Abstract
NmrA is a negative transcriptional regulator involved in the post-translational modulation of the GATA-type transcription factor AreA, forming part of a system controlling nitrogen metabolite repression in various fungi. X-ray structures of two NmrA crystal forms, both to 1.8 Å resolution, show NmrA consists of two domains, including a Rossmann fold. NmrA shows an unexpected similarity to the short-chain dehydrogenase/reductase (SDR) family, with the closest relationship to UDP-galactose 4-epimerase. We show that NAD binds to NmrA, a previously unreported nucleotide binding property for this protein. NmrA is unlikely to be an active dehydrogenase, however, as the conserved catalytic tyrosine in SDRs is absent in NmrA, and thus the nucleotide binding to NmrA could have a regulatory function. Our results suggest that other transcription factors possess the SDR fold with functions including RNA binding. The SDR fold appears to have been adapted for other roles including non-enzymatic control functions such as transcriptional regulation and is likely to be more widespread than previously recognized.
Keywords: crystal structure/NmrA/short-chain dehydrogenase/reductase/transcriptional regulation
Introduction
Neurospora crassa, Aspergillus nidulans and other ascomycetous fungi are able to utilize a wide array of nitrogen sources and many of the pathways involved are regulated at the level of transcription by pathway-specific control proteins. When the preferred nitrogen sources of ammonium or glutamine are present in the growth medium with an alternative nitrogen source, the pathway for the non-preferred source remains inactive. This situation is known as nitrogen metabolite repression and the alternative nitrogen utilization pathway is said to be repressed (Marzluf, 1993). These observations show there is a signal transduction pathway (STP) that responds to the presence of ammonium/glutamine and targets the control of transcription of the genes involved in nitrogen metabolism.
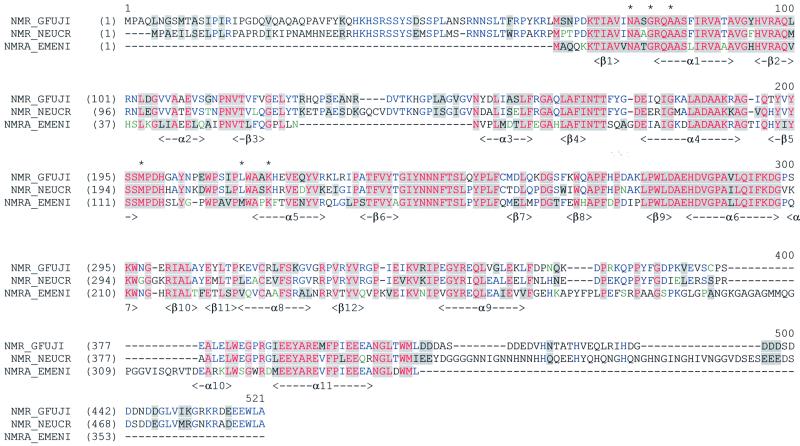
This STP is complex, having elements of positive/negative control, mRNA stability mediated through 3′-untranslated regions (UTRs) of positively acting control genes and the re-modelling of chromatin domains (Fu and Marzluf, 1990; Marzluf, 1993; Platt et al., 1996; Muro-Pastor et al., 1999). The nit-2 and areA-encoded NIT2 and AreA of N.crassa and A.nidulans, respectively, are GATA binding proteins, contain single zinc fingers and are required to stimulate transcription of genes controlled by nitrogen metabolite repression. GATA binding proteins are not just found in fungi but are widely distributed in other eukaryotes. Loss-of-function mutants of nit-2 and areA are unable to use non-preferred nitrogen sources and are said to have a repressed phenotype in contrast to the wild-type repressible phenotype (Fu and Marzluf, 1990; Marzluf, 1993; Platt et al., 1996; Wilson and Arst, 1998; Muro-Pastor et al., 1999). Isolating mutants that had a partially de-repressed phenotype in the presence of the nitrogen metabolite signalling molecule glutamine (Dunn-Coleman et al., 1981) identified the nmr-1 gene in N.crassa. The partially de-repressed phenotype of the mutants implies a negative role for NMR1 in the nitrogen metabolite repression STP. In A.nidulans, searching the expressed sequence tag (EST) database with the nmr-1 sequence identified the equivalent nmrA gene, which codes for a protein of 352 amino acids (Figure 1). The entire nmrA gene was isolated, sequenced, and used to generate deletion mutants (Andrianopoulos et al., 1998). NMR1 and NmrA share ∼60% amino acid sequence identity (Figure 1). Strains deleted for nmrA had a phenotype of partial de-repression of activities subject to nitrogen metabolite repression (Andrianopoulos et al., 1998), confirming the similar observation seen with nmr-1 mutants thereby strengthening the interpretation that NMR1 and NmrA function as repressors. Functionally important domains in NMR1 have been sought by analysis of bisulfite-induced randomly generated mutants. Many of the mutants generated had multiple amino acid substitutions making their analysis difficult, but by eliminating amino acid substitutions which occurred in other mutants that maintained their wild-type phenotype four residues whose alteration apparently abolishes nmr-1 function can be inferred (Jarai and Marzluf, 1990). NMR1 binds directly to the zinc finger and the extreme C-terminal 30 amino acids of NIT2 (Xiao et al., 1995) and given its close similarity it is likely that NmrA is able to bind to the NIT2 homologue AreA.
