Abstract
Notch is a highly conserved transmembrane receptor that determines cell fate. Notch signaling denotes cleavage of the Notch intracellular domain, its translocation to the nucleus, and subsequent activation of target gene transcription. Involvement of Notch signaling in several cancers is well known, but its role in melanoma remains poorly characterized. Here we show that the Notch1 pathway is activated in human melanoma. Blocking Notch signaling suppressed whereas constitutive activation of the Notch1 pathway enhanced primary melanoma cell growth both in vitro and in vivo yet had little effect on metastatic melanoma cells. Activation of Notch1 signaling enabled primary melanoma cells to gain metastatic capability. Furthermore, the oncogenic effect of Notch1 on primary melanoma cells was mediated by β-catenin, which was upregulated following Notch1 activation. Inhibiting β-catenin expression reversed Notch1-enhanced tumor growth and metastasis. Our data therefore suggest a β-catenin–dependent, stage-specific role for Notch1 signaling in promoting the progression of primary melanoma.
Introduction
Notch signaling is critical for developing and maintaining tissue homeostasis (1). Its pathway comprises a family of transmembrane receptors and their ligands, negative and positive modifiers, and transcription factors. To date, 4 mammalian receptors (Notch1 through Notch4) and at least 5 ligands (Delta 1, 3, and 4 and Jagged 1, 2) have been identified. Binding of the ligand renders the Notch receptor susceptible to metalloprotease- and γ-secretase–mediated proteolytic cleavage, which in turn results in the release of the Notch intracellular domain (NIC) from the plasma membrane and its subsequent translocation into the nucleus. Once there, NIC associates with DNA-binding protein recombination signal-binding protein Jκ/CBF1/Su(H)/Lag-1 (RBP-Jκ/CSL) and mastermind-like (MAML) protein, which recruit additional factors with histone acetylase activity, such as p300 and p300/CREB-binding protein-associated factor (PCAF). These proteins form a heteromeric complex that mediates transcription of target genes, including basic helix-loop-helix transcription factors of the hairy and enhancer of split (Hes) family (2) and the Hes-related repressor protein (Herp, also known as Hrt/Hey) family (3). Other targets, such as pre-Tα (expressed in pre-T cells) (4) and the cell cycle regulator p21 (expressed in keratinocytes) (5), are tissue specific, whereas Notch-regulated ankyrin-repeat protein (Nrarp) (6) may play an important role as a negative-feedback modulator of Notch signaling. The stability of the NIC–CSL–MAML complex is regulated by E3 ligases of the suppressor and/or enhancer of the lin-12 10 (Sel10) family (7). In addition, Notch can signal independently of CSL as it does in pathways using the cytoplasmic protein Deltex (DTX) (8).
Aberrant Notch signaling has been linked to a wide variety of tumors (9). Notch can either suppress or promote tumors depending on the cell type and context (10, 11). The first evidence of Notch’s involvement in tumorigenesis came from a chromosomal translocation of the mammalian NOTCH1 gene that resulted in constitutive activation of Notch signaling in a small subset of T cell acute lymphoblastic leukemias (T-ALL) (12). Additionally, a recent study demonstrated that novel types of activating mutations in the NOTCH1 gene occur in more than half of all T-ALL cases (13). Aberrant Notch signaling was also observed in small-cell lung cancer (14), neuroblastoma (15, 16), and cervical (17, 18) and prostate carcinoma (19). Activated Notch can transform primary Schwann cells (20). Experimental mouse models have implicated Notch signaling in skin cancer. Notch signaling drives cell-growth arrest and differentiation in keratinocytes (5, 21). Conditional ablation of Notch1 in murine epidermis results in epidermal hyperplasia, skin carcinoma, and facilitation of chemical-induced skin carcinogenesis (basal and squamous carcinomas), which implies a role for Notch1 as tumor suppressor (22). The antioncogenic effect of Notch1 in murine skin appears to be mediated by p21Waf1/Cip1 induction and repression of sonic hedgehog (Shh) and wingless/integration (Wnt) signaling.
Unlike keratinocyte-derived squamous cell and basal cell carcinomas, melanomas originate from pigment-producing melanocytes. In human skin, melanocytes are positioned at the epidermal-dermal junction and are interspersed every 5 to 10 basal keratinocytes. Using their dendrites, they interact with keratinocytes to distribute the pigment melanin (23). In turn, melanocytes are strictly controlled by keratinocytes and maintain a nonproliferative status. Transformation to melanoma is the pathological consequence of cellular control disruptions, which are environmentally initiated and likely governed by specific genetic aberrations. Melanoma development and progression is a step-wise process that consists of (a) common acquired and congenital nevi with structurally normal melanocytes; (b) dysplastic nevi with structural and architectural atypia; (c) radial growth phase (RGP), nontumorigenic primary melanomas without metastatic competence; (d) vertical growth phase (VGP), tumorigenic primary melanomas with competence for metastasis; and (e) metastatic melanoma (24).
Little is known about the role of Notch signaling in maintaining normal melanocyte homeostasis or in the development and progression of melanoma. Nor is it clear whether Notch signaling inhibits melanoma as it does keratinocyte-derived carcinoma. In this study, we investigated the involvement of Notch signaling in regulating melanoma progression using in vitro and in vivo approaches. We observed that the Notch1 signaling pathway is activated and plays an oncogenic role in melanoma progression. Furthermore, we found this effect to be β-catenin dependent. Our data define a novel role and mechanism for Notch signaling in melanoma progression and suggest that critical members of the Notch pathway are potential targets of therapy.
Results
Notch1 pathway is activated in human melanoma.
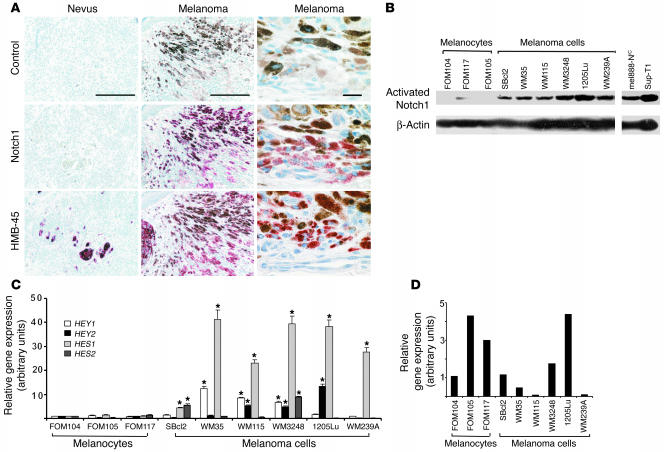
To investigate the potential involvement of Notch1 signaling in melanoma development and progression, we first carried out immunohistochemistry for Notch1 expression in human melanoma lesions. Formalin-fixed, paraffin-embedded melanoma samples from 15 patients were tested and compared with nevus samples derived from 15 individual donors. Notch1 was present in 10 out of 15 lesions but was detectable in only 1 out of 15 nevus samples (Figure 1A). HMB45 staining, which is specific for melanosomes, demonstrated that malignant cells expressing Notch1 are of melanocytic origin. Notch1 appears to be expressed in the cytosol and nucleus, implying an activated status. We also used RT-PCR to examine NOTCH1 expression in 8 melanoma cell lines and 4 normal primary human melanocytes and found an increased expression of NOTCH1 in most melanoma cell lines compared with melanocytes (data not shown). These data suggest a role for Notch1 in melanoma. To determine the functional status of Notch signaling in melanoma, we examined whether the Notch1 pathway is activated in melanoma cell lines. Six melanoma cell lines derived from different stages of progression and 3 normal primary human melanocytes were tested using an antibody specifically recognizing activated Notch1 (ab8925, Abcam). Compared with the level of activated Notch1 in melanocytes, higher levels were observed in all 6 melanoma cell lines (Figure 1B), indicating the activation of Notch1 signaling in melanoma cells. Both SUP-T1 (which are T cell leukemia harboring the t(7;9)(q34;q34.3) translocation and constitutively expressing NIC) and NIC/lenti-transfected mel888 (mel888-NIC) cells were used as positive controls. Notch1 activation in melanoma cell lines versus melanocytes was not caused by differing culture conditions (data not shown). In addition, we analyzed expression of Notch target genes HEY1, HEY2, HES1, and HES2 using quantitative real-time RT-PCR. We found at least 1 Notch target gene upregulated in a given melanoma cell line compared with melanocytes although the type of Notch target gene varied among different melanoma cells (Figure 1C). Taken together, our data suggest that the Notch signaling pathway is activated in melanoma cells. Almost all cell lines expressed the JAGGED1 gene (Figure 1D), but we saw no significant change in levels between melanocytes and melanoma cells. Keratinocytes also expressed high levels of JAGGED1, suggesting that it can act as a ligand to stimulate Notch1 on melanoma cells.
