Abstract
The general secretory pathway (GSP) is a two-step process for the secretion of proteins by Gram-negative bacteria. The translocation across the outer membrane is carried out by the type II system, which involves machinery called the secreton. This step is considered to be an extension of the general export pathway, i.e. the export of proteins across the inner membrane by the Sec machinery. Here, we demonstrate that two substrates for the Pseudomonas aeruginosa secreton, both phospholipases, use the twin-arginine translocation (Tat) system, instead of the Sec system, for the first step of translocation across the inner membrane. These results challenge the previous vision of the GSP and suggest for the first time a mosaic model in which both the Sec and the Tat systems feed substrates into the secreton. Moreover, since P.aeruginosa phospholipases are secreted virulence factors, the Tat system appears to be a novel determinant of bacterial virulence.
Keywords: phospholipase/Pseudomonas aeruginosa/Sec machinery/Tat system/type II secretory pathway
Introduction
Pseudomonas aeruginosa is an opportunistic human pathogen and an important cause of hospital-acquired infections in predisposed or immunocompromised individuals. It is the most lethal Gram-negative pathogen in nosocomial pneumonia (25–50% mortality) and is responsible for lethal chronic infections in the airways of patients with cystic fibrosis (Doring, 1997). Several virulence factors contributing to P.aeruginosa pathogenesis (Liu, 1974) are exoproteins secreted through three distinct secretory pathways, named type I, II and III (Tommassen et al., 1992; Frank, 1997). The type II pathway, also called the main terminal branch of the general secretory pathway (MTB of the GSP) (Pugsley, 1993), is used by exotoxin A and most degradative enzymes of P.aeruginosa (Filloux et al., 1998). These exoproteins carrying a cleavable N-terminal signal peptide are presumed to be exported across the cytoplasmic membrane via the Sec machinery (Douglas et al., 1987; Economou, 2000). In the periplasm, the proteins fold into a (near-)native conformation and are recognized by the Xcp machinery, which allows translocation of the proteins across the outer membrane (Braun et al., 1996; Voulhoux et al., 2000). The Xcp machinery, also called the secreton, thus defines the type II pathway in P.aeruginosa. An intimate interaction between the Sec machinery and the secreton may be important for an efficient secretion, since a temporary arrest of periplasmic intermediates considerably slows down the kinetics of the second step, i.e. the transport across the outer membrane (Poquet et al., 1993).
The Tat pathway was found originally in eukaryotes for proteins transported across the chloroplast thylakoid membrane (Settles et al., 1997). Recently, it has been established, in Escherichia coli, that the Tat pathway operates in parallel to the Sec system in the export of periplasmic proteins. The bacterial Tat system is distinct from the Sec pathway in terms of its remarkable ability to transport folded enzymes (Berks et al., 2000; Wu et al., 2000). Moreover, the signal peptides of the Tat-dependent proteins have a number of peculiar properties. They resemble Sec-dependent signal peptides, considering their overall structures, but possess a twin-arginine motif in the positively charged n-region, a weakly hydrophobic h-region and a positively charged Sec-avoidance signal in the c-region (Bogsch et al., 1997; Cristobal et al., 1999). Most of the known Tat-dependent proteins contain redox factors and are components of various respiratory chains that support the anaerobic growth of the bacteria. Other Tat substrates are involved in the adaptation of bacteria to a particular environment with high osmolarity (Wu et al., 2000). In addition, the Tat machinery is also capable of exporting the green fluorescent protein, which probably folds too tightly to be exported by the Sec system (Feilmeier et al., 2000; Santini et al., 2001).
Here, we demonstrate that the P.aeruginosa Tat system is used for the secretion of at least two Xcp-dependent proteins. Therefore, the Tat system is also involved in protein secretion and covers almost all functions of the Sec pathway. Importantly, this result shows the existence of a mosaic structure in which the Tat system operates in parallel with the Sec machinery for the secretion of proteins via the type II secretory pathway, and thus challenges the established mechanism for type II secretion.
Results
Identification of the Pseudomonas aeruginosa tat genes
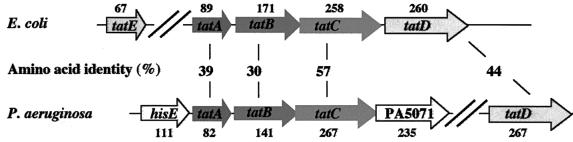
In E.coli, five tat genes located in two loci, tatABCD and tatE, have been identified and characterized (Berks et al., 2000; Wu et al., 2000). Since mutations in the tatC gene lead to mislocation of all the Tat substrates analysed so far, TatC is considered as a crucial component of the Tat translocase. TatA, TatB and TatE share sequence homology at their N-termini, which includes a transmembrane segment and an adjacent amphipathic domain, whereas their C-termini vary in both sequence and length. The functions of these proteins overlap to a certain extent. Recently, it was demonstrated that E.coli TatD is a cytoplasmic protein with DNase activity and is not required for the function of the Tat system (Wexler et al., 2000). The release of the P.aeruginosa genome sequence (Stover et al., 2000) allowed us to identify a tatABC gene cluster in this bacterium (Figure 1). The tatA gene starts at position 5 706 531 of the genome sequence and the tatC gene ends at position 5 708 038. The TatA, TatB and TatC proteins comprise 82, 141 and 267 amino acids, and show 39, 30 and 57% sequence identity to their E.coli counterparts, respectively. The TopPred 2 program (http://www.sbc.su.se/∼erikw/toppred2) predicted that the TatC protein possesses six transmembrane segments between residues 31 and 46, 82 and 100, 123 and 146, 162 and 187, 203 and 221, and 225 and 241, and that the TatA and TatB proteins each have a single putative transmembrane domain at their N-terminus. The gene immediately upstream of tatA encodes a protein highly homologous to the phosphoribosyl ATP pyrophosphatase (HisE) from Azotobacter chroococcum (90% identity), and the one downstream of tatC, PA5071, encodes a protein homologous to a hypothetical protein from Acinetobacter haemolyticus (41% identity) (Figure 1). No tatE homologues were found. Therefore, the Tat system of P.aeruginosa comprises one copy each of tatA, tatB and tatC that form a gene cluster. It should be noted that a tatD gene homologue was found at another chromosomal locus that encoded a 267 amino acid protein 44% identical to E.coli TatD (Figure 1).