Fig. 1. Alignment of amino acid sequences of NmrA with NMR1 homologues from N.crassa and G.fujikuroi. The colour scheme is: black on window default colour, non-similar residues; blue window default colour, consensus residue derived from a block of similar residues at a given position; black on grey, consensus residue derived from the occurrence of >50% of a single residue at a given position; red on grey, consensus residue derived from a completely conserved residue at a given position; green window default colour, residue weakly similar to consensus residue at given position. This alignment was created and visualized via the AlignX Program from the Vector NTI Suite (Informax, Inc.). Regions of secondary structure in NmrA are indicated. Residues structurally equivalent to SDR motifs are indicated by an asterisk.
We have determined crystal structures of NmrA and its complex with NAD in order to characterize this transcriptional regulator at the molecular level.
Results and discussion
Structure determination
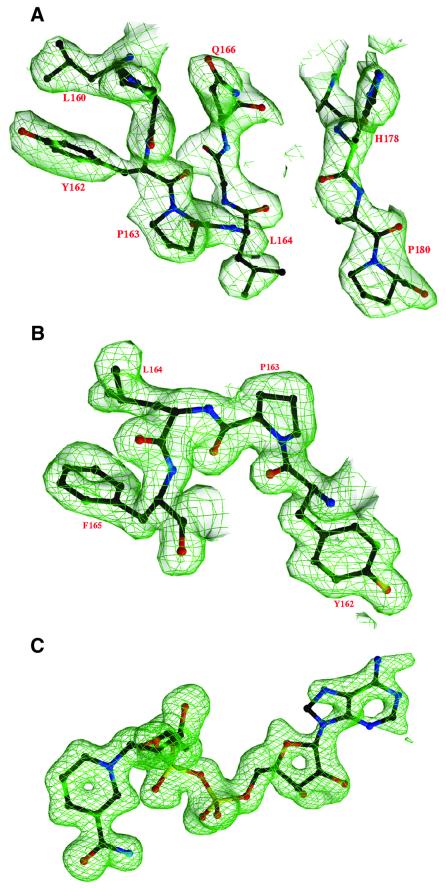
The structure of form A crystals of NmrA was determined by multiple isomorphous replacement with anomalous scattering (MIRAS) using two mercury derivatives (Table I; Figure 2A). The form B NmrA crystal structure was in turn solved by molecular replacement. Both structures were then refined to 1.8 Å resolution with final R factors of 0.205 (form A) and 0.164 (form B) (Table I). Comparison of the three different NmrA structures determined (one from form A and two from form B crystals) showed root mean square (r.m.s.) deviations for α-carbons of 0.38, 0.38 and 0.43 Å. The three structures have between 92 and 93% of residues in the most favoured regions in Ramachandran plots, however Leu164 has energetically unfavourable torsion angles (ϕ = 62°, ψ = –45°) (Figure 2B), these being observed independently in all three NmrA molecules.
Table I. Data collection and refinement statistics for native and derivative NmrA crystals.
| Data set | Cell dimensions |
Resolution limits (Å) | Reflections |
Completion % | Average I/σI | Rmergea | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| a (Å) | b (Å) | C (Å) | β (°) | Unique | Redundancy | |||||
| Form A native | 76.8 | 76.8 | 104.9 | 20.0–1.8 | 29 825 | 10.6 (8.8) | 88.5 (51.6) | 19.4 (9.2) | 0.065 (0.150) | |
| Form A MMCl | 76.8 | 76.8 | 104.8 | 30.0–2.6 | 11 281 | 8.8 (8.1) | 99.3 (99.2) | 45.6 (17.1) | 0.087 (0.245) | |
| Form A HgCl2 | 76.8 | 76.8 | 104.8 | 30.0–2.3 | 16 314 | 18.8 (15.8) | 98.4 (99.4) | 24.0 (7.2) | 0.087 (0.181) | |
| Form B native | 148.8 | 64.3 | 110.2 | 121.8 | 20.0–1.8 | 78 344 | 5.2 (3.5) | 95.5 (73.3) | 12.8 (1.3) | 0.060 (0.369) |
| Form A NAD | 76.7 | 76.7 | 104.5 | 30.0–1.5 | 54 613 | 8.6 (3.1) | 95.0 (68.4) | 29.5 (2.2) | 0.064 (0.247) | |
| MIRAS analysis 20.0–2.5Å: | R isob | Cullis R factorc |
Phasing powerd |
|||||||
| |
|
|
Centric |
Acentric Iso |
Acentric Anom |
Centric |
Acentric Iso |
Acentric Anom |
||
| MMCl | 0.174 | 0.473 | 0.474 | 0.885 | 2.52 | 3.17 | 1.39 | |||
| HgCl2 | 0.169 | 0.609 | 0.630 | 0.860 | 1.93 | 1.73 | 1.42 | |||
| Mean figure of merit | Centric = 0.51 | Acentric = 0.50 | ||||||||
| Refinement statistics | ||||||||||
| |
Resolution range (Å) |
R factore Rwork/Rfree |
R factore(all data) |
No. of atomsf |
R.m.s. bonds deviation (Å) |
R.m.s. angles deviation (°) |
Mean B factorg |
|
|
|
| Form A | 20.0–1.8 | 0.209/0.258 | 0.205 | 2530/381/4/– | 0.0095 | 1.55 | 22.8/21.5/31.2/– | |||
| Form B | 20.0–1.8 | 0.165/0.202 | 0.164 | 5113/867/7/– | 0.0093 | 1.55 | 27.4/25.5/38.2/– | |||
| Form A NAD | 30.0–1.5 | 0.189/0.222 | 0.182 | 2564/599/7/44 | 0.0091 | 1.54 | 22.0/18.7/35.2/28.9 | |||
Figures in brackets are for outer shell data.
