Abstract
Biochemical data have shown that specific, tightly bound phospholipids are essential for activity of the cytochrome bc1 complex (QCR), an integral membrane protein of the respiratory chain. However, the structure and function of such phospholipids are not yet known. Here we describe five phospholipid molecules and one detergent molecule in the X-ray structure of yeast QCR at 2.3 Å resolution. Their individual binding sites suggest specific roles in facilitating structural and functional integrity of the enzyme. Interestingly, a phosphatidylinositol molecule is bound in an unusual interhelical position near the flexible linker region of the Rieske iron–sulfur protein. Two possible proton uptake pathways at the ubiquinone reduction site have been identified: the E/R and the CL/K pathway. Remarkably, cardiolipin is positioned at the entrance to the latter. We propose that cardiolipin ensures structural integrity of the proton-conducting protein environment and takes part directly in proton uptake. Site-directed mutagenesis of ligating residues confirmed the importance of the phosphatidylinositol- and cardiolipin-binding sites.
Keywords: cardiolipin/cytochrome bc1 complex/oxidoreductase/phospholipid/proton transfer
Introduction
Biological membranes are essential for life. They provide a permeability barrier for cells and cell organelles and they form the matrix for membrane-spanning proteins. A general property of the phospholipid bilayer is to hinder diffusion of ions, a prerequisite for the generation of electrochemical potentials that are utilized for synthesis of ATP or active transport. The proportion of putative membrane proteins predicted from sequenced genomes is between 20 and 35% (Stevens and Arkin, 2000). The large number of membrane-embedded proteins covers a broad range of functions in cellular metabolism. A tight interaction of these molecules with the phospholipid bilayer is required to maintain the diffusion barrier. During the past few years, a number of structures of integral membrane proteins at high atomic resolution have emerged. Some of them show protein-associated phospholipid molecules (Fyfe et al., 2001). Biochemical studies have shown that phospholipids are essential for the activity of several membrane proteins (Dowhan, 1997), but the role of structurally resolved phospholipids has not been clear up to now and has not yet been addressed by site-directed mutagenesis (Fyfe et al., 2001). Elucidating the function of specifically bound phospholipid molecules might be essential to understand fully the molecular mechanism of a membrane protein.
The ubiquinol:cytochrome c oxidoreductase (QCR; cytochrome bc1 complex, EC 1.10.2.2) is a multisubunit membrane protein complex, which is one of the fundamental components of the respiratory and photosynthetic electron transfer chains. The enzyme catalyzes electron transfer from ubiquinol to cytochrome c and couples this process to electrogenic translocation of protons across the membrane (Brandt and Trumpower, 1994; Berry et al., 2000). Each monomer of the homodimeric complex contains three essential catalytic subunits with prosthetic groups: cytochrome b (COB) with two b-type hemes, cytochrome c1 (CYT1) with a c-type heme, and the Rieske protein (RIP1) containing a [2Fe–2S] iron–sulfur cluster. Mitochondrial QCRs contain up to eight additional subunits (Schägger et al., 1986). Structures determined by X-ray crystallography have been reported for bovine and chicken mitochondrial QCRs (Xia et al., 1997; Iwata et al., 1998; Zhang et al., 1998) and recently for the complex from the yeast Saccharomyces cerevisiae (Hunte et al., 2000).
The mechanism of the enzyme is known as the protonmotive Q cycle (Mitchell, 1976) and involves separate catalytic sites for quinol oxidation (Qo site) and quinone reduction (Qi site). Protons are taken up from the matrix side while quinone is reduced to quinol and protons are released to the intermembrane side upon quinol oxidation. During quinol oxidation, two electrons are transferred in a bifurcated manner via the [2Fe–2S] cluster to cytochrome c and via heme bL and heme bH to the Qi site. An alternating movement of the extrinsic domain of RIP1 between the redox center domains of COB and CYT1 may facilitate the bifurcation of the electron transfer pathway at the Qo site (Iwata et al., 1998; Zhang et al., 1998; Hunte, 2001). While the main features of electron transfer in the QCR are understood, pathways for proton uptake and release are still hypothetical (Trumpower and Gennis, 1994; Berry et al., 1999; Crofts et al., 1999; Hunte et al., 2000).
The mitochondrial QCR is integrated in the inner mitochondrial membrane. A characteristic phospholipid of this membrane is cardiolipin (CL), which in eukaryotic cells exist primarily in mitochondria (Schlame et al., 2000). Phospholipids are essential for the function of the QCR; increased delipidation of the protein leads to a gradual decrease in enzyme activity up to complete inactivity and destabilization of the multisubunit complex (Yu and Yu, 1980; Schägger et al., 1990). The unique dianionic phospholipid CL (bis-phosphatidylglycerol) seems to be of specific importance, as biochemical data indicate that it is tightly bound to the complex (Schägger et al., 1990; Hayer-Hartl et al., 1992). Digestion of tightly bound phospholipids was shown to inactivate the QCR without disturbing the spectral properties of the enzyme, and reactivation of the enzyme depended on addition of CL (Gomez and Robinson, 1999). These results suggest that CL may have a specific role in the activity of the QCR. A large number of other mitochondrial proteins that reside in the inner membrane interact with CL (Schlame et al., 2000). The presence of this phospholipid molecule is, for example, essential for full activity of the ADP–ATP carrier (Beyer and Nuscher, 1996) and cytochrome c oxidase (Robinson et al., 1990). However, it is not clear how CL contributes to full catalytic activity of these enzymes.
Here we present the assignment of five tightly bound phospholipid molecules in the high-resolution structure of yeast QCR. Their binding sites and conformations are unexpected and remarkable, and provide hints on their function in the mechanism, assembly and stability of the complex. This was supported by site-directed mutagenesis of amino acid residues involved in stabilizing the phosphatidylinositol (PI) and CL molecules in their specific binding pockets. An interhelical PI molecule contributes to the stability of the transmembrane anchor of RIP1. Two proton uptake pathways at the Qi site were identified, and CL is bound at the entrance to one of them, indicating its structural and/or functional role for quinone reduction.