Figure 1.
Expression of Notch pathway components in human melanocytes and melanoma cells. (A) Immunohistochemistry for Notch1 in primary lesions of human malignant melanoma and in nevi. Specimens were stained using standard immunoperoxidase methods. Reddish staining indicates Notch1 positivity whereas brown staining indicates melanin. Negative control was stained with isotype-matched secondary antibodies. HMB45 staining confirmed melanocytic origin. Scale bars: 200 μm. High-magnification images are shown in the right column. Scale bar: 20 μm. (B) Western blot analysis for the levels of activated Notch1 in human melanocytes and melanoma cell lines. Increased levels of activated Notch1 in most melanoma cell lines compared with melanocytes. β-actin was used as loading control. (C) HEY1, HEY2, HES1, and HES2 mRNA expression in normal and malignant melanocytic cells as determined by quantitative real-time RT-PCR. Results are mean ± SD of 3 independent experiments. *P < 0.05, Student’s t test. (D) Relative amounts of JAGGED1 mRNA expression in normal and malignant melanocytic cells as determined by quantitative real-time RT-PCR. Results are mean ± SD of 3 independent experiments.
Suppression of Notch pathway activation inhibits melanoma growth.
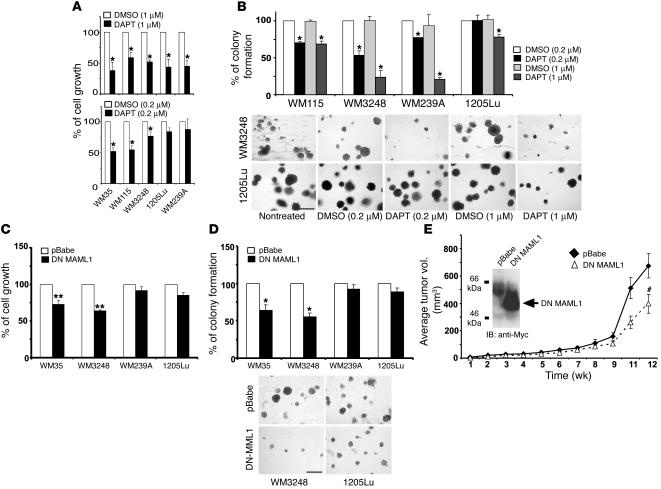
Since our data indicated that Notch1 signaling is activated in melanoma, we therefore determined whether suppression of Notch signaling would inhibit melanoma growth. To inhibit the Notch pathway, we used the γ-secretase inhibitor N-[N-(3, 5-difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester (DAPT) (Calbiochem). Treating melanoma cell lines (WM35, WM115, and WM3248 from primary lesions; 1205Lu and WM239A from metastatic lesions) with a high dose of DAPT (1 μM) significantly reduced their growth rates. However, at a low dose (0.2 μM), DAPT selectively inhibited primary but not metastatic tumor cells (Figure 2A). Similarly, colony formation of primary melanoma cells was selectively inhibited by a low dose of DAPT (Figure 2B). These results suggest a difference in sensitivity to DAPT treatment of primary versus metastatic tumor cells at low dosage. To specifically block the Notch pathway, a dominant-negative (DN) mutant of MAML1 was transfected into WM35, WM3248, WM239a, and 1205Lu melanoma cells using a retroviral vector (pBabe). DN MAML1 expression was confirmed by immunoblotting assays in WM3248 cells (Figure 2E, inner panel). As shown in Figure 2, C and D, cell growth rate — measured by 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide (MTT) assay — and colony formation in soft agar were significantly reduced in DN MAML1–transfected primary but not metastatic melanoma cells compared with those in pBabe-transfected cells. Thus, our data suggest a potential stage-specific manipulation of Notch pathway activation on tumor progression. WM3248 melanoma transfectants were then injected subcutaneously into SCID mice to determine the effects of DN MAML1 on tumor growth in vivo. Tumor growth decreased when Notch signaling was inhibited (Figure 2E). Taken together, our data suggest that Notch signaling activation plays an important role in stimulating primary melanoma proliferation and implicate the Notch pathway as a potential therapeutic target.
Figure 2.
Suppression of Notch signaling inhibits melanoma cell growth. (A) Cells were treated with DAPT (0.2 μM and 1 μM) and compared with equal volume of solvent DMSO. Cell growth was analyzed by MTT. Results indicate percentage of cell growth compared with controls (adjusted to 100%). *P < 0.005, Student’s t test. (B) Colonies formed in soft agar were counted and photographed after 10 days incubation. Graphs and representative photographs from each condition are shown. Scale bar: 500 μm. Results are mean ± SD of 3 independent experiments. (C) Growth of cells transfected with DN MAML1/pBabe was analyzed by MTT. Results are mean ± SD of 3 independent experiments. **P < 0.001, Student’s t test. (D) Number of colonies of cells transfected with DN MAML1/pBabe or pBabe (mock). Results are mean ± SD of 3 independent experiments. Representative photos are shown. Scale bar: 500 μm. (E) Effect of DN MAML1 on WM3248 cell growth was assessed in SCID mice. Tumor size was measured at indicated times after subcutaneous injection. Results are mean ± SD (n = 8). #P = 0.024, Student’s t test. Inner panel, WM3248 cells were transfected with DN MAML1/pBabe or pBabe. Immunoblots confirmed Myc-tagged DN MAML1 expression.
Activated Notch1 enhances human primary melanoma cell growth in vitro.