aRmerge = Σ|I – <I>| /Σ<I>
bRiso = Σ|FPH – FP| /ΣFP where FP and FPH are native and derivative structure amplitudes.
cCullis R factor = Σ|||FPH|±ΣFP||–|FH||/Σ||FPH|±|FP||
dPhasing power = <FH>/ε, where ε is the lack of closure error.
eR factor =Σ|Fo – Fc| / ΣFo
fNo. of atoms (protein/water/ions/ligand).
gFor all atoms/protein/water/ligand.
Fig. 2. (A) Part of the MIRAS electron density map for the trigonal NmrA crystal form (form A). (B) 2Fo – Fc electron density map at 1.8 Å resolution for NmrA form A showing the region containing residue Leu164 with an unusual main-chain geometry. (C) Simulated annealing omit electron density map at 1.5 Å resolution showing NAD in the NmrA–NAD complex.
Overall structure of NmrA
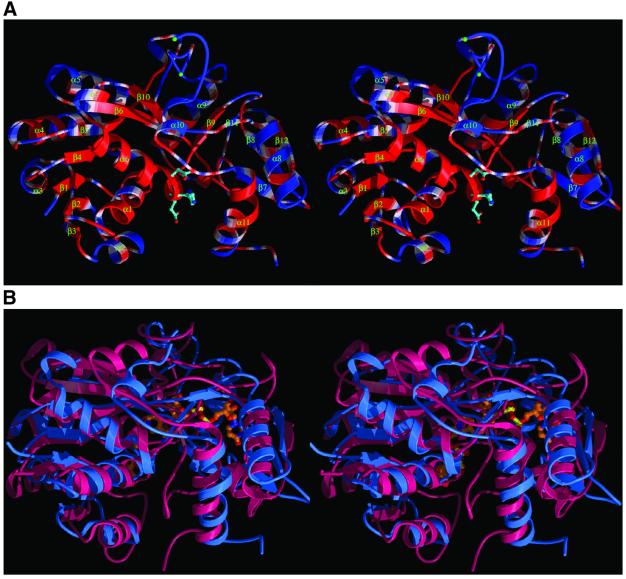
NmrA has a two-domain structure with the most striking feature being a dinucleotide (Rossmann) fold within the N-terminal region (Figure 3A). This domain has a central core of parallel β-sheet (strands β1–β6) interconnected by α-helices (α1–α5, residues 1–147) with a sixth helix (α6) and seventh strand (β10) being contributed by the C-terminal domain. Further secondary structure elements in the C-terminal domain include two β-sheets (β9, β11) and (β7, β8, β12) as well as three α-helices containing 10 or more residues (α8, α9, α11). A loop region of up to 33 amino acids (residues 283–316) is largely disordered in all three NmrA structures, this is positioned between two α-helices (α9 and α10). The two domains of NmrA have a deep cleft dividing them, reminiscent of a buried substrate binding site on an enzyme.
Fig. 3. (A) A ribbon stereodiagram showing the overall fold of NmrA with secondary structure labelled. The conserved residues among the NmrA and NMR1s (Figure 1) are coloured red whilst non-conserved residues are coloured blue. The side chains of three conserved charged residues on helix α6 are shown. (B) Stereodiagram comparing the overall structure of NmrA (blue) and UDP-galactose 4-epimerase (red). The bound NAD and UDP-glucose are shown as ball and stick representations.
Functionally important regions of NmrA
Functionally important regions and residues in NMR1 have been sought by analysis of bisulfite-induced randomly generated mutants (Jarai and Marzluf, 1990). Many of the mutants generated had multiple amino acid substitutions making their analysis difficult, but by eliminating amino acid changes which occurred in other mutants that maintained their wild-type phenotype, four residues (Gly176Ser, Met196Ile, Cys314Tyr and Glu347Lys) whose alteration apparently abolishes nmr-1 function can be inferred (Jarai and Marzluf, 1990). When the NMR1 primary sequence is mapped on to the NmrA three-dimensional structure, Gly176Ser and Met196Ile correspond to position Gly92 (helix α4) and Met113 (β5) in NmrA, suggesting that maintenance of the integrity of the Rossman fold is required for wild-type activity. NMR1 residues 123–147 in N.crassa (which are also present in the Neurospora intermedia and Neurospora sitophila NMR1 homologues) and the Giberrella fujikuroi NMR1 sequence 127–148 have no equivalent in NmrA (Young and Marzluf, 1991), and these sequences map to a loop region connecting β3 and α3 (Figures 1 and 3A), which looks capable of accommodating these additional residues without disrupting the Rossmann fold. The apparently non-functional NMR1 Cys314Tyr and Glu347Lys mutants correspond to positions Cys229 (helix α8) and Glu263 (helix α9) in NmrA and map to its C-terminal domain. Introduction into nmr-1 of a stop codon at position Trp259 encodes a protein approximately one half the size of the wild-type NMR1 and results in a loss of repression (Young et al., 1990; Young and Marzluf, 1991). By analogy to the NmrA structure, this results in the deletion of the majority of the C-terminal domain. These observations imply that the C-terminal domain of NmrA is required for full biological activity.