Fig. 1. Genetic organization of the P.aeruginosa tat gene cluster. The genetic organization of the P.aeruginosa tat genes is compared with that of E.coli. The percentage identity between the corresponding homologous Tat proteins is indicated. The tatD and tatE genes are not clustered with the other tat genes of P.aeruginosa and E.coli, respectively. In P.aeruginosa, the intergenic region between the tatA and tatB genes is 14 bp long, whereas the start codon of tatC overlaps with the stop codon of tatB. The stop codon of the gene (hisE) encoding a 111 amino acid protein homologous to the A.chroococcum phosphoribosyl ATP pyrophosphatase is located 26 bp upstream from the tatA start codon. The start codon of the gene encoding a 235 amino acid protein homologous to a hypothetical protein from A.haemolyticus overlaps with the stop codon of tatC. The deduced size in amino acids of the gene products is indicated above or under each gene for E.coli and P.aeruginosa, respectively.
The RR–pfColA hybrid, and its use as a genetic tool
In order to investigate further whether the Tat pathway was functional in P.aeruginosa, we adapted a previously described genetic selection (Voulhoux et al., 2001). Colicin A (ColA) is a toxin which is produced by and is active against a wide variety of Enterobacteriaceae such as E.coli. The cells are killed as a result of the insertion of the ColA pore-forming domain (pfColA) from the periplasm and within the cytoplasmic membrane of E.coli (Espesset et al., 1994). The producing strain is protected from the action of pfColA by the immunity protein (Cai) while releasing ColA to the extracellular medium (Espesset et al., 1996). The pfColA domain is then brought into the target cells due to the action of the two other ColA domains, called receptor (R) and translocator (T).
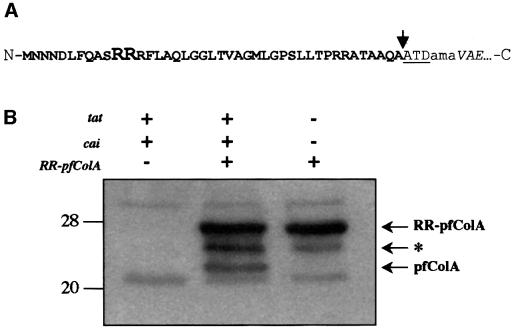
In the previous studies, it was shown that the producing strains might be killed in the absence of Cai, by replacing the ColA-R and ColA-T domains with a signal peptide allowing periplasmic targeting and subsequent membrane insertion of pfColA in a Sec-dependent fashion. Here, we replaced the domain encoding the Sec-dependent signal peptide (sp) with a DNA fragment encoding the Tat-dependent twin-arginine peptide (RR) of the trimethylamine n-oxide (TMAO) reductase (TorA) from E.coli (Santini et al., 1998) (Figure 2A). We subsequently cloned the gene fusion (RR–pfColA) behind the tac promoter of pMMB67EH, yielding pRR-pfColA67. The latter plasmid could only be introduced into E.coli (C600) containing pImTc (Cai), indicating that the hybrid protein RR– pfColA is toxic for E.coli (Table I) and thus transported to the periplasm. In contrast, pRR-pfColA67 could be introduced, in the absence of pImTc (Cai), into an E.coli tat mutant (tatABCDE), indicating that the translocation is Tat dependent and Sec independent.
Fig. 2. (A) Schematic representation of the amino acid sequence at the junction point of the hybrid RR–pfColA. The twin-arginine signal peptide is represented in bold letters, with the RR residues of the twin-arginine motif in a larger font size. The pfColA amino acid sequence is in italics. The added linker region in the hybrid proteins is indicated in lower case letters. The leader peptidase cleavage site is indicated by an arrow and is followed by the first three residues of the mature TorA protein (ATD). N and C indicate the N- and C-termini of the protein, respectively. (B) Tat-dependent processing of RR–pfColA. The presence (+) or absence (–) of the tat, cai and RR–pfcolA genes for each cell sample is indicated. Whole-cell extracts of E.coli wild-type strains containing pImTc and pRR-pfColA or the vector pMMB67EH, and the E.coli tat mutant (tatABCDE) containing pRR-pfColA were loaded on an 11% acrylamide gel containing SDS, and proteins were separated by electrophoresis and blotted onto nitrocellulose. The immunoblot was revealed using antibodies directed against pfColA (diluted 1:1000). The arrow indicates the position of the precursor (RR–pfColA) and mature (pfColA) proteins. The deduced mol. wts of 26.6 and 22.8 kDa are in agreement with the expected size of RR–pfColA and pfColA, respectively. The asterisk indicates the position of a putative degradation product. Molecular weight markers are indicated on the left.