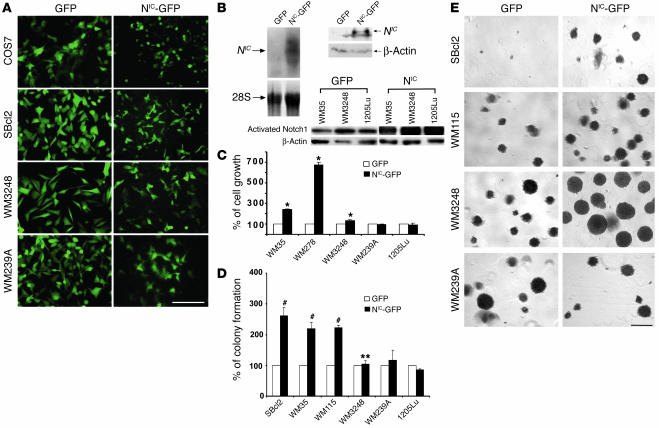
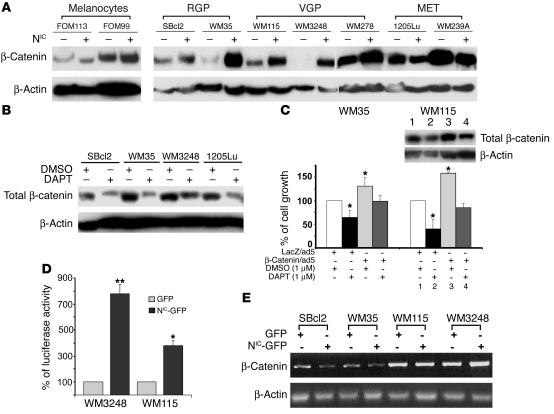
To investigate the role of aberrant Notch1 signaling on cellular properties of human melanoma, we tested the effect of activated Notch1 on melanoma cell growth in vitro by introducing the NIC into cells. First, COS7 cells were transfected with lentiviral vectors encoding either the GFP marker gene or the GFP marker gene linked to the NIC via internal ribosome entry site to express GFP and NIC independently. The NIC–GFP-transfected COS7 cells simultaneously expressed GFP (Figure 3A) and NIC (Figure 3B, upper panel). Note that the band below the NIC shown in the immunoblot appears to be nonspecific since it is detectable in both control and NIC-transfected cells. GFP therefore can be a reliable marker to indicate NIC expression in NIC-GFP/lentiviral vector–transfected cells. Melanoma cell lines were subsequently transfected. At an MOI of 5, more than 95% of transfected cells expressed GFP as observed by fluorescence microscopy (Figure 3A). Expression of total activated Notch1 in several melanoma transfectants was shown in Figure 3B, lower panel. GFP-transfected melanoma cells exhibited the same properties as untransfected parental cells whereas NIC–GFP-transfected primary melanoma cells displayed marginal morphological changes, which might reflect the increased cell adhesion capability. Experiments were carried out using pools of each cell line in which GFP-positive cells accounted for more than 95% of total. Growth rates of NIC–GFP-transfected primary melanoma cells (WM35 [RGP], WM278, and WM3248 [VGP]), significantly increased compared with that of parental cells (data not shown), GFP-transfected control cells, or NIC–GFP-transfected metastatic melanoma cells (WM239A and 1205Lu) (Figure 3C). Similarly, substantially increased colony formation was observed in NIC–GFP-transfected RGP (WM35 and SBcl2) and VGP (WM115) primary melanoma cells but not in metastatic melanoma cells (WM239A and 1205Lu) (Figure 3, D and E). The colony size of NIC–GFP-transfected WM3248 cells enlarged significantly compared with that of controls, although the number of colonies did not increase. This change also represents an increased capability in colony formation. Ectopic expression of activated Notch1 in melanocytes did not enhance their growth rate or colony formation in soft agar (data not shown). These results show that constitutively active Notch1 signaling promotes primary melanoma cell growth in vitro in a stage-specific manner.
Figure 3.
Constitutive activation of Notch1 pathway enhances melanoma growth in vitro. (A) Cells were transfected with either NIC-GFP or GFP. Equal numbers of stable transfectants were plated and photographed after 4 hours under fluorescence microscopy. Scale bar: 20 μm. More than 95% of green cells were observed in all other melanoma cell lines. (B) Upper panel shows COS7 cells transfected with NIC-GFP or GFP lentiviruses were analyzed by Northern (left) and Western (right) blot. NIC gene and protein expression are shown. 28S rRNA and β-actin were used as controls for Northern and Western blots, respectively. Lower panel shows expression of activated Notch1 in melanoma transfectants as detected by Western blot. (C) Cell growth was measured by 3[H]-thymidine incorporation assays. Ectopic expression of NIC accelerated primary melanoma but did not affect metastatic melanoma cell growth. Data were normalized by setting the activities of GFP-transfected control cells at 100. Results are mean ± SD from triplicates in 3 independent experiments. *P < 0.005, Student’s t test. (D) Percentage of colonies formed in soft agar. NIC enhances colony formation of primary but not metastatic melanoma cells. **Note that colonies formed by WM3248-NIC-GFP cells are larger than those of control, although the numbers are not significantly increased. Data were normalized by setting the activities of GFP-transfected control cells as 100. Results are mean ± SD from independent experiments. #P < 0.001, Student’s t test. (E) Representative fields in soft agar plates. Scale bar: 500 μm.
Activated Notch1 promotes human primary melanoma progression in vivo.
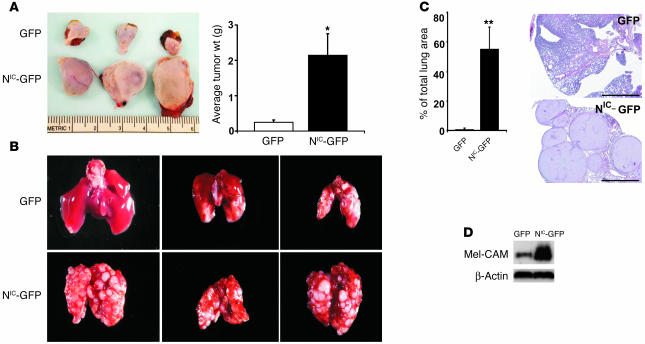
To further investigate the function of activated Notch1 signaling, we examined tumor growth in vivo of 2 stably NIC–GFP-transfected primary melanoma cell lines, WM35 (RGP) and WM3248 (VGP). We subcutaneously injected 3 × 106 WM35-NIC–GFP and WM35-GFP cells into SCID mice, respectively (n = 12 for each group). Tumor samples were harvested and measured after 7 weeks. An approximately 7-fold (by weight) increase in melanoma expansion was achieved in WM35-NIC–GFP versus WM35-GFP (Figure 4A), confirming an oncogenic effect of aberrant Notch1 signaling in melanoma. Similar results were obtained with WM3248 and WM278 cells (data not shown). Whether activated Notch1 signaling promotes progression of primary melanoma was tested by lung colony formation in an experimental metastasis model. We injected intravenously 2 × 105 WM3248-NIC-GFP and WM3248-GFP cells into SCID mice (n = 12 for each group). After 7 weeks, mice were sacrificed and lung samples were collected. Tumor colonies in lungs were macroscopically counted under a dissection microscope. In WM3248-GFP–injected mice, only a few colonies had formed in the lungs. In contrast, a dramatically increased number of colonies was observed in the lungs of WM3248-NIC–GFP-injected mice (Figure 4B), indicating that primary melanoma cells had acquired an enhanced metastatic ability when the Notch1 pathway was constitutively activated. Figure 4C shows H&E staining of lung tumor tissue and the percentage of tumor size accounting for total lung area. Similarly, an increased metastatic ability was observed in WM278-NIC-GFP cells compared with control cells (data not shown). The increased lung metastatic ability suggests that particular adhesion molecules required for 1 or more steps in extravasation or homing/attachment and adhesion of melanoma cells at distant sites may be induced. In studies aimed to identify such adhesion molecules, we found an upregulated expression of melanoma-associated cell adhesion molecule (Mel-CAM) in WM3248-NIC-GFP cells relative to control cells (Figure 4D). Although extensive studies into the biological function of Mel-CAM in mediating tumor metastasis are required, our observation does provide a substantial basis to support our hypothesis. Taken together, these data implicate aberrant Notch1 signaling in oncogenesis and primary melanoma progression.
Figure 4.
Activated Notch1 increases primary melanoma growth and promotes lung metastasis in adult mice. (A) 3 × 106 WM35-NIC-GFP or WM35-GFP cells were injected subcutaneously into mice, respectively. Mice were sacrificed after 7 weeks, and tumor samples were harvested, measured, and photographed. Representative photos of harvested melanoma (with skin) from 3 samples of each group are shown (left panel). Total tumor weight (g) from each group of mice was calculated and shown (right panel). Data are mean ± SD. *P = 0.006, Student’s t test. (B) WM3248-NIC-GFP or WM3248-GFP cells were injected intravenously into SCID mice. Mice were sacrificed after 7 weeks, and lungs were harvested. Representative lungs from each group are shown: GFP (upper panel) and NIC-GFP (lower panel). (C) Section of lungs from WM3248-NIC-GFP– and WM3248-GFP–injected mice (right panel). Percentage of total area occupied by lung tumor was measured using Image-Pro Plus Phase 3 Imaging System (MediaCybernetics) based on H&E stained sections. Data are mean ± SD. **P < 0.005, Student’s t test. Scale bars: 1 mm. (D) Activated Notch1 upregulates Mel-CAM expression on melanoma cells. Immunoblotting analysis of whole-cell lysates of NIC-GFP- or GFP-transfected WM3248 cells demonstrates increased levels of Mel-CAM in NIC–GFP–transfected but not GFP-transfected cells. Levels of β-actin are shown for equal loading conditions.
