Abstract
Dipeptidyl peptidase I (DPPI) or cathepsin C is the physiological activator of groups of serine proteases from immune and inflammatory cells vital for defense of an organism. The structure presented shows how an additional domain transforms the framework of a papain-like endopeptidase into a robust oligomeric protease-processing enzyme. The tetrahedral arrangement of the active sites exposed to solvent allows approach of proteins in their native state; the massive body of the exclusion domain fastened within the tetrahedral framework excludes approach of a polypeptide chain apart from its termini; and the carboxylic group of Asp1 positions the N-terminal amino group of the substrate. Based on a structural comparison and interactions within the active site cleft, it is suggested that the exclusion domain originates from a metallo-protease inhibitor. The location of missense mutations, characterized in people suffering from Haim–Munk and Papillon–Lefevre syndromes, suggests how they disrupt the fold and function of the enzyme.
Keywords: apoptosis/cysteine protease/granzyme/inflammation/zymogen activation
Introduction
Zymogen activation by limited proteolysis is the crucial step in control of the proteolytic activity of most proteases (Neurath, 1984). Dipeptidyl peptidase I (DPPI), also known as cathepsin C, is becoming recognized as one of the most multifaceted protease-processing machines known so far (Nuckolls and Slavkin, 1999; Podack, 1999), having been shown to function beyond its role of a non-specific lysosomal protease (Turk et al., 2000). Cell lines derived from DPPI-deficient mice fail to activate groups of serine proteases from granules of immune (cytotoxic T-lymphocytes, natural killer cells) and inflammatory (neutrophils, mast cells) cells primarily involved in the defense of the organism, demonstrating that DPPI is involved in their activation (Pham and Ley, 1999; Wolters et al., 2001). The current list of unprocessed zymogens of proteases in DDPI null mice includes granzymes A, B and C, cathepsin G, neutrophil elastase and chymase.
Granzyme A and, in particular, granzyme B are best known for their role in apoptotic clearance of virus-infected cells and tumor cells (Shresta et al., 1998). Granzyme B delivers fast cell death by activating the caspase-dependent pathway (Darmon et al., 1995), with caspase-3 being its most likely in vivo target (Kumar, 1999), whereas granzyme A acts with a delay and induces a caspase-independent cell death pathway (Shresta et al., 1999). Cathepsin G, chymases and neutrophil elastase are, on the other hand, involved in the inflammatory response (Travis, 1988). The demonstration that cytoplasmic granules are the major source of DPPI in mast cells in dog airways and macrophages in alveoli led to the suggestion that DPPI may have a role in chronic airway diseases such as asthma (Wolters et al., 2000).
Moreover, genetic studies revealed that loss-of-function mutations in the DPPI gene result in early-onset periodontitis and palmoplantar keratosis, characteristics of Haim–Munk and Papillon–Lefevre syndromes (Toomes et al., 1999; Hart et al., 2000a,b; Allende et al., 2001), quite probably as a result of incomplete processing of some as yet unidentified proteases. Furthermore, Nuckolls and Slavkin (1999) suggested that DPPI may be essential for establishing or maintaining the structural organization of the epidermis of the extremities and the integrity of the tissues surrounding the teeth, and may participate indirectly in the processing of proteins such as keratins. Due to the demonstrated potential of DPPI to activate coagulation cascade enzymes (Lynch and Pfueller, 1988; Nauland and Rijken, 1994), DPPI may play a role as a protease link between inflammation and thrombosis.
DPPI (EC 3.4.14.1) was discovered by Gutman and Fruton in 1948 (Gutman and Fruton, 1948); however, the cDNA of the human enzyme was first described in 1995 (Pariš et al., 1995). DPPI is an abundant lysosomal cysteine protease from the papain superfamily with a mol. wt of 200 kDa and is widely expressed in many tissues of mammals and other animals. It is the only member of this family that is functional as a tetramer, consisting of four identical subunits. Each is composed of an N-terminal fragment, a heavy chain and a light chain (Dolenc et al., 1995). The N-terminal fragment, also named the residual propart domain (Cigič et al., 2000), was suggested to be involved in the formation of the tetramer (Cigič et al., 2000; Dahl et al., 2001).
In the acidic lysosomal milieu, DPPI is primarily an amino dipeptidase, cleaving two-residue units from the N-terminus of a polypeptide chain (B.Turk et al., 1998), although it can also act as a transferase at higher pH and catalyze the reverse reaction (Planta et al., 1964; McGuire et al., 1992). DPPI is the only papain-like enzyme that requires halide ions for its activity in addition to a thiol reducing compound (Fruton and Mycek, 1956; McDonald et al., 1966). The enzyme is not very specific and will progressively cleave two-residue units from protein and peptidyl substrates until either the N-terminus is no longer available or a stop sequence has been reached. DPPI stop sequences are positively charged residues (arginine or lysine) at the N-terminus or proline at either side of the scissile bond of a substrate (McDonald et al., 1966; McGuire et al., 1992).
Although it is the processing enzyme of many proteases (reviewed in B.Turk et al., 1998), DPPI is not capable of autoactivation and requires other proteases, presumably endopeptidases such as cathepsin L or S, for its own activation (Dahl et al., 2001). However, only the initial endopeptidase cleavages are accomplished by these other enzymes, whereas the final trimming to the naturally occurring N-terminus of the heavy chain of DPPI seems to be carried out by DPPI itself (Dahl et al., 2001). The N-terminus of its light chain, however, is naturally protected by a stop sequence. Similarly, DPPI seems to trim the N-termini of other lysosomal cysteine proteases (Turk et al., 2000), as has been demonstrated in vitro for cathepsin B (Rowan et al., 1992). The second residue from the N-terminus of these cathepsins is a proline, the DPPI stop sequence.
As mentioned above, DPPI is unique among proteases within the papain superfamily because of its oligomeric structure. Indeed, it also has a unique structure and mechanism compared with other oligomeric proteolytic complexes such as the proteasome (Lowe et al., 1995; Groll et al., 1997), bleomycin hydrolase (Joshua-Tor et al., 1995) and tryptase (Pereira et al., 1998), which all have their active site located on the inside of the structure. The active sites of DPPI, which can bind four molecules of cystatin per oligomer, were suggested to be on the outside (Dolenc et al., 1996). Our structure confirms this hypothesis.
Our wish to understand the mechanism underlying the unusual protease-processing activity of DPPI, which seems to be tightly controlled and in such widespread use, necessitated determination of its crystal structure. The crystal structure of human DPPI was solved by combined use of molecular replacement, multiple anomolous displacement (MAD) and multiple isomorphous replacement (MIR) phasing techniques and refined against 2.15 Å resolution data. The structure revealed that the N-terminal fragment domain folds into a β-barrel structure, which alters the papain-like protease framework into an enzyme exhibiting dipeptidyl aminopeptidase activity. The carboxylic group of Asp1 controls entry into the S2 binding pocket by docking with the N-terminal amino group of the substrate.
