Short abstract
cDNA microarray-derived expression profiles of MMTV-Wnt-1 and MMTV-Neu transgenic mice reveal several hundred genes to be differentially expressed at each stage of breast tumor development.
Abstract
Background
In human breast cancer normal mammary cells typically develop into hyperplasia, ductal carcinoma in situ, invasive cancer, and metastasis. The changes in gene expression associated with this stepwise progression are unclear. Mice transgenic for mouse mammary tumor virus (MMTV)-Wnt-1 exhibit discrete steps of mammary tumorigenesis, including hyperplasia, invasive ductal carcinoma, and distant metastasis. These mice might therefore be useful models for discovering changes in gene expression during cancer development.
Results
We used cDNA microarrays to determine the expression profiles of five normal mammary glands, seven hyperplastic mammary glands and 23 mammary tumors from MMTV-Wnt-1 transgenic mice, and 12 mammary tumors from MMTV-Neu transgenic mice. Adipose tissues were used to control for fat cells in the vicinity of the mammary glands. In these analyses, we found that the progression of normal virgin mammary glands to hyperplastic tissues and to mammary tumors is accompanied by differences in the expression of several hundred genes at each step. Some of these differences appear to be unique to the effects of Wnt signaling; others seem to be common to tumors induced by both Neu and Wnt-1 oncogenes.
Conclusion
We described gene-expression patterns associated with breast-cancer development in mice, and identified genes that may be significant targets for oncogenic events. The expression data developed provide a resource for illuminating the molecular mechanisms involved in breast cancer development, especially through the identification of genes that are critical in cancer initiation and progression.
Background
Gene expression arrays are being widely used to improve the classification of human cancers and to improve our understanding of the molecular changes associated with carcinogenesis [1,2]. However, their use in defining expression patterns in tumor evolution and in correlating genotypes with phenotypes has been limited because of the poor availability of tissues at different stages in cancer development and because of the great diversity of genetic backgrounds among individuals [3-5]. Mouse models of cancer have advantages for exploring the use of this method: a partially defined neoplastic genotype, relatively uniform genetic background, and ample sources of tissue samples from different stages in mammary tumor evolution. Some features of expression profiles identified in mouse mammary tumors are shared by patterns seen in RNA from human tumors [6]. By comparing expression patterns of mammary tumors in six different transgenic mouse models, Desai and coworkers [7] have shown that the initiating pathway determines a distinctive expression phenotype in tumors. In addition, using proteins as markers of cell phenotypes, we showed that initiating oncogenes determine the developmental status of mammary tumor cells [8].
Members of the Wnt gene family were discovered as proto-oncogenes that are frequently activated in mammary tumors arising in mice infected with mouse mammary tumor virus (MMTV) [9,10]. Wnt genes encode extracellular matrix binding proteins that control many developmental processes, including cell fate specification and stem cell renewal; they are also involved in mammary morphogenesis and progenitor cell renewal [11,12]. Made as secreted glycoproteins, Wnt proteins exert their biologic effects by binding to at least two membrane receptors, namely the frizzled and low-density lipoprotein receptor related proteins. As a result of signaling via the 'canonical' pathway, β-catenin is stabilized, translocates to the nucleus, and transactivates different sets of genes depending on the cellular context [13].
Mice expressing Wnt-1 under the control of the enhancer elements in the MMTV long terminal repeat develop extensive hyperplasias of the mammary glands at prepubertal ages, mammary tumors at a median age of 6 months, and sometimes pulmonary metastases ([14]; Podsypanina K, unpublished observations). Tumors in these MMTV-Wnt-1 transgenic mice appear to arise from progenitor cells in the mammary gland, because many cells in both hyperplastic and neoplastic lesions express putative progenitor cell markers (such as Sca-1 and keratin-6) and efflux fluorescent Hoechst 33342 dye - a property that has been associated with stem cells in the hematopoietic system [8,15]. The resulting tumors also contain tumor cells with myoepithelial as well as epithelial markers, implying that they arise from a progenitor cell that gives rise to both lineages [8,15]. Because at least some human breast cancers are also thought to arise from progenitor cells [16], it is important to define better the molecular events that lead to tumor formation in this line of mice.
Here we report the expression profiles at different steps of tumor evolution in the MMTV-Wnt-1 transgenic model, and we compare these profiles with those in the MMTV-Neu transgenic model. We addressed the following questions. Can we follow progression in MMTV-Wnt-1 transgenic mice from hyperplasia to primary tumor? Are differences apparent between tumors induced by different transgenic oncogenes? Can we distinguish tumors with additional genetic alterations in MMTV-Wnt-1 transgenic mice from those without other known genetic alterations?
Results and discussion
Mammary tumors in MMTV-Wnt-1 transgenic mice have an expression profile distinct from that seen in mammary tumors induced by MMTV-Neu
Comparison of expression profiles of tumors from several transgenic models has led to the identification of expression signatures for different oncogenic pathways [7]. In order to determine whether tumors from MMTV-Wnt-1 transgenic mice also have a distinctive expression profile, we determined the profiles of 23 mammary tumors from MMTV-Wnt-1 transgenic mice and, for comparison, 12 mammary tumors from mice carrying the MMTV-Neu transgene (Tables 1 and 2 provide sample information and a list of all comparisons). Neu (ErbB2/HER2), a proto-oncogene that is amplified in approximately 25% of human breast cancers [17], encodes a member of the epidermal growth factor receptor family of receptor tyrosine kinases [18]. It activates signaling pathways different from those activated by Wnt-1, and the two oncogenes can collaborate in mammary tumorigenesis [19].
Table 1.
Tissue samples
| Tissue type | Abbreviation | Number of samples | Age (weeks) | Array size |
| Normal virgin mammary gland | VMG or V | 5 | 9 | 15k |
| Hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice | WntH | 7 | 9 | 15k |
| Mammary tumors from MMTV-Wnt-1 transgenic mice | WntT | 33 | 9-56 | 15k (23 arrays), 8.7k (10 arrays) |
| Mammary tumors from MMTV-Neu transgenic mice | NeuT | 12 | 32-60 | 15k |
| Normal fat tissue | Fat | 3 | 12 | 15k |
| Mammary tumors from MMTV-Wnt-1 transgenic/P53-/- mice | WntT/p53-/- | 6 | 9-14 | 8.7k |
| Mammary tumors from MMTV-Wnt-1 transgenic/Pten+/- mice with LOH at the Pten locus | WntT/Pten+/-LOH | 3 | 12-21 | 8.7k |
LOH, loss of heterozygosity; MMTV, mouse mammary tumor virus.
Table 2.
Numbers of genes that are differentially expressed
| Comparisons | Number of genes differentially expressed | |||
| Tissue A (number of samples) | Tissue B (number of samples) | Total | Up | Down |
| WntH (7) | VMG (5) | 584 | 121 | 463 |
| WntT (23) | WntH (7) | 388* | 112 | 276 |
| WntT (23) | NeuT (12) | 1,296 | 624 | 672 |
| NeuT (12) | VMG (5) | 1,263* | 419 | 844 |
| WntT, H-ras mutant (12) | WntT, H-ras wild-type (9) | 40 | 31 | 9 |
| WntT/p53-/- | WntT (10) | 113 | 43 | 70 |
| WntT/Pten+/- LOH (3) | WntT (10) | 115 | 45 | 70 |
Expression ratio is computed by dividing the average expression level of the A group by the average expression level of the B group. The numbers of differentially expressed genes were determined by random permutation (P < 0.001), as described in the Materials and method section. *In selected comparisons, to reduce potential false signals due to stromal effects, the genes that were less than three-fold different in expression were filtered out from the listed total number of genes. LOH, loss of heterozygosity.