Results and discussion
Phospholipid-binding sites
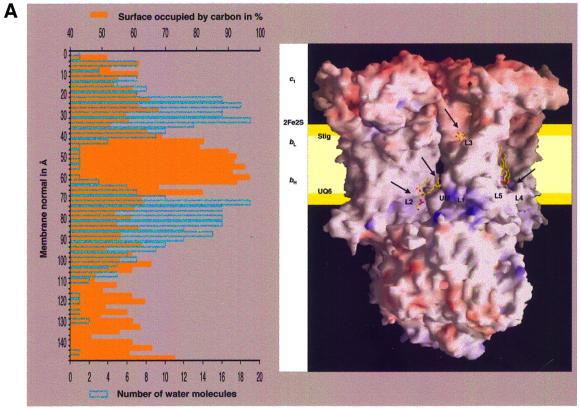
Five phospholipid molecules and one detergent molecule were identified in the 2.3 Å resolution structure of yeast QCR (Hunte et al., 2000) by typical ‘hairpin’-shaped features of the electron density map close to the surface of the transmembrane core. The quality of the electron density map allowed the assignment of individual headgroup moieties. Two phosphatidylethanolamines (PEs), one phosphatidylcholine (PC), one PI, one CL and one detergent molecule (n-undecyl-β-d-maltopyranoside, UM) were introduced and refined. Phospholipid analysis by mass spectrometry confirmed the presence of PI, PE, PC and CL in purified as well as in crystallized samples of yeast QCR (C.Lange and C.Hunte, in preparation). This is in agreement with the phospholipid composition of the inner mitochondrial membrane of S.cerevisiae that consists of 38.4% PC, 24.0% PE, 16.2% PI, 3.8% phosphatidylserine (PS), 16.1% CL and 1.5% phosphatidic acid (PA) (Zinser et al., 1991). All phospholipid molecules described in the structure are well defined in the final 2Fo – Fc electron density map. The molecules are bound to the transmembrane region at the surface of the two monomers and in two large lipophilic clefts, which are present at the dimer interface (Figure 1A and B). Binding of the lipophilic substrates quinol and quinone to the catalytic Qi and Qo sites occurs via diffusion through these clefts. The isoprenoid tails of bound quinone molecules extend into these cavities (Figure 1B). The localization of the acyl chains of the phospholipid bilayer most probably corresponds to a lipophilic core of 27 Å thickness perpendicular to the membrane plane that matches the position of the transmembrane helices (Figure 1A). This is in accordance with the experimentally determined thickness of pure PC bilayers with 18:1 acyl chains, where 38 Å were measured between the phosphodiester groups and 27 Å for the hydrophobic core (Lewis and Engelman, 1983). The deduced interface layer that resembles the position of the phospholipid headgroups is flanked by a segment of the structure with the highest number of ordered water molecules. Headgroups of all phospholipids except L3 are positioned on the matrix-oriented side. Their headgroups are bound to the protein surface in regions with positive electrostatic potential (Figure 1A).
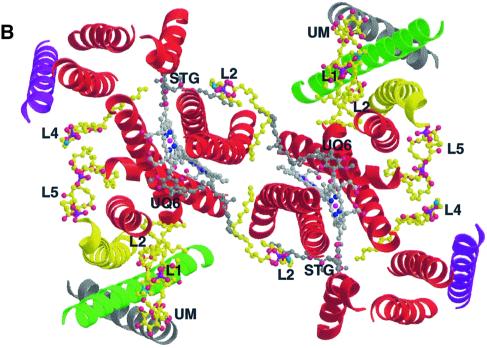
Fig. 1. Binding of lipid molecules (L1–L5) and one detergent molecule (UM) to the yeast QCR. (A) View of the homodimeric QCR parallel to the membrane plane with the intermembrane space at the top and the matrix side at the bottom. The molecule is shown in a surface representation colored according to the electrostatic potential, with positive and negative charges in blue and red, respectively. Arrows indicate the regions of bound phospholipids and detergent, which are presented as stick models. The relative orientation of the inner mitochondrial membrane is depicted in yellow. It was determined by analysis of the percentage of solvent-exposed carbon atoms (orange) and of the number of water molecules (blue) in segments along the dimer axis. The relative positions of the heme cofactors (c1, bL and bH) and the iron–sulfur cluster (2Fe–2S) as well as of ubiquinone bound at the Qi site (UQ6) and stigmatellin bound at the Qo site (Stig) are indicated. (B) Binding of phospholipids (yellow) and detergent (yellow) in the transmembrane region of QCR viewed from the matrix side on the membrane plane. The core of the dimeric complex is made up of eight helices of COB (red) per monomer. Single helices of RIP1 (green), CYT1 (yellow), QCR8 (purple) and QCR9 (gray) are attached on each side at the periphery. The heme b cofactors (light gray) as well as UQ6 and Stig (dark gray) are shown. The helices are depicted as ribbon drawings, and further molecules as ball-and-stick representation. Standard colors are used for all non-carbon atoms.
Binding in the lipophilic clefts
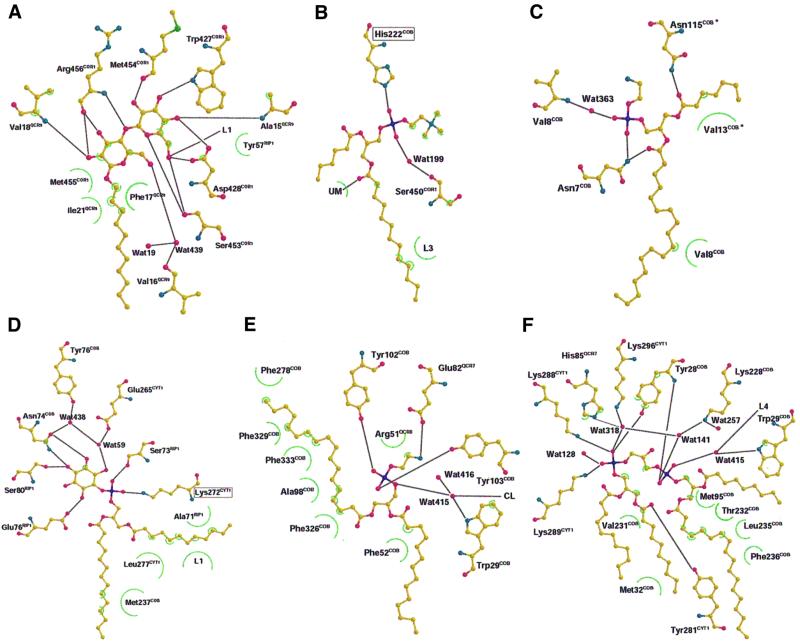
Three well-defined lipid molecules and one detergent molecule coat each of the lipophilic clefts at the dimer interface (Figure 1B). The large maltopyranoside headgroup of the detergent molecule is tightly stacked between residues of subunit COR1, subunit RIP1 and subunit 9 of the QCR (QCR9). The binding is stabilized by several hydrogen bonds between hydroxyl oxygen atoms of UM and neighboring amino acid residues (Figure 2A). Furthermore, one hydrogen bond is present between a hydroxyl oxygen atom and the neighboring phospholipid molecule (L1). The hydrocarbon chain extends into the large lipophilic cleft and the shape of the electron density map perfectly fits the length of an undecyl chain. We assume that this site is naturally occupied by a phospholipid molecule.