A glycine-rich loop (residues between 283 and 316) situated between α9 and α10 is disordered to varying degrees in the NmrA structures we have determined. Interestingly NmrA residues 299–320 have no counterpart in NMR1 (Figure 1). The loop sequence is similar in length (21 amino acids) to the N-terminal non-homologous sequence in NMR1, but the two sequences are unrelated and thus a common function appears unlikely. Mapping regions of conserved residues onto the three-dimensional structure of NmrA reveals some clustering (Figure 3A). As would be expected, some of the conserved residues are internal whilst a particularly long surface region is apparent on helix α6 (Figure 3A), which is slightly recessed but is solvent accessible. The conserved residues on the outer surface of α6 include a cluster of charged residues Glu193, His 194 and Asp 195 (Figure 3A), which could have a functional property such as the interaction with AreA.
NmrA and short-chain dehydrogenase/reductases form a structural superfamily
DALI searches (Holm and Sander, 1993) revealed that the structures most closely related to NmrA are members of the short-chain dehydrogenase/reductase (SDR) family (Jornvall et al., 1995), the highest match being with UDP-galactose 4-epimerase (UDP-GE) (Figure 3B). Using SHP analysis (Stuart et al., 1979) against human UDP-GE in complex with NADH and UDP-glucose (Thoden et al., 2000) showed that for 251 equivalent α-carbons out of 315 for NmrA and 346 for UDP-GE the r.m.s. deviation was 2.7 Å. The C-terminal domain of NmrA (a total of 175 amino acids comprising residues 155–283 and 314–352) was also most closely matched to UDP-GE in DALI searches (r.m.s. deviation of 3.8 Å for 126 equivalent residues). NmrA is more distantly related to the TrpE subunit of anthranilate synthase than to UDP-GE (r.m.s. deviation of 3.5 Å for 101 equivalenced α-carbons out of 195 for TrpE), thereby not providing support for the earlier hypothesis that NMR1 (and by implication NmrA) had evolved from this protein (Hawkins et al., 1994).
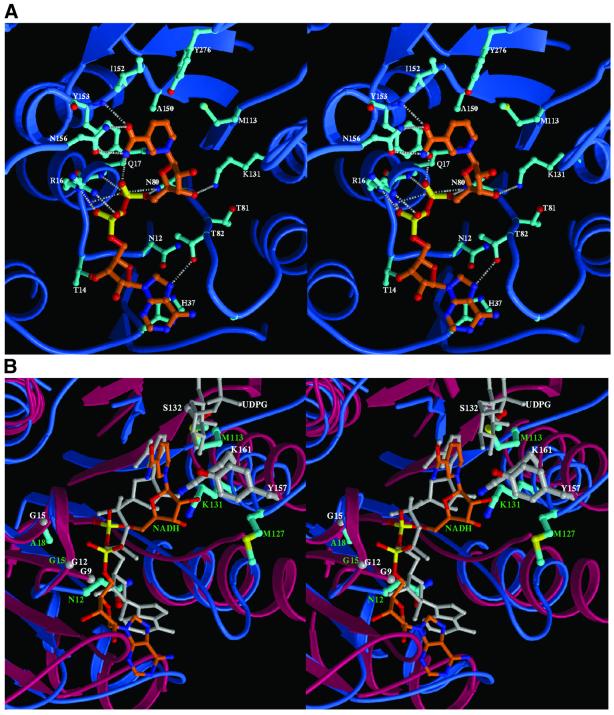
The SDRs form a large protein family, >1600 examples have been identified from a range of genomes (Oppermann et al., 2001). The relationship of NmrA to the SDR family could not be determined from sequence data base searching, however the identification of the three-dimensional structural similarity between NmrA and UDP-GE enables us to compare functionally important regions of these two proteins. SDRs generally contain at least two characteristic motifs that are involved in either the catalytic mechanism or nucleotide binding. The TyrXXXLys motif (residues 157–161 in UDP-GE) is associated with the enzyme active site and is situated on an α-helix such that the Tyr and Lys residues are pointing in a similar direction (Thoden et al., 2000). The tyrosine is thought to catalyse hydride transfer from the substrate to the C4 of the nicotinamide ring and is an absolute requirement for enzyme activity (Jornvall et al., 1995). The lysine is able to lower the pKa of the tyrosine and also hydrogen bonds to the ribose moiety of the nicotinamide mononucleotide portion of NAD (Thoden et al., 2000). For NmrA the structural equivalent to the TyrXXXLys of UDP-GE is MetXXXLys (residues 127–131) positioned on helix α5 (Figure 4B). Replacement of the Tyr by Met would strongly suggest that NmrA cannot be an active dehydrogenase of the SDR type. A less conserved residue (usually Ser/Thr) appears able to form, in conjunction with the conserved Tyr and Lys residues, a catalytic triad in some SDRs (Thoden et al., 2000). This residue is upstream from the TyrXXXLys motif and in the case of UDP-GE is Ser132 which can form a hydrogen bond to the 4′-hydroxyl of the sugar substrate (Thoden et al., 2000). For NmrA the structurally equivalent residue to Ser132 is Met113 (Figure 4B), which obviously cannot fulfil the same side-chain hydrogen bonding function as serine.