The expression profiles of these two sets of tumors were clearly separated into two groups by unsupervised average linkage hierarchical clustering analysis (Figure 1), suggesting that the global expression patterns of these two sets of tumors differ significantly. This finding extends previous reports of significant divergence in histopathobiology, cellular composition, and possibly the cell types of origin between these two groups of tumors [8,20,21].
Figure 1.
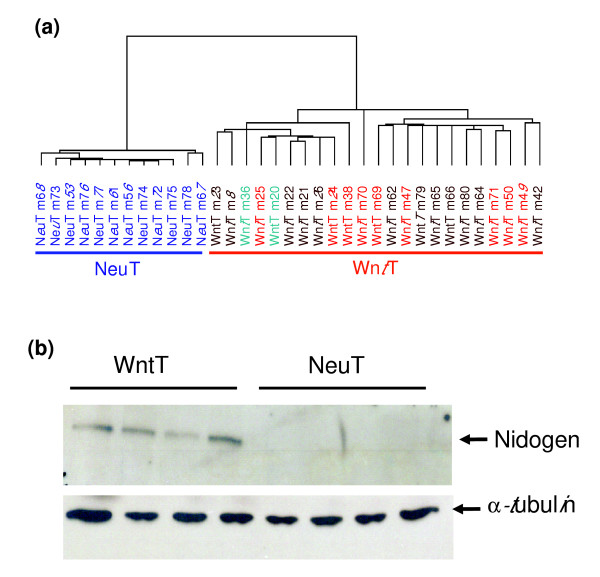
Gene expression in mammary tumors from MMTV-Wnt-1 versus MMTV-Neu transgenic mice. (a) Dendrogram of 35 mammary tumors analyzed by average linkage hierarchical clustering analysis using 1,932 genes selected for high variability across all tumors. 15k arrays were used. The status of Ha-Ras on MMTV-Wnt-1-induced tumors is color coded: red, wild-type; brown, mutant; green, unknown. (b) Western blot analysis for nidogen protein expression on representative mammary tumors from MMTV-Wnt-1 and MMTV-Neu transgenic mice. MMTV, mouse mammary tumor virus; NeuT, mammary tumors from MMTV-Neu transgenic mice; WntT, mammary tumors from MMTV-Wnt-1 transgenic mice.
In an effort to identify genes that are specifically dysregulated in tumors induced by MMTV-Wnt-1, we performed a permutation t-test (see Materials and methods, below, for details) on these two groups of array data. In total, 1,296 genes were differentially expressed between MMTV-Wnt-1-induced and MMTV-Neu-induced tumors (P < 0.001; Table 2 and Additional data file 1). Among the 1,296 genes that we found to be differentially expressed between Wnt-1-induced and Neu-induced tumors, 842 genes are represented in the 8.7k chips used in the previous report [7]. In that study, 672 genes were found to be differentially expressed among tumors from MMTV-Neu, MMTV-Ha-Ras, MMTV-c-Myc, MMTV-polyoma middle T antigen, C3(1)/simian virus 40 T/t antigen, and Wap-simian virus 40 T/t antigen transgenic mice using the 8.7k chips. Comparing the 842 differentially expressed genes in the present study with the 672 genes from the earlier study, we found that 165 genes were present in both lists (Additional data file 1), including 91 of the 178 genes (51%) reported as the Neu-Ras-polyoma middle T antigen cluster [7]. Examples of these 91 genes include Rap1-GTPase activating protein 1, matrix metalloproteinase 15, and CD81 (Additional data file 1).
It should be noted that the MMTV-Wnt-1 transgenic mice had a mixed genetic background that was mostly FVB (>75%), whereas MMTV-Neu transgenic mice were on a pure FVB background. Although this small variation in genetic background between these two groups of mice is unlikely to account for the differences in expression profiles we detected, we cannot exclude the possibility that some of the genes identified by this analysis might be due to variation in genetic background.
A panel of 652 genes were reported to be differentially expressed between MMTV-Neu-induced tumors and normal virgin mammary glands in the study of Desai and coworkers [7] using the 8.7k chips (> two-fold). In the present study comparing 12 tumors from MMTV-Neu transgenic mice and five nontransgenic normal virgin mammary glands using the 15k chips, 1,263 genes were differentially expressed (P < 0.001, more than three-fold; Table 2). Among these 1,263 genes, 626 genes were represented in the 8.7k arrays used by Desai and coworkers. Of these 626 genes, 225 (35%) overlapped with the 652 genes reported to be differentially expressed between MMTV-Neu-induced tumors and normal virgin mammary glands in the study conducted by Desai and colleagues. We consider this to be an acceptable level of reproducibility, considering the multiple differences in the generation of the two data sets (including differences in reference RNAs, array prints, age of the virgin mammary glands, and sample size).
Genes that were more highly expressed (P < 0.001) in MMTV-Wnt-1-induced tumors than in MMTV-Neu-induced tumors include genes reported to be transcriptional targets of Wnt signaling [22-26] such as cyclin D1 (2.0-fold), c-Myc (2.0-fold), frizzled 7 (2.1-fold), and Wnt-5a (9.2-fold; Additional data file 1). Wnt-5b, another member of the Wnt family, was also more highly expressed (3.7-fold) in tumors from MMTV-Wnt-1 transgenic mice than in tumors from MMTV-Neu transgenic mice; it remains to be determined whether this Wnt member is also a transcriptional target of Wnt signaling.
Retinoic acid signaling has been reported to synergize with Wnt signaling to induce gene expression [27,28]. Retinoic acid receptor and Stra6, a gene activated by the addition of retinoids to cultured cells [29], have also been suggested to be targets of Wnt signaling [27]. Consistent with these reports, we found higher level of Stra6 (P < 0.001, 9.0-fold) in MMTV-Wnt-1-induced tumors than in MMTV-Neu-induced tumors. In addition, cellular retinol binding protein (RBP)1, a gene related to retinoic acid signaling, was also more highly expressed (P < 0.001, two-fold) in MMTV-Wnt-1-induced tumors than in MMTV-Neu-induced tumors, which is consistent with our recent report that RBP1 is induced by β-catenin [30].
MMTV-Wnt-1-induced tumors contain both epithelial and myoepithelial cells in approximately equal numbers, unlike tumors induced by the MMTV-Neu transgene, which contain only epithelial tumor cells [8,21,31]. Consistent with these reports, we observed higher expression levels (P < 0.001) of myoepithelial markers, including calponin 1 (12.5-fold) and calponin 2 (2.5-fold and 4.0-fold for two separate clones), in tumors from MMTV-Wnt-1 transgenic than in tumors from MMTV-Neu transgenic mice (Additional data file 1). Consistent with earlier reports that tumors may arise from mammary progenitor cells in MMTV-Wnt-1 transgenic mice [8,15], we found that RNA encoding the candidate progenitor cell markers keratin 6 (13-fold), tenascin (3.1-fold), osteoblast specific factor 2 (2.0-fold), insulin-like growth factor binding protein 7 (2.0-fold), and nidogen 1 (1.8-fold) [8,32] were more abundant (P < 0.001) in tumors from MMTV-Wnt-1 transgenic mice. Using immunoassays, we demonstrated that keratin 6 and nidogen proteins are expressed at higher level in MMTV-Wnt-1-induced tumors than in MMTV-Neu-induced tumors (Figure 1b) [8].