Fig. 2. Binding interactions between the individual phospholipids, detergent and stabilizing amino acid residues: (A) detergent (UM), (B) L1 (PC), (C) L2 (PE), (D) L3 (PI), (E) L4 (PE) and (F) L5 (CL). Apparent hydrogen bonds are indicated with black lines, and hydrophobic interactions with green semi-circles (see Materials and methods). Molecules are shown in ball-and-stick presentation using standard colors. They have been rearranged for clarity after flattening to a two-dimensional plane.
The electron density of the neighboring phospholipid (L1) is compatible with PC. The stabilizing ligand of the phosphodiester moiety is His222 of COB (Figure 2B). A stabilizing hydrogen bond is mediated by a water molecule to the backbone oxygen atom of Ser450 of subunit COR1. The acyl chains of L1 extend along helix E of COB. His222 is highly conserved in mitochondrial COB (Degli Esposti et al., 1993). Interestingly, the side chain of the neighboring residue Met221 stabilizes the orientation of the ubiquinone headgroup bound in the Qi site (Hunte et al., 2000; Hunte, 2001). Mutational analysis has to show if the L1 binding site is of functional or structural importance.
The lipid molecule L2, a PE molecule, is located at the dimer interface near the matrix side in a V-shaped opening, which is formed by helix D and D* of both COB subunits (Figures 1B and 2). Remarkably, stabilization of its binding is achieved towards one monomer by hydrogen bond interactions between Asn7 of COB and the oxygen atom of the ester-carbonyl group of the acyl chain 2 as well as the phosphodiester group (Figure 2C). Towards the second monomer (*), Asn115 of COB* is within hydrogen bonding distance of the ester-carbonyl oxygen of acyl chain 1. The acyl chains are embedded in hydrophobic grooves of COB helices D and A*, thereby closing the gap between the helices at the dimer interface. Asn115 is highly conserved in mitochondrial COB and Asn7 is only replaced by histidine or glutamine, indicating a conserved lipid-binding site. Since this lipid interacts with both monomers, it might stabilize the dimer interface and it could be involved in dimer formation. Close to L2, elongated features in the electron density map indicate the presence of additional phospholipid molecules, but an assignment has not been possible.
In general, the boundary lipids of the hydrophobic clefts cover local grooves and depressions and provide a smooth hydrophobic surface, which might support diffusion exchange of the substrates ubiquinol and ubiquinone between the Qi and Qo site and the bulk quinol pool of the inner mitochondrial membrane. The position of L2 at the dimer interface could promote a direct quinol–quinone exchange between the catalytic sites within one cleft, i.e. between the quinol-producing Qi site of one monomer and the quinol-oxidizing Qo site of the second monomer (Figure 1B).
Interhelical phospholipid
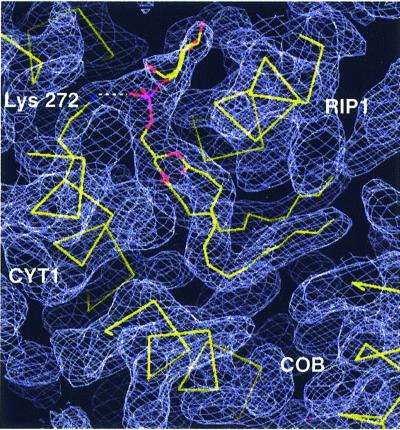
The third lipid (L3), a PI molecule, has an unusual interhelical position (Figure 3). It is wrapped around the transmembrane helix of RIP1 with the ends of the acyl chains reaching into the hydrophobic cleft at the dimer interface. Whereas all other visible lipids are located on the matrix side of the phospholipid bilayer, the headgroup of the interhelical PI points towards the intermembrane space. Remarkably, the position is ∼8 Å away from the zone where the phospholipid headgroups of the intermembrane leaflet of the phospholipid bilayer would be expected (Figure 1A). Furthermore, the headgroup is surrounded by COB helices A, B and E, as well as by the transmembrane helices of RIP1, CYT1 and subunit QCR9. The phosphodiester group of L3 forms a stabilizing ion pair with Lys272, which is conserved in eukaryotic CYT1, with the exception of Neurospora crassa. Furthermore, several hydrogen bonds between the hydroxyl oxygen atoms of the inositol moiety and side chain atoms stabilize the binding of the headgroup (Figure 2D). It is noteworthy that side chains of all catalytic subunits contribute to the stabilization of the headgroup. Beside Lys272 of CYT1, Ser73, Glu76 and Ser80 of RIP1 and Asn74 of COB are involved. In two cases, water molecules mediate the stabilizing contact. The acyl chains of PI fit tightly in a groove between the helices nearly parallel to the membrane plane and their binding is stabilized by hydrophobic interactions with residues of all catalytic subunits. The specific binding mode implies that this PI molecule stabilizes the helix packing between the transmembrane anchor of RIP1 and the transmembrane core of the protein complex and that it might be of importance for the assembly of the complex, as it is located at a point where four out of five membrane-spanning subunits of a monomer come together. Furthermore, the PI headgroup is present at the border between the transmembrane helix (Ser80) and the flexible linker region of RIP1. The flexibility of the latter ensures the movement of the extrinsic, catalytic RIP1 domain that is essential for the activity of the enzyme (Zhang et al., 1998; Nett et al., 2000). L3 might stabilize the transmembrane anchor of RIP1 close to the pivot point of movement by dissipating torsion forces generated by the fast movement.
Fig. 3. Close packing of the interhelical phospholipid L3 (PI) between the transmembrane helices of the catalytic subunits COB, CYT1 and RIP1. The phospholipid molecule, the trace of α-carbon atoms of the polypeptide chains as well as the side chain of Lys272 of CYT1 are shown. The latter forms an ion pair with the phosphodiester group of PI. The 2Fo – Fc electron density map (blue–gray) is contoured at 1.0σ. Molecules are shown in stick presentation.
When Lys272 of CYT1 is mutated to alanine, the QCR activity of the respective membranes is essentially the same as that of the membranes from the wild-type strain. When QCR is isolated from this mutant, the enzyme activity is only ∼1% compared with that of the enzyme from the wild-type strain (data not shown). The reason for this is an almost complete loss of RIP1, as can be seen by western blots of mitochondrial membranes and isolated enzyme (Figure 4). Whereas there are normal amounts of both mature and intermediate size RIP1 detectable in mitochondrial membranes, only trace amounts of RIP1 can be detected after isolation of QCR from the mutant strain. This loss of RIP1 could not be attributed to proteolysis, since the antibodies did not detect low molecular weight proteolytic fragments from RIP1 in either the membranes or the purified enzyme complex. The most likely explanation for the presence of RIP1 in the membranes and its absence in the isolated enzyme is that the destabilization of the interhelical phospholipid-binding site results in extraction of RIP1 from the enzyme during purification of QCR. Lacking the major binding interaction, i.e. the ion pair between Lys272 and the phosphodiester of L3, detergent molecules might compete with PI binding and displace the lipid molecule. This supports the assumption that the interhelical phospholipid has an important role in stabilization of RIP1 in the complex. The described mutation does not necessarily prevent binding of PI in the specific site. Further mutations are required to find out whether the interhelical PI is important for assembly of the QCR.