Activation of the Notch1 pathway increasesβ-catenin stability, specifically in primary melanoma cells.
To understand how activated Notch1 promotes primary melanoma progression, we investigated several potential target molecules. When determining β-catenin expression in human melanocytes and melanoma cells, we found increased levels of β-catenin in both the NIC–GFP-transfected RGP and VGP primary melanoma cells when compared with GFP-transfected (Figure 5A) and nontransfected cell lines. This increase was not observed in melanocytes or metastatic melanoma cell lines, suggesting a stage-specific effect of Notch signaling on β-catenin expression. To confirm the regulatory effect of Notch signaling on β-catenin expression, we tested whether inhibiting the endogenous Notch pathway by DAPT treatment affects β-catenin expression in melanoma cells. DAPT (1 μM) indeed suppressed β-catenin expression in melanoma cells (Figure 5B), supporting our hypothesis. We further examined whether overexpression of exogenous β-catenin overrides the inhibitory effect of DAPT on primary melanoma cell growth. We tested 2 melanoma cell lines (WM35 and WM115) by infecting them with β-catenin/adenovirus (25) (kindly provided by T. Force, Tufts University School of Medicine, Boston, Massachusetts, USA), then treating them with DAPT (1 μM). As shown in Figure 5C, tumor cells overexpressing β-catenin grew faster. Although DAPT was able to suppress cell proliferation, the cell growth rate was comparable to that of parental cells not treated with DAPT. These data further support an important role for β-catenin in promoting melanoma cell proliferation.
Figure 5.
Activation of Notch1 signaling stabilizes β-catenin in primary melanoma cell lines. (A) Immunoblotting analysis demonstrates increased levels of β-catenin in NIC–GFP-transfected RGP and VGP primary melanomas but not in NIC–GFP-transfected melanocytes or metastatic melanoma cells. Levels of β-actin are shown for equal loading conditions. (B) DAPT treatment inhibits β-catenin expression in melanoma cells. Cells were treated with 1 μM DAPT for 24 hours, and whole-cell lysates were analyzed by immunoblotting assays. Levels of β-actin are shown for equal loading conditions. (C) Introducing exogenous β-catenin overrides the inhibitory effect of DAPT on primary melanoma cell growth. β-catenin/adenovirus–transfected melanoma cells were treated with DAPT (1 μM) or DMSO. Overexpression of β-catenin accelerated cell growth as measured by MTT assay. Inner panel shows detection of β-catenin expression by immunoblot. Although DAPT suppressed cell proliferation, the growth rate was comparable to that of parental cells not treated with DAPT. Results are mean ± SD of 3 independent experiments. *P < 0.01, Student’s t test. (D) Luciferase assay demonstrates an enhanced TCF-mediated transcription activity in NIC–GFP-transfected cells compared with control cells. Data were normalized by setting the luciferase activities of control cells as 100. Results are mean ± SD of 3 independent experiments. **P < 0.001, Student’s t test. (E) RT-PCR analysis demonstrates that RNA levels of β-catenin are not concurrently upregulated in the NIC–GFP-transfected RGP and VGP cell lines, suggesting a posttranscriptional mechanism in stabilizing β-catenin. β-actin was used as control.
Moreover, a functional analysis of β-catenin activity by T cell–specific transcription factor–luciferase (TCF-luciferase) assays confirmed the enhanced β-catenin activity in NIC–GFP-transfected WM3248 and WM115 cells compared with controls (Figure 5D). To determine whether the upregulation of β-catenin by activated Notch1 occurred at the gene transcription level or posttranscriptionally, we performed RT-PCR assays to assess the level of β-catenin mRNA in transfectants. As shown in Figure 5E, transcripts of β-catenin were not increased in NIC–GFP-transfected primary melanoma cells, indicating that the increased level of β-catenin protein was due to enhanced stability. The stage-specific role of activated Notch1 in regulating β-catenin stability is coincident with its stage-specific effect on primary melanoma cell growth in vitro, thus suggesting that the oncogenic effect of activated Notch1 is mediated through β-catenin.
The oncogenic role of activated Notch1 in promoting primary melanoma progression isβ-catenin dependent.
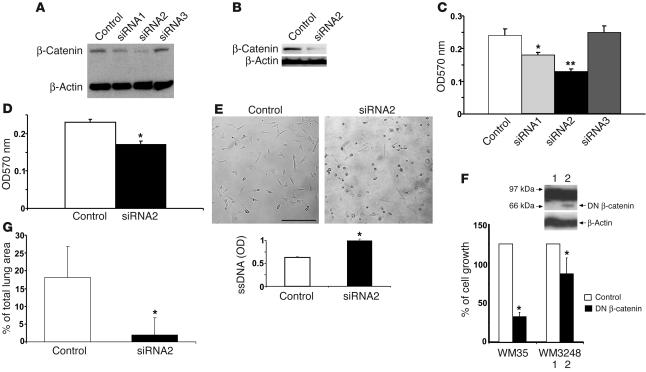
To further address the above-mentioned issue, we used a lentiviral vector–mediated RNAi to stably knock down β-catenin levels. Based on the recently described siRNA technique for persistent gene silencing in mammalian cells (26–28), we designed a mammalian expression vector, H1UG. Three RNAi vectors were constructed by cloning DNA sequences encoding short hairpin RNA (shRNA) that targets β-catenin mRNA into BamH1 and XhoI sites to render them under control of the H1 promoter. Recombinant lentiviruses were generated to infect melanoma cells at 5 MOI per cell. One out of 3 shRNA vectors displayed high-efficiency knockdown of β-catenin in melanoma cell lines and was selected for subsequent experiments. The original DNA sequences encoding this shRNA that targets β-catenin mRNA was: 5′-GCAAGCTCATCATACTGGC-3′. The C at the 3′ end of sense was changed to T to create a “G:U wobble” base pair upon pairing with its antisense RNA complement in the hairpin. This modification enhances the efficiency of specific gene silencing (29). WM35-NIC and WM3248-NIC cells were transfected with a lentiviral vector encoding shRNA targeting sequence to establish sublines (termed β-cat-siRNA/WM35-NIC and β-cat-siRNA/WM3248-NIC). Immunoblotting analyses showed that, compared with parental WM35-NIC and WM3248-NIC cells (data not shown) and H1UG lentivirus–transfected control cells (called H1UG/WM35-NIC and H1UG/WM3248-NIC), there was a substantial reduction of β-catenin in these sublines (approximately 78% in WM35 and more than 90% in WM3248 cells) whereas the β-actin control remained unaltered (Figure 6, A and B).
Figure 6.
Suppressing β-catenin reverses Notch1-induced tumor growth and metastasis. (A and B) β-catenin was examined by immunoblotting in WM35–NIC-GFP cells expressing β-cat–siRNA1, β-cat–siRNA2, β-cat–siRNA3 and control vector (A) and in WM3248–NIC-GFP cells expressing β-cat–siRNA2 (B). β-cat–si–RNA2 efficiently suppressed expression of β-catenin. β-actin was used as loading control. (C and D) MTT assay shows a decreased growth rate induced by β-cat–siRNA1 (*P < 0.01, Student’s t test) and β-cat–siRNA2 (**P < 0.001, Student’s t test) in WM35–NIC-GFP cells and β-cat–siRNA2 in WM3248-NIC-GFP cells when compared with control. Results are mean ± SD from triplicates of 3 independent experiments. (E) Induction of cell apoptosis by β-cat–siRNA2 in WM35–NIC-GFP cells. Round shape indicates cell death. Scale bar: 20 μm. Cell apoptosis was quantitatively measured by ELISA. Results are mean ± SD from triplicates of 3 independent experiments. Similar results were observed in WM3248–NIC-GFP cells. (F) 3[H]-Thymidine incorporation assay shows a decreased growth rate of WM3248 cells transfected with DN β-catenin compared with the control. Results are mean ± SD from 3 independent experiments. Inner panel shows expression of DN β-catenin protein. β-actin was used as loading control. (G) In vivo lung colony–formation assay. β-cat–siRNA-WM3248-NIC-GFP or H1UG-WM3248-NIC-GFP cells were injected intravenously into mice. After 12 weeks, lung samples were harvested and analyzed. Tumor colonies formed on the lung surface were macroscopically counted under a dissection microscope. Data are mean ± SD.