Results
Overall structure: tetrahedron is dimer of dimers
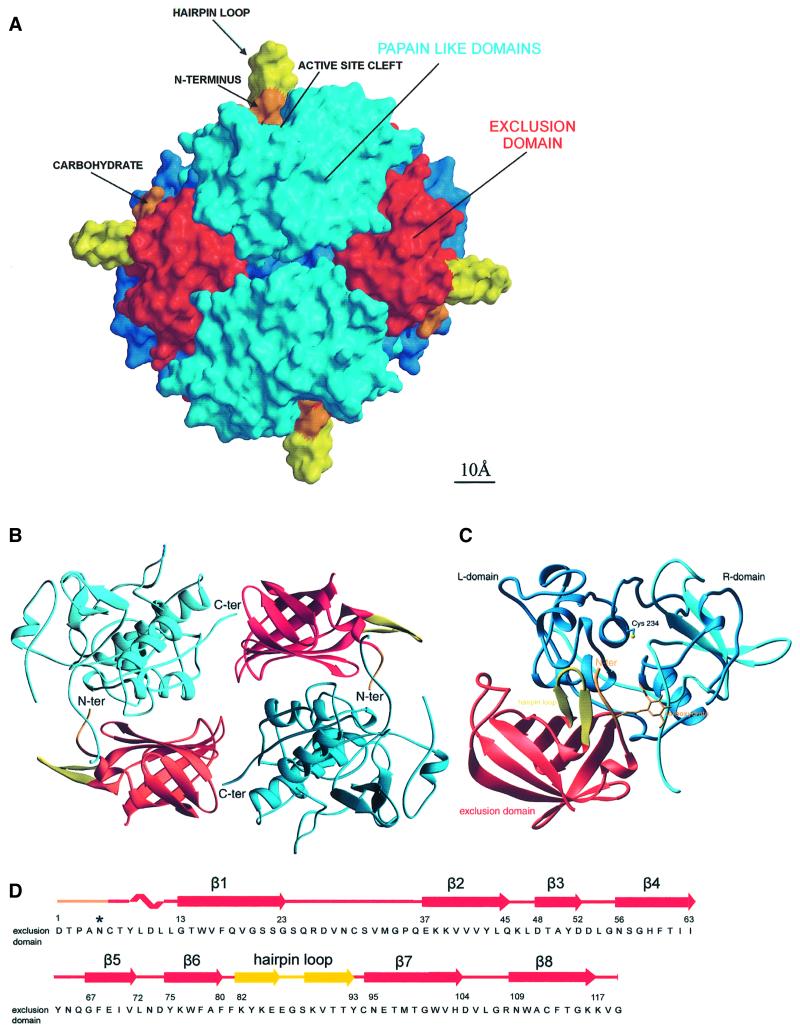
The tetrameric molecule of DPPI has theshape of a slightly flattened sphere with a diameter of ∼80 Å and a spherical cavity with a diameter of ∼20 Å in the middle. The molecule has tetrahedral symmetry. The molecular symmetry axis coincides with the crystal symmetry axis of the I222 space group. The asymmetric unit of the crystal thus contains a monomer. Each monomer consists of three domains, the two domains of the papain-like structure containing the catalytic site and an additional domain. This additional domain, with no analogy within the family of papain-like proteases, contributes to the tetrahedral structure and creates an extension of the active site cleft, providing features that endow DPPI with dipeptidyl aminopeptidase activity (Figure 1). Therefore, we term this additional domain the ‘exclusion’ domain instead of the less specific ‘residual propart’ domain, which was used to describe a domain with an unknown function (Dahl et al., 2001).
Fig. 1. Tetrahedral structure of DPPI. (A) Molecular surface of the tetrahedral structure of DPPI. The surface of papain-like domains is shown in blue, whereas the surface of exclusion domains is shown in red, orange and yellow. The view is along two active sites towards the exclusion domain hairpin loop (Lys82–Tyr93), shown in yellow, building a wall behind the active site cleft and five N-terminal residues, shown in orange. The left and right molecules are shown from the back towards the exclusion domain. The molecular surface was generated with GRASP (Nicholls et al., 1991); the figure was prepared in MAIN (Turk, 1992) and rendered with RENDER (Merritt and Bacon, 1997). (B) DPPI dimer. Head-to-tail arrangement of two pairs of papain-like and exclusion domains. Color coding is the same as in (A). The view is from the inside of the tetramer along the dimer 2-fold axis. The figure was created with RIBBONS (Carson, 1991). (C) Ribbon plot of the functional monomer of DPPI. The view shows the structure from the top, down the central α-helix. It is perpendicular to the view used in (A). The color coding is the same as in (A) except that here the heavy and light chains are shown in cyan and blue, respectively. The side chain of catalytic Cys234 and disulfides are shown with yellow sticks. The figure was created with RIBBONS (Carson, 1991). (D) Sequence of the exclusion domain with its secondary structure assignment.
The residues of a monomer are numbered consecutively according to the zymogen sequence (Pariš et al., 1995). The observed crystal structure of the mature enzyme contains 119 residues of the exclusion domain from Asp1 to Gly119 and 233 residues of the two papain-like domains from Leu207 to Leu439. The papain-like structure is composed of N-terminal heavy and C-terminal light chains generated by cleavage of the peptide bond between Arg370 and Asp371. The 87 propeptide residues from Thr120 to His206, absent in the mature enzyme structure, were removed during proteolytic activation of the proenzyme. The structure confirms the cDNA sequence (Pariš et al., 1995) and is in agreement with the amino acid sequence of the mature enzyme (Cigič et al., 1998; Dahl et al., 2001). With the exception of Arg26, all residues are well resolved in the final 2Fo – Fc electron density map. The conformations of the regions Asp27–Asn29 within the exclusion domain and Gly317–Arg320 at the C-terminus of the heavy chain are partially ambiguous.
During activation, the structure of DPPI undergoes a series of transformations. From the presumably monomeric form of preproenzyme (Muno et al., 1993), via a dimeric form of proenzyme (Dahl et al., 2001), the tetrameric form of the mature human enzyme is assembled (Dolenc et al., 1995). Visual inspection along each of the three molecular 2-fold axes showed that one of the axes reveals a head-to-tail arrangement of a pair of papain-like and exclusion domains (Figure 1B). The N-terminus of the exclusion domain of one dimer binds into the active site cleft of the papain-like domain of the next, while the C-terminus of one papain-like domain binds into the β-barrel groove of the adjacent exclusion domain of its symmetry partner. The N-termini of the heavy and light chains are, however, each arranged around one of the two remaining 2-fold axes. Interestingly, both chain termini result from proteolytic cleavages that appear during proenzyme activation, whereas the head-to-tail arrangement involves chain termini, already present in the zymogen. This suggests that the head-to-tail arrangement observed in the crystal structure originates from the zymogen form, whereas the N-termini contacts are suggested to be formed during tetramer formation. The 87 residue propeptide, cleaved off during activation, not only blocks access to the active site of the enzyme, but also prevents formation of the tetramer. This is in contrast to the proenzymes of related structures (Cygler et al., 1996; Turk et al., 1996; Podobnik et al., 1997). A similar role is given to the ∼8 residue insertion from Asp371 to Leu378 (Figure 2), cleavage of which breaks the single polypeptide chain of the papain-like domain region into heavy and light chains.
Fig. 2. Structure-based sequence alignment and sequences of exopeptidases cathepsins C (or DPPI), B (1HUC), H (8PCH), X (1EF7) and endopetidase cathepsin L (1ICF). Occluding loop residues of cathepsin B and mini-chain residues of cathepsin X are marked with blue and green, respectively.
The surface area of the complete tetramer is 57 530 Å2, that of the head-to-tail dimer 29 410 Å2, and those of the other two dimers 29 690 and 30 250 Å2. This indicates that the head-to-tail dimer is the most compact of all the three possible dimers found in the crystal and is thus in agreement with other data, giving further support to the suggestion that this is the zymogen dimer. The surface areas of the monomer and its constituents the exclusion domain and papain-like structure are 15 120, 6210 and 10 060 Å2, respectively. The monomer constituents interact along 570 Å2. A head-to-tail dimer includes interaction surface contained within each of the two monomer subunits (1140 Å2) plus an additional 420 Å2, whereas the interaction surface area between two head-to-tail dimers is 650 Å2.
The positioning of the exclusion domain at the end of the active site cleft and the extended contact surface with the papain-like domain leave no doubt as to which three domain units form the functional monomer (Figure 1). However, this functional monomer is only a putative substructure, presumably not existing in solution, i.e. no enzymatic activity has been associated with a monomeric form of DPPI to date. It is thus not unlikely that the exclusion domain swapping in the dimeric form of the zymogen is a mandatory intermediate leading to the tetrameric form of the mature enzyme. The answer to the question of whether the domains of the functional monomer originate from the same polypeptide chain, as would be assumed, is thus not obvious. The disconnected termini of the head-to-tail dimer (C-termini of the exclusion domains and N-termini of heavy chains) are 45 Å apart, and visual inspection of the structure of the cathepsin B propeptide (Podobnik et al., 1997) superimposed on the structure of DPPI provides no clear hints. Therefore, the resolution of this question must await crystal structure determination of a zymogen.