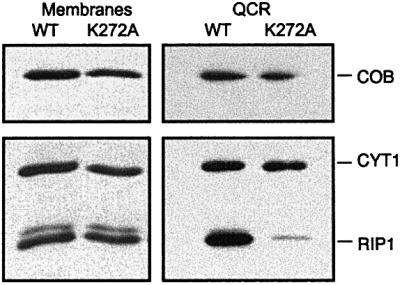
Fig. 4. Western blot analysis after SDS–PAGE separation of mitochondrial membranes (20 µg) and isolated QCR (2 pmol) from yeasts expressing wild-type and mutated CYT1 (K272A). K272 is the stabilizing ligand of the interhelical phospholipid (L3). RIP1, COB and CYT1 were detected using specific antibodies.
Binding sites at the monomer surface
Two lipid molecules (L4 and L5) are bound in a depression formed by non-polar residues of the helices A, B, E and G of COB and the transmembrane helices of CYT1 and RIP1 (Figure 1A and B). The phosphodiester group of L4, a PE molecule, is within hydrogen-bonding distance of Tyr102 and Tyr103 of COB (Figure 2E). Additional hydrogen bonding is mediated via a water molecule towards Trp29 and the headgroup of L5. Glu82 of QCR7 is within hydrogen-bonding distance of the nitrogen atom of the PE headgroup. Both tyrosine residues and Trp29 are highly conserved in mitochondrial COB. One acyl chain of L4 is kinked and inserts in a narrow bore between the transmembrane helices of COB (Figure 1B). The phospholipids bound to the monomer surface should be part of the matrix leaflet of the phospholipid bilayer. They form a mediating layer between the membrane-spanning part of the protein and help to position the complex vertically in the bilayer.
Cardiolipin
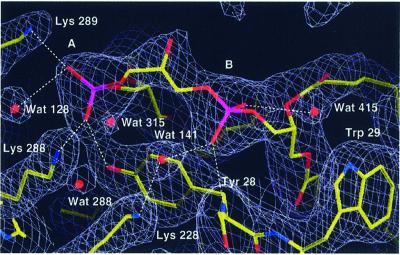
Electron density for a CL molecule (L5) is present close to lipid L4. The electron density for the two individual phosphodiester headgroups (A and B) is well defined. A continuous tube-like density is present for the bridging glycerol moiety (Figure 5). Phosphodiester group A of L5 is within hydrogen-bonding distance of Lys288 and Lys289 of CYT1 and Tyr28 of COB (Figure 2F). These residues are highly conserved in mitochondrial QCR. In addition, Tyr281 of CYT1 is within hydrogen-bonding distance of one of the ester-carbonyl oxygen atoms. Phosphodiester group B has a single hydrogen bond with the backbone nitrogen atom of Tyr28 of COB. On one side, the phosphodiester is interacting with Lys228 of COB via a hydrogen-bonded water molecule; on the other side there is a connection by a hydrogen-bonded water molecule with the headgroup of the neighboring PE molecule. The latter water molecule is within hydrogen-bonding distance of the side chain nitrogen atom of the conserved Trp29 of COB. The aromatic ring of this residue sticks out of the protein surface at the level of the ester moieties of the acyl chains. A favorable positioning of tryptophan indole rings in small peptides close to the ester carbonyls of phospholipids at the membrane water interface region has been shown experimentally (Yau et al., 1998). Trp29 is located between lipids L4 and L5 with the indole ring parallel to the dimer axis. This lamellar separation restricts lateral mobility of the phospholipids and stabilizes their binding to the protein. A similar tryptophan arrangement was found to separate purple membrane lipids bound to bacteriorhodopsin (Essen et al., 1998). The central hydroxyl group of CL most probably points towards His85 of subunit QCR7.
Fig. 5. Section of the yeast QCR model showing L5 (cardiolipin), neighboring amino acid residues and water molecules. Hydrogen bonds or ion pairs with the oxygen atoms of the phosphodiester groups A and B of the cardiolipin headgroup are indicated as dashed lines (Lys288, Lys289 of CYT1, Tyr28 of COB). Water molecules are shown as balls, and other molecules in stick presentation. The final 2Fo – Fc electron density map (blue–gray) is contoured at 1.0σ. Atoms are shown in standard colors.
To our knowledge, this is the first structural characterization of CL binding to a mitochondrial integral membrane protein. A preliminary report indicates binding of CL molecules in the structure of bovine cytochrome c oxidase (Mizushima et al., 1999). So far, the only known high-resolution structure of CL bound to a membrane protein is that of the reaction center from Rhodobacter sphaeroides (McAuley et al., 1999). The described stabilizing interactions involving the two phosphodiester groups are similar to those described here: one phosphodiester group interacts with arginine and tyrosine, and the second interacts via a water molecule with lysine. The role of CL binding for the structure and function of the reaction center is not known yet (Fyfe et al., 2001). The importance of this dianionic phospholipid may result from its unique large and charged headgroup, requiring a specific and tightly interacting binding site, which might result in a clamp-like stabilizing effect for a portion of the protein.
To analyze the importance of the specific CL-binding site in yeast QCR, each of the residues Lys288, Lys289 and Lys296 of CYT1 were changed to alanine, methionine or leucine by site-directed mutagenesis. None of these single mutations had a measurable effect on QCR activity (results not shown). Double and triple replacements of the lysines with leucines, however, resulted in a slow growth phenotype of the K289L/K296L mutant on non-fermentable carbon sources. The triple mutant grew even more slowly and was petite on solid media (Table I). Determination of turnover numbers of QCR in mitochondrial membranes revealed that the QCR of mutant K289L/K296L retained almost 90% of wild-type activity, which was inconsistent with the observed slow growth phenotype. The slow growth can be explained by reduced amounts of QCR in the mitochondrial membranes, as indicated by a low specific enzyme activity, expressed on the basis of mitochondrial membrane protein (Table I). This was confirmed by absorption difference spectra quantification of heme b in the mitochondrial membranes (Table I). Western blot analysis showed the loss of COB and CYT1 in mutant K288L/K289L, as well as in mutants K289L/K296L and K288L/K289L/K296L (Figure 6). No stable and homogenous protein preparations of QCR of these mutants could be obtained (data not shown). These results suggest a structural role for the intact CL-binding site, in either assembly or stability of the enzyme. For CL-deficient yeast mutant strains, a decreased mitochondrial membrane potential and a reduced mitochondrial function were shown (Schlame et al., 2000). It appears that elevated levels of the anionic phospholipid phosphatidylglycerol found in these mutants can only partially compensate for the CL requirement, thus supporting the need for this unique dianionic phospholipid. CL may have, in addition to its structural role, a specific function for activity of the QCR.