Expression profiles are similar among mammary tumors with additional genetic alterations in MMTV-Wnt-1 transgenic mice
The distinct patterns of genes expressed in MMTV-Wnt-1-induced and MMTV-Neu-induced tumors described in the preceding section suggest that initiating oncogenes strongly influence gene expression in the tumors arising in these two models. We have observed that other genetic events accelerate tumorigenesis in MMTV-Wnt-1 transgenic mice [14,33]. We next evaluated whether the events that mediate acceleration are reflected in the gene expression patterns.
We recently reported that approximately 50% of mammary tumors in MMTV-Wnt-1 transgenic mice have activating mutations in the Ha-Ras locus [19]. Thus, we first considered whether tumors carrying mutant Ha-Ras have an expression profile distinct from that observed in tumors that are wild-type at the Ha-Ras locus. We sequenced Ha-Ras cDNA to seek mutations in 21 out of the 23 MMTV-Wnt-1-induced tumors: 12 tumors carry Ha-Ras mutations and nine have only Ha-Ras wild-type alleles (Figure 1a). Tumors with and without Ha-Ras mutations did not have distinct global expression profiles (Figure 1a). Independent multidimensional scaling (MDS) and hierarchical clustering of these 21 tumors based on expression profiles also did not separate them according to Ha-Ras status (data not shown). Nevertheless, permutation t test identified 40 genes differentially expressed between tumors bearing wild-type Ha-Ras and those carrying a mutant Ha-Ras (Table 2 and Additional data file 2). This is more than expected (P < 0.001) but many fewer than we saw in our earlier comparison between MMTV-Wnt-1-induced and MMTV-Neu-induced tumors. In addition, the average fold difference is much smaller (Additional data file 2) than that in the earlier comparison.
We previously determined that loss of either p53 or Pten accelerates mammary tumorigenesis in MMTV-Wnt-1 transgenic mice [34,35]. To further investigate the influence of these genetic alterations on expression patterns in MMTV-Wnt-1-induced tumors, we determined the expression profiles of six tumors from MMTV-Wnt-1 transgenic mice that were p53 null and three tumors from MMTV-Wnt-1 transgenic/Pten+/- mice that had lost the wild-type allele of Pten (i.e. loss of heterozygosity). When genes in the 8.7k array data set from these two groups of tumors and those from 10 tumors from MMTV-Wnt-1 transgenic mice that were otherwise wild-type were subjected to analysis by MDS or unsupervised hierarchical clustering, the three groups of samples could not be separated from each other (Figure 2 and data not shown). These findings suggest that the global expression profiles of MMTV-Wnt-1 tumors carrying different additional genetic alterations cannot be distinguished.
Figure 2.
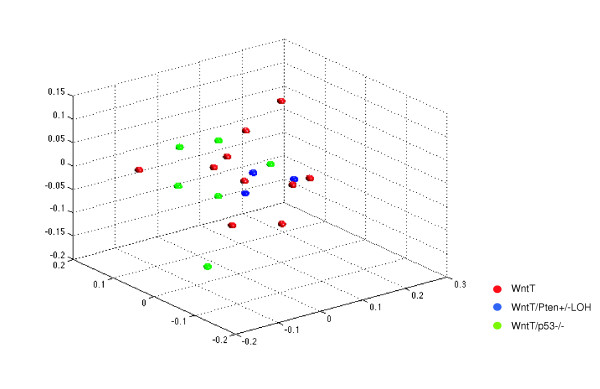
Multidimensional scaling analysis of 18 tumor samples of indicated genotypes. 8.7k arrays were used. MMTV, mouse mammary tumor virus; WntT, mammary tumors from MMTV-Wnt-1 transgenic mice; WntT/Pten+/- loss of heterozygosity (LOH), mammary tumors from MMTV-Wnt-1 transgenic/Pten+/- mice with Pten loss of heterozygosity; WntT/P53-/-, mammary tumors from MMTV-Wnt-1/P53-/- mice.
Permutation t test identified 113 genes that were differentially expressed (p < 0.001) between tumors from MMTV-Wnt-1 transgenic mice and those from MMTV-Wnt-1 transgenic/p53-/- mice (Table 2). Among the 113 genes, 43 were upregulated, and 70 were downregulated in the latter set of tumors (Additional data file 3). Examples of the upregulated genes are cyclin D2 (3.7-fold), Myb (2.9-fold and 2.8-fold for two separate clones), Bcl11a (1.5-fold), and Pbx3 (1.8-fold average), which promote proliferation or survival. Examples of the downregulated genes are CD59a antigen (two-fold), a potential p53 target [36], the Rb1 tumor suppressor gene, and the Met proto-oncogene. Using similar analyses, we found that 115 genes were differentially expressed (P < 0.001) between tumors from MMTV-Wnt-1 transgenic mice and those from MMTV-Wnt-1/Pten+/- mice with Pten loss of heterozygosity (Table 2). Forty-five were upregulated, and 70 of them were downregulated in the latter set of tumors (Additional data file 4). Interestingly, among the downregulated genes is tensin (two-fold), a cell adhesion molecule that is related to Pten. Similar to the comparison between Ha-Ras mutant and Ha-Ras wild-type tumors in MMTV-Wnt-1 transgenic mice, the number of genes differentially expressed and the average fold difference were much smaller in the above two comparisons than in the comparison between MMTV-Wnt-1-induced and MMTV-Neu-induced tumors (Additional data files 1, 3, and 4).
Collectively, these findings suggest that tumors from MMTV-Wnt-1 transgenic mice are similar to each other in their global expression profiles, regardless of whether the tumors have additional genetic alterations. It is not known whether the modest differences in RNA levels we identified among these groups of tumors explain the accelerating effects of these alterations on tumorigenesis in MMTV-Wnt-1 transgenic mice. We plan to test some of these changes by expressing cDNAs in mammary glands in Wnt-1 transgenic mice using TVA-mediated somatic gene transfer technology [37].
Distinct changes in gene expression accompany the evolution from normal mammary glands to hyperplasias and to tumors in MMTV-Wnt-1 transgenic mice
Hyperplastic lesions are widespread in MMTV-Wnt-1 transgenic mice before the development of mammary tumors [14]. To determine whether unique gene expression patterns accompany the evolution from normal mammary cells to hyperplasias and then to tumors, we compared expression profiles among mammary glands of nontransgenic virgin mice, hyperplastic mammary glands, and mammary tumors from MMTV-Wnt-1 transgenic mice. Unsupervised hierarchical clustering analysis and MDS showed that expression profiles from these three groups of tissues were separated from each other (Figure 3a and data not shown). The difference between hyperplastic and normal glands is unlikely to be due to decreased contribution of stromal RNA in the preparation of RNAs from the hyperplastic glands from MMTV-Wnt-1 transgenic mice, because the expression levels of epithelial and myoepithelial marker genes (keratin 19, calponin 1, and calponin 2) were not significantly statistically different between hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice and mammary glands from age-matched nontransgenic virgins.
