl,G. (1988) Cis-acting sequences responsible for anterior localization of bicoid mRNA in Drosophila embryos. Nature, 336, 595–598. [DOI] [PubMed] [Google Scholar]
- Micklem D.R., Dasgupta,R., Elliott,H., Gergely,F., Davidson,C., Brand,A., González-Reyes,A. and St Johnston,D. (1997) The mago nashi gene is required for the polarisation of the oocyte and the formation of perpendicular axes in Drosophila. Curr. Biol., 7, 468–478. [DOI] [PubMed] [Google Scholar]
- Micklem D.R., Adams,J., Grünert,S. and St Johnston,D. (2000) Distinct roles of two conserved Staufen domains in oskar mRNA localisation and translation. EMBO J., 19, 1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuman-Silberberg F. and Schüpbach,T. (1993) The Drosophila dorsoventral patterning gene gurken produces a dorsally localized RNA and encodes a TGFα-like protein. Cell, 75, 165–174. [PubMed] [Google Scholar]
- Nüsslein-Volhard C. and Wieschaus,E. (1980) Mutations affecting segment number and polarity in Drosphila. Nature, 287, 795–801. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard C., Frohnhöfer,H.G. and Lehmann,R. (1987) Determination of anteroposterior polarity in Drosophila. Science, 238, 1675–1681. [DOI] [PubMed] [Google Scholar]
- Padgett R.W., St Johnston,R.D. and Gelbart,W.M. (1987) A transcript from a Drosophila pattern gene predicts a protein homologous to the transforming growth factor-β family. Nature, 325, 81–84. [DOI] [PubMed] [Google Scholar]
- Palacios I. and St Johnston,D. (2001) Getting the message across: the intracellular localisation of mRNAs in higher eukaryotes. Annu. Rev. Cell Dev. Biol., 17, 569–614. [DOI] [PubMed] [Google Scholar]
- Pelham H.R. (1989) Heat shock and the sorting of luminal ER proteins. EMBO J., 8, 3171–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pokrywka N.J. and Stephenson,E.C. (1991) Microtubules mediate the localization of bicoid RNA during Drosophila oogenesis. Development, 113, 55–66. [DOI] [PubMed] [Google Scholar]
- Ramos A., Grunert,S., Adams,J., Micklem,D.R., Proctor,M.R., Freund,S., Bycroft,M., St Johnston,D. and Varani,G. (2000) RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J., 19, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S., Stein,D. and Nüsslein-Volhard,C. (1989) A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell, 59, 1189–1202. [DOI] [PubMed] [Google Scholar]
- Roth S., Neuman-Silberberg,F.S., Barcelo,G. and Schüpbach,T. (1995) cornichon and the EGF receptor signaling process are necessary for both anterior–posterior and dorsal–ventral pattern formation in Drosophila. Cell, 81, 967–978. [DOI] [PubMed] [Google Scholar]
- Ruohola H., Bremer,K.A., Baker,D., Sedlow,J.R., Jan,L.Y. and Jan,Y.N. (1991) Role of neurogenic genes in establishment of follicle cell fate and oocyte polarity during oogenesis in Drosophila. Cell, 66, 433–449. [DOI] [PubMed] [Google Scholar]
- Rushlow C.A., Han,K., Manley,J.L. and Levine,M. (1989) The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell, 59, 1165–1177. [DOI] [PubMed] [Google Scholar]
- Schnorrer F., Bohmann,K. and Nusslein-Volhard,C. (2000) The molecular motor dynein is involved in targeting Swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nature Cell Biol., 2, 185–190. [DOI] [PubMed] [Google Scholar]
- Schuldt A.J., Adams,J.H.J., Davidson,C.M., Micklem,D.R., St Johnston,D. and Brand,A. (1998) Miranda mediates the asymmetric protein and RNA localisation in the developing nervous system. Genes Dev., 12, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schüpbach T. (1987) Germ line and soma cooperate during oogenesis to establish the dorsoventral pattern of egg shell and embryo in Drosophila melanogaster. Cell, 49, 699–707. [DOI] [PubMed] [Google Scholar]
- Schüpbach T. and Wieschaus,E. (1986) Maternal-effect mutations altering the anterior–posterior pattern of the Drosophila embryo. Wilhelm Roux’s Arch. Dev. Biol., 195, 302–317. [DOI] [PubMed] [Google Scholar]
