Abstract
The branch site of group II introns is typically a bulged adenosine near the 3′-end of intron domain 6. The branch site is chosen with extraordinarily high fidelity, even when the adenosine is mutated to other bases or if the typically bulged adenosine is paired. Given these facts, it has been difficult to discern the mechanism by which the proper branch site is chosen. In order to dissect the determinants for branch-point recognition, new mutations were introduced in the vicinity of the branch site and surrounding domains. Single mutations did not alter the high fidelity for proper branch-site selection. However, several combinations of mutations moved the branch site systematically to new positions along the domain 6 stem. Analysis of those mutants, together with a new alignment of domain 5 and domain 6 sequences, reveals a set of structural determinants that appear to govern branch-site selection by group II introns.
Keywords: catalysis/group II introns/ribozyme/spliceosome/splicing
Introduction
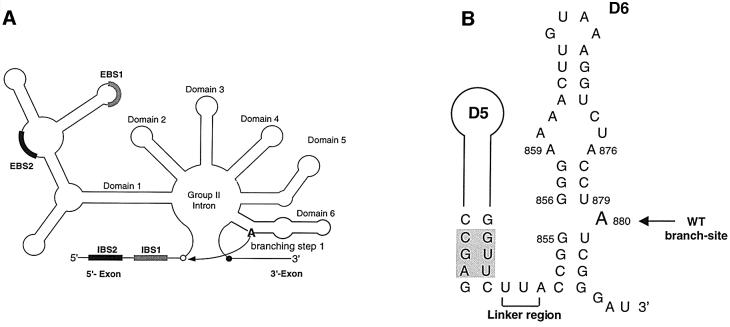
Group II introns are a class of self-splicing and transposable RNA molecules that adopt a conserved secondary, and presumably tertiary, structure (Michel and Ferat, 1995; Qin and Pyle, 1998; Bonen and Vogel, 2001). The intron sequence is arranged in a series of six domains with specific roles in catalysis, folding or the encoding of proteins (Michel et al., 1989; Qin and Pyle, 1998). While group II introns generally interrupt the organellar genes of plants, fungi and yeast, they are also common in eubacteria (Martinez-Abarca and Toro, 2000). The self-splicing reaction of group II introns differs from that of group I introns in that the nucleophile for the first step of splicing is contained within the intron itself. Specifically, the 2′-hydroxyl of a bulged adenosine (the branch point) within domain 6 (D6) is the nucleophile during the first step of splicing through transesterification (Figure 1) (Peebles et al., 1986; van der Veen et al., 1986). This results in a characteristic lariat structure which is remarkably similar to that observed during excision of nuclear introns by the eukaryotic spliceosome (Padgett et al., 1984; Konarska et al., 1985). This parallel has led to many comparisons of the two systems and an interest in a hypothetical common ancestor that may have preceded them (Cech, 1986; Madhani and Guthrie, 1992; Sun and Manley, 1995; Yu et al., 1995).
Fig. 1. Secondary structural elements from group II intron ai5γ. (A) Schematic secondary structure of the ai5γ group II intron. The 5′ and 3′ splice sites are indicated as open and closed circles, respectively. The exon/intron pairing interactions (EBS1 and EBS2 pairing with IBS1 and IBS2) are shown as bold shaded boxes. Branching occurs during the first step of splicing, when the 2′-hydroxyl of A880 in D6 (bold) attacks the 5′-splice site (curved arrow). (B) The secondary structure of D56 is shown, with the branch site indicated by an arrow and the linker region by a bracket. Catalytically important residues in D5 are shaded.
Spliceosomal branch sites have been studied intensively and, while a number of specificity determinants have been shown to be important (Newman et al., 1985; Query et al., 1994, 1996), the precise location of the branch site is not always tightly fixed. Mammalian introns in particular have been observed to react at several possible functional branch sites (Ruskin et al., 1985; Hornig et al., 1986; Query et al., 1994; Lund et al., 2000). Given the relative promiscuity of branch-site selection by the spliceosome, it has been of interest to determine the means by which group II introns select their branch sites.
The specificity determinants for branch-site choice by group II introns have been very difficult to discern. Paradoxically, the intronic region containing the branch site (D6, Figure 1B) is not highly conserved (Michel et al., 1989). It is therefore difficult to infer which structural elements might designate the proper branch site. Although cross-links between domain 5 (D5) and its linker with D6 have been observed (Podar and Perlman, 1999), there have been no tertiary contacts identified to help position the branch site for attack. These findings would seem to suggest that there is something particularly important about the conserved, bulged adenosine at the common branch site, and that this bulge structure marks the nucleotide chosen for branching. However, this notion has been contradicted by results from mutational studies in which the morphology of the branch site has been radically altered. For example, effective removal of the bulge by pairing the adenosine to another base (such as guanosine) has only minor effects on efficiency and no effect on proper choice of the branch site (Chu et al., 1998). Furthermore, the bulged nucleotide need not be an adenosine, as branching (at a reduced rate) occurs at the proper site for both guanosine and uridine branch points (Liu et al., 1997; Chu, 2000). These results indicate that the bulged adenosine does not designate the site of branching. Studies of branching are further complicated by the fact that, unlike the spliceosome, group II introns have a second pathway for ensuring that splicing occurs. Water readily serves as the nucleophile during the first step of splicing by group II introns both in vitro and in vivo (Jarrell et al., 1988b; Daniels et al., 1996; Podar et al., 1998).