Figure 3.
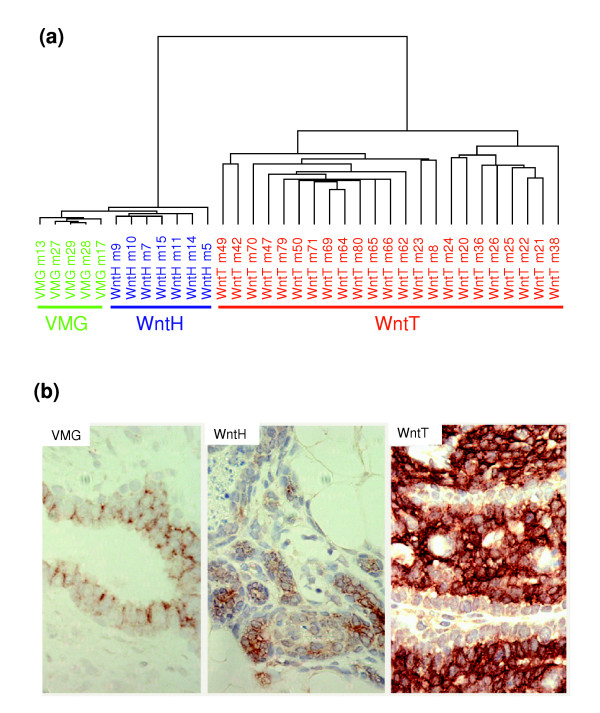
Gene expression in mammary tumor evolution in MMTV-Wnt-1 transgenic mice. (a) Dendrogram of 35 samples analyzed by average linkage hierarchical clustering analysis, using 3,359 genes selected for high variability across all samples. 15k arrays were used. (b) Immunohistochemical staining for c-Kit in the indicated tissue sections. A 20× objective was used. MMTV, mouse mammary tumor virus; VMG, virgin mammary glands from nontransgenic mice; WntT, mammary tumors from MMTV-Wnt-1 transgenic mice; WntH, hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice.
In total, 584 genes were differentially expressed (P < 0.001, Table 1) between hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice and normal mammary glands from nontransgenic littermates. Among these 584 genes, 121 were more highly expressed in the hyperplastic glands (Additional data file 5), which includes some of the known transcriptional targets of Wnt signaling such as c-Myc (3.6-fold) and frizzled 7 (2.1-fold). This list may therefore provide an important starting point for confirming mammary-specific target genes and for discovering novel in vivo targets of Wnt signaling. In fact, two genes in this list, namely RBP1 (2.9-fold) and tumor-associated calcium signal transducer (3.5-fold), were shown to be upregulated by β-catenin in 293 cells in our recent studies [30].
One of the greatest challenges in identifying specific genes and expression patterns associated with the evolution from hyperplastic glands to tumors is the change in cellular composition. Normal and MMTV-Wnt-1-induced hyerplastic ductal trees are embedded in stroma, but tumors often contain much less stroma. Thus, the differential contribution of RNA from the stromal cells may skew array analysis, which is based on total RNA content. However, stromal cells are mostly large adipocytes whose RNA to mass ratio is small; thus, the relative contribution of RNA from these cells is probably much less than it appears to be from histologic assessment. The average expression level of epithelial and myoepithelial markers (keratin 19, calponin 1, and calponin 2) was 2.6-fold higher in the tumors (which contain very few stromal cells) than in the hyperplastic tissues, suggesting that 38% (1/2.6 = 38%) of the RNA in the hyperplastic tissues might come from the ducts and alveoli. Thus, to eliminate genes that were not truly differentially expressed, we filtered out any genes that were less than three-fold different in our comparison between tumors and hyperplastic glands in Table 2.
Based on the above calculation, approximately 62% of the RNAs from hyerplastic mammary glands might come from adipocyte-rich stroma. Thus, tumor samples, which have very little contribution from adipocytes, may appear to have downregulated the genes that are associated with adipocytes. In order to identify these genes, we compared the expression profiles of a set of three fat samples with those of the 35 mammary tumors from MMTV-Wnt-1 and MMTV-Neu transgenic mice. Expression of 741 genes was at least three-fold or higher (P < 0.001) in fat than in the mammary tumors (Table 2 and Additional data file 6). These include published fat-specific genes (Additional data file 6), such as fat-specific gene 27, lipoprotein lipase, CD36, carbonic anhydrases, and solute carrier family members [7]. We note these genes in our table comparing hyperplastic mammary glands with tumors (Additional data file 7).
In total, 1,372 genes were differentially expressed (P < 0.001) between tumors and hyperplastic glands from MMTV-Wnt-1 transgenic mice. Among them, expression levels for 388 differed by at least three-fold (Additional data file 7). This subgroup is likely to contain genes that are important for the evolution of hyperplastic lesions into tumors, including genes that are required for tumor cell proliferation and survival. One such candidate is c-Kit, a proto-oncogene that is frequently overexpressed in cancers and that encodes a receptor that activates both Ras and Akt pathways. The expression of c-Kit was 3.6-fold higher in tumors than in hyperplastic lesions (Additional data file 7), although it was similarly expressed in normal virgin glands and hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice. This is consistent with a recent report that c-Kit is highly expressed in the basal group of human breast tumors compared to other groups [38]. Using immunohistochemical staining, c-Kit protein was barely detectable in normal mammary glands from nontransgenic mice and in hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice, but was readily and widely detected in the tumor samples from MMTV-Wnt-1 transgenic mice (Figure 3b).
Some of the genes that were differentially expressed between mammary tumors and hyperplastic glands in MMTV-Wnt-1 transgenic mice may be needed for evolution of tumors induced by both Wnt-1 and other oncogenes. Other genes may be uniquely important for induction of tumors from hyperplastic glands in MMTV-Wnt-1 transgenic mice. For example, certain signaling pathways may need to be activated in hyperplastic cells in MMTV-Wnt-1 transgenic mice before a tumor will form, but they may be optional for tumorigenesis initiated by other oncogenes. To discover genes that might be uniquely important for tumors to develop in hyperplastic glands in MMTV-Wnt-1 transgenic mice, we compared the 388 genes that we found to be differentially expressed between tumors and hyperplastic glands from MMTV-Wnt-1 transgenic mice with the 1,296 genes that we found to be differentially expressed between tumors from MMTV-Wnt-1 transgenic and MMTV-Neu transgenic mice. Fifty-six genes corresponding to 59 cDNA clones in the former group were shared in the latter group (Table 3), suggesting they might be specifically involved in neoplastic progression in MMTV-Wnt-1 transgenic mice. Among these 56 genes, 23 were more highly expressed and 33 were expressed at lower level in MMTV-Wnt-1-induced tumors than in either MMTV-Wnt-1-induced hyperplasia or MMTV-Neu-induced tumors (Table 3). The upregulated genes (P < 0.001) include TNFRSF19 (3.5-fold), NGFR (3.6-fold), apolipoprotein D (4.7-fold), and Wnt5a (4.4-fold), and the downregulated genes (P < 0.001) include BNIP3 (2.5-fold) and caveolin (2-, 3.3-, and 3.3-fold for three separate clones). Of note, apolipoprotein D has been reported to be upregulated in a subset of human breast cancers [39], and caveolin 1, a negative regulator of the Ras-p42/p44 mitogen-activated protein kinase cascade, has been reported to inhibit growth in human breast cancer cells [40].