We then assessed whether β-catenin suppression was sufficient to affect Notch1-induced melanoma cell growth and metastatic capacity. As shown in Figure 6, C and D, β-catenin suppression significantly reversed Notch1-induced melanoma cell proliferation. This is due to the induction of melanoma cell apoptosis since more dead cells were observed in apoptosis assays (Figure 6E). To confirm the effect of β-catenin suppression on melanoma cell growth, we also examined the effect of DN β-catenin mutant. NIC–GFP-WM3248 cells were transfected with DN β-catenin/pcDNA4 (30) (kindly provided by J.B. Trepel, National Cancer Institute, Bethesda, Maryland, USA) using Lipofectamine 2000 (Invitrogen Corp.). DN β-catenin inhibited cell proliferation compared with controls (Figure 6F). Expression of DN β-catenin in transfectants was confirmed by immunoblotting analysis (Figure 6F, inner panel).
Next, we tested β-cat-siRNA/WM3248-NIC and H1UG/WM3248-NIC cells for their ability to form metastatic lung colonies in mice. We injected 4 × 105 cells into SCID mice (n = 4) via tail vein. After 12 weeks, mice were sacrificed and lungs were collected. Compared with the H1UG/WM3248-NIC cell–injected group, a significantly decreased number of colonies had developed in lungs of mice injected with β-cat-siRNA/WM3248-NIC cells (Figure 6G). These results indicate that β-catenin mediates, at least partly, the oncogenic effect of activated Notch1 and that β-catenin places as a functional downstream target of Notch1 signaling in primary melanoma cells.
Discussion
In this study, we demonstrate that Notch1 signaling is activated in human melanoma. Activated Notch1 plays a β-catenin–dependent and stage-specific oncogenic role in promoting primary melanoma progression. Notch signaling has varied roles in cancer that reflect its multiple functions in development and tissue homeostasis. In contrast to its oncogenic role in melanoma progression, Notch1 signaling exerts an antioncogenic effect in mouse basal cell and squamous cell carcinomas of the skin. Correspondingly, Notch receptors are downregulated in human skin carcinomas (31). The versatile effects of Notch signaling on cell differentiation, proliferation, survival, and tumorigenesis have been clearly demonstrated, so it is not surprising that Notch signaling plays differential roles in 2 types of skin cancers. For example, in B cell development, Notch1 induces growth arrest and apoptosis (32) whereas in T cells, it induces cell proliferation (33). Notch signaling provides cell proliferation cues and elicits oncogenic activities in acute lymphoblastic T cell leukemias (12), mouse mammary tumors (34), and transformed kidney epithelial cells (35). On the other hand, it acts as a growth antagonist or tumor suppressor not only in murine basal cell carcinoma but also in small cell lung cancer (14), hepatocellular carcinoma (36), and prostate cancer (19). Notch signaling also counteracts HPV-induced transformation (18), and its antioncogenic activity in cervical cancer might be mediated by MAML (37). Although the precise mechanism for the dual action of Notch remains to be explored, it is now well accepted that the consequences of Notch signaling depend on signal strength, timing, and especially cellular context (11).
We focused on Notch1 signaling in this study because Notch1 is upregulated in melanoma cell lines and lesions relative to normal melanocytes and nevi. Our data point to Notch1 as a critical player in melanoma progression but do not exclude the potential involvement of other Notch family members. Consistent with our observation is the recent report that intracellular forms of all 4 Notch receptors, including Notch1, are expressed in primary lesions of human malignant melanoma (38). Yet it is not clear whether several Notch receptor–mediated signaling pathways are simultaneously activated in melanoma or whether a specific Notch receptor is preferentially activated in melanoma lesions or cell lines. Using cDNA microarray analyses, Hoek et al. found the NOTCH2 gene upregulated in melanomas relative to melanocytes (39). They also found the expression of the HEY1 gene upregulated, which is consistent with our observation and supports our assertion that Notch signaling is activated in melanoma.
β-catenin has been suggested as a functional target gene for Notch1 signaling that mediates the tumor-suppressive effect in murine skin carcinoma (22, 40). In melanoma, however, β-catenin mediates the oncogenic role of Notch1 signaling. Thus, given the role of Notch signaling in various skin cancers, β-catenin exerts differential roles on tumorigenesis that depend on Notch activation. This suggests that differential roles for Notch signaling in skin cancer are determined upstream of β-catenin. Aberrant activation of β-catenin caused by mutation (41), elevation in wild-type β-catenin nuclear content (42), or stabilization by IGF-1 signaling (43) has been linked to melanoma progression. β-catenin plays a fundamental role in regulating E- or N-cadherin–mediated cell adhesion. It also plays a role in the Wnt signaling pathway by activating TCF/lymphoid enhancer–binding factor–regulated (TCF/LEF-regulated) gene transcription. Indeed, increased expression of β-catenin protein, but not mRNA, and enhanced TCF-luciferase activity implicate β-catenin stabilization. Regulation of β-catenin by Notch activation in melanoma may be mediated by crosstalk with the Wnt pathway or by regulation of N-cadherin, which may also account for its increased growth and antiapoptotic and metastatic activities (44).
The oncogenic role of Notch signaling in melanoma displays a stage-specific characteristic. Notch signaling advances primary melanoma but has little effect on metastatic melanoma and appears insufficient to transform melanocytes on its own (enforced ectopic activation of Notch1 signaling in normal melanocytes neither promotes growth rate nor induces colony formation in soft agar). This agrees with published reports that collaborating genetic events are required for tumorigenic transformation via Notch signaling (45, 46). Such a stage-specific effect of Notch signaling in melanoma agrees with the well-known temporal/spatial properties of Notch during development. However, the mechanism remains obscure. Upregulation of both NOTCH1 and targeting genes occurs in metastatic melanoma cell lines whereas enforced activation of the Notch1 pathway fails to upregulate β-catenin expression or promote cell proliferation. The status of other pathways involved in crosstalk or collaboration with the Notch pathway may likely vary in primary and metastatic melanoma cells. Or potentially all required oncogenic signals have been triggered, and downstream elements such as β-catenin or cell cycle machinery are already maximally activated in metastatic cells. In this case, further activation could not enhance downstream pathways. Alternatively, unknown negative feedback regulators could be activated in metastatic cells to antagonize the effect of Notch signaling.
Tumor progression/metastasis is a complicated process that requires multiple cellular events including cell proliferation, survival, migration, invasion, etc. It appears that the primary role of β-catenin in mediating tumor progression/metastasis is to support cell survival. This is consistent with our observation that β-catenin overexpression accelerates cell proliferation. The metastatic cascade involves a series of steps: local invasion by tumor cells, intravasation, circulation through blood and lymphoid vascular systems, extravasation, homing/attachment and adhesion at distant sites, and growth as a new lesion (47). The artificial metastatic assay employed in this study reflects only the last steps of this cascade. Notch signaling does not seem to affect the initial steps of the metastatic cascade, i.e., local invasion and intravasation of tumor cells. We subcutaneously injected WM3248-NIC transfectants into SCID mice and were unable to detect lung metastases after 12 weeks. This suggests that Notch signaling might regulate particular cell adhesion/migration molecules required for 1 or more steps in extravasation or homing/attachment and adhesion of melanoma cells at distant sites during tumor dissemination. The increased expression of Mel-CAM provides us with one such candidate for being a cell adhesion molecule of interest. Further studies elucidating the functional responsibility of Mel-CAM and other cell surface molecules that mediate multiple steps in tumor metastasis will help us not only understand how Notch signaling regulates melanoma metastasis but also reveal potential therapeutic targets for preventing metastasis.
Methods
Reagents.