Table I. Viability of E.coli and P.aeruginosa strains upon pfColA production.
| Wild type | E.coli | P.aeruginosa | |||
|---|---|---|---|---|---|
| Wild type (cai)a | tatA-E | Wild type | tatC | ||
| pMMB67EH | + | + | + | + | + |
| pRR-pfColA67 | – | + | + | – | + |
The + and – indicate the strains that are able to support efficient growth when transformed with the pMM67EH vector or the plasmid pRR-pfColA67 encoding RR–pfColA. Wild type indicates the wild-type strains of E.coli and P.aeruginosa in contrast to the tat mutant derivatives.
aThe strain carries the pImTc encoding the immunity protein Cai.
We then assessed the production and maturation of RR–pfColA. The wild-type E.coli strain and the tat mutant-containing pMMB67EH or its derivative pRR- pfColA67 were grown at 37°C in L broth to an OD600 of 0.6. Expression of RR-pfColA was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 30 min. Whole-cell extracts were prepared and proteins separated on gels containing 11% acrylamide. Immunoblotting was performed subsequently using antibodies directed against pfColA. The three bands identified are specific for the RR–pfcolA gene fusion since they are not detected when the strain containing the empty vector is analysed (Figure 2B). Moreover in the tatA-E mutant, the lowest band is not detected, which is in agreement with the fact that in the tat mutant no processing is expected. The calculated molecular mass confirms that the higher and lower bands corresponded to precursor and mature forms, respectively. This result clearly confirmed that the processing of the RR–pfColA chimera and the translocation across the cytoplasmic membrane of the pfColA domain are Tat dependent.
The Tat pathway is functional in P.aeruginosa
We previously showed that ColA toxicity could also be observed in P.aeruginosa when pfColA is synthesized from pRV300 (sp–pfColA) and translocated into the periplasm via the Sec pathway (Voulhoux et al., 2001). Using a similar strategy, we showed here that pfColA can be translocated into the E.coli periplasm via the Tat pathway and kill the cells. Thus, we reasoned that P.aeruginosa should not support RR–pfColA expression if it has a functional Tat system.
The pRR-pfColA67 plasmid was electroporated into the wild-type P.aeruginosa strain PAK. Recombinant bacteria were allowed to grow for 48 h at 30°C, after plating on Pseudomonas isolation agar (PIA) supplemented with 300 µg/ml carbenicillin and 2 mM IPTG. The number of colonies that could be obtained, in these conditions, was 1 × 105 less than the control experiment in which P.aeruginosa was electroporated with the cloning vector pMMB67EH (Table I). Moreover, the few transformants obtained appeared not to produce pfColA and were spontaneous carbenicillin-resistant clones. Such an observation was reported previously with the use of the sp–pfColA chimera (Voulhoux et al., 2001).
We constructed a tatC mutant using a recombination-interruption approach. Briefly, a fragment of 450 bp covering the central region of tatC was amplified by PCR and cloned into the suicide vector pCR2.1. The resulting construct was introduced into P.aeruginosa PAK and the tatC mutant (PAK-TC) was isolated and characterized (see Materials and methods). In contrast to the PAK wild-type strain, the P.aeruginosa tatC mutant was fully able to support growth upon electroporation with pRR-pfColA67 (Table I). These results indicated that, as in E.coli, the Tat-dependent signal peptide (RR) is able to target pfColA to the periplasmic side of the P.aeruginosa cytoplasmic membrane in a Tat-dependent manner. Therefore, it is likely that the Tat pathway is functional in P.aeruginosa.
Identification of P.aeruginosa Tat substrates
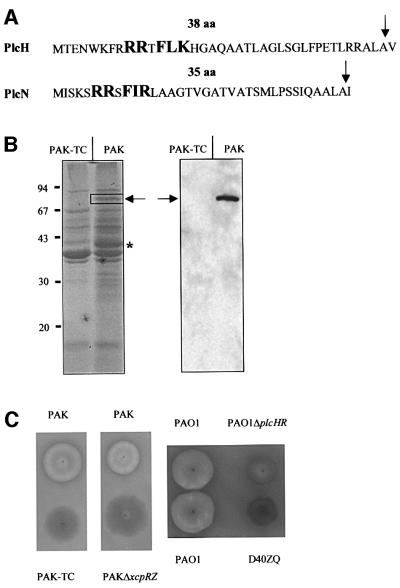
We attempted to identify putative substrates of the P.aeruginosa Tat machinery by screening the genome at the PEDANT website (http://pedant.mips.biochem. mpg.de) for proteins containing an RRxF motif. Among many candidates, we identified the nitrous oxide reductase encoded by nosZ, the homologue of which is translocated by the Tat pathway in Ralstonia eutropha (Bernhard et al., 2000). In addition, two phospholipases C particularly attracted our attention since they are secreted by the Xcp pathway (Martinez et al., 1998) and their signal peptides fitted perfectly the criteria of the twin-arginine signal peptide (Figure 3A). These enzymes are also known as haemolytic (PlcH) and non-haemolytic (PlcN) phospholipases and both are capable of cleaving phosphatidylcholine which is present in the outer leaflet of mammalian erythrocytes (Ostroff et al., 1990). In addition, PlcH cleaves outer leaflet-located sphingomyelin, whereas PlcN shows specificity for phosphatidylserine, which is found in the inner leaflet (Ostroff et al., 1990). These two proteins are encoded by non-tandem genes and are 40% identical. Finally, their synthesis is regulated by the concentration of inorganic phosphate (Pi) in the medium (Gray et al., 1981).