Table I. Doubling times and enzyme activities of yeast QCR with mutations in cardiolipin-ligating residues.
| Strain | Wild type | K288L/K289L | K288L/K296L | K289L/K296L | K288L/K289L/K296L |
|---|---|---|---|---|---|
| Doubling time (h) | 3.30 | 3.37 | 3.21 | 4.21 | 9.93 |
| Specific enzyme activity (% of WT)a | 100 | 67.6 | 79.8 | 28.6 | 12.7 |
| Turnover number of QCR (% of WT)a | 100 | 98.5 | 99.3 | 88.6 | 37.8 |
| COB content of mitochondrial membranes (% of WT) | 100 | 62.6 | 65.5 | 37.0 | 26.7 |
The turnover number of QCR in the wild-type membranes was 65/s. The COB content of the wild-type membranes was 0.095 nmol/mg.
aEnzyme activities measured in membrane preparations (see Materials and methods).
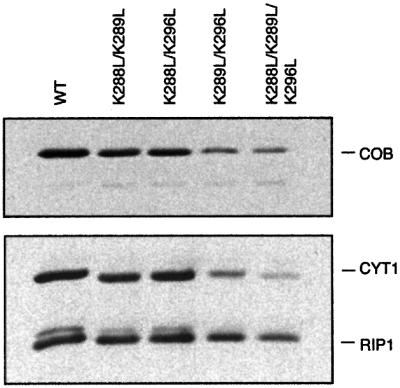
Fig. 6. Western blot analysis after SDS–PAGE separation of mitochondrial membranes (20 µg) from yeasts expressing wild-type or double and triple mutants of CYT1. Residues ligating the L5 (CL) phosphodiester group A have been mutated: (K288L/K289L, K288L/K296L, K289L/K296L and K288L/K289L/K296L). RIP1, COB and CYT1 were detected using specific antibodies.
Cardiolipin positioned at the entrance of a suggested proton uptake pathway at the Qi site
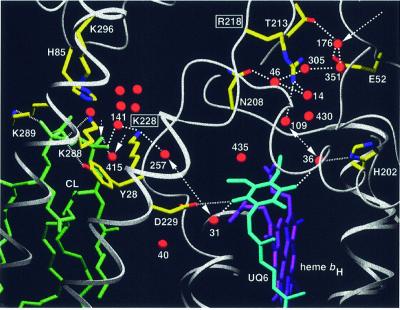
The headgroup of CL is bound in close proximity to the site of quinone reduction (Qi site). The natural substrate coenzyme Q6 (UQ6) is bound to this site in the yeast QCR structure with the ubiquinone headgroup located in a binding pocket oriented towards the matrix (Figure 1A; Hunte et al., 2000). One carbonyl oxygen atom of UQ6 is ligated by Asp229 of COB and the second via a water molecule by His202 of COB. These highly conserved residues were proposed to stabilize the protein–substrate complex and to act directly and/or indirectly via water as primary proton donors, which have to be reprotonated from the matrix (Berry et al., 1999; Hunte et al., 2000). The location of the binding pocket at the border of the transmembrane hydrophobic core as well as the lack of a direct opening to the bulk solvent (Hunte et al., 2000) require the uptake of protons by a short proton-‘wire’ or a hydrogen-bonded ‘network’ from the protein surface. A close examination of the high-resolution structure reveals two possible pathways capable of conducting protons from the bulk aqueous phase to the quinone headgroup. Interestingly, cardiolipin is positioned at the entrance to one of them (Figure 7). Well-defined water molecules are present in the spacious ubiquinone-binding pocket and each of the carbonyl groups of the quinone ring is within hydrogen-bonding distance of a water molecule (Wat31 and Wat36). Further disordered water molecules are likely to be present in the two lobes of the binding pocket, which extend to the matrix side. Reduction of quinone by heme bH most probably takes place without major reorientation of the quinone headgroup. Therefore, protonation pathways for both sides seem to be favorable.
Fig. 7. Putative proton uptake pathways at the Qi site of yeast QCR via two arrays of hydrogen-bonded water molecules, which connect the bulk solvent at the matrix side with the site of ubiquinone reduction. The entrance to the E/R pathway is formed by Glu52 of QCR7 and Wat176. The gating residue towards the quinone-binding pocket is Arg218 of COB. Cardiolipin (L5) is positioned at the entrance to the CL/K pathway, for which Lys228 of COB is the gating residue. Arrows indicate the access sites from the bulk solvent, and double arrows show proton transfer between the key residues Arg218 or Lys228 of COB and UQ6. Side chains of amino acid residues that are involved in hydrogen bond interactions or ion pair formation are shown (standard colors). Dashed lines indicate hydrogen bond interactions. Dotted lines are used for hydrogen bond interactions of UQ6 and CL ligands (His202, Asp229 and Tyr28 of COB, and Lys288 and Lys289 of CYT1). Water molecules in the cavity above the cardiolipin headgroup are stabilized by interactions with side chains of Lys228 of COB, Lys296 of CYT1 and His85 of QCR7. A surrounding layer of non-polar residues (not shown) encloses the water-filled cavity. Transmembrane helices are shown as ribbon presentation and other polypeptide backbones as rope presentation. UQ6, heme bH and CL are represented as stick models.
On the His202 oriented side of the ubiquinone headgroup, Arg218 connects the water molecules of the binding pocket with an array of several water molecules, which are connected by hydrogen bonds and stabilized by interactions with surrounding residues: Wat14, Wat46, Wat305, Wat351 and Wat176 (Figure 7). Glu52 of QCR7 stabilizes the water molecule Wat176, which has access to the bulk solvent at the dimer interface, where a hydrophilic cavity is formed by COB, subunit QCR7 and subunit QCR8. We suggest that upon reduction of ubiquinone, a proton is abstracted from Arg218, and that proton transfer occurs directly via the hydrogen-bonded water molecules, thereby allowing replenishment of this proton from the matrix side: the E/R pathway. The importance of residue Arg218 with its presumably gating role is supported by the fact that it is highly conserved and only replaced by lysine in mitochondrial COB. Further support derives from mutations of Asn223 of COB, the prokaryotic homolog of Asn208. The side chain of this residue stabilizes water molecule Wat46 of the suggested proton-conducting array. A change of this residue to valine in a prokaryotic organism led to a disturbance of the Qi kinetics (Brasseur et al., 1996).