Papain-like domains structure
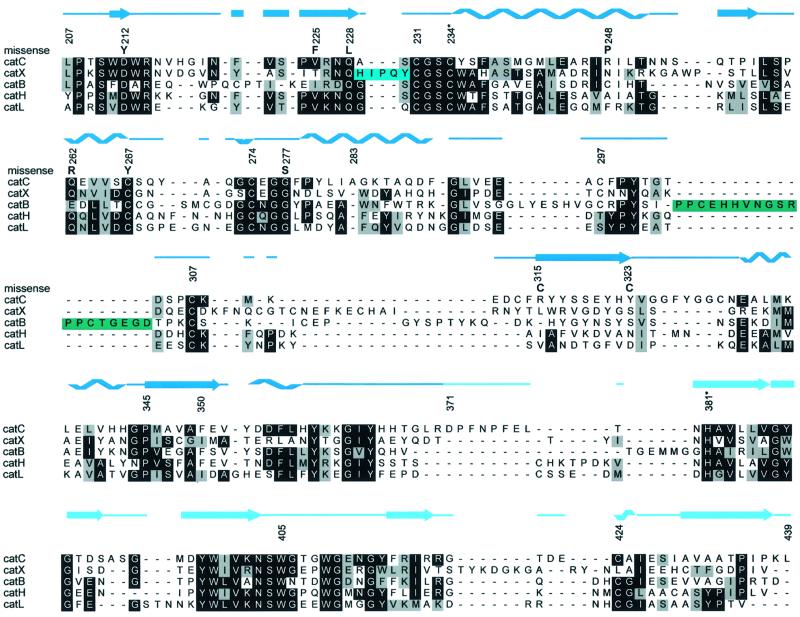
The two domains of the papain-like structure are termed left (L-) and right (R-) domains according to their position seen in Figure 1C. The L-domain contains several α-helices, the most pronounced being the structurally conserved 28 residue central α-helix with catalytic Cys234 on its N-terminus. The R-domain is a β-barrel with a hydrophobic core. The interface of the two domains is quite hydrophobic, in contrast to the interface of the cathepsin B structure (Musil et al., 1991), which is stabilized by numerous salt bridges. The interface opens in front, forming the active site cleft, in the middle of which is the catalytic ion pair Cys234 and His381. The papain-like domains contain nine cysteines, six of them being involved in disulfide bridges (231–274, 267–307 and 297–313) and three being free (catalytic Cys234, Cys331 and Cys424). The side chain of Cys424 is exposed to the solvent and was the major binding site for osmium and the only binding site for the gold derivative, whereas the side chain of Cys331 is buried in the hydrophobic environment of the side chains of Met336, Met346, Val324 and Ala430.
To demonstrate the similarity of the structures among homologous proteins, several structures were superimposed on the DPPI papain-like domains and their sequences aligned (Figure 2).
Exclusion domain structure
The exclusion domain forms an enclosed structure allowing it to fold independently from the rest of the enzyme (Cigič et al., 2000). This domain folds as an up-and-down β-barrel composed of eight antiparallel β-strands wrapped around a hydrophobic core formed by tightly packed aromatic and branched hydrophobic side chains. The strands are numbered consecutively as they follow each other in the sequence. The exclusion domain contains four cysteine residues, which form two disulfide bridges (Cys6–Cys94 and Cys30–Cys112). The N-terminal residues from Asp1 to Gly13 seal one end of the β-barrel, whereas there is a broad groove filled with solvent molecules and a sulfate ion at the other end (Figure 1C and D).
Two long loops project out of the β-barrel. The first (Ser24–Gln36) is a broad loop from β-strand 1, shielding the first and the last strands from solvent. This loop additionally stabilizes the barrel structure via the disulfide Cys30–Cys112, which fastens the loop to strand 8. The second loop (Lys82–Tyr93), termed the hairpin loop, is a two-stranded β-sheet structure with a tight β-hairpin at its end. The loop comes out of strands 7 and 8 and encloses the structure by the disulfide Cys6–Cys94, which connects the loop to the N-terminus of the exclusion domain. This loop stands out of the tetrameric structure (Figure 1A and C) and is reminiscent of the cathepsin X 110–123 loop (Gunčar et al., 2000) in terms of its pronounced form and charged side chains, indicating a possible common role for these structural features.
Interface of papain-like domains and the exclusion domain
All three domains make contacts along the edges of the two papain-like domains and form a large binding surface of predominantly hydrophobic character. The wall formed by β-strands 4–7 of the exclusion domain attaches to the surface of the papain-like domains. There are three stacks of parallel side chains from each of the strands of the β-sheet mentioned above interacting in a zipper-like manner with the side chains of a short three-turn α-helix between Phe278 and Phe290. This feature is a conserved structural element in all homologous enzymes (Figure 2). The middle turn of this helix contains an additional residue, Ala283, thus forming a π-helical turn, which is a unique feature of DPPI. The branched side chain of Leu281 is the central residue of a small hydrophobic core formed at the interface of the three domains. Only the side chain of Glu69 escapes the usual β-sheet side chain stacking and forms a salt bridge with Lys285. The exchange of electrostatic interactions continues from Lys285 towards the side chains of His103 and Asp289.
The active site cleft
The four active site clefts are positioned approximately at the tetrahedral corners of the molecule, ∼50–60 Å apart, and are exposed to the solvent. Each active site cleft is formed by features of all three domains of a functional monomer of DPPI (Figure 3); the papain-like domains form the sides of the monomer, which is closed at one end by the exclusion domain.
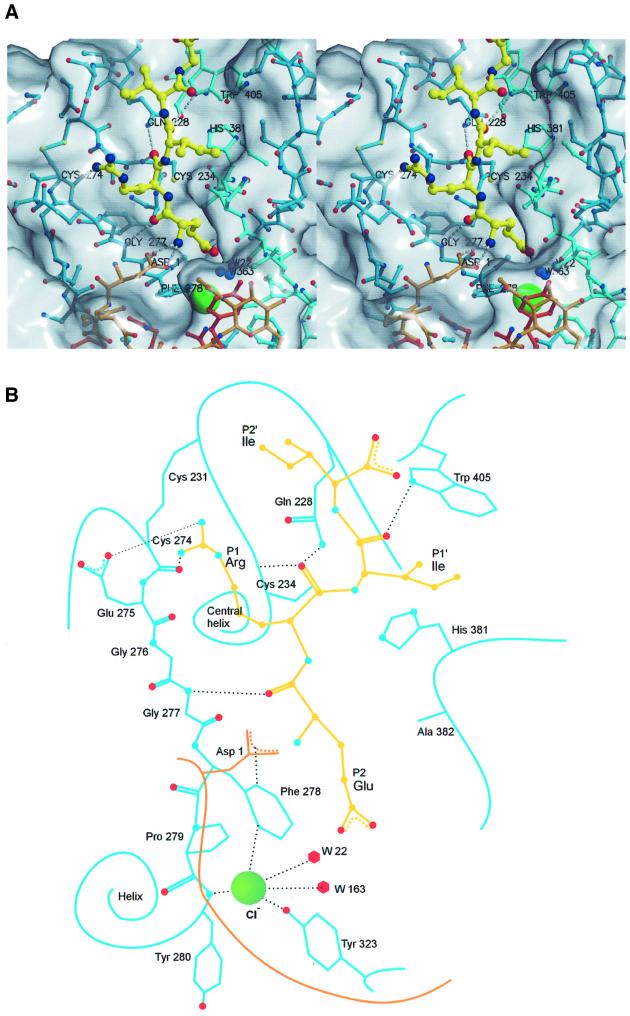
Fig. 3. Active site cleft of DPPI with a bound model of the N-terminal sequence ERIIGG from the biological substrate, granzyme A. (A) Stereo view: covalent bonds of papain-like domains and the exclusion domain are shown in the colors used in Figure 1C. Covalent bonds of the substrate model are shown as yellow sticks. Corresponding carbon atoms are shown as balls using the covalent bond color scheme. The chloride ion is shown as a large green sphere. Oxygen, nitrogen and sulfur atoms are shown as red, blue and yellow spheres, respectively. The residues relevant for substrate binding are marked and hydrogen bonds are shown as white broken lines. The molecular surface was generated with GRASP (Nicholls et al., 1991); the figure was prepared in MAIN (Turk, 1992) and rendered with RENDER (Merritt and Bacon, 1997). (B) Schematic presentation. The color codes are the same as in (A).