- Sen J., Goltz,J.S., Stevens,L. and Stein,D. (1998) Spatially restricted expression of pipe in the Drosophila egg chamber defines embryonic dorsal–ventral polarity. Cell, 95, 471–481. [DOI] [PubMed] [Google Scholar]
- Shen C.-P., Jan,L.-Y. and Jan,Y.-N. (1997) Miranda is required for the asymmetric localization of Prospero during mitosis in Drosophila. Cell, 90, 449–458. [DOI] [PubMed] [Google Scholar]
- Shulman J.M., Benton,R. and St Johnston,D. (2000) The Drosophila homolog of C.elegans PAR-1 organizes the oocyte cytoskeleton and directs oskar mRNA localisation to the posterior pole. Cell, 101, 1–20. [DOI] [PubMed] [Google Scholar]
- Smith J.C. (1993) Mesoderm-inducing factors in early vertebrate development. EMBO J., 12, 4463–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer F.A., Hoffman,F.M. and Gelbart,W.M. (1982) Decapentaplegic: a gene complex affecting morphogenesis in Drosophila melanogaster. Cell, 28, 451–461. [DOI] [PubMed] [Google Scholar]
- Spradling A. (1993) Developmental genetics of oogenesis. In Bate,M. and Martinez-Arias,A. (eds), The Development of Drosophila melanogaster. Vol. 1. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 1–70.
- Sprenger F., Stevens,L.M. and Nüsslein-Volhard,C. (1989) The Drosophila gene torso encodes a putative receptor tyrosine kinase. Nature, 338, 478–483. [DOI] [PubMed] [Google Scholar]
- Stein D., Roth,S., Vogelsang,E. and Nüsslein-Volhard,C. (1991) The polarity of the dorsoventral axis of the Drosophila embryo is defined by an extracellular signal. Cell, 65, 725–735. [DOI] [PubMed] [Google Scholar]
- Stevens L.M., Frohnhöfer,H.G., Klingler,M. and Nüsslein-Volhard,C. (1990) Localized requirement for torso-like expression in follicle cells for development of terminal anlagen of the Drosophila embryo. Nature, 346, 660–663. [DOI] [PubMed] [Google Scholar]
- St Johnston D. and Nüsslein-Volhard,C. (1992) The origin of pattern and polarity in the Drosophila embryo. Cell, 68, 201–219. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Driever,W., Berleth,T., Richstein,S. and Nüsslein-Volhard,C. (1989) Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development, Suppl. 107, 13–19. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Hoffmann,F.M., Blackman,R.K., Segal,D., Grimaila,R., Padgett,R.W., Irick,H.T. and Gelbart,W.M. (1990) Molecular organization of the decapentaplegic gene of Drosophila melanogaster. Genes Dev., 4, 1114–1127. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Beuchle,D. and Nüsslein-Volhard,C. (1991) Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell, 66, 51–63. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Brown,N.H., Gall,J.G. and Jantsch,M. (1992) A conserved double-stranded RNA-binding domain. Proc. Natl Acad. Sci. USA, 89, 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston R.D. and Gelbart,W.M. (1987) Decapentaplegic transcripts are localized along the dorsal–ventral axis of the Drosophila embryo. EMBO J., 6, 2785–2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G. (1982) Decapentaplegic—hopes held out. Nature, 298, 13–14. [DOI] [PubMed] [Google Scholar]
- Theurkauf W.E., Smiley,S., Wong,M.L. and Alberts,B.M. (1992) Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development, 115, 923–936. [DOI] [PubMed] [Google Scholar]
- Treisman R. (1995) Journey to the surface of the cell: Fos regulation and the SRE. EMBO J., 14, 4905–4913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eeden F.J.M., Palacios,I.M., Petronczki,M., Weston,M.J.D. and St Johnston,D. (2001) Barentsz is essential for the posterior localization of oskar mRNA and colocalizes with it to the posterior. J. Cell Biol., 154, 511–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. and Lehmann,R. (1991) Nanos is the localized posterior determinant in Drosophila. Cell, 66, 637–647. [DOI] [PubMed] [Google Scholar]
- Wharton K., Ray,R. and Gelbart,W. (1993) An activity gradient of decapentaplegic is necessary for the specification of dorsal pattern elements in the Drosophila embryo. Development, 117, 807–822. [DOI] [PubMed] [Google Scholar]