On the Asp229 oriented side of ubiquinone, Lys228, which is highly conserved in mitochondrial COB, is the key residue for the second possible pathway for proton uptake: the CL/K pathway (Figure 7). This residue is within hydrogen-bonding distance of the water molecule Wat257 of the ubiquinone-binding pocket. Towards the protein surface, Lys228 forms a hydrogen bond with water molecule Wat141. Most remarkably, this water molecule is within hydrogen-bonding distance of the phosphodiester group B of CL. We propose that during reduction of ubiquinone, a proton is abstracted from Lys228. This consequently will be replenished from the matrix side either by reversible deprotonation of the CL phosphodiester group B, as this group is in contact with the bulk solvent and it interacts with Lys228 via a hydrogen-bonded water molecule (Wat141), or by proton exchange via Wat415, which is located between phosphodiester group B of CL and the headgroup of the neighboring phospholipid L4. It is remarkable that phosphodiester group B of CL and the phosphodiester group of L4 both lack a charge-compensating residue. This might enforce protonation of Wat415, providing a proton uptake gate. The CL headgroup and loops of CYT1, subunits QCR7 and QCR8 as well as the upper part of helix E and the ef-helix of COB form a cavity, which encloses a group of water molecules including Wat141 and Wat415. This water network and the surrounding cluster of protonable residues (Lys228, Lys296, Lys288, Lys289 and His85) may act as a buffer system.
The entrance to the CL/K pathway for proton uptake is formed by the boundary phospholipid CL. This allows the direct coupling of ubiquinone reduction with the proton source at the membrane surface of the matrix side. It was proposed that the high surface charge density of bilayer anionic phospholipids acts as a trap or buffer that concentrates protons and passes them directly to a proton-translocating pore (Haines, 1983). CL might serve as a reservoir of protons at relatively high pH due to the specific electrostatic properties of its headgroup, as in aqueous bilayer suspensions one of the two pKa values is close to the physiological pH (Kates et al., 1993). The effective pKa of the CL phosphodiester group B of the ligated CL and the electrostatic potential of the local environment needs to be analyzed to clarify, whether the proton is more likely to be taken up directly via the phosphodiester group B of CL or via the bound water molecule.
The above-described mutations have shown that CL binding at the described site is responsible for stability and integrity of the complex. Further evidence for the importance of this specific binding site derives from the observation that CL can restore binding of the Qi site-specific inhibitor antimycin in delipidated yeast QCR (Tsai and Palmer, 1986). It is likely that less tightly bound CL molecules are present in the phospholipid annulus surrounding the QCR. However, the specific CL position described here may already explain the results from biochemical studies on CL dependence of the QCR enzyme activity. Further studies including electrostatic calculations are required to prove the functional role of CL in gating proton uptake for quinone reduction in addition to its importance for the structural integrity of the Qi site architecture.
Conclusions
The described phospholipid-binding sites reflect general principles of stabilizing the interaction between membrane protein and boundary phospholipid. (i) The strongest interaction is found between the negatively charged phosphodiester moiety and a positive countercharge of lysine and histidine side chains. (ii) Depending on the individual headgroup moiety, several hydrogen bonds stabilize the binding site. (iii) Acyl chains fit tightly into hydrophobic grooves present at the protein surface and this binding is stabilized by non-polar interactions with amino acid residues. It is noteworthy that each of the described tightly bound phospholipid molecules interacts with several subunits of QCR, thus implying that these molecules act as additional stabilizing elements for the structural integrity of the multisubunit complex. It has been stressed that fluctuations of boundary phospholipids occur on a microsecond time scale and that this has to be considered for functional implications (Pebay-Peyroula and Rosenbusch, 2001). However, exchange rates for boundary lipids cannot be generalized. The described binding sites are specific for the headgroup moieties, a feature related to higher binding constants (Jost and Griffith, 1980). Furthermore, several highly conserved residues contribute to the headgroup stabilization. The species and orientation of the described phospholipid molecules including the orientation of acyl chains are astonishingly consistent between yeast QCR structures calculated from different X-ray data sets (unpublished data). It is obvious that only a small number of the array of boundary phospholipids are present in the highly delipidated complex and therefore detected in the yeast QCR structure. We assume that the highly ordered phospholipid molecules are tightly bound and are thus able to fulfill the proposed functions.
Materials and methods
X-ray crystallography
The structure of QCR from the yeast S.cerevisiae at 2.3 Å resolution was determined with a bound antibody Fv-fragment [Hunte et al., 2000; Protein Data Bank (PDB) entry 1EZV]. Elongated electron density features in the transmembrane region, which were not accounted for by the initial model, clearly indicated the presence of phospholipid molecules and one detergent molecule. PA molecules with fully saturated C18 acyl chains and one UM were positioned manually using Fo – Fc, 2Fo – Fc and omit maps of the starting model (1EZV) using the program O (Jones et al., 1991). The model was refined applying positional and B-factor refinement using the program package CNS (Brünger et al., 1998). Topology and parameter files were generated using the program XPLO2D (Kleywegt and Jones, 1997). According to Fo – Fc and 2Fo – Fc electron density maps, headgroups were identified and complete models of the specific phospholipid molecules introduced. Acyl chains were cut at the end of continuous electron density. Additional water molecules were identified close to the phospholipid headgroups. They were introduced into the model and refined. The final crystallographic R-factor of the model is 21.8% (15–2.3 Å). The free R-factor within the same resolution range is 24.9% (2.5% of the data). Introduction and refinement of the additional molecules lowered the R-factor and Rfree by 0.4 and 0.5%, respectively. Coordinates have been deposited in the PDB (accession No. 1KB9).
The location of the inner mitochondrial membrane relative to the protein was determined by analyzing the protein coordinates for solvent-exposed atoms along the membrane normal in segments of 2.5 Å thickness. The percentage of carbon atoms in each segment was used to determine the lipophilic zone of the transmembrane region. A segment of 27 Å thickness perpendicular to the membrane plane has the highest hydrophobic parameter values. This corresponds to the localization of the transmembrane helices. The thickness of the hydrophobic core is in accordance with the experimentally determined thickness of pure PC bilayers with 18:1 acyl chains, where 27 Å were measured for the lipophilic center and 38 Å for the total thickness of the bilayer (Lewis and Engelman, 1983). From the latter, the position of the two phospholipid headgroup layers was deduced. Additionally, the number of water molecules in each segment was determined and yielded two peaks separated by 40 Å that flank the two headgroup regions of the bilayer.
Analysis of neighboring atoms and hydrogen bond interactions was performed using the programs HBP (McDonald and Thornton, 1994) and LIGPLOT (Wallace et al., 1995).
Sequence analysis
Comparison of CYT1 sequences from S.cerevisiae, Kluyveromyces lactis, Neurospora crassa, Homo sapiens, Bos taurus, Solanum tuberosus, Euglena gracilis, Rhodospirillum rubrum, Rhodobacter capsulatus, Rhodobacter sphaeroides, Paracoccus denitrificans and Rhodopseudo monas viridis was performed using the program Clustal_W (Thompson et al., 1994). Sequences were taken from the Swiss Prot Database. Residues Lys272, Lys288, Lys289 and Lys296 are conserved in mitochondrial CYT1.