The present study examines the branch-site choice and catalytic activity of a large family of mutations in D6 of intron ai5γ. While single mutations failed to alter branch-site selectivity, more elaborate constructs allowed us to move the branch site systematically from one position to the next. This information was combined with phylogenetic analysis to deduce a set of rules that appear to govern branch-site selection by group II introns.
Results
In order to dissect the molecular determinants for branch-site selection, mutations were made in the conserved structural features that surround the branch site and adjacent regions. The mutants can be grouped into several families that contain alterations of the bulge structure at the branch site, the length of the linker that connects D5 to D6, or the helical register of the bulged adenosine (Figure 2). Mutant RNA precursors were allowed to self-splice under standard reaction conditions (see Materials and methods), reaction kinetics were evaluated and lariat products were isolated. In each case, the site of branching was determined by exploiting new high-resolution mapping procedures (Figure 3).
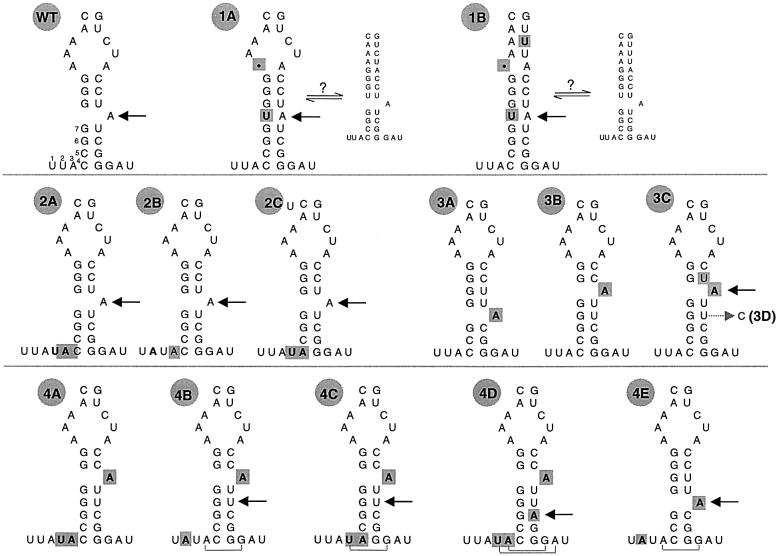
Fig. 2. Schematic secondary structure of D6 mutants. Mutations to the abbreviated D6 sequence are shown in bold and highlighted in gray. The name of each variant is indicated to the upper left. The branch site of each mutant is indicated with an arrow. For the WT variant, the 7 nt downstream from D5 are shown. For mutants 1A and 1B, the two possible equilibrium conformations of D6 are indicated. For mutants 4B–E, brackets indicate additional base pairs that may serve to extend the D6 stem. RNA 3D is shown as a mutant of 3C, from which it is derived, with an arrow indicating the site of uridine to cytosine mutation.
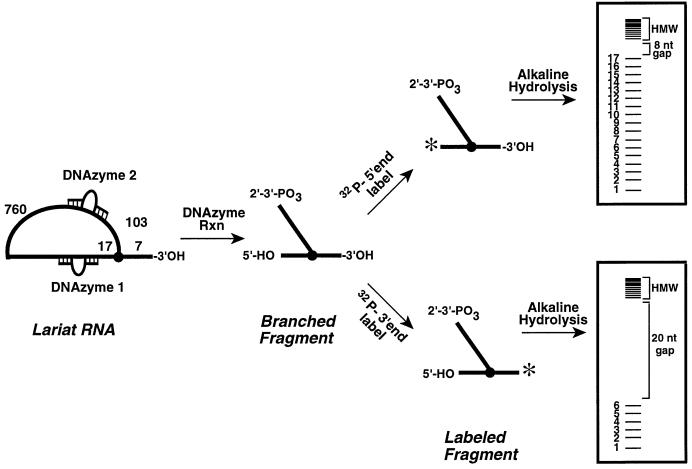
Fig. 3. Schematic of the DNAzyme method for mapping group II intron branch points from both the 5′ and 3′ ends.
The contribution of a bulged structure at the branch site
Given previous results with the spliceosome (Ruskin et al., 1985), one might expect that cryptic branching in group II introns could be activated by eliminating the bulged structure at the branch site. One of the first mutations designed to investigate the role of the bulge was prA–U, in which the branch-point adenosine (A880) was paired to a uridine inserted between G855 and G856. Although the rate of hydrolytic splicing was unaffected by this mutation, the rate of branching was reduced by three orders of magnitude (van der Veen et al., 1987; Chu et al., 1998). In parallel studies, the prA–U mutant was transformed into yeast mitochondrial DNA. In vivo revertants of the prA–U strain were isolated and included several suppressor mutations that restored branching activity in vivo (Podar, 1997).