Table 3.
List of genes potentially specifically involved in neoplastic progression in MMTV-Wnt-1 transgenic mice
| Expression ratio | |||||
| Image ID | Gene name | Symbol | WntH/V | WntT/WntH | WntT/NeuT |
| 474107 | Tumor necrosis factor receptor superfamily, member 19 | Tnfrsf19 | 2.56 | 3.48 | 6.23 |
| 406897 | Retinol binding protein 1, cellular | Rbp1 | 2.93 | 3.02 | 1.96 |
| 572428 | Cell cycle cyclin D1 Mm.35804 Cyclin D1 | Ccnd1 | = | 5.53 | 1.97 |
| 536306 | Hormone/GF growth/diff 1 Mm.22621 Procollagen type I α1 | Col1a1 | = | 3.04 | 3.27 |
| 536526 | ESTs | = | 8.8 | 5.95 | |
| 329780 | Cadherin 3 | Cdh3 | = | 3.74 | 3.96 |
| 427360 | Mus musculus, clone IMAGE:3590270, mRNA, partial cds | LOC192176 | = | 3.5 | 2.44 |
| 638805 | Expressed sequence AI504637 | = | 3.3 | 2.27 | |
| 355990 | Chondroitin sulfate proteoglycan 2 | Cspg2 | = | 3.62 | 5.05 |
| 574500 | ESTs | = | 4.13 | 1.65 | |
| 476431 | Nerve growth factor receptor | Ngfr | = | 3.61 | 6.34 |
| 680894 | Glial cell line derived neurotrophic factor family receptor α1 | Gfra1 | = | 4.74 | 4.05 |
| 482943 | Odd-skipped related 1 (Drosophila) | Osr1 | = | 4.1 | 3.65 |
| 695687 | Calponin 2 | Cnn2 | = | 3.49 | 2.54 |
| 335572 | Dihydropyrimidinase-like 3 | = | 3.27 | 7.61 | |
| 621246 | Interferon concensus sequence binding protein | Icsbp | = | 3.38 | 4.91 |
| 482170 | Four jointed box 1 (Drosophila) | Fjx1 | = | 4.13 | 3.78 |
| 468019 | IL-17b | Il17b | = | 4.27 | 5.37 |
| 373716 | Tumor-associated calcium signal transducer 2 | Tacstd2 | = | 4.16 | 3.69 |
| 479405 | ESTs | = | 3.01 | 4.95 | |
| 722262 | Wingless-related MMTV integration site 5A | Wnt5a | = | 4.44 | 9.23 |
| 719592 | GATA binding protein 2 | Gata2 | = | 6.06 | 10.54 |
| 1247541 | Apolipoprotein D | Apod | = | 4.67 | 3.86 |
| 331186 | Caveolin, caveolae protein, 22 kDa | Cav1 | 0.31 | 0.29 | 3.67 |
| 596968 | Caveolin, caveolae protein, 22 kDa | Cav1 | 0.3 | 0.31 | 3.05 |
| 948509 | Caveolin, caveolae protein, 22 kDa | Cav1 | 0.52 | 0.18 | 3 |
| 386555 | CD36 antigen | Cd36 | 0.21 | 0.06 | 0.08 |
| 832585 | ESTs | 0.21 | 0.06 | 0.21 | |
| 620819 | transcription elongation factor A (SII) 1 | AI158848 | 0.24 | 0.07 | 0.25 |
| 831701 | ESTs | 0.2 | 0.09 | 0.25 | |
| 775253 | ESTs | 0.16 | 0.07 | 0.22 | |
| 334182 | Unknown | 0.24 | 0.06 | 0.21 | |
| 493675 | Riken cDNA 2700018N07 gene | Scd1 | 0.18 | 0.1 | 0.21 |
| 571367 | Riken cDNA 2410012F02 gene | 0.24 | 0.11 | 0.27 | |
| 463388 | Epoxide hydrolase 2, cytoplasmic | Ephx2 | 0.3 | 0.11 | 0.41 |
| 374030 | EST | 0.25 | 0.11 | 0.33 | |
| 1067881 | Fc receptor, IgG, low affinity III | Fcgr3 | 0.36 | 0.16 | 0.26 |
| 474184 | Expressed sequence AW554339 | 0.3 | 0.15 | 0.33 | |
| 579349 | Expressed sequence AI593221 | 2410127E18Rik | 0.29 | 0.18 | 0.23 |
| 874232 | Riken cDNA 1110025G12 gene | 1110025G12Rik | 0.33 | 0.19 | 0.56 |
| 891453 | Expressed sequence AI315208 | AI315208 | = | 0.2 | 0.16 |
| 1399595 | Riken cDNA 2810422B09 gene | 1810061M12Rik | 0.3 | 0.18 | 0.4 |
| 737745 | ESTs | 0.34 | 0.17 | 0.37 | |
| 876063 | BCL2/adenovirus E1B 19 kDa-interacting protein 1, NIP3 | Bnip3 | 0.38 | 0.27 | 0.62 |
| 850642 | Epoxide hydrolase 2, cytoplasmic | Ephx2 | 0.18 | 0.22 | 0.44 |
| 1248105 | Expressed sequence AI595343 | AI595343 | 0.42 | 0.21 | 0.37 |
| 330661 | Mus musculus golli-interacting protein mRNA, complete cds | Nif3 | 0.66 | 0.21 | 0.22 |
| 1349720 | Apoptosis NIP3, Bcl-2-binding protein homolog (Nip3) mRNA | Bnip3 | 0.37 | 0.29 | 0.52 |
| 1067414 | Expressed sequence AI413399 | 1110001E17Rik | = | 0.24 | 0.52 |
| 832584 | ESTs, Weakly similar to STHM MOUSE STATHMIN [M. musculus] | 0.31 | 0.21 | 0.46 | |
| 876369 | Amyotrophic lateral sclerosis 2 (juvenile) homolog (human) | Als2 | 0.41 | 0.29 | 0.39 |
| 764542 | ESTs, Weakly similar to GPRY_mouse probable G protein-coupled receptor GPR34 [M. musculus] | Gpr43 | 0.49 | 0.24 | 0.43 |
| 1248075 | Transcription factor 1 | Tcf1 | 0.29 | 0.31 | 0.36 |
| 949592 | ESTs, Moderately similar to hypothetical protein [H. sapiens] | = | 0.32 | 0.22 | |
| 1195295 | Actinin α3 | Actn3 | = | 0.29 | 0.43 |
| 820307 | A kinase (PRKA) anchor protein (gravin) 12 | Akap12 | = | 0.31 | 0.08 |
| 864409 | Riken cDNA 1200013I08 gene | 1200013I08Rik | 0.47 | 0.33 | 0.39 |
| 947659 | Riken cDNA 2310016E22 gene | 2310016E22Rik | = | 0.33 | 0.37 |
| 1396547 | Myosin binding protein H | = | 0.25 | 0.45 | |
This list displays genes that are differentially expressed between mammary tumors (WntT) and hyperplastic mammary gland (WntH) from MMTV-Wnt-1 transgenic mice, and that are also differentially expressed between WntT and mammary tumors from MMTV-Neu transgenic mice (NeuT). Genes are sorted according to the average ratio of WntT versus WntH. A numeric ratio is displayed if the gene expression meets the criteria (statistical significance and fold cutoff) described in Table 2; otherwise, it is marked as '=', indicating that there is no significant difference between the two sets of samples in comparison. EST, expressed sequence tag; MMTV, mouse mammary tumor virus.