Plasmid construction and site-directed mutagenesis
The gene for CYT1, including 1.1 kb of the 5′- and 3′-untranslated regions, was cloned from genomic DNA with flanking AvaI and SacI sites by PCR and ligated into pGem3 and the expression vector pFL39 (CEN, Trp) to create plasmids pJN202 and pJN203, respectively. Plasmid pJN202 was used as the template for construction of the mutants using the Clontech Transformer Mutagenesis Kit. After mutagenesis, the fragments were excised with HpaI and NsiI and subcloned into pJN203. The plasmids were then transformed into the yeast strain LLD6, in which the CYT1 gene is deleted.
Western blot analysis of mitochondrial membranes and QCRs
Mitochondrial membranes (Nett et al., 1994) or QCRs (Brandt et al., 1994) were resolved on 15% SDS–polyacrylamide gels (Laemmli, 1970) and blotted to nitrocellulose membranes. RIP1, COB and CYT1 were detected by western blotting (Towbin et al., 1979), using monoclonal antibodies to RIP1 or CYT1, or polyclonal antibodies to COB.
QCR activity measurements
QCR activities of mitochondrial membranes and purified QCR were assayed in 50 mM potassium phosphate pH 7.0, 250 mM sucrose, 0.2 mM EDTA, 1 mM NaN3, 0.1% (w/v) and 0.01% Tween-20 at 23°C, using 50 µM 2,3-dimethoxy-5-methyl-6n-decyl-1,4-benzoquinol as substrate and 50 µM cytochrome c. Reduction of cytochrome c was monitored in an Aminco DW-2a™ spectrophotometer at 550 versus 539 nm in dual wavelength mode. Data were collected and analyzed using an Online Instrument Systems Inc. computer interface and software. Turnover numbers of QCR in membranes and of the purified enzyme were calculated on the basis of the concentration of COB, which was determined from optical spectra of the dithionite-reduced minus ferricyanide-oxidized samples (Ljungdahl et al., 1989). For turnover numbers, membranes from two individual isolations were assayed in triplicate; specific enzyme activities are based on the protein content of membranes. They were determined from four individual isolations and assayed in triplicate.
Acknowledgments
Acknowledgements
We are grateful to H.Michel for continuous support and we thank W.Grabarse for helpful discussions. The gift of antibodies against cytochrome b from Brian H.Robinson is acknowledged. This work was supported by the DFG (SFB472), the Fonds der Chemischen Industrie and NIH grant GM 20379.
References
- Berry E.A., Zhang,Z., Huang,L.S. and Kim,S.H. (1999) Structures of quinone-binding sites in bc complexes: functional implications. Biochem. Soc. Trans., 27, 565–572. [DOI] [PubMed] [Google Scholar]
- Berry E.A., Guergova-Kuras,M., Huang,L. and Crofts,A.R. (2000) Structure and function of cytochrome bc complexes. Annu. Rev. Biochem., 69, 1005–1075. [DOI] [PubMed] [Google Scholar]
- Beyer K. and Nuscher,B. (1996) Specific cardiolipin binding interferes with labeling of sulfhydryl residues in the adenosine diphosphate/adenosine triphosphate carrier protein from beef heart mitochondria. Biochemistry, 35, 15784–15790. [DOI] [PubMed] [Google Scholar]
- Brandt U. and Trumpower,B.L. (1994) The protonmotive Q cycle in mitochondria and bacteria. Crit. Rev. Biochem. Mol. Biol., 29, 165–197. [DOI] [PubMed] [Google Scholar]
- Brandt U., Uribe,S., Schägger,H. and Trumpower,B.L. (1994) Isolation and characterization of QCR10, the nuclear gene encoding the 8.5-kDa subunit 10 of the Saccharomyces cerevisiae cytochrome bc1 complex. J. Biol. Chem., 269, 12947–12953. [PubMed] [Google Scholar]
- Brasseur G., Saribas,A.S. and Daldal,F. (1996) A compilation of mutations located in the cytochrome b subunit of the bacterial and mitochondrial bc1 complex. Biochim. Biophys. Acta, 1275, 61–69. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite of macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Crofts A.R., Hong,S.J., Ugulava,N., Barquera,B., Gennis,R., Guergova Kuras,M. and Berry,E.A. (1999) Pathways for proton release during ubihydroquinone oxidation by the bc1 complex. Proc. Natl Acad. Sci. USA, 96, 10021–10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DegliEsposti M., De Vries,S., Crimi,M., Ghelli,A., Patarnello,T. and Meyer,A. (1993) Mitochondrial cytochrome b: evolution and structure of the protein. Biochim. Biophys. Acta, 1143, 243–271. [DOI] [PubMed] [Google Scholar]
- Dowhan W. (1997) Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu. Rev. Biochem., 66, 199–232. [DOI] [PubMed] [Google Scholar]
- Essen L.-O., Siegert,R., Lehmann,W.D. and Oesterhelt,D. (1998) Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin–lipid complex. Proc. Natl Acad. Sci. USA, 95, 11673–11678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe P.K., McAuley,K.E., Roszak,A.W., Isaacs,N.W., Cogdell,R.J. and Jones,M.R. (2001) Probing the interface between membrane proteins and membrane lipids by X-ray crystallography. Trends Biochem. Sci., 26, 106–112. [DOI] [PubMed] [Google Scholar]
- Gomez B. Jr, and Robinson,N.C. (1999) Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry, 38, 9031–9038. [DOI] [PubMed] [Google Scholar]
- Haines T.H. (1983) Anionic lipid headgroups as a proton-conducting pathway along the surface of membranes: a hypothesis. Proc. Natl Acad. Sci. USA, 80, 160–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayer-Hartl M., Schägger,H., von Jagow,G. and Beyer,G.K. (1992) Interactions of phospholipids with the mitochondrial cytochrome-c reductase studied by spin-label ESR and NMR spectroscopy. Eur. J. Biochem., 209, 423–430. [DOI] [PubMed] [Google Scholar]
- Hunte C. (2001) Insights from the structure of the yeast cytochrome bc1 complex: crystallization of membrane proteins with antibody fragments. FEBS Lett., 504, 126–132. [DOI] [PubMed] [Google Scholar]
- Hunte C., Koepke,J., Lange,C., Roßmanith,T. and Michel,H. (2000) Structure at 2.3 Å resolution of the cytochrome bc1 complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Structure, 8, 669–684. [DOI] [PubMed] [Google Scholar]