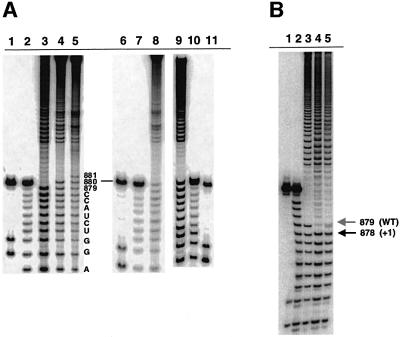
One of the most active suppressors contained guanosine, in place of uridine, paired with the branch-site adenosine. The self-splicing efficiency of this mutant is almost indistinguishable from that of the wild-type (WT) intron, and branching occurs at the correct position (Chu et al., 1998) (Figure 4B, lane 7). In vivo, this mutant splices efficiently, although a modest second-step defect is indicated by accumulation of intron-3′-exon RNA (Podar et al., 1998). Another suppressor mutation retains the uridine that can pair with the branch site adenosine, but has a deletion in the internal loop of D6 (A859, RNA 1A; Figure 2). Yeast containing 1A respire well and accumulate lariat-3′-exon molecules (Podar, 1997). The self-splicing of 1A is much faster than that of the prA–U mutant [kbr = 0.00012 min–1 (Chu et al., 1998)], having a branching rate (kbr = 0.017 min–1) that is only 10-fold slower than that of the WT intron (0.14 min–1; Table I). To determine whether efficient reaction by 1A could be attributed to the formation of a new branch site, particularly at position A876 (Figure 2), branched fragments were mapped (Figure 4). This analysis revealed that branching occurs at the normal branch-site (A880, Figure 4A, lane 4; Figure 4B, lane 6).
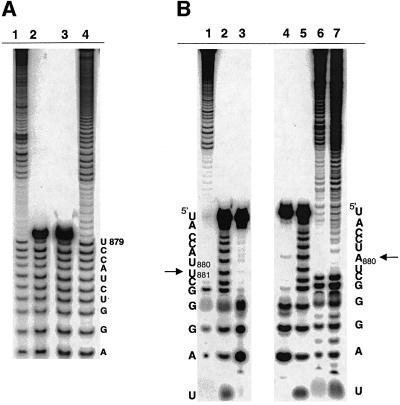
Fig. 4. Analysis of branch-site choice for intron revertants. (A) The branch points for mutants 1A (lane 4) and 1B (lane 1) were mapped by limited alkaline hydrolysis of branched fragments labeled at their 5′-termini. These were subjected to electrophoresis next to alkaline hydrolysis ladders of an oligonucleotide marker corresponding to the 17 nt immediately upstream of the WT branch-point adenosine (lanes 2 and 3). For the mapping procedure employed (Figure 3), the last band before the gap indicates the nucleotide 5′ of the selected branch point (A880, in each of these cases). (B) Partial alkaline hydrolysis of 3′-labeled branched fragments. For the mapping procedure employed (Figure 3), the band directly beneath the gap will correspond to a position 2 nts downstream from the branch point. Thus, the gap beginning at position C882 (lanes 6 and 7) corresponds to a WT branch-site choice at position A880 for mutants 1A (lane 6) and the prA–G (lane 7; Chu et al., 1998). Partial alkaline hydrolysis of these fragments is shown next to corresponding T1 (lane 4) and hydroxide (lane 5) ladders on an oligonucleotide marker corresponding to the terminal 13 nt of the intron. Partial alkaline hydrolysis of a cryptic branching mutant (RNA 4B, lane 1) is shown next to T1 (lane 3) and hydroxide (lane 2) ladders, and corresponds to branching at position U881 (arrow).
Table I. Kinetic analysis of branch-site mutants.
| Mutant | Branch site | Hydrolytic splicing rate khy (min–1 × 10–2) | Branching rate kbr (min–1 × 10–2) | Relative rate–1 1/krel |
|---|---|---|---|---|
| WT | WT | 0.479 ± 0.010 | 13.8 ± 0.9 | 1.0 |
| 1A | WT | 1.70 ± 0.088 | 1.72 ± 0.15 | 8.0 |
| 1B | WT | 1.89 ± 0.093 | 0.0585 ± 0.008 | 240 |
| 2A | WT | 1.14 ± 0.13 | 3.02 ± 0.239 | 4.6 |
| 2B | WT | 0.342 ± 0.27 | 8.97 ± 0.37 | 1.5 |
| 2C | WT | 0.898 ± 0.159 | 13.4 ± 0.62 | 1.0 |
| 3A | – | 1.01 ± 0.14 | 0.00613 ± 0.0029 | 2200 |
| 3B | – | 2.1 ± 0.151 | 0.068 ± 0.0093 | 203 |
| 3C | +1 | 1.9 ± 0.26 | 2.8 ± 0.26 | 4.9 |
| 3D | +1 | 2.1 ± 0.25 | 0.54 ± 0.068 | 26 |
| 4A | – | 1.56 ± 0.15 | * | * |
| 4B | –1 | 1.55 ± 0.096 | 0.083 ± 0.015 | 170 |
| 4C | –1 | 1.69 ± 0.12 | 0.0345 ± 0.0089 | 400 |
| 4D | –2 | 2.58 ± 0.30 | 0.0224 ± 0.0026 | 616 |
| 4E | –1 | 2.34 ± 0.24 | 0.44 ± 0.046 | 31 |
Splicing kinetics of all variants were analyzed (Daniels et al., 1996; Chu et al., 1998), resulting in parameters describing the rate of splicing by hydrolysis (khy) and by transesterification (branching, kbr). The relative rate of branching (1/krel) represents the relative degree to which transesterification has been inhibited by mutation. The site of branching (second column) is normal for most mutants (WT), but has moved downstream (–1 or –2 nt) for mutants 4B–E and upstream for mutants 3C and 3D (+1 nt). The latter mutants were readily mapped (Figure 6), despite their low branching efficiency, which is comparable to that of mutant 3B. By contrast, all attempts to map mutant 3B failed (after four independent trials), potentially due to instability or heterogeneity of the branch site.