Conclusion
Our analysis of different stages of tumorigenesis in mouse models identified changes in gene expression accompanying tumor initiation and evolution. We also extended the report by Desai and coworkers [7] that the initiating oncogene determines the expression profiles of primary mammary tumors. In addition, we observed that the tumors from MMTV-Wnt-1 transgenic mice are similar to each other in their global expression profiles, regardless of whether the tumors have additional genetic alterations. These data may be useful for elucidation of oncogenic signaling pathways in breast cancer initiation and evolution.
Materials and methods
cDNA microarray slides
The mouse 15k slides and 8.7k slides used in this study were arrayed at the National Cancer Institute microarray facility. All slides of each array type were printed in a single batch. The 8.7k slides contain the 8700 Incyte GEM1 clone set, which are mapped to 6,877 Unigene cluster IDs, among which 2,953 are named genes, 2,206 are expressed sequence tags, and 1,628 are Riken cDNAs. The 15k slides contain the 8700 Incyte GEM1 clone set and the mammary 6000 clone set; a total of 1,444 clones do not map to a Unigene cluster ID, whereas the rest of the clones map to 10,062 unique genes as defined by Unigene cluster ID. Among the 10,062 Unigene clusters, 3,750 are named genes, 3,922 are expressed sequence taqs, and 2,390 are Riken cDNAs.
Sample information
All nontransgenic and MMTV-Neu transgenic animals used in this study were on the FVB background. All MMTV-Wnt-1 transgenic mice [14] were a mixture of FVB (>75%), SJL, and C57BL/6 strains. MMTV-Neu transgenic mice [41] were purchased from Jackson Laboratories (Bar Harbor, ME, USA). This transgenic line carries a rat cDNA encoding the wild-type Neu protein. Fat tissues were collected from intestinal fat in virgin FVB mice. All samples were collected fresh and snap-frozen in liquid nitrogen. RNA was extracted by Trizol (Invitorgen, Carlsbad, CA, USA). Reference RNA was a mixture of ovarian RNA (Ambion, Austin, TX, USA; Cat number 7824) and RNA extracted from tissues of liver, spleen, kidney, thymus, pancreas, lung, and normal lactating mammary gland of FVB mice of 6 months of age. All reference RNA used in this study is from a single preparation.
cDNA microarray hybridization and data extraction
The cDNA probes were prepared from a total of 35-50 μg reference RNA and 50-75 μg sample RNA from normal, hyperplastic, or tumor tissues, as described [42,43]. The cDNA from reference RNA was labeled with cyanine 3-dUTP, and that from sample RNA was labeled with cyanine 5-dUTP. Fluorescent images of hybridized microarrays were obtained by using a GenePix 4000 scanner (Axon Instruments, Foster City, CA, USA). Microarray images were analyzed using the ArraySuite software [44,45] based on the Scanalytics IPlab platform (Scanalytics, Fairfax, VA, USA). For each cDNA probe location, fluorescence intensity ratio and its associated measurement quality (q) were calculated. The evaluation of measurement quality is based on spot size, signal to noise ratio, background uniformity, and saturation pixel percentage [45]. The range of measurement quality is from 1.0 to 0, with higher measurements reflecting better quality. Areas of the array with obvious blemishes were automatically given a low quality value.
Statistical analysis of cDNA microarray data
Normalized log test to reference ratios and their corresponding quality measurements in each experiment were calculated as described previously [45]. A gene was excluded from further analyses (see below for description) if the average quality measurement was under 0.5 across samples in that specific comparison. Approximately 14,000 genes are suitable for analyzing on the 15k chips and 8,000 genes on the 8.7k chips.
Two methods were used to visualize the expression patterns among samples. Both used the Pearson correlation as a similarity measure. In average linkage hierarchical clustering, the distances between samples are represented on a tree called a dendrogram. In MDS, samples with similar expression ratios were placed closer to each other in three dimensional space. Average linkage hierarchical clustering analysis was implemented using the CLUSTER program, and the results were displayed using TREEVIEW [46]. MDS was developed in the MATLAB (Natick, MA, USA) environment.
A permutation t-test was used to select genes significantly differentially expressed between any two groups [47]. Here, a standard t-statistic was computed between two groups on the log-transformed ratios of each gene. The group labels were randomly permuted and the t-statistic for each gene in the permuted data set was calculated. The process was repeated 10,000 times. A P value was reported for each gene by comparing the observed statistic with the permutation statistics. To control for multiple comparisons, only genes with P values less than 0.001 were considered differentially expressed. The distribution of the significant differences expected by chance and the probability of observing as many or more differentially expressed genes were calculated from the permuted data. This latter probability is the P value reported in the Results and discussion section.
Western blot analysis and immunohistochemical staining
Antibodies used include rabbit IgG directed against nidogen and c-Kit (Santa Cruz, CA, USA). Tissues were fixed in 10% neutral formalin and processed as previously described [34] to obtain paraffin sections of 4 μm in thickness. For immunohistochemistry, the sections were boiled for 15 minutes in 10 mmol/l citrate buffer of pH 6.0 (to unmask antigen epitopes). Endogenous peroxidase activity was inactivated by 10 minute incubation in 3% hydrogen peroxide, and subsequent steps were performed using Vector ABC kits and the Nova-Red substrate (Vector Laboratories, Burlingame, CA, USA) following the manufacturer's recommendations.
For Western blotting, tumors were ground to powder in liquid nitrogen and lysed in the M-PER tissue lysis solution (Pierce, Rockford, IL, USA) with gentle shaking overnight at 4°C. Proteins in resulting supernatant (25 μg protein) were denatured using 2-mercaptoethanol, resolved on 10% polyacrylamide mini-gels containing 10% sodium dodecyl sulfate, and transferred to nitrocellulose membranes. The membranes were then incubated with primary antibodies and peroxidase-conjugated secondary antibodies (Jackson Laboratories) in tris-borate buffer (1 mmol/l Tris and 13.7 mmol/l NaCl, pH 7.6)/0.05% Tween 20/5% nonfat dried milk. Proteins recognized by specific antibodies were visualized using a chemiluminescent substrate (Supersignal; Pierce).
Accession number
The GEO accession number for the series of array data-sets is GSE2860.
Additional data files
The following additional data are available with the online version of this article: a table listing genes differentially expressed between mammary tumors from MMTV-Wnt-1 and MMTV-Neu transgenic mice (Additional data file 1); a table listing genes differentially expressed between Ha-Ras mutant and Ha-Ras wild-type tumors in MMTV-Wnt-1 mice (Additional data file 2); a table listing genes differentially expressed between MMTV-Wnt-1-induced tumors in p53-null and in p53-wild-type background (Additional data file 3); a table listing genes differentially expressed between tumors from MMTV-Wnt-1 transgenic/Pten+/- mice with loss of heterozygosity at the Pten locus and tumors from Wnt-1 transgenic/Pten+/+ mice (Additional data file 4); a table listing genes differentially expressed between virgin mammary glands from nontransgenic mice and hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice (Additional data file 5); a table listing genes expressed three-fold or higher in fat tissues than in mammary tumors from MMMTV-Wnt-1 and MMTV-Neu transgenic mice (Additional data file 6); and a table listing genes differentially expressed between mammary tumors and hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice (Additional data file 7).