SDS-polyacrylamide gels were obtained from Invitrogen Corp., 3[H]-thymidine (1 mCi/ml) was obtained from Amersham Biosciences, and DAPT was obtained from Calbiochem. The MTT cell proliferation kit was obtained from ATCC, and the ssDNA apoptosis ELISA kit was purchased from Chemicon International. All other chemicals and solutions were obtained from Sigma-Aldrich, unless otherwise indicated.
Cell culture.
Human melanoma cell lines (WM35 [RGP], WM115, WM278, WM3248 [VGP], WM239A, and 1205Lu [metastatic]) derived from different progression stages were isolated and cultured as described (48). SBcl2 RGP-like cells were a gift from B. Giovanelli (Stehlin Foundation for Cancer Research, St. Joseph Hospital, Houston, Texas, USA). Normal human primary melanocytes FOM104, FOM105, and FOM117 were isolated from human epidermis and cultured as described (49). We cultured 293T, Phoenix (AMPHO), and COS7 cells in DMEM (Invitrogen Corp.) supplemented with 10% FBS. SUP-T1 cells were propagated in RPMI 1640 medium supplemented with 10% FBS, 2 mM L-glutamine.
Northern blotting, immunoblotting, and immunohistochemistry.
Blotting assays were performed as described (50). For Northern blotting, the probe was prepared by random amplification of a 1.2 kb DraIII fragment from MigR1-FLN1 containing full-length human NOTCH1 gene (kindly provided by T. Kadesch, University of Pennsylvania, Philadelphia, Pennsylvania, USA). For immunoblotting, membranes were probed with Abs to activated Notch1 (ab8925, Abcam), Notch1 (925, a rabbit polyclonal antiserum directly against residues 1759–2095) (51), β-catenin (Cell Signaling Technology), Myc-Tag 9B11 (New England Biolabs Inc.), Mel-CAM (P1H12, Abcam), or β-actin (AC-15, Sigma-Aldrich). For immunohistochemistry, 7-μm paraffin sections were processed by deparaffinization and rehydration followed by endogenous peroxidase blocking (1% H2O2 in methanol for 20 minutes) and antigen retrieval (boiled in 10 mM citrate buffer for 10 minutes). Tissue sections were blocked with 2% goat or horse serum (Vector Laboratories) and incubated with rat monoclonal antibody against Notch1, bTAN20 (obtained from Developmental Studies Hybridoma Bank), or mouse monoclonal HMB45 antibody (DakoCytomation) for 1 hour at room temperature, then with biotinylated secondary antibodies (Vector Laboratories). Immunoreactivity was detected using the ABC Elite kit (Vector Laboratories). We used AEC as final chromogen and hematoxylin as the nuclear counterstain. Negative controls for all antibodies were made by replacing the primary antibody with nonimmunogen IgG.
Real-time RT-PCR and semiquantitative RT-PCR.
Total RNA was isolated by Trizol reagent (Invitrogen Corp.). For real-time RT-PCR, cDNA was synthesized from 500 ng of total RNA using TaqMan Gold RT-PCR Kit (Applied Biosystems) according to the manufacturer’s protocol. The cDNA samples were diluted 20-fold, and real-time PCR reaction was carried out using SYBR green JumpStart Taq ReadyMix (Sigma-Aldrich) with 100 μM of primer. Amplifications were performed in an ABI PRISM 7000 Sequence Detection System (Applied Biosystems). Thermal cycler conditions were 50°C for 2 minutes and 95°C for 10 minutes to activate/inactivate different enzymes, then 40 cycles of 15 seconds at 95°C (denaturation) followed by 1 minute at 59°C (annealing and extension). The β-actin plasmid was used as standard cDNA. All standards and samples were assayed in triplicate. The threshold cycle (Ct) values were used to plot a standard curve. All samples were normalized to the relative levels of β-actin and results expressed as fold increase in relative levels of all cells to FOM104 by setting the values of each gene in FOM104 to 1. Primers were designed using PrimerExpress (Applied Biosystems) software as follows: HEY1, 5′-AGCCGAGATCCTGCAGATGA-3′ and 5′-GCCGTATGCAGCATTTTCAG-3′; HEY2, 5′-AGATGCTTCAGGCAACAGGG-3′ and 5′-CAAGAGCGTGTGCGTCAAAG-3′; HES1, 5′-GTGCATGAACGAGGTGACCC-3′ and 5′-GTATTAACGCCCTCGCACGT-3′; HES2, 5′-AGAACTCCAACTGCTCGAAGCT-3′ and 5′-CGGTCATTTCCAGGACGTCT-3′; Jagged1, 5′-GAGCTATTTGCCGACAAGGC-3′ and 5′-GGAGTTTGCAAGACCCATGC-3′; and β-actin, 5′- CCTCACCCTGAAGTACCCCA-3′ and 5′- TCGTCCCAGTTGGTGACGAT-3′. Semiquantitative RT-PCR was performed as described (52). The following primer pairs were used: β-catenin, 5′-GTTCGTGCACATCAGGATAC-3′ and 5′-CGATAGCTAGGATCATCCTG -3′, which amplify a 429-bp fragment; and β-actin, 5′-TCTACAATGAGCTGCGTGTG-3′ and 5′-CAACTAAGTCATAGTCCGCC-3′, which amplify an 878-bp fragment.
Cell growth and apoptosis assays.
Cell proliferation was measured by either MTT or 3[H]-thymidine uptake assays. MTT assay was performed according to the manufacturer’s protocol and 3[H]-thymidine uptake assay was carried out as described (52). Cell apoptosis was detected using an ssDNA apoptosis ELISA kit (Chemicon International) according to manufacturer’s protocol.
Colony formation assay.
Colony formation in soft agar was carried out as described (53). In brief, 3 × 103 melanoma cells per well were embedded into 0.33% agar gel containing FBS in 6-well plates precoated with 0.5% of agar solution in triplicate and covered with tumor medium. Colonies (defined as a minimum of 4 cells) were counted after 10 days of incubation.
Recombinant lenti-, retro-, and adeno-viruses.
For gene transfer, we constructed various viral vectors. GFP/lenti plasmid (pHX’-CMV-GFP) was obtained from the Gene Therapy Program, Division of Medical Genetics, University of Pennsylvania School of Medicine, Philadelphia, Pennsylvania, USA (54). To construct NIC-GFP/lenti (pHX’-CMV-NIC-IRES-GFP), a gene fragment encoding NIC (52) was inserted into pIRES2-EGFP (BD Biosciences — Clontech) between EcoRI and BamHI sites. The NIC-IRES-GFP gene fragment was then digested out with XhoI and DraI and blunted. The GFP gene in vector pHX’-CMV-GFP was replaced by the NIC-IRES-GFP gene fragment. To do this, pHX’-CMV-GFP vector was digested with NotI and BamHI. After removal of the small fragment containing the GFP gene, the remaining 2 fragments were ligated, along with a linker (5′-GGCCGCCCGGG-3′ annealed with 5′-GATCCCCGGGC-3′) to generate a SmaI site between NotI and BamHI sites. The blunted NIC-IRES-GFP gene fragment was subsequently ligated into the modified pHX’-CMV backbone vector at a digested SmaI site. H1UG was derived from the FG12 lentiviral vector (55) by inserting a human H1 promoter (HuH1) with an XbaI-EcoRI-HuH1-BamHI-HindIII-ClaI-SalI-XhoI fragment into XbaI and XhoI sites between the Flap element and the UbiC promoter. All generated plasmids were confirmed by restriction enzyme digestion and DNA sequencing.
Production of pseudotyped lentivirus was achieved by cotransfecting 293T cells with 3 plasmids (54), a packaging-defective helper construct, pCMVΔR8.2, pMD.G (Gene Therapy Program, Division of Medical Genetics, University of Pennsylvania, Philadelphia, Pennsylvania, USA) that codes for a vesicular stomatitis virus glycoprotein (VSV-G) envelope, and a vector containing the transgene. In brief, 48 hours after transient transfection of 293T cells via the calcium phosphate method, viral supernatants were collected, filtered through 0.45-μm low protein–binding filters (NALGENE Labware; Nalge Nunc International), and stored in aliquots at –70°C. The titers were determined by transducing NIH/3T3 cells. Green cells were counted under fluorescence microscope. Lentiviruses collected 48 hours after transfection displayed titers of around 108 transducing units/ml in NIH/3T3 cells.