*No branching observed.
To determine whether conformational flexibility in the upstream internal loop of D6 contributes to the efficiency and fidelity of branching by 1A, RNA 1B was constructed in which an A–C pair (positions 861 and 874) was changed to an A–U, although the deletion of A859 was maintained (Figure 2). Unlike the suppressor mutants described previously, branching by mutant 1B is strongly inhibited (kbr = 0.000585 min–1). This RNA reacts ∼200-fold slower than WT and ∼20-fold slower than 1A (Table I), although it reacts at the proper branch site (Figure 4A, lane 1). The high fidelity of branching by RNA 1A and 1B suggests that a bulge structure is not required for branch-site choice in a group II intron (Chu et al., 1998). However, it is possible that the secondary structures of these mutants rearrange in a way that permits the normal branch-site adenosine to be bulged; possible D6 isomers are indicated in Figure 2 for RNAs 1A and 1B. Regardless of the mechanism, these experiments underscore the high fidelity of branch-site choice and the primacy of the normal branch site.
Identity of the branch-site nucleotide
Changing the branch-point adenosine (A880) to other nucleotides results in a dramatic decrease in the rate of branching (Liu et al., 1997). Although A880C does not branch detectably, A880G and A880U react with a branching rate that is ∼100× the WT, and branch at the correct position (Liu et al., 1997). These results were confirmed using the new DNAzyme mapping procedure (Chu et al., 1998; Pyle et al., 2000) on branched fragments labeled at the 3′ or 5′ ends (Chu, 2000). Together with the collective data on the fidelity of branch-site mutants (see below), these results are inconsistent with a report of cryptic branching of the A880G mutant at intron positions C877 and U879 (Gaur et al., 1997). Branch-site choice therefore appears to be independent of base identity, suggesting that adenosine is not involved in specification of the branch site. However, adenosine makes an important contribution to branching efficiency, which is consistent with its high level of conservation (Michel et al., 1989) and the importance of its functional groups (Liu et al., 1997). This example highlights a feature that was observed throughout this study: reduced branching efficiency does not translate into a loss of fidelity for branch-site choice. Branching efficiency and fidelity appear to be uncoupled and are likely to be governed by different determinants.
Length of the D56 linker region
Another feature that might influence the choice of branch site is the length of the linker between D5 and D6. Previous studies have suggested that the 3 nucleotide (nt) D56 linker in ai5γ (Figure 1B) is important for positioning the branch-point nucleophile in the catalytic core of group IIB introns (Dib-Hajj et al., 1993; Boulanger et al., 1996; Podar and Perlman, 1999). In that work, a 5 nt linker (see Figure 2, mutant 2A) strongly inhibited branching in vivo, although some branching was observed in vitro (Boulanger et al., 1996). Mutants with 4 nt linkers were significantly more reactive in vivo and in vitro (Boulanger et al., 1996) (see Figure 2, mutants 2B and 2C) and accurate branching in vivo was demonstrated for mutant 2C (Podar et al., 1998).
To further characterize the effects of linker length on the efficiency and accuracy of branch-site selection, three D56 linker mutants were examined. RNA 2A (Figure 2) contains a 5 nt linker and branched with a reduced rate (0.0302 min–1; Table I). Mutant 2B was obtained as a revertant of 2A; it contains a 4 nt linker due to a deletion of a uridine (either U849 or U850; see Figure 2). Mutant 2C is another revertant of 2A; it contains a deletion of C852 or C853 at the base of D6. In both mutants 2B and 2C, the rate of branching is similar to that of the WT intron (2B, kbr = 0.090 min–1; 2C, kbr = 0.13 min–1; Table I), consistent with previous results (Boulanger et al., 1996). The remarkable parity in rates of RNAs 2B and 2C strongly suggests that they have similar structures (see below).
To determine whether alterations in linker length displace the site of branching in vitro, the lariat branch points of these linker mutants were mapped from the 5′ end (Figure 5, lanes 3–5). Cryptic branch sites were not observed, as confirmed by mapping RNA 2B from the 3′-end (data not shown). In each case, the branch point was chosen correctly, occurring at the bulged A in D6 (position A880 based on the WT numbering). Therefore, data obtained in vitro and in vivo confirm that these alterations in linker length do not activate cryptic branching.