The reactive site residues Cys234(25)–His381(159) form an ion pair and are at their usual positions above the oxyanion hole formed by the amides of the Gln228(19) side chain and Cys234(25) main chain. The HE1 hydrogen atom from a ring of Trp405(177) is in the correct orientation to bind the substrate carbonyl atom of a P1′ residue, and the extended stretch of conserved Gly276(65)–Gly277(66) is in the usual place to bind substrate P2 residue with an antiparallel hydrogen bond ladder (D.Turk et al., 1998). The resulting hydrogen bonds are indicated in Figure 3. (For easier sequence comparison, the papain numbering is given in parentheses.)
As expected, the substrate-binding area beyond the S2 binding site is blocked. DPPI utilizes the exclusion domain to build a wall, which prevents formation of a binding surface beyond the S2 substrate-binding site. This wall spans across the active site cleft as well as away from it. A broad loop made of the five N-terminal residues surrounds the S2 binding site and forms a layer across the active site cleft. The blockade of the cleft is enhanced additionally by carbohydrate rings attached to Asn5. (The first carbohydrate ring is well resolved by the electron density map.) Behind the N-terminal loop, there is an upright β-hairpin (Lys82–Tyr93), which protrudes far into the solvent.
Substrate-binding sites
Surprisingly, the anchor for the N-terminal amino group of a substrate is not the C-terminal carboxylic group of a peptide chain, as expected based on analogy with cathepsin H (Gunčar et al., 1998) and bleomycin hydrolase (Joshua-Tor et al., 1995), but, instead, it is the carboxylic group of the Asp1 side chain, the N-terminal residue of the exclusion domain (Figure 3). The N-terminal amino group of Asp1 is fixed with two hydrogen bonds between the main chain carbonyl of Glu275 and the side chain carbonyl of Gln272. The Asp1 side chain reaches towards the entrance of the S2 binding site, where it interacts with the electrostatically positive edge of the Phe278 ring (Figure 3).
The side chains of Ile429, Pro279, Tyr323 and Phe278 form the surface of the S2 binding site. This site has the shape of a pocket, and is the deepest known thus far. The bottom of the pocket is filled with an ion and two solvent molecules. The high electron density peak, chemical composition of the coordinated atoms and the requirement of DPPI for chloride ions lead to the conclusion that this ion is chloride. It is positioned at the N-terminal end of the three-turn helix (Phe278–Phe290) and is coordinated by the main chain amide group of Tyr280 (3.2 Å) and is 3.3 Å away from the hydroxyl group of Tyr323 and two solvent molecules (Figure 3). The ring of Phe278 is thus positioned with its electropositive edge between the negative charges of chloride and the Asp1 carboxylic group.
The surfaces of the other substrate binding sites (S1, S1′ and S2′) show no features unique for DPPI when compared with other members of the family (D.Turk et al., 1998). The S1 binding site is placed between the active site loops Gln272–Gly277 and Gln228–Cys234, beneath the disulfide 274–231 and Glu275. The S1′ substrate-binding site is rather shallow, with a hydrophobic surface contributed by Val352 and Leu357, and the S2′ binding site surface is placed within the Gln228–Cys234 loop. The molecular surface along the active site cleft beyond the S2′ binding area is wide open, indicating that there is no particular site defined for binding of substrate residues.
Discussion
Mechanisms of exopeptidases: peptide patches and the exclusion domain
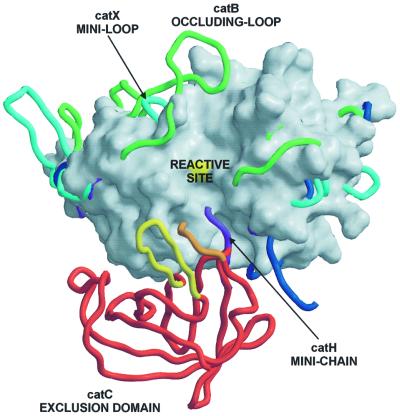
Elucidation of the structure of DDPI explains its unique exopeptidase activity. Figure 4 clearly shows that converting endo- to exopeptidase activity of a papain-like protease is achieved by features added to the structure of a typical papain-like endopeptidase framework on either side of the active site cleft (D.Turk et al., 1998; McGrath, 1999). The carboxypeptidases cathepsin B (Musil et al., 1991) and cathepsin X (Gunčar et al., 2000) utilize loops that block access along the primed side and provide histidine residues to anchor the C-terminal carboxylic group of a substrate. In contrast, the aminopeptidases cathepsin H (Gunčar et al., 1998) and a more distant homolog bleomycin hydrolase (Joshua-Tor et al., 1995) utilize a polypeptide chain in an extended conformation that blocks access along the non-primed binding sites and provides its C-terminal carboxylic group as the anchor for the N-terminal amino group of a substrate. DPPI recognizes the N-terminal amino group of a substrate in a unique way. The anchor is a charged side chain group of the N-terminal residue Asp1, folded as a broad loop on the surface. However, this loop is not a part of the polypeptide chain of papain-like domains, but belongs to an additional domain. It has an independent origin that adds to the framework of a papain-like endopeptidase and turns it into an exopeptidase. As it excludes endopeptidase activity of the enzyme, we named it the exclusion domain.
Fig. 4. Features of papain-like exopeptidases. A view towards the active site clefts of superimposed papain-like proteases. The underlying molecular surface of cathepsin L, shown in white, is used to demonstrate an endopeptidase active site cleft, which is blocked by features of the exopeptidase structures. The surface of the catalytic cysteine is colored in yellow. Chain traces of cathepsins B, X and H are shown in green, cyan and purple, respectively. Chain traces of papain-like domains of DPPI are shown in dark blue, whereas for the chain trace of the exclusion domain the color code is the same as in Figure 1. The bleomycin hydrolase chain trace is not shown for reasons of clarity, although its C-terminal residues superimpose almost perfectly with the C-terminal residues of the cathepsin H mini-chain (purple).
Substrate-excluding specificity of DPPI
The selectivity of DPPI is best described by exclusion rules, and the structure provides some clues to understanding their mechanism.
DPPI shows no endopeptidase activity, in contrast to cathepsins B and H. It is, however, inhibited by cystatin-type inhibitors, non-selective protein inhibitors of papain-like cysteine proteases (Turk et al., 2000), as are the other papain-like exopeptidases cathepsins B, H and X. The patches on the papain-like endopeptidase structure framework responsible for the exopeptidase activity of cathepsins B and H are relatively short polypeptide fragments, which lie on the surface (Musil et al., 1991; Gunčar et al., 1998). It was shown for the cathepsin B occluding loop (Illy et al., 1997; Podobnik et al., 1997) that these rather flexible structural features compete with substrates and inhibitors for the same binding sites within the active site cleft. A similar function has been suggested for the cathepsin H mini-chain (Gunčar et al., 1998). Analogously, the flexibility of the five N-terminal residues of the exclusion domain can explain the complex formation of DPPI with cystatin-type inhibitors. However, proximal to this short region is the massive body of the exclusion domain with its extended binding surface for the papain-like domain and its projecting feature β-hairpin Lys82–Tyr93 tightly fastened within the tetrameric structure. Therefore, it is highly unlikely that the exclusion domain could be pushed away by an approaching polypeptide. This indicates the robust mechanism by which the endopeptidase activity of DPPI is excluded. Control at the micro level is then achieved by the carboxylate group of the Asp1 side chain, which is oriented towards the active site cleft to rule out the approach of a substrate without an N-terminal amino group (McGuire et al., 1992), as demonstrated in Figure 3.
DPPI, similarly to other papain-like proteases, does not cleave substrates with proline at the P1 or P1′ position. A simple modeling study (not shown) suggested that proline residues at these positions would disturb the hydrogen bonding network and may produce clashes within the S1 substrate-binding site. Prolines, in contrast to other amino acid residues, do not have the amide proton. The peptide bond nitrogen instead is bound to the side chain carbon, forming a ring structure, which is therefore not capable of forming the same type of hydrogen bonding patterns as other amino acids, whereas the ring structure reduces the available conformational space of the residue. Cleaving peptide bonds of prolines inside a peptidyl substrate thus imposes additional constraints on the active site of a protease, and special classes of proteolytic enzymes, named prolyl endo- and oligopeptidases, have evolved to perform this cleavage effectively (Barrett et al., 1998).