Fig. 3. N-terminal signal sequences of PlcH and PlcN, protein profiles of the supernatants and haemolytic activities of the wild type and the tatC mutant (PAK-TC). (A) The twin-arginine motif R-R-x-F-(I/L)-(K/R) is represented in bold and upper case letters. The number of residues (aa) in each signal peptide is indicated. The signal peptide cleavage site is indicated by an arrow. (B) Proteins from culture supernatants of PAK and PAK-TC strains equivalent to 1 absorption unit of cells were separated on a denaturing SDS–gel containing 11% acrylamide and stained with Coomassie Blue (left panel), or immunoblotted and revealed by using monoclonal antibodies directed against PlcH (right panel). The expected migration position of PlcH and PlcN is indicated by an arrow. The bands analysed by mass spectrometry and missing in the tatC mutant are boxed (PlcN) or indicated with an asterisk (GlpQ). The positions of molecular weight markers (kDa) are indicated on the left. (C) Haemolytic activity on blood agar plates of PAK or PAO1, and the derivative xcpR-Z or plcHR and xcpZ-Q (D40ZQ) mutant strains, respectively.
Pseudomonas aeruginosa phospholipases C are secreted Tat dependently
To identify proteins secreted efficiently by the Tat pathway, we compared the protein profiles of the culture supernatants from the wild-type strain PAK and its tatC derivative (PAK-TC), both grown to an optical density at 600 nm of 2, in proteose peptone broth, which is limiting for Pi. The protein profiles of both strains were rather similar. However, we particularly noticed the absence of a band at ∼77 kDa in the profile of the tatC mutant (Figure 3B). The migration position of the 77 kDa band corresponds well with the expected sizes of the mature forms of both PlcH (78.2 kDa) and PlcN (73.5 kDa), which showed similar mobilities in denaturing SDS–PAGE (Ostroff et al., 1990).
To determine the identity of the 77 kDa band, it was excised from a gel, treated with trypsin and analysed by MALDI-TOF mass spectrometry (see Materials and methods). The peptide mapping was achieved by using appropriate programs at http://www.matrixscience.com and http://www.expasy.ch/cgi-bin/peptident.p1. Among the 15 monoisotopic peptide peaks identified, we succeeded in assigning nine peptides with a mass difference <100 p.p.m. and two peptides with a mass difference <250 p.p.m. to the PlcN. Therefore, this result unambiguously reveals the presence of PlcN in this sample. Even though PlcH was not identified in the excised 77 kDa band by mass spectrometry analysis, immunoblotting analysis unambiguously confirmed the presence of PlcH in this band (Figure 3B).
We then analysed the haemolytic activity of P.aeruginosa strains on blood agar plates, to determine whether the tatC mutation affects the secretion of PlcH (Figure 3C). The haemolytic activity of the tatC mutant (PAK-TC) was drastically reduced as compared with that of the wild-type strain, and was similar to that of xcpR-Z or plcHR (devoid of PlcH activity) (Shortridge et al., 1992) mutant strains (Figure 3C). Moreover, full haemolytic activity was recovered upon introduction of pTATC, a pLAFR3 derivative that carries the PAK tatC gene, into the PAK-TC strain (data not shown). We confirmed that the haemolytic activity observed on plates is correlated with the presence of PlcH in the extracellular medium of the bacterial culture, by immunoblotting analysis using monoclonal antibodies directed against PlcH (Figure 3B). Finally, we observed that PlcH is blocked within the cytoplasmic membrane fraction of the tatC mutant, by looking at phospholipase C activity as measured using ρ-nitrophenylphosphorylcholine (NPPC) as substrate (Table II).
Table II. Phospholipase C activitya in P.aeruginosa cell fractions.
| Strain | Extracellular medium | Outer membrane periplasm | Cytoplasm | Cytoplasmic membrane |
|---|---|---|---|---|
| PAK | 2.80 | 0.50 | 0.10 | 0.68 |
| PAKΔxcpR-Z | 0.03 | 4.74 | 0.16 | 0.57 |
| PAK-TC (tatC) | 0.01 | <0.01 | 0.04 | 4.21 |
aAssayed by hydrolysis of ρ-nitrophenylphosphorylcholine (NPPC), which is expressed in relative units: ΔOD410/min/100 µl × 100. Cultures were adjusted to the same OD590 before fractionation. OD590 of the original cultures was 1.05 for PAK wild type; 0.99 for PAKΔxcpR-Z; and 1.01 for PAK-TC. Since protein concentration varies between each fraction from the same organism (e.g. supernatant versus cytoplasm), comparisons can only be made between the same fraction (e.g. cytoplasmic membrane or supernatant) of each of the strains.
To confirm that the secretion deficiency in the tatC mutant is indeed specific for the phospholipases, we also analysed the secretion of Sec-dependent elastase and exotoxin A in this mutant. The secretion efficiency of these proteins in the tatC mutant strain and its parental strain PAK appeared to be comparable as revealed by immunoblotting (data not shown). Therefore, we conclude that the Tat pathway mediates the first step of secretion of the Xcp-dependent exoproteins PlcN and PlcH.
Interestingly, a protein with a molecular mass of ∼42 kDa is also missing in the extracellular growth medium of the PAK-TC mutant culture (Figure 3B). MALDI-TOF mass spectrometry revealed that the corresponding band, excised from the gel, contained a protein coded by the open reading frame (ORF) annotated PA0347 in the P.aeruginosa genome (http://www.pseudomonas.com). The protein presents a high level of similarity (49–52%) to the glycerophosphoryl diester phosphodiesterase (GlpQ) from E.coli or Bacillus subtilis (Tommassen et al., 1991; Antelmann et al., 2000). Analysis of the deduced amino acid sequence showed that the P.aeruginosa GlpQ protein possesses a putative 27 amino acid N-terminal signal peptide containing a twin-arginine motif. This result supports the observed Tat dependence of the P.aeruginosa GlpQ.