- Iwata S., Lee,J.W., Okada,K., Lee,J.K., Iwata,M., Rasmussen,B., Link,T.A., Ramaswamy,S. and Jap,B.K. (1998) Complete structure of the 11-subunit bovine mitochondrial cytochrome bc1 complex. Science, 281, 64–71. [DOI] [PubMed] [Google Scholar]
- Jones A., Zou,J.-Y, Cowan,S. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron-density maps and the location of error in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jost P.C. and Griffith,O.H. (1980) The lipid–protein interface in biological membranes. Ann. N. Y. Acad. Sci., 348, 391–407. [DOI] [PubMed] [Google Scholar]
- Kates M., Syz,J.-Y., Gosser,D. and Haines,T.H. (1993) pH-dissociation characteristics of cardiolipin and its 2′-deoxy analogue. Lipids, 28, 877–882. [DOI] [PubMed] [Google Scholar]
- Kleywegt G.J. and Jones,T.A. (1997) Model building and refinement practice. Methods Enzymol., 277, 208–230. [DOI] [PubMed] [Google Scholar]
- Laemmli U.K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lewis B.A. and Engelman,D.M. (1983) Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J. Mol. Biol., 166, 211–217. [DOI] [PubMed] [Google Scholar]
- Ljungdahl P.O., Beckmann,J.D. and Trumpower,B.L. (1989) Mutational analysis of the mitochondrial Rieske iron–sulfur protein of Saccharomyces cerevisiae. II. Biochemical characterization of temperature-sensitive RIP1– mutations. J. Biol. Chem., 264, 3723–3731. [PubMed] [Google Scholar]
- McAuley K.E., Fyfe,P.K., Ridge,J.P., Isaacs,N.W., Cogdell,R.J. and Jones,M.R. (1999) Structural details of an interaction between cardiolipin and an integral membrane protein. Proc. Natl Acad. Sci. USA, 96, 14706–14711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald I.K. and Thornton,J.M. (1994) Satisfying hydrogen bonding potential in proteins. J. Mol. Biol., 238, 777–793. [DOI] [PubMed] [Google Scholar]
- Mitchell P. (1976) Possible molecular mechanisms of the proton motive function of cytochrome systems. J. Theor. Biol., 62, 327–367. [DOI] [PubMed] [Google Scholar]
- Mizushima T. et al. (1999) Structure of phospholipids in a membrane protein complex, bovine heart cytochrome c oxidase. Acta Crystallogr. A (Suppl.), 55, P06.04.069. [Google Scholar]
- Nett J.H., Schägger,H. and Trumpower,B.L. (1994) Processing of the presequence of the Schizosaccharomyces pombe Rieske iron–sulfur protein occurs in a single step and can be converted to two-step processing by mutation of a single proline to serine in the presequence. J. Biol. Chem., 273, 8652–8658. [DOI] [PubMed] [Google Scholar]
- Nett J.H., Hunte,C. and Trumpower,B.L. (2000) Changes to the length of the flexible linker region of the Rieske protein impair the interaction of ubiquinol with the cytochrome bc1 complex. Eur. J. Biochem., 267, 5777–5782. [DOI] [PubMed] [Google Scholar]
- Pebay-Peyroula E. and Rosenbusch,J.P. (2001) High-resolution structures and dynamics of membrane protein–lipid complexes: a critique. Curr. Opin. Struct. Biol., 11, 427–432. [DOI] [PubMed] [Google Scholar]
- Robinson N.C., Zborowski,J. and Talbert,L.H. (1990) Cardiolipin-depleted bovine heart cytochrome c oxidase: binding stoichiometry and affinity for cardiolipin derivatives. Biochemistry, 29, 8962–8969. [DOI] [PubMed] [Google Scholar]
- Schägger H., Link,T.A., Engel,W.D. and von Jagow,G. (1986) Isolation of the eleven subunits of the bc1 complex from beef heart. Methods Enzymol., 126, 224–237. [DOI] [PubMed] [Google Scholar]
- Schägger H., Hagen,T., Roth,B., Brandt,U., Link,T.A. and von Jagow,G. (1990) Phospholipid specificity of bovine heart bc1 complex. Eur. J. Biochem., 190, 123–130. [DOI] [PubMed] [Google Scholar]
- Schlame M., Rua,D. and Greenberg,M.L. (2000) The biosynthesis and functional role of cardiolipin. Prog. Lipid Res., 39, 257–288. [DOI] [PubMed] [Google Scholar]
- Stevens T.J. and Arkin,I.T. (2000) Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins, 39, 417–420. [DOI] [PubMed] [Google Scholar]
- Thompson J.D., Higgins,D.G. and Gibson,T.J. (1994) CLUSTAL_W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res., 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H., Staehhelin,T. and Gordon,J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl Acad. Sci. USA, 76, 4350–4354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trumpower B.L. and Gennis,R. (1994) Energy transduction by cytochrome complexes in mitochondrial and bacterial respiration: the enzymology of coupling electron transfer reactions to transmembrane proton translocation. Annu. Rev. Biochem., 63, 675–716. [DOI] [PubMed] [Google Scholar]
- Tsai A. and Palmer,G. (1986) The role of phospholipids in the binding of antimycin to yeast complex III. Biochim. Biophys. Acta, 852, 100–105. [DOI] [PubMed] [Google Scholar]
- Wallace A.C., Laskowski,R.A. and Thornton,J.M. (1995) LIGPLOT: a program to generate schematic diagrams of protein ligand interactions. Protein Eng., 8, 127–134. [DOI] [PubMed] [Google Scholar]
- Xia D., Yu,C.A., Kim,H., Xia,J.Z., Kachurin,A.M., Zhang,L., Yu,L. and Deisenhofer,J. (1997) Crystal structure of the cytochrome bc1 complex from bovine heart mitochondria. Science, 277, 60–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yau W.-M., Wimley,W.C., Gawrisch,K. and White,S.H. (1998) The preference of tryptophan for membrane interfaces. Biochemistry, 37, 14713–14718. [DOI] [PubMed] [Google Scholar]
- Yu C.A. and Yu,L. (1980) Structural role of phospholipids in ubiquinol-cytochrome c reductase. Biochemistry, 19, 5715–5720. [DOI] [PubMed] [Google Scholar]
- Zhang Z.L., Huang,L.S., Shulmeister,V.M., Chi,Y.I., Kim,K.K., Hung,L.W., Crofts,A.R., Berry,E.A. and Kim,S.H. (1998) Electron transfer by domain movement in cytochrome bc1. Nature, 392, 677–684. [DOI] [PubMed] [Google Scholar]
- Zinser E., Sperka-Gottlieb,C.D.M., Fasch,E.-V., Kohlwein,S.D., Paltauf,F. and Daum,G. (1991) Phospholipid synthesis and lipid composition of subcellular membranes in the unicellular eukaryote Saccharomyces cerevisiae. J. Bacteriol., 173, 2026–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]