Supplementary Material
A table listing genes differentially expressed between mammary tumors from MMTV-Wnt-1 and MMTV-Neu transgenic mice
A table listing genes differentially expressed between Ha-Ras mutant and Ha-Ras wild-type tumors in MMTV-Wnt-1 mice
A table listing genes differentially expressed between MMTV-Wnt-1-induced tumors in p53-null and in p53-wild-type background
A table listing genes differentially expressed between tumors from MMTV-Wnt-1 transgenic/Pten+/- mice with loss of heterozygosity at the Pten locus and tumors from Wnt-1 transgenic/Pten+/+ mice
A table listing genes differentially expressed between virgin mammary glands from nontransgenic mice and hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice
A table listing genes expressed three-fold or higher in fat tissues than in mammary tumors from MMMTV-Wnt-1 and MMTV-Neu transgenic mice
A table listing genes differentially expressed between mammary tumors and hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice
Acknowledgments
Acknowledgements
We thank Patrick O Brown, Michael B Eisen, Vishy Iyer, Nick Socci, Larry Donehower, Xinbin Chen, and Alfonso Bellacosa for advice and members of the Varmus laboratory for helpful discussion. We thank Xiaomei Zhang for immunohistochemical staining and Gary Chamness for assistance in the preparation of this manuscript. We thank Raju Chaganti for providing access to the scanner and Veronique Bourdon for assistance in its use. S.H. was supported by Department of Defense Breast Cancer Research Program awards. This work was supported in part by a National Institutes of Health Grant P01 CA94060-02 (to H.E.V.) and funds from the Martell Foundation (to H.E.V.) and Department of Defense (USAMRMC) BC030755 (to Y.L.).
Contributor Information
Shixia Huang, Email: shixiah@bcm.tmc.edu.
Yi Li, Email: liyi@bcm.tmc.edu.
Yidong Chen, Email: yidong@mail.nih.gov.
Katrina Podsypanina, Email: podsypak@mskcc.org.
Mario Chamorro, Email: chamorrm@mskcc.org.
Adam B Olshen, Email: olshena@mskcc.org.
Kartiki V Desai, Email: kartiki@hopkins.edu.sg.
Anne Tann, Email: anne_tann@mail.utexas.edu.
David Petersen, Email: petersed@mail.nih.gov.
Jeffrey E Green, Email: jegreen@mail.nih.gov.
Harold E Varmus, Email: varmus@mskcc.org.
References
- Fuller AP, Palmer-Toy D, Erlander MG, Sgroi DC. Laser capture microdissection and advanced molecular analysis of human breast cancer. J Mammary Gland Biol Neoplasia. 2003;8:335–345. doi: 10.1023/B:JOMG.0000010033.49464.0c. [DOI] [PubMed] [Google Scholar]
- Perez EA, Pusztai L, Van de Vijver M. Improving patient care through molecular diagnostics. Semin Oncol. 2004;31(5 Suppl 10):14–20. doi: 10.1053/j.seminoncol.2004.07.017. [DOI] [PubMed] [Google Scholar]
- Ma XJ, Salunga R, Tuggle JT, Gaudet J, Enright E, McQuary P, Payette T, Pistone M, Stecker K, Zhang BM, et al. Gene expression profiles of human breast cancer progression. Proc Natl Acad Sci USA. 2003;100:5974–5979. doi: 10.1073/pnas.0931261100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter D, Lahti-Domenici J, Keshaviah A, Bae YK, Argani P, Marks J, Richardson A, Cooper A, Strausberg R, Riggins GJ, et al. Molecular markers in ductal carcinoma in situ of the breast. Mol Cancer Res. 2003;1:362–375. [PubMed] [Google Scholar]
- Porter DA, Krop IE, Nasser S, Sgroi D, Kaelin CM, Marks JR, Riggins G, Polyak K. A SAGE (serial analysis of gene expression) view of breast tumor progression. Cancer Res. 2001;61:5697–5702. [PubMed] [Google Scholar]
- Hu Y, Sun H, Drake J, Kittrell F, Abba MC, Deng L, Gaddis S, Sahin A, Baggerly K, Medina D, Aldaz CM. From mice to humans: identification of commonly deregulated genes in mammary cancer via comparative SAGE studies. Cancer Res. 2004;64:7748–7755. doi: 10.1158/0008-5472.CAN-04-1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai KV, Xiao N, Wang W, Gangi L, Greene J, Powell JI, Dickson R, Furth P, Hunter K, Kucherlapati R, et al. Initiating oncogenic event determines gene-expression patterns of human breast cancer models. Proc Natl Acad Sci USA. 2002;99:6967–6972. doi: 10.1073/pnas.102172399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci USA. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- Nusse R, Varmus HE. Wnt genes. Cell. 1992;69:1073–1087. doi: 10.1016/0092-8674(92)90630-U. [DOI] [PubMed] [Google Scholar]
- Brennan KR, Brown AM. Wnt proteins in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2004;9:119–131. doi: 10.1023/B:JOMG.0000037157.94207.33. [DOI] [PubMed] [Google Scholar]
- Hatsell S, Rowlands T, Hiremath M, Cowin P. Beta-catenin and Tcfs in mammary development and cancer. J Mammary Gland Biol Neoplasia. 2003;8:145–158. doi: 10.1023/A:1025944723047. [DOI] [PubMed] [Google Scholar]
- Nelson WJ, Nusse R. Convergence of Wnt, beta-catenin, and cadherin pathways. Science. 2004;303:1483–1487. doi: 10.1126/science.1094291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto AS, Grosschedl R, Guzman RC, Parslow T, Varmus HE. Expression of the int-1 gene in transgenic mice is associated with mammary gland hyperplasia and adenocarcinomas in male and female mice. Cell. 1988;55:619–625. doi: 10.1016/0092-8674(88)90220-6. [DOI] [PubMed] [Google Scholar]
- Liu BY, McDermott SP, Khwaja SS, Alexander CM. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci USA. 2004;101:4158–4163. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalley M, Ashworth A. Stem cells and breast cancer: a field in transit. Nat Rev Cancer. 2003;3:832–844. doi: 10.1038/nrc1212. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Olayioye MA, Neve RM, Lane HA, Hynes NE. The ErbB signaling network: receptor heterodimerization in development and cancer. EMBO J. 2000;19:3159–3167. doi: 10.1093/emboj/19.13.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podsypanina K, Li Y, Varmus H. Evolution of somatic mutations in mammary tumors in transgenic mice is influenced by the inherited genotype. BMC Med. 2004;2:24. doi: 10.1186/1741-7015-2-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff RD, Anver MR, Gusterson BA, Hennighausen L, Jensen RA, Merino MJ, Rehm S, Russo J, Tavassoli FA, Wakefield LM, et al. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene. 2000;19:968–988. doi: 10.1038/sj.onc.1203277. [DOI] [PubMed] [Google Scholar]