Discussion
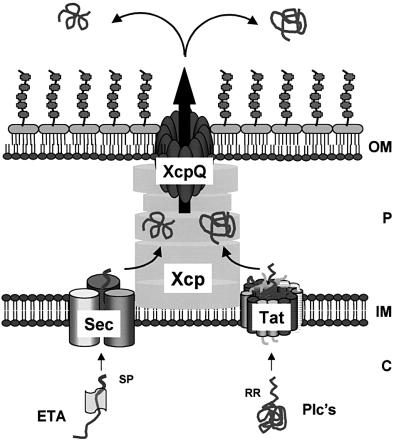
The term general secretory pathway (GSP) was used originally to define a two-step secretion process, which was considered to be an extended version of the general export pathway (GEP) (Pugsley, 1993). The first step in this process was proposed to be carried out exclusively by the Sec machinery, which translocates signal peptide-bearing precursors across the cytoplasmic membrane. The Sec complex comprises the membrane-embedded SecYEG translocon, which functions in concert with SecA, an ATP-hydrolysing protein that pushes precursors through the translocon by successive cycles of ATP hydrolysis (Economou and Wickner, 1994). An important feature of this export process is that the precursors are translocated in a delineated fashion (Manting et al., 2000). Once released into the periplasm, the proteins acquire a folded conformation, which is a prerequisite for the subsequent translocation across the outer membrane (Braun et al., 1996; Voulhoux et al., 2000). This second step is carried out by specialized machinery, the secreton, which is composed of at least 12 components, designated XcpA and XcpP–Z in P.aeruginosa (Filloux et al., 1998). One of these components, the secretin XcpQ, forms a membrane-embedded multimeric complex with a large central cavity that allows the passage of folded proteins (Bitter et al., 1998; Brok et al., 1999) (Figure 4).
Fig. 4. Mosaic model of protein secretion via the type II pathway. The Xcp-dependent exoproteins exotoxin A (ETA), containing a Sec-dependent signal peptide (SP), or the phospholipases C (Plc’s), bearing a twin-arginine (RR) signal peptide, are exported across the inner membrane through the Sec and the Tat pathway, respectively. After cleavage of the signal peptides, the exoproteins are recognized in the periplasm by the Xcp system, directed to the secretin XcpQ, and released into the external medium. C = cytoplasm; IM = inner membrane; P = periplasm; OM = outer membrane. Different shades in the Sec and Tat translocons correspond to different subunits.
The recently discovered Tat system is distinct from the Sec pathway by its unusual ability to transport folded enzymes often containing redox cofactors across the cytoplasmic membrane. In our study, we reveal for the first time that the Tat pathway can feed substrates into the Xcp system and we show a mosaic structure in which the Tat system operates in parallel with the Sec machinery in the secretion of type II-dependent proteins (Figure 4). This observation conceptually affects several important issues.
First, it challenges well established dogmas in protein secretion. As previously mentioned, it is generally accepted that the type II secretory pathway is an extension of the general export pathway (Sec), i.e. a terminal branch of the GSP (Pugsley, 1993). Moreover, folding of the secretion intermediates in the periplasm is required for the subsequent translocation across the outer membrane via specialized machinery, the secreton. Here, we demonstrate that this concept is wrong, and we provide compelling evidence for a mosaic mechanism for type II secretion. From our observations, the Tat system appears to be used as an alternative Sec-independent step to feed the secreton and, since Tat substrates may be folded in the cytoplasm, subsequent periplasmic folding is not necessarily required. Our data thus reveal a general scheme for a network of protein transport systems, i.e. the signal recognition particle (SRP) and Sec routes converge at the Sec translocon for inner membrane transport (Manting and Driessen, 2000), whereas the Tat and Sec routes converge at the secreton for outer membrane translocation. Now, the type II pathway should be defined as the secreton-dependent pathway (SDP) rather than the GSP. It is thus a unique protein secretion mechanism mediated by the secreton, which is fed by multiple pipelines including both the Tat and Sec systems.
Secondly, the spectrum of the Tat substrates is definitely broader than anticipated, and not dedicated exclusively to the export of metalloenzymes, since PlcN and PlcH do not belong to these categories of proteins.
Thirdly, the Tat pathway appears to be a new virulence determinant of P.aeruginosa, since evidence that the phospholipases C are significant virulence factors in bacterial pathogenesis is accumulating. These extracellular proteins may be particularly relevant to virulence of P.aeruginosa in the lung, where the substrate for these enzymes, phosphatidylcholine, is abundant in lung surfactant. Moreover, it was shown that a plcH/plcN double mutant has a reduced virulence (54%) in the lethal paralysis model of Caenorhabditis elegans (Darby et al., 1999), and a plcH mutant has a reduced mortality in a mouse burn model (40%) (Rahme et al., 2000).
Finally, in this study, we identified a Tat-dependently secreted protein that is the putative glycerophosphoryl diester phosphodiesterase GlpQ protein. Interestingly, this protein is phosphate starvation-inducible and secreted in B.subtilis (Antelmann et al., 2000). GlpQ is involved in the hydrolysis of deacylated phospholipids to glycerol 3-phosphate (G3P). It is thus interesting to speculate that G3P subsequently could be a substrate for secreted phosphate starvation-induced alkaline phosphatases that will liberate the inorganic phosphate.
We will investigate the virulence of a tatC mutant in these different models further as well as the role of the Tat system in P.aeruginosa pathogenesis. Since the Tat system is absent from all sequenced animal genomes, the Tat system requirement for the secretion of virulence factors paves a new way for developing new-generation safe antibiotics with the Tat system as the therapeutic target.