Fig. 4. (A) Stereodiagram showing the interaction between NAD and NmrA. All side chains of the protein which have any atoms ≤3.8 Å to the NAD (orange) are shown (cyan). The grey broken lines indicate hydrogen bonds. (B) Stereodiagram showing the superimposition of the NAD binding site of NmrA (blue) and the NADH site of UDP-GE (red). The NAD in NmrA is shown in orange and the NADH in UDP-GE is shown in grey. Part of the UDP-glucose molecule bound to UDP-GE is also shown in grey. The side chains of the TyrXXXLys and GlyXXGlyXXGly motifs as well as Ser132 of UDP-GE are shown as thicker grey bonds; the structurally equivalent residues in NmrA are shown in cyan.
For bi-domain SDRs the characteristic nucleotide binding motif is GlyXXGlyXXGly (Mulichak et al., 1999). The structural overlap of NmrA with UDP-GE reveals that this motif is AsnXXGlyXXAla in NmrA (Figure 1), Asn12 and Ala18 replacing two of the glycines in UDP-GE. One result of this change is that the side chain of Asn12 in NmrA protrudes into part of the equivalent region of UDP-GE containing the binding region for the adenosine moiety of NADH.
SDRs are generally homodimeric or homotetrameric (Jornvall et al., 1995), whilst NmrA is monomeric (Nichols et al., 2001). Use of the SDR-type subunit contacts might seem a plausible means of NmrA interacting with AreA, however the putative AreA binding site on NmrA involving helix α6 is not structurally equivalent to such regions.
NAD binding to NmrA
The identification of the structural relationship of NmrA to the SDR fold lead us to investigate whether NAD could bind to NmrA, using co-crystallization of NAD with NmrA and structure determination. A difference map at 1.5 Å resolution clearly reveals the NAD binding site in NmrA (Figure 2C), which is located in an overall similar position to that in UDP-GE (Figure 4B). There are a number of hydrogen bonding interactions between NmrA and NAD particularly with the NMN moiety (Figure 4A): the side-chain amide of Asn156 to the nicotinamide group, the main-chain nitrogen of Tyr153 to the nicotinamide O, the side-chain NH2 of Lys131 to the ribose 3′-OH, the main-chain carbonyl of Thr81 to the ribose 3′-OH; the side-chain hydroxyl of Tyr153 to P1 O and this also contacts the ribose O; the side-chain nitrogen of Gln80 to P1 O; the main-chain nitrogen of Gln17 to P1 O. There are also some van der Waals contacts including the side chains of Met113 and Tyr153 with the nicotinamide ring and the side-chain NH2 of Gln17 to the ribose O. In contrast, the AMP portion of NAD has fewer contacts with the protein (Figure 4A), which include hydrogen bonds from the side-chain hydroxyl of Thr82 to the adenine ring, from the side-chain hydroxyl of Thr14 to the ribose 3′-OH and from the side-chain guanidinium group of Arg16 to P2 O. The effect of the Asn12 side chain is to reorientate the adenosine group of NAD relative to its position in UDP-GE (Figure 4B). The adenine in NmrA is then able to form a ring-stacking interaction with His37 (Figure 4A). The changes in conformation of NmrA on binding NAD are mainly limited to reorientation of side chains, including Tyr153 (50° rotation about χ2) to stack with the nicotinamide ring, His37 (20° rotation about χ2) to stack with the adenine ring and the movement of Arg16 NE by 1.3 Å to interact with the phosphate group of the AMP fragment.