Retroviral vectors MAML1/pBabe, DN MAML1/pBabe, and empty pBabe vector (mock) were described previously (56). Recombinant retroviruses were generated by transfection of vector into Phoenix (AMPHO) helper-free retrovirus producer lines, obtained from G.P. Nalon (Stanford University, Palo Alto, California, USA) using the calcium phosphate method. In general, retroviruses displayed titers of approximately 108 transducing units/ml in NIH/3T3 cells. Recombinant adenovirus encoding β-catenin was described previously (25).
Viral infection of targeting cells.
To infect target cells by lentiviruses or retroviruses, we exposed cells overnight to viruses with an MOI ranging from 2 to 10 in the presence of 4 μg/ml polybrene (Sigma-Aldrich). To infect cells by adenoviruses, we exposed cells to virus (100 PFUs/cell) in serum-free medium for 4 hours. Cells were washed, cultured with regular complete medium for 2 additional days, and analyzed for protein expression by immunoblot or pooled for subsequent analysis as indicated in individual experiments.
Animal experiments.
Six-week-old SCID CB-17 mice were purchased from Charles River Laboratories. Animal experiments were approved by the Wistar Institute’s Institutional Animal Care and Use Committee. For subcutaneous injection, 3 × 106 cells per mouse were injected with 8 mice in each group. Subcutaneous tumor growth was measured once per week, and tumors were harvested after 7 weeks and weighed. For the lung colony formation assay, cells were injected (2 × 105 in 100 μl PBS) into 12 SCID mice in each group via tail vein. The experiment was terminated at week 7; the lungs were harvested, tumor colonies were macroscopically counted under a Leica MZ125 dissecting microscope, and samples were subjected to histological examination. For the metastasis inhibition study, 4 × 105 cells were injected into SCID mice via tail vein. Lung samples were harvested after 12 weeks and examined as described above.
Luciferase assay.
Cells grown in 24-well plates were transiently transfected using Lipofectamin 2000 (Invitrogen Corp.) and 1 μg/well of either the TOPflash or FOPflash reporter plasmids (Promega). Cells were incubated for 24 hours at 37°C in 2 ml of serum-free culture medium. Cell lysates were prepared and luciferase activity was measured following the manufacturer’s protocol (Enhanced Luciferase Assay kit; BD Biosciences — Pharmingen). Measurements were corrected for background activity by subtraction of the FOPflash values from the corresponding TOPflash values and were then normalized by setting values from GFP-transfected cells at 100.
Statistics.
Data are presented as mean ± SD and were analyzed by 2-tailed Student’s t test. A P value of less than 0.05 was considered significant.
Acknowledgments
We thank J.M. Wilson for providing lentiviral vector for gene expression study, T. Force for adenovirus, G.P. Nalon for Phoenix cells, J.B. Trepel for DN β-catenin/pcDNA4, and T. Kadesch for MigR1-FLN1 plasmid. We also thank W.S. Pear and R. Marchelle for helpful discussion, J. Hayden for imaging analyses, M.E. Herbaly for immunohistochemical assistance, and G. Ascione for preparing the manuscript. This work was supported by grants from the NIH (CA47159, CA76674, CA25874, and CA10815) and partially supported by funds from the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health.
Footnotes
Nonstandard abbreviations used: CSL, CBF1/Su(H)/Lag-1; DAPT, N-[N-(3, 5-difluorophenacetyl-L-alanyl)]-S-phenylglycine t-butyl ester; DN, dominant negative; Hey, Hes-related repressor protein; LEF, lymphoid enhancer–binding factor; MAML, mastermind-like; Mel-CAM, melanoma-associated cell adhesion molecule; MTT, 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide; NIC, Notch intracellular domain; RGP, radial growth phase; shRNA, short hairpin RNA; TCF, T cell–specific transcription factor; VGP, vertical growth phase; Wnt, wingless/integration.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- 1.Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Davis RL, Turner DL. Vertebrate hairy and enhancer of split related proteins: transcriptional repressors regulating cellular differentiation and embryonic patterning. Oncogene. 2001;20:8342–8357. doi: 10.1038/sj.onc.1205094. [DOI] [PubMed] [Google Scholar]
- 3.Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the notch signaling pathway. J. Cell. Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 4.Reizis B, Leder P. Direct induction of T lymphocyte-specific gene expression by the mammalian notch signaling pathway. Genes Dev. 2002;16:295–300. doi: 10.1101/gad.960702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rangarajan A, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krebs LT, Deftos ML, Bevan MJ, Gridley T. The Nrarp gene encodes an ankyrin-repeat protein that is transcriptionally regulated by the notch signaling pathway. Dev. Biol. 2001;238:110–119. doi: 10.1006/dbio.2001.0408. [DOI] [PubMed] [Google Scholar]
- 7.Wu G, et al. SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 2001;21:7403–7415. doi: 10.1128/MCB.21.21.7403-7415.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez Arias A, Zecchini V, Brennan K. CSL-independent notch signalling: a checkpoint in cell fate decisions during development? Curr. Opin. Genet. Dev. 2002;12:524–533. doi: 10.1016/s0959-437x(02)00336-2. [DOI] [PubMed] [Google Scholar]
- 9.Allenspach EJ, Maillard I, Aster JC, Pear WS. Notch signaling in cancer. Cancer Biol. Ther. 2002;1:466–476. doi: 10.4161/cbt.1.5.159. [DOI] [PubMed] [Google Scholar]
- 10.Radtke F, Raj K. The role of Notch in tumorigenesis: oncogene or tumour suppressor? Nat. Rev. Cancer. 2003;3:756–767. doi: 10.1038/nrc1186. [DOI] [PubMed] [Google Scholar]
- 11.Maillard I, Pear WS. Notch and cancer: best to avoid the ups and downs. Cancer Cell. 2003;3:203–205. doi: 10.1016/s1535-6108(03)00052-7. [DOI] [PubMed] [Google Scholar]
- 12.Ellisen LW, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 13.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 14.Sriuranpong V, et al. Notch signaling induces cell cycle arrest in small cell lung cancer cells. Cancer Res. 2001;61:3200–3205. [PubMed] [Google Scholar]
- 15.Gestblom C, et al. The basic helix-loop-helix transcription factor dHAND, a marker gene for the developing human sympathetic nervous system, is expressed in both high- and low-stage neuroblastomas. Lab. Invest. 1999;79:67–79. [PubMed] [Google Scholar]
- 16.Grynfeld A, Pahlman S, Axelson H. Induced neuroblastoma cell differentiation, associated with transient HES-1 activity and reduced HASH-1 expression, is inhibited by notch1. Int. J. Cancer. 2000;88:401–410. [PubMed] [Google Scholar]
- 17.Zagouras P, Stifani S, Blaumueller CM, Carcangiu ML, Artavanis-Tsakonas S. Alterations in notch signaling in neoplastic lesions of the human cervix. Proc. Natl. Acad. Sci. U. S. A. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Talora C, Sgroi DC, Crum CP, Dotto GP. Specific down-modulation of notch1 signaling in cervical cancer cells is required for sustained HPV-E6/E7 expression and late steps of malignant transformation. Genes Dev. 2002;16:2252–2263. doi: 10.1101/gad.988902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shou J, Ross S, Koeppen H, de Sauvage FJ, Gao WQ. Dynamics of notch expression during murine prostate development and tumorigenesis. Cancer Res. 2001;61:7291–7297. [PubMed] [Google Scholar]
- 20.Li Y, et al. Notch and Schwann cell transformation. Oncogene. 2004;23:1146–1152. doi: 10.1038/sj.onc.1207068. [DOI] [PubMed] [Google Scholar]
- 21.Lowell S, Jones P, Le Roux I, Dunne J, Watt FM. Stimulation of human epidermal differentiation by delta-notch signalling at the boundaries of stem-cell clusters. Curr. Biol. 2000;10:491–500. doi: 10.1016/s0960-9822(00)00451-6. [DOI] [PubMed] [Google Scholar]
- 22.Nicolas M, et al. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet. 2003;33:416–421. doi: 10.1038/ng1099. [DOI] [PubMed] [Google Scholar]
- 23.Liu, Z.J., and Herlyn, M. 2005. Molecular biology of cutaneous melanoma. In Cancer: principles and practice of oncology. 7th edition. V.T. DeVita Jr., S. Hellman, and S.A. Rosenberg, editors. Lippincott Williams & Wilkins. Philadelphia, Pennsylvania, USA. 1745–1753.