Materials and methods
Bacterial strains, plasmids and growth conditions
Escherichia coli C600, MC4100 and the isogenic tatABCDE mutant (Wexler et al., 2000) were used to probe the Tat-dependent processing of RR–pfColA. The E.coli TG1 strain was used for propagating plasmids. The P.aeruginosa PAK strain was used for construction of the tatC mutant derivative PAK-TC. Bacterial cells were grown in Luria broth at 30 or 37°C with aeration. Plasmids containing genes encoding pfColA or derivatives were prepared systematically and propagated together with pImTc encoding the immunity protein (Cai), which protects the cells from pfColA toxicity (Espesset et al., 1994). The recombinant plasmids were introduced into E.coli or P.aeruginosa by transformation or electroporation (Smith and Iglewski, 1989), respectively, and clones selected on L-agar or PIA (Difco Laboratories) plates containing antibiotics, respectively. The following antibiotic concentrations were used. For E.coli: ampicillin (Ap), 100 µg/ml; and tetracycline (Tc), 15 µg/ml. For P.aeruginosa: carbenicillin (Cb), 300 µg/ml; Tc, 200 µg/ml; and kanamycin (Km), 2000 µg/ml. Blood agar plates were purchased from Bio-Rad.
Construction of plasmid encoding RR-pfColA
Construction of pRR-pfColA67 encoding RR–pfColA was as follows. A DNA fragment encoding the TorA signal sequence was amplified by PCR. The resulting fragment was digested using EcoRI and NcoI, and ligated into the pfcolA-containing pCT1 plasmid (Espesset et al., 1996) digested with the same enzymes. The gene fusion subsequently was re-cloned into the broad host range vector pMMB67EH (Fürste et al., 1986) using the EcoRI and HindIII restriction sites, and placed under the control of the IPTG-inducible tac promoter.
In vivo expression in E.coli
In E.coli, expression of the genes cloned under control of the tac promoter was monitored by induction with IPTG. Cells were grown at 37°C up to an OD600 of 0.6. IPTG (1 mM) was added to the culture and cells were grown further for 30 min.
SDS–PAGE and immunoblotting
The E.coli and P.aeruginosa cells were grown as described. For E.coli, total cell samples were resuspended in SDS–PAGE buffer. For P.aeruginosa, cells and supernatants were separated by centrifugation. Cell fractionations were performed exactly as previously described (Wilderman et al., 2001). Proteins from the supernatant were precipitated with 12.5% trichloroacetic acid (TCA) for 1 h at 4°C and washed twice with 90% acetone. Pellets were subsequently resuspended in SDS–PAGE buffer and heated for 10 min at 95°C. The equivalent of 0.1 units of OD600 was used for protein separation by electrophoresis in polyacrylamide gels containing SDS. Immunoblotting was used for the detection of pfColA and PlcH with specific antibodies, diluted 1:1000 and 1:500, respectively, and anti-rabbit or anti-mouse peroxidase-conjugated antibodies as secondary antibodies, respectively. Revelation was performed using the ECL chemiluminescence system (Pierce).
Phospholipase C activity
Phospholipase C activity was measured as previously described (Cota-Gomez et al., 1997) using the synthetic phospholipase C substrate NPPC (Sigma-Aldrich, St Louis, MO).
Computer analysis of tat genes and Tat-dependent proteins
The identification of the tat genes was performed using E.coli TatA, B, C, D and E proteins as queries against P.aeruginosa genome and using the BLAST (http://www.ncbi.nlm.nih.gov/BLAST/unfinishedgenome.html) and ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) programs. The tatA gene starts at position 5 706 531 of the genome sequence and the tatC gene ends at position 5 708 038 (http://www.pseudomonas.com). The transmembrane domains in the Tat proteins were predicted using the TopPred 2 program (http://www.sbc.su.se/∼erikw/toppred2). Screening of the P.aeruginosa genome for twin-arginine signal peptide-containing proteins was performed at the PEDANT website (http://pedant.mips.biochem.mpg.de) using the RRxF motif.
Engineering of the tatC mutant strain and cloning of the tatC gene
A 450 bp internal DNA fragment of the tatC gene was amplified by PCR using a P.aeruginosa colony as template and two oligonucleotides: TATC1, 5′-CACCTGACCGAACTGCGTACG-3′; and TATC2, 5′-GTC AGGACGAAGTCCAGGTAG-3′. The DNA fragment was then cloned into plasmid PCR2.1 (Invitrogen), previously deleted for the gene encoding β-lactamase. The resulting plasmid, pTATC, which does not replicate in P.aeruginosa, was introduced in the PAK strain by electroporation in order to interrupt the tatC gene. The chromosomal integration of the plasmid was selected by plating on PIA containing 2000 µg/ml kanamycin. The insertion was confirmed by PCR using the oligonucleotide couple TATC1 and the M13 reverse primer.
The tatC gene was PCR amplified using a P.aeruginosa colony as template and two oligonucleotides: TATC5, 5′-TACCCACGCCGC CGACCAGCAC-3′; and TATC3, 5′-GCACGATCCGCCGACACA CGAAGT-3′. The 860 bp DNA fragment obtained was cloned at the EcoRI site of the pLAFR3 cosmid (Staskawicz et al., 1987), yielding pTATC.