There is as yet no known biological requirement for NmrA to bind NAD and as discussed above the absence of the catalytic Tyr means NmrA is unlikely to have dehydrogenase activity. The fact that NmrA has many hydrogen bonding groups in positions to form a nucleotide binding site and also that the structurally analogous Lys from the SDR TyrXXXLys motif retains a similar hydrogen bonding role in the NmrA–NAD complex (i.e. interaction with the ribose 3′-hydroxyl), strongly suggests that a nucleotide binding function is associated with NmrA. That this may be of functional significance is indicated by the fact that mutation of the homologous residue to Met113 in NMR1 (Met196) gives an apparently inactive protein. In contrast, the structurally equivalent region to the UDP-glucose site of UDP-GE looks unlikely to be able to provide specific interactions to bind a further nucleotide in NmrA since it contains a number of hydrophobic residues: Ile152, Phe165, Trp177, Phe181 as well as the conformationally strained residue Leu164. Thus, the SDR fold in NmrA as well as providing a platform for interacting with the positive regulator AreA appears to have maintained a nucleotide binding function albeit in an adapted form from an SDR, whilst in contrast the dehydrogenase activity has apparently been deleted. Assuming the binding of nucleotides to NmrA does not merely represent a vestigial property which is the result of an evolutionary origin from the SDR family, the question arises as to the possible biological role for such a binding site on NmrA. It has been shown that null mutants of nit-2 are deficient in purine catabolic enzymes, implying that NIT2 is required for the expression of the genes encoding these enzymes (Jarai and Marzluf, 1990). It is thus conceivable that there is a control mechanism whereby nucleotide binding to NmrA can in turn modulate the activity of AreA. The structural effects of NAD binding to NmrA are small and thus a regulatory mechanism could depend on subtle side-chain rearrangements. Further work is currently in progress to test such ideas.
Structural relationship of other transcription factors to the SDR/NmrA fold
It has been proposed that certain proteins having functions other than dehydrogenase activity could be related to SDRs. Some transcription factors contain conserved motifs common to SDRs and it has been suggested that they are structurally related to this protein family. Thus TIP30, a host protein involved in the activation of transcription by the HIV-1 tat protein, has the GlyXXGlyXXGly associated with the Rossmann fold in certain SDRs as well as the active site TyrXXXLys motif preceded by a Ser (Xiao et al., 1998). It would thus be expected that TIP30 would be capable of functioning as a dehydrogenase (Baker et al., 2000). An alternative suggestion is that the GlyXXGlyXXGly sequence in TIP30 is an ATP binding motif and it has been reported that TIP30 is capable of autophosphorylation in vitro (Xiao et al., 2000), perhaps implying that the SDR fold can be further adapted to a kinase function. There are also cases of non-SDR dehydrogenases having additional functions involving the binding of RNA. The Rossmann fold of GAPDH can bind AU-rich RNA (Nagy and Rigby, 1995) and dihydrofolate reductase is able to bind RNA and act as its own repressor (Chu et al., 1993). It thus appears that some enzymes with NAD(P) binding domains have evolved additional functions such that the di-nucleotide site can also interact with RNA (Hentze, 1994). There is evidence that the SDR fold can be adapted to allow RNA binding as well as to give distinctive catalytic properties. For example the transcription factor CSP41 from chloroplasts (Yang et al., 1996) has been shown by multiple sequence alignment searches to be related to the SDRs but more distantly than is TIP30 (Baker et al., 1998). CSP41 retains two glycines of the GlyXXGlyXXGly motif of the bi-domain SDR Rossmann fold whilst the TyrXXXLys motif is lost. CSP41 can bind the 3′-UTR of the petD precursor mRNA, has an exoribonuclease activity that cleaves this 3′-UTR within the stem–loop structure, and thus has a role in affecting mRNA stability (Yang and Stern, 1997).
Thus, a picture is emerging of proteins with the SDR fold having a range of functional properties other than dehydrogenase or reductase activity. Proteins that apparently have lost the primary SDR function are adapted to purely transcriptional regulation properties by virtue of either interacting with another protein or RNA or having an RNase activity. In the case of NmrA, the SDR fold is adapted for use in protein–protein interaction, involving the positive regulator and GATA binding protein AreA. It also looks plausible that the nucleotide binding properties of the SDR fold have been adapted in NmrA, in this case from an enzymatic activity to another function, possibly regulatory.
This report provides the first experimental demonstration of the relatedness at the three-dimensional structural level of a transcription factor to an SDR fold and the identification of a structural superfamily. It also raises some further possibilities. Given that the NmrA/SDR relationship could not be detected by alignments of amino acid sequences, then it might be the case that the SDR fold is even more prevalent than previously realized. The SDR protein fold is already widely distributed and although largely recognized as a dehydrogenase/reductase appears to be adaptable to other functions such as those associated with transcriptional regulation including protein–protein interaction as well as RNA binding and RNase activity.
Materials and methods
Crystallization and data collection
NmrA from the A.nidulans strain R153 was expressed in Escherichia coli and purified as described previously (Nichols et al., 2001). NmrA crystals of form A (space group P3221, one molecule per asymmetric unit) and form B (space group C2, two molecules per asymmetric unit) were grown as described previously (Nichols et al., 2001). Crystals of the complex of NmrA with NAD (10 mM) were grown under form A conditions.
X-ray data for native form A and form B crystals were collected at SBS, APS Chicago, IL on beamline SBC-2 at a wavelength of 0.9537 Å. Data for the NmrA–NAD complex were collected at ESRF, Grenoble, France on beamline ID29 using a wavelength of 0.9919 Å.
Heavy atom derivatives were produced for form A crystals by soaking for 18 h in either 2 mM mercuric chloride (HgCl2) or 2 mM methyl mercury chloride (MMCl). Data were collected for both derivatives using an in-house system consisting of a MAR345 image plate and a Rigaku generator. Indexing and integration of data images were carried out with HKL2000, and data were merged using SCALEPACK (Otwinowski and Minor, 1996). X-ray data statistics are given in Table I.