The mechanism regarding the selectivity of the S2 binding pocket seems puzzling at first sight. The selectivity of the S2 binding site appears to be dominated by two negative charges; the Asp1 side chain at the entry of the pocket and the chloride ion at the very bottom of the deep hydrophobic pocket. Yet residues with positively charged groups, such as lysines and arginines, are not cleaved, whereas residues with a negative charge, such as glutamate and aspartate, appear in the sequences of the biological (Pham and Ley, 1999) and synthetic (McGuire et al., 1992) substrates of DPPI. (The chloride ion seems strongly bound, as it was not removed by extended dialysis during sample preparation.) The strongest oligopeptide-based inhibitors with an arginine at their N-terminus were even shown to be weak competitive inhibitors (Ki = 10–5) of DPPI (Horn et al., 2000). The weak inhibition constant suggests an assembly of binding geometries, indicating that the side chain of arginine does not even enter the S2 binding pocket, but probably only interacts with the carboxylic group of Asp1 at the entrance. This mechanism is consistent with the biological role of DPPI, which should not be inhibited when it reaches a stop sequence, but rather move on to process the next molecule.
The exclusion domain is a structural homolog of a protease inhibitor
No sequence homolog is known for the exclusion domain; however, 44 similar structural folds were found using DALI (Holm and Sander, 1996). The highest similarity scores were obtained with the structures of streptavidin (PDB accession No. 1SWU) and Erwinia chrysanthemi inhibitor (1SMP), whose structure was determined in complex with the Serratia marcescens metallo-protease (Baumann et al., 1995).
The large number of structural homologs is not surprising as the eight-stranded antiparallel β-barrels are quite a common folding pattern. However, the geometry of binding of the E.chrysanthemi inhibitor to metallo-protease also indicates a functional similarity. The N-terminal tail of E.chrysanthemi inhibitor binds into the active site cleft of the S.marcescens metallo-protease along the substrate-binding sites towards the active site cleft. Even the chain traces of the N-terminal parts are similar, i.e. an extended chain, which continues into a short helical region (Figure 5). In contrast to the exclusion domain of DPPI, which enters the active site cleft from the non-primed region (in a substrate-like direction), the N-terminal tail of E.chrysanthemi inhibitor binds along the primed substrate-binding sites (in the direction opposite to that of a substrate). It is thus intriguing to suggest that the exclusion domain is an adapted inhibitor, which does not abolish the catalytic activity of the enzyme, but prevents its endopeptidase activity by blocking access to only a portion of the active site cleft.
Fig. 5. Superposition of Erwinia chrysanthemi metallo-protease inhibitor on the exclusion domain. All structurally homologous Cα atoms from seven out of eight β-strands form 56 pairs, which after superposition yield an r.m.s.d. of 1.70 Å. After applying a 1.5 Å cut-off, the remaining 48 pairs yield an r.m.s.d. of 1.04 Å. Only the strand β-7 does not superimpose well, as in the exclusion domain it connects the hairpin loop and C-terminal strand β-8. The inhibitor is shown in red and the exclusion domain with the same color codes as used in Figure 1. The figure was prepared with MAIN (Turk, 1992) and rendered with RENDER (Merritt and Bacon, 1997).
Genetic disorders located on the DPPI structure
Quite a few of the genetic disorders of DPPI described are nonsense mutations resulting in truncation of the expressed sequence (Hart et al., 1999; Toomes et al., 1999). However, there is a series of missense mutations (D212Y, V225F, Q228L, R248P, Q262R, C267Y, G277S, R315C and Y323C) in the sequence of the heavy chain (Figure 6A) (Toomes et al., 1999; Hart et al., 2000a,b; Allende et al., 2001). Their structure-based interpretation suggests that not all missense mutations necessarily result in complete loss of DPPI activity.
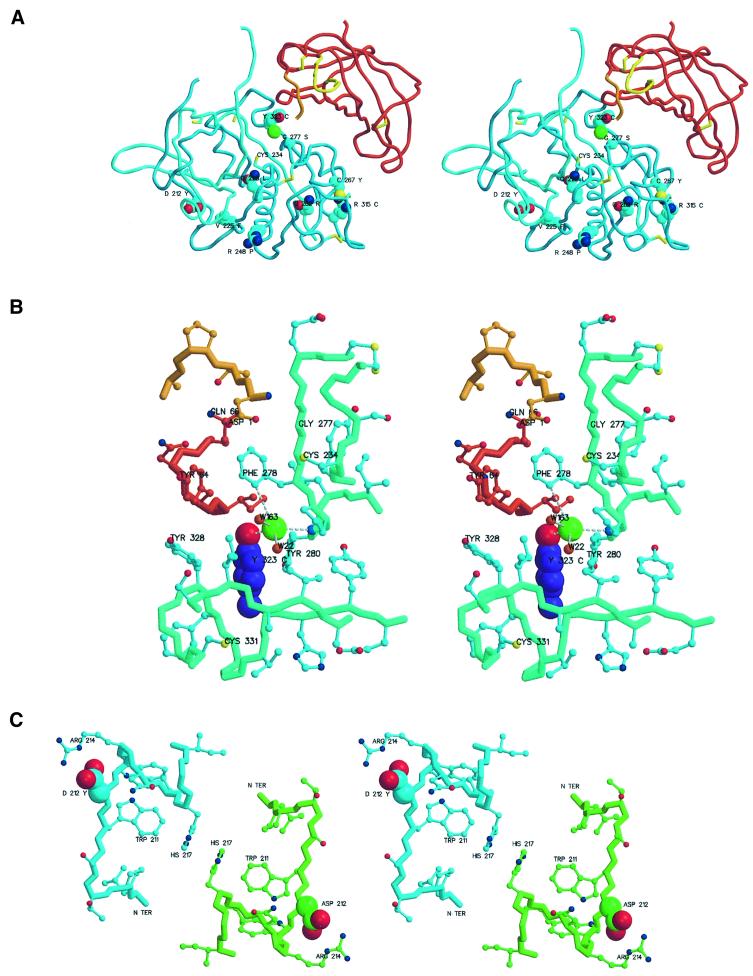
Fig. 6. Regions with missense mutations resulting in genetic diseases. The figures were prepared with MAIN (Turk, 1992) and rendered with RENDER (Merritt and Bacon, 1997). (A) Overview of missense mutations. The chain trace of the DPPI domain is shown in the colors used in Figure 1. Side chains of mutated residues are shown as cyan, red and dark blue balls representing carbon, oxygen and nitrogen atoms, respectively. All cysteine residues are shown as sticks. Mutated residues are marked with their sequence IDs and residue names in one-letter code. The catalytic cysteine is also marked. (B) Y323C mutant with chloride ion coordination. A side view towards the S2 binding pocket containing the chloride ion and its coordination with the active site residues Asp1 and Cys234 at the top. The color scheme is as in (A). The exceptions are the side chain atoms of Tyr323, shown as balls with carbon atoms colored purple, and the papain-like domains residues of the main and side chain trace, which are shown in greenish and cyan, respectively. The main chain bonds are thicker. Oxygens of the main chain carbonyls are omitted for clarity. The chloride ion is a large green ball, and the small red balls adjacent to it are solvent molecules. Chloride coordination is shown with white disconnected sticks. Relevant residues are marked with their sequence IDs and residue names. (C) D212Y mutant: view along a molecular 2-fold axis. Carbon atoms and covalent bonds of chain trace and side chains are differentiated by color: cyan for the left and green for the right molecule. Asp212 side chain atoms are highlighted as larger balls.
Gln228 and Gly277 are two of the key residues involved in substrate binding. Mutation of Q228L disrupts the oxyanion hole surface and consequently severely affects productive binding of the carbonyl oxygen of the scissile bond of the substrate. The G277S mutation presumably disrupts the main chain–main chain interactions with the P2 residue, as the glycine conformation cannot be preserved (see Figure 3).