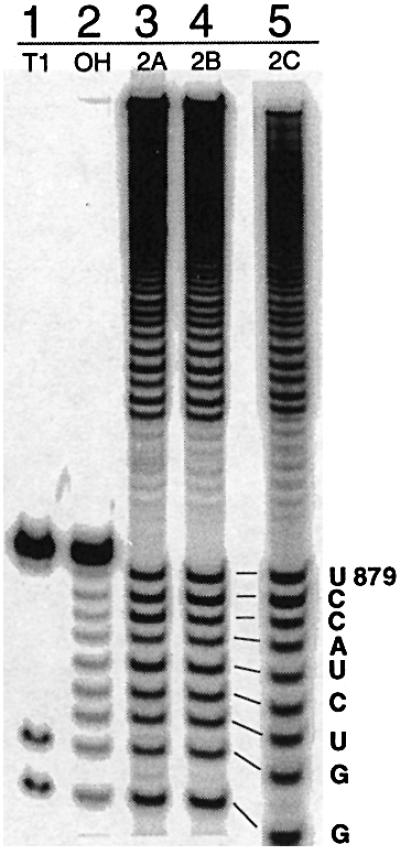
Fig. 5. Mapping the branch points of linker mutants 2A–C. Partial alkaline hydrolysis of branched fragments labeled at the 5′-end indicates that, like the WT intron, these mutants all branch at position A880 (lanes 3–5). The T1 digest (T1) and alkaline hydrolysis (OH) ladders on the 13 nt oligonucleotide serve as markers.
Spatial positioning of the branch-site in D6
Given this remarkable fidelity, which contrasts with behavior of the mammalian spliceosome (see below), it was of interest to determine whether a group II intron can be induced to choose an incorrect branch-site. To evaluate this possibility, mutants were constructed in which the bulged adenosine (A880) was displaced either 1 nt down (RNA 3A) or 1 nt up (RNA 3B) from its original register (Figure 2). These mutations resulted in such a radical reduction of branching efficiency that insufficient quantities of lariat product were available for mapping (Table I).
Systematic alteration of branch-site choice through multiple mutations
Single changes in branch-site morphology or positioning failed to disrupt the fidelity of branch-point choice, suggesting that multiple redundant determinants are involved in maintaining fidelity. This notion is consistent with previous structural studies on group II introns in which multiple weak tertiary interactions support a particular RNA architectural element (Chanfreau and Jacquier, 1996; Costa et al., 1997; Boudvillain and Pyle, 1998). In these cases, disrupting any one interaction is insufficient to completely eliminate a particular activity. To explore this possibility, two types of mutations were simultaneously incorporated into D6 and their effects were assessed.
The lack of branching by mutants 3A and 3B suggested that branch-site positioning is particularly important for reaction. To test this notion, a number of double mutants were analyzed in which the register of the bulged adenosine was shifted while simultaneously altering the length of the D56 linker. In these mutants, the branch-point adenosine was shifted upstream by 1 nt while the D56 linker was extended to 4 (RNA 4B, 4C, Figure 2) or 5 nt (RNA 4A, Figure 2).
In marked contrast to the behavior of mutants 3A and 3B, reactivity was sufficient to map all variants except mutant 4A, which did not branch at all. Remarkably, for mutants in which the linker length was effectively increased by 1 nt, mapping of 5′-labeled fragments indicated that the branch site shifted 1 nt 3′ of its normal position to nt 881 (RNAs 4B, 4C, Figure 2; Figure 6A, lanes 9 and 8, respectively). In accordance with this, mapping of mutant 4B from the 3′ end demonstrates a concomitant shift in position of the branch-site (Figure 4B, lane 1). It is notable that RNA 4C (like RNA 2C) actually contains a 5 nt linker, and would only contain a 4 nt linker if an A–G pair forms at the terminus of D6 (Figure 2). Like mutants 2B and 2C, this would result in very similar structures for RNAs 4B and 4C, which is supported by their almost indistinguishable branching rates (kbr = 0.000830 min–1 for 4B and kbr = 0.000345 min–1 for 4C). Another intriguing aspect of these mutants is that they both branch from a uridine, with reaction rates that are ∼100× slower than WT. Because uridine substitution at A880 is known to reduce the branching rate by 100× (Liu et al., 1997), mutants 4B and 4C may branch more slowly due to the presence of a uridine nucleophile rather than due to morphological changes at the branch site.