Structure solution and refinement
Crystal form A of NmrA was solved by multiple isomorphous replacement with anomalous scattering. The CCP4 program suite was used for difference Patterson calculations and initial heavy atom refinement (CCP4, 1994). Isomorphous and anomalous difference Patterson maps could be easily interpreted as distinct single sites for the HgCl2 and MMCl derivatives. Refinement of heavy atom parameters was carried out using SHARP (De la Fourtelle and Bricogne, 1997); the final statistics from this are shown in Table I. A native Fourier map to 2.5 Å resolution was calculated and solvent flattened with SOLOMON (Abrahams and Leslie, 1996) assuming a solvent content of 43%, which resulted in a correlation coefficient of 0.21. The final map was of good quality (Figure 2A) and largely continuous main-chain electron density was observed starting at residue 3 apart from two breaks at 78–79 and 283–318. Alpha carbon positions were assigned to the map for 315 amino acids. Starting at Cys229 (the position of the binding sites for the mercury derivatives), a high proportion of the amino acid side chains could be fitted to the electron density map (Figure 2A). CNS (version 1.0) (Brunger et al., 1998) was used for the refinement of structures. A bulk solvent correction and anisotropic B-factor scaling was applied, rounds of positional and individual B-factor refinement were alternated with manual model rebuilding. Simulated annealing was used at the first round of refinement only. Model building was carried out using O running on an SGI O2 workstation (Jones et al., 1991). Refined co-ordinates of NmrA from form A crystals were then used to solve the B crystal form by molecular replacement with CNS.
Structure alignments
The NmrA coordinates were aligned with the protein structures available in the Protein Data Bank using the distance matrix alignment algorithm DALI (Holm and Sander, 1993). For more detailed comparisons of protein structures, SHP (Stuart et al., 1979) was used to align NmrA and UDP-GE.
Coordinates
The coordinates and structured factors for the NmrA structures have been deposited in the Protein Data Bank with access codes 1K6I, 1K6J and 1K6X.
Acknowledgments
Acknowledgements
We would like to thank the staff of the APS, Chicago, IL and ESRF, Grenoble, France for help with the data collection. We thank Dr R.Esnouf and Ms J.Dong for computer support and Dr K.Harlos for help with the in-house data collection. This work is funded by the BBSRC and The Wellcome Trust. D.K.S. is supported by the MRC.
References
- Abrahams J.P. and Leslie,A.W.G. (1996) Methods used in the structure determination of bovine mitochondrial F1 ATP-ase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Andrianopoulos A., Kourambas,S., Sharp,J.A., Davis,M.A. and Hynes,M.J. (1998) Characterization of the Aspergillus nidulans nmrA gene involved in nitrogen metabolite repression. J. Bacteriol., 180, 1973–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M.E., Grundy,W.N. and Elkan,C.P. (1998) Spinach CSP41, an mRNA-binding protein and ribonuclease, is homologous to nucleotide-sugar epimerases and hydroxysteroid dehydrogenases. Biochem. Biophys. Res. Commun., 248, 250–254. [DOI] [PubMed] [Google Scholar]
- Baker M.E., Yan,L. and Pear,M.R. (2000) Three-dimensional model of human TIP30, a coactivator for HIV-1 Tat-activated transcription and CC3, a protein associated with metastasis suppression. Cell Mol. Life Sci., 57, 851–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr., D54, 905–921. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chu E., Takimoto,C.H., Voeller,D., Grem,J.L. and Allegra,C.J. (1993) Specific binding of human dihydrofolate reductase protein to dihydrofolate reductase messenger RNA in vitro. Biochemistry, 32, 4756–4760. [DOI] [PubMed] [Google Scholar]
- De la Fourtelle E. and Bricogne,G. (1997) Maximum-likelihood heavy atom refinement for multiple isomorphous replacement and multiwavelength anomalous dispersion methods. Methods Enzymol., 276, 472–494. [DOI] [PubMed] [Google Scholar]
- Dunn-Coleman N.S., Tomsett,A.B. and Garrett,R.H. (1981) The regulation of nitrate assimilation in Neurospora crassa: biochemical analysis of the nmr-1 mutants. Mol. Gen. Genet., 182, 234–239. [DOI] [PubMed] [Google Scholar]
- Fu Y.H. and Marzluf,G.A. (1990) nit-2, the major positive-acting nitrogen regulatory gene of Neurospora crassa, encodes a sequence-specific DNA-binding protein. Proc. Natl Acad. Sci. USA, 87, 5331–5335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins A.R., Lamb,H.K., Radford,A. and Moore,J.D. (1994) Evolution of transcription-regulating proteins by enzyme recruitment: molecular models for nitrogen metabolite repression and ethanol utilisation in eukaryotes. Gene, 146, 145–158. [DOI] [PubMed] [Google Scholar]