The most frequent missense mutation appears to be Y323C (Toomes et al., 1999; Hart et al., 2000b). Normally, the hydroxyl group of Tyr323 is involved in the binding of the chloride ion, which seems to stabilize the S2 substrate-binding site (Figure 6B). The mutation into a cysteine may disrupt not only chloride binding but also positioning of Phe278 and, consequently, Asp1. The change to a cysteine residue has even more of an impact. It may alter the structure of the short segment of the chain towards Cys331 by forming a new disulfide bond. Even the binding surface for the exclusion domain may be disrupted, and it is possible that this mutant may not form an oligomeric structure at all and may thus even exhibit endopeptidase activity.
The mutations C267Y, R315C and Q262R are located around the surface loop enclosed by the disulfide Cys297–Cys313. In the observed structure, the side chains of Gln262 and Phe298 form the center around which the loop is folded (Figure 6A). Cys267 is located in the vicinity of Gln262 and fastens the structure of the loop via the disulfide Cys267–Cys307. Arg315 is involved in a salt bridge with Glu263, the residue following the central loop residue Gln262, and is adjacent to Cys13. Either of these mutations may thus prevent proper folding of the loop and disrupt formation of the two disulfides. Free cysteines may thus result in non-native disulfide connectivity, which has the potential to aggregate the improperly folded DPPI monomers.
The R248P mutant presumably leads to folding problems as a proline at this position quite probably breaks the central helix at the second turn from its C-terminus. A phenylalanine ring at the position of Val225 is too large to form the basis of the short loop Asn403–Gly413 and thereby disrupts the primed substrate-binding sites, in particular the positioning of the conserved Trp405 involved in P1′ residue binding (see Figure 3).
The mutation D212Y, however, seems a special case. It does not seem to be linked to the active site structure or aggregation problems. Asp212, the sixth residue from the N-terminus of the papain-like domain, is exposed to the surface where it forms a salt bridge with Arg214. Disruption of the salt bridge structure may result in a different positioning of the N-terminus and, since the N-terminal region is involved in molecular symmetry contacts, this mutation may prevent tetramer formation (Figure 6C).
DPPI is a protease-processing machine
Oligomeric proteolytic machineries such as the 20S proteasome (Lowe et al., 1995; Groll et al., 1997), bleomycin hydrolase (Joshua-Tor et al., 1995) and tryptase (Pereira et al., 1998) restrict access of substrates to their active sites. Proteasomes are barrel-like structures composed of four rings of α- and β-subunits, which cleave unfolded proteins captured in the central cavity into short peptides. Tryptases are flat tetramers with a central pore in which the active sites reside. The pore restricts the size of accessible substrates and inhibitors. In addition, the active sites of bleomycin hydrolase are also located within the hexameric barrel cavity. In contrast, the active sites of DPPI are located on the external surface and the tetrahedral architecture introduces a long distance between them and allows them to behave independently. This makes DPPI a protease, capable of hydrolysis of protein substrates in their native state regardless of their size. Its robust design, supported by the oligomeric structure, confines the activity of the enzyme to an aminodipeptidase and thereby makes it suitable for use in many different environments, where DPPI can selectively activate quite a large group of chymotrypsin-like proteases.
Materials and methods
Protein purification and crystallization
DPPI was expressed in the insect cell/bacullovirus system (Dahl et al., 2001). The purified DPPI was concentrated to 10 mg/ml in a spin concentrator (Centricon, Amicion). Crystals were grown using the sitting drop vapor diffusion method. The reservoir contained 1 ml of 2.0 M ammonium sulfate solution with 0.1 M sodium citrate and 0.2 M potassium/sodium tartrate at pH 5.6 (Hampton screen II, solution 14). The drop was composed of 2 µl of reservoir solution and 2 µl of protein solution. Acetic acid and sodium hydroxide were used to adjust the pH.
The crystals of DPPI belong to the orthorhombic space group I222 with cell dimensions a = 87.15 Å, b = 88.03 Å, c = 114.61 Å. Native crystals diffracted to 2.15 Å resolution on the XRD1 beamline at Elettra. Before data collection, crystals of DPPI were soaked in 30% glycerol solution before being dipped into liquid nitrogen and frozen. All data sets were processed using the program DENZO (Otwinowski and Minor, 1997).
Phasing and structure solution
The position of the enzymatic domain was determined by molecular replacement implemented in the EPMR program (Kissinger et al., 1999) using various cathepsin structures. The partial model did not enable us to proceed with the structure determination, therefore a heavy atom derivative screen was performed. Two soaks proved successful (K2Cl6Os3 and AuCl3). A three-wavelength MAD data set of osmium derivative was measured at the Max-Planck beamline at DESY Hamburg. We had to use the native data set as a reference to solve the heavy atom positions and treat the MAD data as MIR data. The RSPS program (Knight, 1989) suggested a single heavy atom position. The derived map was not of sufficient quality to enable model building. It did, however, show that the molecular replacement solution and MAD/MIR map were consistent. Phasing based on a single gold heavy atom site and an additional five minor osmium heavy atom sites located from the residual maps, refined and solvent flattened with SHARP (de La Fortelle and Bricogne, 1997) using data to 3.0 Å, resulted in an interpretable electron density map (Table I).
Refinement and structure validation
This structure was then refined to an R-value of 0.184 (Rfree = 23.8 using 5% of reflections) against 2.15 Å resolution data (Table I). When using 2.6 Å data, individual B-value refinement was included, and with 2.4 Å resolution data and an R-value ∼0.24, the inclusion of solvent molecules was initiated using an automated procedure. The chloride ion was identified from a water molecule which, after positional and B-value refinement, returned a B-value for oxygen at the minimum boundary. It was still positioned within a 4.5σ positive peak of the Fo – Fc difference electron density map. Three sulfate ions were found by visual inspection of large clouds of positive density contoured at 3.0σ in the vicinity of already built solvent molecules. The only carbohydrate ring observed was attached to Asn5 in the exclusion domain. It was recognized from a cluster of solvent molecules and peaks of positive density in the Fo – Fc map and positioned among them. The position of the chloride ion was also confirmed by an iodine derivative, which resulted in the electron difference density peak of 28σ. The center of the iodine peak was found to be 0.4 Å away from the native chloride ion position.
Table I. Diffraction data and refinement statistics.
| Data set | Native | Os 1.13987 Å | Os 1.139205 Å | Os 1.04591 Å | Au | Iodine |
|---|---|---|---|---|---|---|
| Space group | I222 | |||||
| Unit cell parameters | ||||||
| a | 87.154 | |||||
| b | 88.031 | |||||
| c | 114.609 | |||||
| Resolution range | 20–2.15 | 20–2.81 | 20–2.82 | 20–2.68 | 20–3.0 | 20–2.23 |
| Total measurements | 96 833 | 71 728 | 80 430 | 79 013 | 11 889 | 39 337 |
| Unique reflections | 23 553 | 18 594 | 19 651 | 21 720 | 3511 | 21 279 |
| Completeness | 0.976 | 0.90 | 0.95 | 0.81 | 0.78 | 95.3 |
| Last shell | 0.99 | 0.70 | 0.76 | 0.76 | 59.4 | |
| Anomalous completeness | 0.75 | 0.84 | 0.75 | |||
| Rsym | 0.070 | 0.055 | 0.063 | 0.056 | 0.053 | 0.043 |
| Last shell | 0.249 | 0.184 | 0.175 | 0.483 | 0.109 | 0.336 |
| Phasing: isomorphous acentric (centric) | 0.57 (0.38) | 0.59 (0.39) | 0.64 (0.52) | 0.52 | ||
| Power: anomalous acentric | 0.23 | 0.31 | 0.23 | |||
| Figure of merit acentric (centric) | 0.51 (0.24) | |||||
| Protein atoms | 2749 | |||||
| Solvent | 464 | |||||
| Sulfate ions | 3 | |||||
| Chloride ion |
1 |
|
|
|
|
|
| Refinement | ||||||
| Resolution | 10.0–2.15 | |||||
| Reflections | 23 353 | |||||
| R-factor | 0.186 | |||||
| Rfree | 0.231 | |||||
| Mean B-factor (bonded) | 23.8 (1.72) | |||||
| R.m.s.d. bonds | 0.0090 Å | |||||
| R.m.s.d. angles | 1.54° | |||||
All model building steps, structure refinement and map calculations were carried out using MAIN (Turk, 1992) running on Compaq Alpha workstations. The Engh and Huber force field parameter set was used (Engh and Huber, 1991). Structure analysis was performed with MAIN during the whole course of model building and refinement: particularly useful were averaged kicked-maps, which, in cases of doubt, pointed to the correct electron density interpretation. The final model was inspected and validated with the program WHAT CHECK (Hooft et al., 1996).