- Rosner A, Miyoshi K, Landesman-Bollag E, Xu X, Seldin DC, Moser AR, MacLeod CL, Shyamala G, Gillgrass AE, Cardiff RD. Pathway pathology: histological differences between ErbB/Ras and Wnt pathway transgenic mammary tumors. Am J Pathol. 2002;161:1087–1097. doi: 10.1016/S0002-9440(10)64269-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398:422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Simcha I, Albanese C, D'Amico M, Pestell R, Ben-Ze'ev A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc Natl Acad Sci USA. 1999;96:5522–5527. doi: 10.1073/pnas.96.10.5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson JS, Leder P. Beta-catenin is a downstream effector of Wnt-mediated tumorigenesis in the mammary gland. Oncogene. 2001;20:5093–5099. doi: 10.1038/sj.onc.1204586. [DOI] [PubMed] [Google Scholar]
- Willert J, Epping M, Pollack JR, Brown PO, Nusse R. A transcriptional response to Wnt protein in human embryonic carcinoma cells. BMC Dev Biol. 2002;2:8. doi: 10.1186/1471-213X-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto W, Jiang W, Tice DA, Rubinfeld B, Hollingshead PG, Fong SE, Dugger DL, Pham T, Yansura DG, Wong TA, et al. Overexpression of the retinoic acid-responsive gene Stra6 in human cancers and its synergistic induction by Wnt-1 and retinoic acid. Cancer Res. 2001;61:4197–4205. [PubMed] [Google Scholar]
- Tice DA, Szeto W, Soloviev I, Rubinfeld B, Fong SE, Dugger DL, Winer J, Williams PM, Wieand D, Smith V, et al. Synergistic induction of tumor antigens by Wnt-1 signaling and retinoic acid revealed by gene expression profiling. J Biol Chem. 2002;277:14329–14335. doi: 10.1074/jbc.M200334200. [DOI] [PubMed] [Google Scholar]
- Bouillet P, Sapin V, Chazaud C, Messaddeq N, Decimo D, Dolle P, Chambon P. Developmental expression pattern of Stra6, a retinoic acid-responsive gene encoding a new type of membrane protein. Mech Dev. 1997;63:173–186. doi: 10.1016/S0925-4773(97)00039-7. [DOI] [PubMed] [Google Scholar]
- Chamorro MN, Schwartz DR, Vonica A, Brivanlou AH, Cho KR, Varmus HE. FGF-20 and DKK1 are transcriptional targets of beta-catenin and FGF-20 is implicated in cancer and development. EMBO J. 2005;24:73–84. doi: 10.1038/sj.emboj.7600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui XS, Donehower LA. Differential gene expression in mouse mammary adenocarcinomas in the presence and absence of wild type p53. Oncogene. 2000;19:5988–5996. doi: 10.1038/sj.onc.1203993. [DOI] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Hively WP, Varmus HE. Use of MMTV-Wnt-1 transgenic mice for studying the genetic basis of breast cancer. Oncogene. 2000;19:1002–1009. doi: 10.1038/sj.onc.1203273. [DOI] [PubMed] [Google Scholar]
- Li Y, Podsypanina K, Liu X, Crane A, Tan LK, Parsons R, Varmus HE. Deficiency of Pten accelerates mammary oncogenesis in MMTV-Wnt-1 transgenic mice. BMC Mol Biol. 2001;2:2. doi: 10.1186/1471-2199-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donehower LA, Godley LA, Aldaz CM, Pyle R, Shi YP, Pinkel D, Gray J, Bradley A, Medina D, Varmus HE. Deficiency of p53 accelerates mammary tumorigenesis in Wnt-1 transgenic mice and promotes chromosomal instability. Genes Dev. 1995;9:882–895. doi: 10.1101/gad.9.7.882. [DOI] [PubMed] [Google Scholar]
- Gazouli M, Kokotas S, Zoumpourlis V, Zacharatos P, Mariatos G, Kletsas D, Perunovic B, Athanasiou A, Kittas C, Gorgoulis V. The complement inhibitor CD59 and the lymphocyte function-associated antigen-3 (LFA-3, CD58) genes possess functional binding sites for the p53 tumor suppressor protein. Anticancer Res. 2002;22:4237–4241. [PubMed] [Google Scholar]
- Fisher GH, Orsulic S, Holland E, Hively WP, Li Y, Lewis BC, Williams BO, Varmus HE. Development of a flexible and specific gene delivery system for production of murine tumor models. Oncogene. 1999;18:5253–5260. doi: 10.1038/sj.onc.1203087. [DOI] [PubMed] [Google Scholar]
- Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET. Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci USA. 2003;100:10393–10398. doi: 10.1073/pnas.1732912100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgroi DC, Teng S, Robinson G, LeVangie R, Hudson JR, Jr, Elkahloun AG. In vivo gene expression profile analysis of human breast cancer progression. Cancer Res. 1999;59:5656–5661. [PubMed] [Google Scholar]
- Lee SW, Reimer CL, Oh P, Campbell DB, Schnitzer JE. Tumor cell growth inhibition by caveolin re-expression in human breast cancer cells. Oncogene. 1998;16:1391–1397. doi: 10.1038/sj.onc.1201661. [DOI] [PubMed] [Google Scholar]
- Guy CT, Webster MA, Schaller M, Parsons TJ, Cardiff RD, Muller WJ. Expression of the neu protooncogene in the mammary epithelium of transgenic mice induces metastatic disease. Proc Natl Acad Sci USA. 1992;89:10578–10582. doi: 10.1073/pnas.89.22.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Guo QM, Malek RL, Kim S, Chiao C, He M, Ruffy M, Sanka K, Lee NH, Dang CV, Liu ET. Identification of c-myc responsive genes using rat cDNA microarray. Cancer Res. 2000;60:5922–5928. [PubMed] [Google Scholar]
- Chen Y, Dougherty ER, Bittner ML. Ratio-based decusuibs abd quantitative analysis of cDNA microarray images. J Biomed Opt. 1997;2:364–374. doi: 10.1117/1.429838. [DOI] [PubMed] [Google Scholar]
- Chen Y, Kamat V, Dougherty ER, Bittner ML, Meltzer PS, Trent JM. Ratio statistics of gene expression levels and applications to microarray data analysis. Bioinformatics. 2002;18:1207–1215. doi: 10.1093/bioinformatics/18.9.1207. [DOI] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedenfalk I, Duggan D, Chen Y, Radmacher M, Bittner M, Simon R, Meltzer P, Gusterson B, Esteller M, Kallioniemi OP, et al. Gene-expression profiles in hereditary breast cancer. N Engl J Med. 2001;344:539–548. doi: 10.1056/NEJM200102223440801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A table listing genes differentially expressed between mammary tumors from MMTV-Wnt-1 and MMTV-Neu transgenic mice
A table listing genes differentially expressed between Ha-Ras mutant and Ha-Ras wild-type tumors in MMTV-Wnt-1 mice
A table listing genes differentially expressed between MMTV-Wnt-1-induced tumors in p53-null and in p53-wild-type background
A table listing genes differentially expressed between tumors from MMTV-Wnt-1 transgenic/Pten+/- mice with loss of heterozygosity at the Pten locus and tumors from Wnt-1 transgenic/Pten+/+ mice
A table listing genes differentially expressed between virgin mammary glands from nontransgenic mice and hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice
A table listing genes expressed three-fold or higher in fat tissues than in mammary tumors from MMMTV-Wnt-1 and MMTV-Neu transgenic mice
A table listing genes differentially expressed between mammary tumors and hyperplastic mammary glands from MMTV-Wnt-1 transgenic mice