Mass spectrometry
After crushing and washing the excised gel, the proteinaceous material was reduced and alkylated, respectively, by dithiothreitol and iodoacetamide in 100 mM NH4HCO3. Proteolytic digestion by trypsin was then carried out overnight at 37°C. The supernatant was collected, the salts were removed by flow through a R2 Poros column and the sample was analysed by mass spectrometry. The peptide mapping was achieved by using appropriate programs at http://www.matrixscience.com and http://www.expasy.ch/cgi-bin/peptident.p1.
Acknowledgments
Acknowledgements
We thank D.Moinier for mass spectrometry analysis, A.Vasil for her excellent technical assistance, and S.Lory for the gift of the PAKΔxcpR-Z strain. This work was supported by a grant to A.F. from the PICS program no. 848, grant QLK3-CT-1999 (Quality of Life and Management of Living Resources) to L.-F.W. from the European Union, and grant no. HL62608 to M.L.V. from the National Institute of Heart, Lung and Blood. R.V. is supported by a grant from the Fondation pour la Recherche Médicale (FRM) and B.I. by a grant from the Ministry of Research and Technology.
References
- Antelmann H., Scharf,C. and Hecker,M. (2000) Phosphate starvation-inducible proteins of Bacillus subtilis: proteomics and transcriptional analysis. J. Bacteriol., 182, 4478–4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berks B.C., Sargent,F. and Palmer,T. (2000) The Tat protein export pathway. Mol. Microbiol., 35, 260–274. [DOI] [PubMed] [Google Scholar]
- Bernhard M., Friedrich,B. and Siddiqui,R.A. (2000) Ralstonia eutropha TF93 is blocked in tat-mediated protein export. J. Bacteriol., 182, 581–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitter W., Koster,M., Latijnhouwers,M., de Cock,H. and Tommassen,J. (1998) Formation of oligomeric rings by XcpQ and PilQ, which are involved in protein transport across the outer membrane of Pseudomonas aeruginosa. Mol. Microbiol., 27, 209–219. [DOI] [PubMed] [Google Scholar]
- Bogsch E., Brink,S. and Robinson,C. (1997) Pathway specificity for a ΔpH-dependent precursor thylakoid lumen protein is governed by a ‘Sec-avoidance’ motif in the transfer peptide and a ‘Sec-incompatible’ mature protein. EMBO J., 16, 3851–3859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P., Tommassen,J. and Filloux,A. (1996) Role of the propeptide in folding and secretion of elastase of Pseudomonas aeruginosa. Mol. Microbiol., 19, 297–306. [DOI] [PubMed] [Google Scholar]
- Brok R., Van Gelder,P., Winterhalter,M., Ziese,U., Koster,A.J., de Cock,H., Koster,M., Tommassen,J. and Bitter,W. (1999) The C-terminal domain of the Pseudomonas secretin XcpQ forms oligomeric rings with pore activity. J. Mol. Biol., 294, 1169–1179. [DOI] [PubMed] [Google Scholar]
- Cota-Gomez A., Vasil,A.I., Kadurugamuwa,J., Beveridge,T.J., Schweitzer, H.P. and Vasil,M.L. (1997) PlcR1 and PlcR2 are putative calcium binding proteins required for secretion of the hemolytic phospholipase C of Pseudomonas aeruginosa. Infect. Immun., 65, 2904–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristobal S., de Gier,J.W., Nielsen,H. and von Heijne,G. (1999) Competition between Sec- and TAT-dependent protein translocation in Escherichia coli. EMBO J., 18, 2982–2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby C., Cosma,C.L., Thomas,J.H. and Manoil,C. (1999) Lethal paralysis of Caenorhabditis elegans by Pseudomonas aeruginosa. Proc. Natl Acad. Sci. USA, 96, 15202–15207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doring G. (1997) Cystic fibrosis respiratory infections: interactions between bacteria and host defence. Monaldi Arch. Chest Dis., 52, 363–366. [PubMed] [Google Scholar]
- Douglas C.M., Guidi-Rontani,C. and Collier,R.J. (1987) Exotoxin A of Pseudomonas aeruginosa: active, cloned toxin is secreted into the periplasmic space of Escherichia coli. J. Bacteriol., 169, 4962–4966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Economou A. (2000) Bacterial protein translocase: a unique molecular machine with an army of substrates. FEBS Lett., 476, 18–21. [DOI] [PubMed] [Google Scholar]
- Economou A. and Wickner,W. (1994) SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell, 78, 835–843. [DOI] [PubMed] [Google Scholar]
- Espesset D., Corda,Y., Cunningham,K., Benedetti,H., Lloubes,R., Lazdunski,C. and Geli,V. (1994) The colicin A pore-forming domain fused to mitochondrial intermembrane space sorting signals can be functionally inserted into the Escherichia coli plasma membrane by a mechanism that bypasses the Tol proteins. Mol. Microbiol., 13, 1121–1131. [DOI] [PubMed] [Google Scholar]
- Espesset D., Duche,D., Baty,D. and Geli,V. (1996) The channel domain of colicin A is inhibited by its immunity protein through direct interaction in the Escherichia coli inner membrane. EMBO J., 15, 2356–2364. [PMC free article] [PubMed] [Google Scholar]
- Feilmeier B.J., Iseminger,G., Schroeder,D., Webber,H. and Phillips,G.J. (2000) Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol., 182, 4068–4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filloux A., Michel,G. and Bally,M. (1998) GSP-dependent protein secretion in Gram-negative bacteria: the Xcp system of Pseudomonas aeruginosa. FEMS Microbiol. Rev., 22, 177–198. [DOI] [PubMed] [Google Scholar]