Fig. 6. Mapping the branch point of cryptic branching mutants. (A) Partial alkaline hydrolysis of branched fragments labeled at the 5′-end indicates that, unlike the WT intron (lane 3), mutants 4E (lane 4), 4C (lane 8) and 4B (lane 9) all branch at the nucleotide 3′ of the normal branch site (U881). Mutant 4D (lane 5) branches at a position 2 nt 3′ of the normal branch-site (U882). T1 nuclease (lanes 1, 6 and 11) and alkaline hydroxide (lanes 2, 7 and 10) ladders are shown as markers. (B) The 3C mutant (lane 4) and the related 3D mutant both react at a bulged adenosine that has been shifted by one nt upstream in contrast to WT (lane 3), and relative to the T1 (lane 1) and alkaline hydrolysis (lane 2) ladders. The solid and gray arrows indicate sites of upstream and WT branch-point selection, respectively. For mapping of mutants 3C and 3D, DNAzyme 1 (which cleaves 3′ of A862) was replaced by DNAzyme 3 (which cleaves 3′ of G866). Accordingly, the 17 nt marker oligonucleotide (lanes 1 and 2) spans intronic sequences U867 to G883.
Although mutants 4B and 4C differ from the WT in several ways, their branch points share a feature in common with the WT intron: there are at least 7 nt between D5 and the base pair that is located directly beneath the selected branch site (see WT, Figure 2). This suggests that there is a measuring function that detects the distance between the D5 active site and the branch-point nucleophile in D6. Based on this model, one can envision a rationale for the inactivity of mutant 4A, which contains two extra linker nucleotides: there is a cytidine located at the predicted branch site (above nt 7 from the base of D5, Figure 2) and it is known that cytosine is unreactive for branching (Liu et al., 1997). To test this, the predicted branch site of 4A was mutated from cytosine to adenosine, resulting in a triple mutation (RNA 4D). Remarkably, 4D supported branching at the engineered adenosine branch site, which is now located 2 nt downstream from the normal branch site (Figure 6, lane 5).
As a further test of this model, it was of interest to move the bulged adenosine downstream (3′) by 1 nt while simultaneously increasing the linker length by 1 nt (RNA 4E). Consistent with activities of mutants 4B–D, an increase in linker length restored branching activity, allowing RNA 4E to react from a bulged adenosine residue that had been shifted downstream by 1 bp (Figure 6, lane 4). Taken together, results with four double mutants (RNA 4B–E) show that branching can be directed to a new site, provided that certain recognition elements are maintained.
In order to evaluate the structural basis for branch-point specification, the sequences of the double mutants were inspected carefully. In each case, one can draw a 3 nt linker between D5 and D6, and a 4 bp stem beneath the branch-site. In some cases (as in 4B and 4E), the closing base pair of D6 is a mismatch; but in each case, it is a mismatch known to be stabilizing at helical termini (Gautheret et al., 1994; Burkard et al., 1999). Another prominent feature of all the mutants capable of branching is that the selected branch-site lies beneath a G–U wobble pair.
It was therefore of interest to determine whether branching activity of mutant 3B could be restored by placing a G–U pair above the bulged adenosine (RNA 3C, Figure 2). RNA 3C forms abundant lariat and branches with relatively high efficiency (0.028 min–1; Table I). Mapping of the branch-site indicates that 3C reacts at the shifted bulged adenosine (Figure 6B, lane 4), which has been activated by the placement of an adjacent G–U pair (Figure 2). Unlike the other engineered branch sites, the branch site of 3C is shifted upstream of the normal position. It is notable that the 3C mutant contains an unusual run of three G–U pairs (Figure 2), which may enable the bulged adenosine to function as a sort of ‘super branch-site’. Interestingly, when the lowest of these wobble pairs is mutated to G–C (corresponding to G855–C881 in D6, RNA 3D, Figure 2), the shifted branch-site is still chosen properly (Figure 6B, lane 5), but the mutant branches less efficiently (0.0054 ± 0.00068 min–1). In both cases, the restoration of branching at a shifted adenosine underscores the importance of an upstream G–U wobble pair adjacent to the chosen branch-site.
Phylogenetic analysis of branch-site recognition determinants
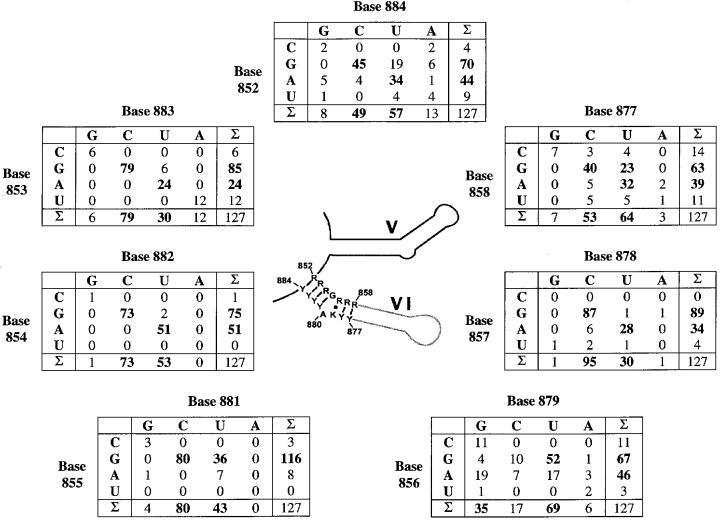
In an effort to establish a phylogenetic understanding of branch-site selection, we carried out a comprehensive search for group II intron sequences available up to November 2000. The resulting sequence set was then reduced by applying criteria designed to help ensure that only functional introns were included in the subsequent analysis. If at least one of the following criteria were met, the sequence was added to our final database: (i) the intron has been reported to splice in vitro or in vivo; (ii) it is possible to define signatures for all six secondary structural domains of the intron (to eliminate group III introns); and (iii) the intron interrupts an essential open reading frame (ORF), thereby suggesting that splicing is obligate for survival of an organism. We excluded a family of group II introns found in genes for tRNAVal and in a chloroplastid protease gene clpP, which lack a branch-site. The tRNAVal introns have been demonstrated to splice exclusively through a hydrolytic pathway in vivo (J.Vogel, personal communication). There are cases in which identical introns are observed at the same (and sometimes different) loci in different organisms as a result of lateral gene transfer (Ehara et al., 2000). In order to ensure the diversity of the database, we included only one example of a particular intron and excluded related examples that were identical throughout the intron sequence. The resulting collection of 127 distinct sequences includes introns from diverse organisms (i.e. bacteria, chloroplasts, plant and fungal mitochondria) and diverse gene families (i.e. tRNA and rRNA genes, cytochrome oxidase, NADH dehydrogenase and other protein genes).