All surfaces were calculated using MAIN (Turk, 1992) without taking into account solvent molecules and ions.
The substrate model using the N-terminal sequence of granzyme A ERIIGG was generated on the basis of crystal structures of papain family enzymes complexed with substrates mimicking inhibitors as described (Turk et al., 1995). Binding of substrate residues P2 and P1 into the S2 and S1 binding sites was indicated by chloromethylketone substrate analog inhibitors bound to papain (Drenth et al., 1976), and the binding of P1′ and P2′ residues into the S1′ and S2′ binding sites was suggested by CA030 in complex with cathepsin B (Turk et al., 1995). The model was built manually on superimposed structures and then minimized energetically under additional distance constraints that preserved the consensus hydrogen bonding network between the substrate and underlying enzymatic surface. The binding geometry of the P3′ and P4′ residues was generated in an extended conformation and minimized with no additional distance restraints.
The structure has been deposited in Protein Data Bank under accession code 1K3B.
Acknowledgments
Acknowledgements
This publication is dedicated to the memory of Vojko Janjić, a PhD student, who left us so abruptly. The authors wish to thank the CNR staff for assistance during native and iodine derivative data set collection on the XRD1 beam line at Elettra, Trieste, Hans Bartunik and Gleb Bourenkov for their assistance with the collection of MAD data sets on the MPG/GBF wiggler beamline BW6/DORIS at DESY, Gregor Gunčar for help with some figure preparations and lively discussions, Anže Slosar for taking part in data processing and initial heavy atom positioning attempts during his summer practice, and Iztok Dolenc and, in particular Tim Mather, for their suggestions and critical reading of the manuscript. The Slovenian Ministry of Schools, Science and Sports and ICGEB are gratefully acknowledged for financial support.
References
- Allende L.M., Garcia-Perez,M.A., Moreno,A., Corell,A., Corasol,M., Martinez-Canut,P. and Arnaiz-Villena,A. (2001) Cathepsin C gene: first compound heterozygous patient with Papillon–Lefevre syndrome and novel symptomless mutation. Hum. Mutat., 17, 152–153. [DOI] [PubMed] [Google Scholar]
- Barrett A.J., Rawlings,N.D. and Woessner,J.F. (1998) Handbook of Proteolytic Enzymes. Academic Press, London, UK. [Google Scholar]
- Baumann U., Bauer,M., Letoffe,S., Delepelaire,P. and Wandersman,C. (1995) Crystal structure of a complex between Serratia marcescens metallo-protease and an inhibitor from Erwinia chrysanthemi. J. Mol. Biol., 248, 653–661. [DOI] [PubMed] [Google Scholar]
- Carson M. (1991) Ribbons 2. J. Appl. Crystallogr., 24, 283–291. [Google Scholar]
- Cigič B., Krizaj I., Kralj,B., Turk,V. and Pain,R.H. (1998) Stoichiometry and heterogeneity of the pro-region chain in tetrameric human cathepsin C. Biochim. Biophys. Acta, 1382, 143–150. [DOI] [PubMed] [Google Scholar]
- Cigič B., Dahl,S.W. and Pain,R.H. (2000) The residual pro-part of cathepsin C fulfills the criteria required for an intramolecular chaperone in folding and stabilizing the human proenzyme. Biochemistry, 39, 12382–12390. [DOI] [PubMed] [Google Scholar]
- Cygler M., Sivaraman,J., Grochulski,P., Coulombe,R., Storer,A.C. and Mort,J. (1996) Structure of rat procathepsin B: model for inhibition of cysteine protease activity by the proregion. Structure, 4, 405–416. [DOI] [PubMed] [Google Scholar]
- Dahl S.W., Halkier T., Lauritzen,C., Dolenc,I., Pedersen,J., Turk,V. and Turk,B. (2001) Human recombinant pro-dipeptidyl peptidase I (cathepsin C) can be activated by cathepsins L and S but not by autocatalytic processing. Biochemistry, 40, 1671–1678. [DOI] [PubMed] [Google Scholar]
- Darmon A.J., Nicholson,D.W., Bleackley,R.C. (1995) Activation of the apoptotic protease CPP32 by cytotoxic T-cell-derived granzyme B. Nature, 377, 446–448. [DOI] [PubMed] [Google Scholar]
- de La Fortelle E. and Bricogne,G. (1997) Maximum-likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction methods. Methods Enzymol. Macromol. Crystallogr., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Dolenc I., Turk,B., Pungerčič,G., Ritonja,A. and Turk,V. (1995) Oligomeric structure and substrate induced inhibition of human cathepsin C. J. Biol. Chem., 270, 21626–21631. [DOI] [PubMed] [Google Scholar]
- Dolenc I., Turk,B., Kos,J. and Turk,V. (1996) Interaction of human cathepsin C with chicken cystatin. FEBS Lett., 392, 277–280. [DOI] [PubMed] [Google Scholar]
- Drenth J., Kalk,K.H. and Swen,H.M. (1976) Binding chloromethyl ketone substrate analogues to crystalline papain. Biochemistry, 15, 3731–3738. [DOI] [PubMed] [Google Scholar]
- Engh R.A. and Huber,R. (1991) Accurate bond and angle parameters for X-ray protein structure refinement. Acta Crystallogr. A, 47, 392–400. [Google Scholar]
- Fruton J.S. and Mycek,M.J. (1956) Studies of beef spleen cathepsin C. Arch. Biochem. Biophys., 65, 11–20. [DOI] [PubMed] [Google Scholar]
- Groll M., Ditzel,L., Lowe,J., Stock,D., Bochtler,M., Bartunik,H.D. and Huber,R. (1997) Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature, 386, 463–471. [DOI] [PubMed] [Google Scholar]
- Gunčar G., Podobnik,M., Pungerèar,J., Štrukelj,B., Turk,V. and Turk,D. (1998) Crystal structure of porcine cathepsin H determined at 2.1 Å resolution: location of the mini-chain C-terminal carboxyl group defines cathepsin H aminopeptidase function. Structure, 6, 51–61. [DOI] [PubMed] [Google Scholar]
- Gunčar G., Klemenčič,I., Turk,B., Turk,V., Karaoglanovic-Carmona,A., Juliano,L. and Turk D. (2000) Crystal structure of cathepsin X: a flip–flop of the ring of His23 allows carboxy-monopeptidase and carboxy-dipeptidase activity of the protease. Structure, 29, 305–313. [DOI] [PubMed] [Google Scholar]
- Gutman H.R. and Fruton,J. (1948) On the proteolytic enzymes of animal tissues VIII: an intracellular enzyme related to chymotrypsin. J. Biol. Chem., 174, 851–858. [PubMed] [Google Scholar]
- Hart T.C., Hart,P.S., Bowden,D.W., Michalec,M.D., Callison,S.A., Walker,S.J., Zhang,Y. and Firatli,E. (1999) Mutations of the cathepsin C gene are responsible for Papillon–Lefevre syndrome. J. Med. Genet., 36, 881–887. [PMC free article] [PubMed] [Google Scholar]
- Hart T.C. et al. (2000a) Haim–Munk syndrome and Papillon–Lefevre syndrome are allelic mutations in cathepsin C. J. Med. Genet., 37, 88–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T.C., Hart,P.S., Michalec,M.D., Zhang,Y., Marazita,M.L., Cooper,M., Yassin,O.M., Nusier,M. and Walker,S. (2000b) Localisation of a gene for prepubertal periodontitis to chromosome 11q14 and identification of a cathepsin C gene mutation. J. Med. Genet., 37, 95–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1996) Mapping the protein universe. Science, 273, 595–602. [DOI] [PubMed] [Google Scholar]
- Hooft R.W.W., Vriend,G., Sander,C. and Abola,E.E. (1996) Errors in protein structures. Nature, 381, 272–272. [DOI] [PubMed] [Google Scholar]
- Horn M., Pavlik,M., Doleèkova,L., Baudyš,M. and Mareš,M. (2000) Arginine-based structures are specific inhibitors of cathepsin C. Eur. J. Biochem., 267, 3330–3336. [DOI] [PubMed] [Google Scholar]
- Illy C., Quraishi,O., Wang,J., Purisima,E., Vernet,T. and Mort,J.S. (1997) Role of the occluding loop in cathepsin B activity. J. Biol. Chem., 272, 1197–1202. [DOI] [PubMed] [Google Scholar]
- Joshua-Tor L., Xu,H.E., Johnston,S.A. and Rees,D.C. (1995) Crystal structure of a conserved protease that binds DNA: the bleomycin hydrolase, Gal6. Science, 269, 945–950. [DOI] [PubMed] [Google Scholar]
- Kissinger C.R., Gehlhaar,D.K. and Fogel,D.B. (1999) Rapid automated molecular replacement by evolutionary search. Acta Crystallogr. D, 55, 484–491. [DOI] [PubMed] [Google Scholar]
- Knight S. (1989) Ribulose 1,5-bisphosphate carboxylase/oxygenase—a structural study. PhD Thesis, Swedish University of Agricultural Sciences, Uppsala.