- Frank D.W. (1997) The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol., 26, 621–629. [DOI] [PubMed] [Google Scholar]
- Fürste J.P., Pansegrau,W., Frank,R., Blocker,H., Scholz,P., Bagdasarian,M. and Lanka,E. (1986) Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene, 48, 119–131. [DOI] [PubMed] [Google Scholar]
- Gray G.L., Berka,R.M. and Vasil,M.L. (1981) A Pseudomonas aeruginosa mutant non-derepressible for orthophosphate-regulated proteins. J. Bacteriol., 147, 675–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P.V. (1974) Extracellular toxins of Pseudomonas aeruginosa. J. Infect. Dis., 130, S94–S99. [DOI] [PubMed] [Google Scholar]
- Manting E.H. and Driessen,A.J.M. (2000) Escherichia coli translocase: the unraveling of a molecular machine. Mol. Microbiol., 37, 226–238. [DOI] [PubMed] [Google Scholar]
- Manting E.H., van Der Does,C., Remigy,H., Engel,A. and Driessen,A.J. (2000) SecYEG assembles into a tetramer to form the active protein translocation channel. EMBO J., 19, 852–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez A., Ostrovsky,P. and Nunn,D.N. (1998) Identification of an additional member of the secretin superfamily of proteins in Pseudomonas aeruginosa that is able to function in type II protein secretion. Mol. Microbiol., 28, 1235–1246. [DOI] [PubMed] [Google Scholar]
- Ostroff R.M., Vasil,A.I. and Vasil,M.L. (1990) Molecular comparison of a nonhemolytic and a hemolytic phospholipase C from Pseudomonas aeruginosa. J. Bacteriol., 172, 5915–5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poquet I., Faucher,D. and Pugsley,A.P. (1993) Stable periplasmic secretion intermediate in the general secretory pathway of Escherichia coli. EMBO J., 12, 271–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugsley A.P. (1993) The complete general secretory pathway in gram-negative bacteria. Microbiol. Rev., 57, 50–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme L.G. et al. (2000) Plants and animals share functionally common bacterial virulence factors. Proc. Natl Acad. Sci. USA, 97, 8815–8821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini C.L., Ize,B., Chanal,A., Muller,M., Giordano,G. and Wu,L.F. (1998) A novel sec-independent periplasmic protein translocation pathway in Escherichia coli.EMBO J., 17, 101–112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santini C.L., Bernadac,A., Zhang,M., Chanal,A., Ize,B., Blanco,C. and Wu,L.F. (2001) Translocation of jellyfish green fluorescent protein via the TAT system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem., 276, 8159–8164. [DOI] [PubMed] [Google Scholar]
- Settles A.M., Yonetani,A., Baron,A., Bush,D.R., Cline,K. and Martienssen,R. (1997) Sec-independent protein translocation by the maize Hcf106 protein. Science, 278, 1467–1470. [DOI] [PubMed] [Google Scholar]
- Shortridge V.D., Lazdunski,A. and Vasil,M.L. (1992) Osmoprotectants and phosphate regulate expression of phospholipase C in Pseudomonas aeruginosa. Mol. Microbiol., 6, 863–871. [DOI] [PubMed] [Google Scholar]
- Smith A.W. and Iglewski,B.H. (1989) Transformation of Pseudomonas aeruginosa by electroporation. Nucleic Acids Res., 17, 10509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck,D., Keen,N. and Napoli,C. (1987) Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J. Bacteriol., 169, 5789–5794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stover C.K. et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature, 406, 959–964. [DOI] [PubMed] [Google Scholar]
- Tommassen J., Eiglmeier,K., Cole,S.T., Overduin,P., Larson,T.J. and Boos,W. (1991) Characterization of two genes, glpQ and ugpQ, encoding glycerophosphoryl diesterphosphodiesterases of Escherichia coli. Mol. Gen. Genet., 226, 321–327. [DOI] [PubMed] [Google Scholar]
- Tommassen J., Filloux,A., Bally,M., Murgier,M. and Lazdunski,A. (1992) Protein secretion in Pseudomonas aeruginosa. FEMS Microbiol. Rev., 9, 73–90. [DOI] [PubMed] [Google Scholar]
- Voulhoux R., Taupiac,M.P., Czjzek,M., Beaumelle,B. and Filloux,A. (2000) Influence of deletions within domain II of exotoxin A on its extracellular secretion from Pseudomonas aeruginosa. J. Bacteriol., 182, 4051–4058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voulhoux R., Lazdunski,A. and Filloux,A. (2001) Colicin A hybrids: a genetic tool for the selection of type II secretion-proficient Pseudomonas strains. EMBO Rep., 2, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wexler M., Sargent,F., Jack,R.L., Stanley,N.R., Bogsch,E.G., Robinson,C., Berks,B.C. and Palmer,T. (2000) TatD is a cytoplasmic protein with DNase activity. No requirement for TatD family proteins in sec-independent protein export. J. Biol. Chem., 275, 16717–16722. [DOI] [PubMed] [Google Scholar]
- Wilderman P.J., Vasil,A.I., Johnson,Z. and Vasil,M.L. (2001) Genetic and biochemical analyses of a eucaryotic-like phospholipase D of Pseudomonas aeruginosa suggest horizontal acquisition and a role for persistence in a chronic pulmonary infection model. Mol. Microbiol., 39, 291–303. [DOI] [PubMed] [Google Scholar]
- Wu L.F., Ize,B., Chanal,A., Quentin,Y. and Fichant,G. (2000) Bacterial twin-arginine signal peptide-dependent protein translocation pathway: evolution and mechanism. J. Mol. Microbiol. Biotechnol., 2, 179–189. [PubMed] [Google Scholar]