Having established a set of group II intron sequences, we then folded and aligned the D56 region of each representative using a combination of manual and algorithmic approaches for calculating secondary structural stability (Mathews et al., 1998). The final database of 127 sequences includes 62 group IIA and group IIB sequences that had been aligned and analyzed previously (Michel et al., 1989) (see Supplementary data available at The EMBO Journal Online).
Co-variations in the resultant alignment were computed and are shown as a series of matrices (Figure 7). This analysis reveals a persistent set of structural features in D6, surrounding the branch point. The most notable feature of the consensus (center, Figure 7) is the presence of an almost invariant 4 bp stem between the base of D6 and the branch-site, regardless of intron subgroup (IIA or IIB). This stem is composed of Watson–Crick (W–C) base pairs, although there is some variation in the terminal base pair of the stem. In 83% of cases, this terminal pair is a W–C or G–U pair. However, the terminal pair can also be G–A (9%), A–C (5%) or U–U (3%).
Fig. 7. Covariation of D6 nucleotides. The secondary structure consensus of D5 and D6 are shown in the center. The numbering of nucleotides is derived from the ai5γ nomenclature. Each table displays the co-variation of 2 nt putatively involved in base pairing around the branching adenosine. The D56 alignment from which these matrices were calculated contains 127 sequences belonging to fungal, plant and algae organelles, and bacteria. All were downloaded from DDBJ/EMBL/GenBank and aligned combining the comparative approach with thermodynamic methods (Mfold server, Zuker, 1995–2000, Rensselaer Polytechnic Institute). Introns were classified by gene family and parsed into groups IIA and IIB according to standardized parameters (Michel et al., 1989). This resulted in the inclusion of 68 group IIB introns, 45 IIA introns and 14 unclassified bacterial introns that adopt features from both subgroups. Seven introns belonging to group IIA were excluded due to the lack of a bulged adenosine. Five of these are inserted into chloroplastid tRNAVal genes (trnV, J.Vogel, personal communication) and two into chloroplastic protease genes [clpP/2 or ORF203/2, sequence numbers 24 and 25 in Michel et al. (1989)]. Alignment of all sequences is provided as Supplementary data available at The EMBO Journal Online.
A second conserved feature of D6 is the polypurine tract that begins at the base of D6 (nt 852–855, Figure 7). As described in a previous phylogenetic analysis (Michel et al., 1989), this conserved tract pairs with a polypyrimidine stretch to form the 4 bp stem beneath the branch site. In most cases, the polypurine tract is composed of guanosines, which can be considered semi-conserved at most positions and fully conserved at position 855. Analysis of the entire database (including the tRNAval genes) reveals that this polypurine tract is one of the most conserved features of group II introns. Because the D6 stem is not merely G–C rich, but predominantly consists of poly(G)–poly(C), it is likely that the motif is involved in an important molecular interaction such as an extended triple helix with other regions of the intron. This type of contact may play an important role in defining the branch site or interacting with D5.
A third feature that is evident among the group IIB introns is the prevalence of a 3 or 4 nt linker between D5 and D6. Analysis of the database reveals that among the 45 group IIB introns, there are 26 with a 3 nt linker, 17 with a 4 nt linker, and two with a 6 nt linker. Thus, linker length is relatively constrained and likely to have significance for function.
Fourthly, the alignment reveals significant trends in the composition of base pairs that flank the branch point in group IIA and group IIB introns (Figure 7, lower panels). The base pair that lies beneath the bulged adenosine contains a conserved guanosine (91%, G855 and Y881 in the ai5γ nomenclature), which interacts either with uridine or cytosine. Above the bulged adenosine (R856–K879), G–U pairing is also predominant, although other non-Watson–Crick base pairs also occur. Thus, it is likely that neighboring wobble or other mispairing orientations contribute strongly to branch-site function and selection.