- Hentze M.W. (1994) Enzymes as RNA-binding proteins: a role for (di)nucleotide-binding domains? Trends Biochem. Sci., 19, 101–103. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander,C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol., 233, 123–138. [DOI] [PubMed] [Google Scholar]
- Jarai G. and Marzluf,G.A. (1990) Analysis of conventional and in vitro generated mutants of nmr, the negatively acting nitrogen regulatory gene of Neurospora crassa.Mol. Gen. Genet., 222, 233–240. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Jornvall H., Persson,B., Krook,M., Atrian,S., Gonzalez-Duarte,R., Jeffery,J. and Ghosh,D. (1995) Short-chain dehydrogenases/reductases (SDR). Biochemistry, 34, 6003–6013. [DOI] [PubMed] [Google Scholar]
- Marzluf G.A. (1993) Regulation of sulfur and nitrogen metabolism in filamentous fungi. Annu. Rev. Microbiol., 47, 31–55. [DOI] [PubMed] [Google Scholar]
- Mulichak A.M., Theisen,M.J., Essigmann,B., Benning,C. and Garavito,R.M. (1999) Crystal structure of SQD1, an enzyme involved in the biosynthesis of the plant sulfolipid headgroup donor UDP-sulfoquinovose. Proc. Natl Acad. Sci. USA, 96, 13097–13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muro-Pastor M.I., Gonzalez,R., Strauss,J., Narendja,F. and Scazzocchio,C. (1999) The GATA factor AreA is essential for chromatin remodelling in a eukaryotic bidirectional promoter. EMBO J., 18, 1584–1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy E. and Rigby,W.F. (1995) Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD(+)-binding region (Rossmann fold). J. Biol. Chem., 270, 2755–2763. [DOI] [PubMed] [Google Scholar]
- Nichols C.E., Cocklin,S., Dodds,A., Ren,J., Lamb,H., Hawkins,A.R. and Stammers,D.K. (2001) Expression purification and crystallisation of Aspergillus nidulans NmrA—a negative regulatory protein involved in nitrogen metabolite repression. Acta Crystallogr. D, 57, 1722–1725. [DOI] [PubMed] [Google Scholar]
- Oppermann U.C., Filling,C. and Jornvall,H. (2001) Forms and functions of human SDR enzymes. Chem. Biol. Interact., 130–132, 699–705. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1996) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Platt A., Langdon,T., Arst,H.N., Kirk,D., Tollervey,D., Sanchez,J.M.M. and Caddick,M.X. (1996) Nitrogen metabolite signalling involves the C-terminus and the GATA domain of the Aspergillus transcription factor AreA and the 3′ untranslated region of its mRNA. EMBO J., 15, 2791–2801. [PMC free article] [PubMed] [Google Scholar]
- Stuart D.I., Levine,M., Muirhead,H. and Stammers,D.K. (1979) The crystal structure of cat pyruvate kinase at a resolution of 2.6Å. J. Mol. Biol., 134, 109–142. [DOI] [PubMed] [Google Scholar]
- Thoden J.B., Wohlers,T.M., Fridovich-Keil,J.L. and Holden,H.M. (2000) Crystallographic evidence for Tyr157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry, 39, 5691–5701. [DOI] [PubMed] [Google Scholar]
- Wilson R.A. and Arst,H.N. (1998) Mutational analysis of AreA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the ‘streetwise’ GATA family of transcription factors. Microbiol. Mol. Biol. Rev., 62, 586–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Tao,Y., Greenblatt,J. and Roeder,R.G. (1998) A cofactor, TIP30, specifically enhances HIV-1 Tat-activated transcription. Proc. Natl Acad. Sci. USA, 95, 2146–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H., Palhan,V., Yang,Y. and Roeder,R.G. (2000) TIP30 has an intrinsic kinase activity required for up-regulation of a subset of apoptotic genes. EMBO J., 19, 956–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Fu,Y.H. and Marzluf,G.A. (1995) The negative-acting NMR regulatory protein of Neurospora crassa binds to and inhibits the DNA-binding activity of the positive-acting nitrogen regulatory protein NIT2. Biochemistry, 34, 8861–8868. [DOI] [PubMed] [Google Scholar]
- Yang J. and Stern,D.B. (1997) The spinach chloroplast endoribonuclease CSP41 cleaves the 3′-untranslated region of petD mRNA primarily within its terminal stem–loop structure. J. Biol. Chem., 272, 12874–12880. [DOI] [PubMed] [Google Scholar]
- Yang J., Schuster,G. and Stern,D.B. (1996) CSP41, a sequence-specific chloroplast mRNA binding protein, is an endoribonuclease. Plant Cell, 8, 1409–1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young J.L. and Marzluf,G.A. (1991) Molecular comparison of the negative acting nitrogen control gene, nmr, in Neurospora crassa and other Neurospora and fungal species. Biochem. Genet., 29, 447–459. [PubMed] [Google Scholar]
- Young J.L., Jarai,G., Fu,Y.H. and Marzluf,G.A. (1990) Nucleotide sequence and analysis of NMR, a negative-acting regulatory gene in the nitrogen circuit of Neurospora crassa. Mol. Gen. Genet., 222, 120–128. [DOI] [PubMed] [Google Scholar]