- Kumar S. (1999) Mechanisms mediating caspase activation in cell death. Cell Death Differ., 6, 1060–1066. [DOI] [PubMed] [Google Scholar]
- Lowe J., Stock,D., Jap,B., Zwickl,P., Baumeister,W. and Huber,R. (1995) Crystal structure of the 20S proteasome from the archaeon T.acidophilum at 3.4 Å resolution. Science, 268, 533–539. [DOI] [PubMed] [Google Scholar]
- Lynch G.W. and Pfueller,S.L. (1988) Thrombin-independent activation of platelet factor XIII by endogenous platelet acid protease. Thromb. Haemostasis, 59, 372–377. [PubMed] [Google Scholar]
- McDonald J.K., Reilly,T.J., Zeitman,B.B. and Ellis,S. (1966) Cathepsin C: a chloride-requiring enzyme.Biochem. Biophys. Res. Commun., 8, 771–775. [DOI] [PubMed] [Google Scholar]
- McGuire M.J., Lipsky,P.E. and Thiele,D.L. (1992) Purification and characterization of dipeptidyl peptidase I from human spleen. Arch. Biochem. Biophys., 295, 280–288. [DOI] [PubMed] [Google Scholar]
- McGrath M.E. (1999) The lysosomal cysteine proteases. Annu. Rev. Biophys. Biomol. Struct., 28, 1818–1204. [DOI] [PubMed] [Google Scholar]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Muno D., Ishidoh,K., Ueno,T. and Kominami,E. (1993) Processing and transport of the precursor of cathepsin C during its transfer into lysosomes. Arch. Biochem. Biophys., 306, 103–110. [DOI] [PubMed] [Google Scholar]
- Musil D. et al. (1991) The refined 2.15 Å X-ray crystal structure of human liver cathepsin B: the structural basis for its specificity. EMBO J., 10, 2321–2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nauland U. and Rijken,D.C. (1994) Activation of thrombin-inactivated single-chain urokinase-type plasminogen activator by dipeptidyl peptidase I (cathepsin C). Eur. J. Biochem., 223, 497–501. [DOI] [PubMed] [Google Scholar]
- Neurath H. (1984) Evolution of proteolytic enzymes. Science, 224, 350–357. [DOI] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–376. [DOI] [PubMed] [Google Scholar]
- Nuckolls G.H. and Slavkin,H.C. (1999) Paths of glorious proteases. Nature Genet., 23, 378–380. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,V. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pariš A., Štrukelj,B., Pungerčar,J., Renko,M., Dolenc,I. and Turk,V. (1995) Molecular cloning and sequence analysis of human preprocathepsin C. FEBS Lett., 369, 326–330. [DOI] [PubMed] [Google Scholar]
- Pereira P.J., Bergner A., Macedo-Ribeiro,S., Huber,R., Matschiner,G., Fritz,H., Sommerhoff,C.P. and Bode,W. (1998) Human β-tryptase is a ring-like tetramer with active sites facing a central pore. Nature, 392, 306–311. [DOI] [PubMed] [Google Scholar]
- Pham C.T. and Ley,T.J. (1999) Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc. Natl Acad. Sci. USA, 96, 8627–8632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planta R.J., Gorter,J. and Gruber,M. (1964) The catalytic properties of cathepsin C. Biochim. Biophys. Acta, 89, 511–519. [DOI] [PubMed] [Google Scholar]
- Podack E.R. (1999) How to induce involuntary suicide: the need for dipeptidyl peptidase I. Proc. Natl Acad. Sci. USA, 96, 8312–8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podobnik M., Kuhelj,R., Turk,V. and Turk,D. (1997) Crystal structure of the wild-type human procathepsin B at 2.5 Å resolution reveals the native active site of a papain-like cysteine protease zymogen. J. Mol. Biol., 271, 774–788. [DOI] [PubMed] [Google Scholar]
- Rowan A.D., Mason,P., Mach,L. and Mort,J.S. (1992) Rat procathepsin B. Proteolytic processing to the mature form in vitro. J. Biol. Chem., 267, 15993–15999. [PubMed] [Google Scholar]
- Shresta S., Pham,C.T., Thomas,D.A., Graubert,T.A. and Ley,T.J. (1998) How do cytotoxic lymphocytes kill their targets? Curr. Opin. Immunol., 10, 581–587. [DOI] [PubMed] [Google Scholar]
- Shresta S., Graubert,T.A., Thomas,D.A., Raptis,S.Z. and Ley,T.J. (1999) Granzyme A initiates an alternative pathway for granule-mediated apoptosis. Immunity, 10, 595–605. [DOI] [PubMed] [Google Scholar]
- Travis J. (1988) Structure, function and control of neutrophil proteinases. Am. J. Med., 84, 37–42. [DOI] [PubMed] [Google Scholar]
- Toomes C. et al. (1999) Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nature Genet., 23, 421–424. [DOI] [PubMed] [Google Scholar]
- Turk B., Dolenc,I. and Turk,V. (1998) Dipeptidyl-peptidase I. In Barrett,A.J., Rawlings,N.D. and Woessner,J.F.,Jr (eds), Handbook of Proteolytic Enzymes. Academic Press, London, UK, pp. 631–634.
- Turk B., Turk,D. and Turk,V. (2000) Lysosomal cysteine proteases: more than scavengers. Biochim. Biophys. Acta, 1477, 98–111. [DOI] [PubMed] [Google Scholar]
- Turk D. (1992) Weiterentwicklung eines Programms für Molekuelgraphik und Elektrondichte-Manipulation und seine Anwendung auf verschiedene Protein-Strukturaufklärungen. PhD Thesis, Technische Universität, München.
- Turk D., Podobnik,M., Popovič,T., Katunuma,N., Bode,W., Huber,R. and Turk,V. (1995) Crystal structure of cathepsin B inhibited with CA030 at 2 Å resolution: a basis for the design of specific epoxysuccinyl inhibitors. Biochemistry, 34, 4791–4797. [DOI] [PubMed] [Google Scholar]
- Turk D., Podobnik,M., Kuhelj,R., Dolinar,M. and Turk,V. (1996) Crystal structures of human procathepsin B at 3.2 and 3.3 Å resolution reveal an interaction motif between a papain like cysteine protease and its propeptide. FEBS Lett., 384, 211–214. [DOI] [PubMed] [Google Scholar]
- Turk D., Gunčar,G., Podobnik,M. and Turk,B. (1998) Revised definition of substrate binding sites of papain-like cysteine proteases. Biol. Chem., 379, 137–147. [DOI] [PubMed] [Google Scholar]
- Wolters P.J., Laig-Webster,M. and Caughey,G.H. (2000) Dipeptidyl peptidase I cleaves matrix-associated proteins and is expressed mainly by mast cells in normal dog airways. Am. J. Respir. Cell Mol. Biol., 22, 183–190. [DOI] [PubMed] [Google Scholar]
- Wolters P.J., Pham,C.T.N., Muilenburg,D.J., Ley,T.J. and Caughey,G.H. (2001) Dipeptidyl peptidase I is essential for activation of mast cell chymases, but not tryptases, in mice. J. Biol. Chem., 276, 18551–18556. [DOI] [PubMed] [Google Scholar]