- 24.Clark WH. Tumour progression and the nature of cancer. Br. J. Cancer. 1991;64:631–644. doi: 10.1038/bjc.1991.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haq S, et al. Stabilization of beta-catenin by a Wnt-independent mechanism regulates cardiomyocyte growth. Proc. Natl. Acad. Sci. U. S. A. 2003;100:4610–4615. doi: 10.1073/pnas.0835895100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brummelkamp TR, Bernards R, Agami R. Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell. 2002;2:243–247. doi: 10.1016/s1535-6108(02)00122-8. [DOI] [PubMed] [Google Scholar]
- 27.Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 28.Paddison PJ, Hannon GJ. RNA interference: the new somatic cell genetics? Cancer Cell. 2002;2:17–23. doi: 10.1016/s1535-6108(02)00092-2. [DOI] [PubMed] [Google Scholar]
- 29.Schwarz DS, et al. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 30.Chung EJ, et al. Regulation of leukemic cell adhesion, proliferation, and survival by beta-catenin. Blood. 2002;100:982–990. doi: 10.1182/blood.v100.3.982. [DOI] [PubMed] [Google Scholar]
- 31.Thelu J, Rossio P, Favier B. Notch signalling is linked to epidermal cell differentiation level in basal cell carcinoma, psoriasis and wound healing. BMC Dermatol. 2002;2:7. doi:10.1186/1471–5945-2-7. doi: 10.1186/1471-5945-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morimura T, et al. Cell cycle arrest and apoptosis induced by notch1 in B cells. J. Biol. Chem. 2000;275:36523–36531. doi: 10.1074/jbc.M006415200. [DOI] [PubMed] [Google Scholar]
- 33.Pui JC, et al. Notch1 expression in early lymphopoiesis influences B versus T lineage determination. Immunity. 1999;11:299–308. doi: 10.1016/s1074-7613(00)80105-3. [DOI] [PubMed] [Google Scholar]
- 34.Uyttendaele H, et al. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- 35.Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S, Bishop JM. Neoplastic transformation by truncated alleles of human NOTCH1/TAN1 and NOTCH2. Mol. Cell. Biol. 1997;17:6265–6273. doi: 10.1128/mcb.17.11.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qi R, et al. Notch1 signaling inhibits growth of human hepatocellular carcinoma through induction of cell cycle arrest and apoptosis. Cancer Res. 2003;63:8323–8329. [PubMed] [Google Scholar]
- 37.Wu L, et al. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 2000;26:484–489. doi: 10.1038/82644. [DOI] [PubMed] [Google Scholar]
- 38.Nickoloff BJ, Osborne BA, Miele L. Notch signaling as a therapeutic target in cancer: a new approach to the development of cell fate modifying agents. Oncogene. 2003;22:6598–6608. doi: 10.1038/sj.onc.1206758. [DOI] [PubMed] [Google Scholar]
- 39.Hoek K, et al. Expression profiling reveals novel pathways in the transformation of melanocytes to melanomas. Cancer Res. 2004;64:5270–5282. doi: 10.1158/0008-5472.CAN-04-0731. [DOI] [PubMed] [Google Scholar]
- 40.Kang DE, et al. Presenilin couples the paired phosphorylation of beta-catenin independent of axin: implications for beta-catenin activation in tumorigenesis. Cell. 2002;110:751–762. doi: 10.1016/s0092-8674(02)00970-4. [DOI] [PubMed] [Google Scholar]
- 41.Rubinfeld B, et al. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- 42.Demunter A, Libbrecht L, Degreef H, De Wolf-Peeters C, van den Oord JJ. Loss of membranous expression of beta-catenin is associated with tumor progression in cutaneous melanoma and rarely caused by exon 3 mutations. Mod. Pathol. 2002;15:454–461. doi: 10.1038/modpathol.3880546. [DOI] [PubMed] [Google Scholar]
- 43.Satyamoorthy K, Li G, Vaidya B, Patel D, Herlyn M. Insulin-like growth factor-1 induces survival and growth of biologically early melanoma cells through both the mitogen-activated protein kinase and beta-catenin pathways. Cancer Res. 2001;61:7318–7324. [PubMed] [Google Scholar]
- 44.Li G, Satyamoorthy K, Herlyn M. N-cadherin-mediated intercellular interactions promote survival and migration of melanoma cells. Cancer Res. 2001;61:3819–3825. [PubMed] [Google Scholar]
- 45.Fitzgerald K, Harrington A, Leder P. Ras pathway signals are required for notch-mediated oncogenesis. Oncogene. 2000;19:4191–4198. doi: 10.1038/sj.onc.1203766. [DOI] [PubMed] [Google Scholar]
- 46.Nam Y, Aster JC, Blacklow SC. Notch signaling as a therapeutic target. Curr. Opin. Chem. Biol. 2002;6:501–509. doi: 10.1016/s1367-5931(02)00346-0. [DOI] [PubMed] [Google Scholar]
- 47.Bogenrieder T, Herlyn M. Axis of evil: molecular mechanisms of cancer metastasis. Oncogene. 2003;22:6524–6536. doi: 10.1038/sj.onc.1206757. [DOI] [PubMed] [Google Scholar]
- 48.Satyamoorthy K, et al. Melanoma cell lines from different stages of progression and their biological and molecular analyses. Melanoma Res. 1997;7(Suppl. 2):S35–S42. [PubMed] [Google Scholar]
- 49.Berking C, Takemoto R, Satyamoorthy K, Elenitsas R, Herlyn M. Basic fibroblast growth factor and ultraviolet B transform melanocytes in human skin. Am. J. Pathol. 2001;158:943–953. doi: 10.1016/S0002-9440(10)64041-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu ZJ, Haleem-Smith H, Chen H, Metzger H. Unexpected signals in a system subject to kinetic proofreading. Proc. Natl. Acad. Sci. U. S. A. 2001;98:7289–7294. doi: 10.1073/pnas.121171998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jeffries S, Capobianco AJ. Neoplastic transformation by notch requires nuclear localization. Mol. Cell. Biol. 2000;20:3928–3941. doi: 10.1128/mcb.20.11.3928-3941.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu ZJ, et al. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol. Cell. Biol. 2003;23:14–25. doi: 10.1128/MCB.23.1.14-25.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nesbit M, et al. Basic fibroblast growth factor induces a transformed phenotype in normal human melanocytes. Oncogene. 1999;18:6469–6476. doi: 10.1038/sj.onc.1203066. [DOI] [PubMed] [Google Scholar]
- 54.Kobinger GP, Weiner DJ, Yu QC, Wilson JM. Filovirus-pseudotyped lentiviral vector can efficiently and stably transduce airway epithelia in vivo. Nat. Biotechnol. 2001;19:225–230. doi: 10.1038/85664. [DOI] [PubMed] [Google Scholar]
- 55.Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc. Natl. Acad. Sci. U. S. A. 2003;100:183–188. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jeffries S, Robbins DJ, Capobianco AJ. Characterization of a high-molecular-weight Notch complex in the nucleus of Notch(ic)-transformed RKE cells and in a human T-cell leukemia cell line. Mol. Cell. Biol. 2002;22:3927–3941. doi: 10.1128/MCB.22.11.3927-3941.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]