The role of branch-site location in 3′ splice site choice
It is feasible that activation of a cryptic branch point (a first-step reaction) may have an effect upon subsequent reactions in splicing. To examine this relationship, we characterized the 3′-end of a lariat intron derived from a mutant that branched at a cryptic site (RNA 4B). When the branched fragment of 4B was mapped from the 3′-end, the last band before the gap migrated as a 5 mer (Figure 4B, lane 1), indicating that a normal 3′-splice site had been selected. This result suggests that 3′-splice site choice can be uncoupled from branch-site selection.
Discussion
High fidelity of branch-point selection
Group II introns have a remarkably robust mechanism for choosing and reacting at the proper branch site. Single mutations that alter branch-point sequence, structure or context all failed to activate cryptic branch sites. High resolution mapping studies revealed that, in most cases, branching either occurs at the correct site or it does not occur at all. This behavior contrasts with that of the spliceosome, in which ectopic branching is commonly observed as part of a normal, productive splicing pathway. The fact that single types of mutations failed to alter branch-site selection suggests that group II introns have multiple determinants for ensuring the proper choice of branch site and for enhancing the efficiency of reaction. These determinants become clear only upon incorporation of multiple mutations, which relax the branch-site selection system and permit systematic alteration of branch-site choice.
Molecular determinants for branch-site selection
Previous work had already shown what a branch point is not: it need not be bulged and it need not be an adenosine (Liu et al., 1997; Chu et al., 1998). It therefore remained to define what a branch point is and to establish the molecular determinants that cause an intron to react at a particular nucleotide. The results of this study suggest a collection of partially redundant determinants that serve to ensure that the proper branch site is chosen.
Four base pair stem in D6. The single mutants with the lowest branching efficiency were 3A and 3B, in which the bulged adenosine was shifted downstream or upstream. Shortening of the stem (3A) caused particularly radical branching defects (Table I). The low reactivity of these mutants suggested that an important determinant for branching is the spatial positioning of the branch-site relative to the base of the D6 stem. This notion is supported by another line of reasoning. All of the mutants that were active for branching have at least 7 nt between the base of D5 and the nucleotide paired to the residue beneath the branch-site (see WT, Figure 2). This suggests that a measuring function aligns the catalytic residues of D5 (spanning a major groove section that includes G817 and residues of the D5 bulge) with the 2′-hydroxyl of the branch-point nucleophile. The sequences of the active mutants (Figure 2), together with phylogenetic analysis of D56 sequences (Figure 7), suggest that the intervening nucleotides are organized in a common structural motif that would rigidify the region, permitting it to be recognized and oriented by the ribozyme core. These data suggest that an active branch-site lies immediately adjacent to a 4 bp stem at the base of D6. This stem usually consists entirely of Watson–Crick base pairs, although it can also contain a stable mispair such as G–A. What appears to be of primary importance is the ability to form a set of four stacked pairs, which provide a rigid structure of constant length that can be used to measure the distance from the active site to the branch site in D6.
A 3 nt linker in IIB introns. The behavior of the double mutants and the triple mutant (RNA 4D) indicates that, in cases where the branch-site location becomes ambiguous, the linker plays an essential role in measuring the distance between D5 and D6. In each of these cases, linker expansion restored the minimal 7 nt distance between D5 and a potential branch site, possibly allowing the D6 stem to lengthen to 4 bp. The data presented herein therefore suggest that three nts is an optimal linker length for this intron, although it is possible for the linker to be longer (see mutants 2A–C) or shorter (Boulanger et al., 1996). In cases where the 4 bp stem in D6 is exceptionally stable and well defined, a sub-class of group II introns tolerates very long D56 linkers (Michel et al., 1989). The measuring functions of IIA and IIB introns are therefore likely to have diverged in a way that accommodates the shorter linkers typical of IIA introns.
The importance of the D56 linker is underscored by studies indicating that D6 may not form an abundant network of tertiary contacts that help to align it properly relative to D5. For example, an appended D6 does not significantly increase the binding affinity of D5 molecules (Chin and Pyle, 1995), suggesting that D5 drags D6 into the folded intron. Furthermore, nucleotide analog interference mapping studies have not revealed an abundance of sensitive functional groups in D6 (Boudvillain and Pyle, 1998). Given this apparent paucity of interactions between D6 and other domains of the intron, the linker between D6 and D5 would appear to be a critical element for ensuring that these domains coordinate their function. This is supported by crosslinking studies, which indicate that the branch point and adjacent nucleotides are in spatial proximity to linker nucleotides (Podar and Perlman, 1999).
The G–U wobble pair above the branch-site. Based on the above reasoning, one can explain the branch-site choice and the reaction efficiency of all mutants except for one, RNA 3B, in which the branch-site adenosine has been shifted upstream by one nt. This mutant can adopt a 3 nt linker and a 4 bp stem that should place the branch-site at U880, immediately beneath the bulged adenosine (now located at position 879). Given the rules spelled out previously, it should behave in a manner similar to that of RNA 4B or 4C, and choose a paired uridine as the nucleophile. However, close inspection of mutants 4B and 4C reveals a related paradox: if a rigid 4 bp stem were so important, why do these mutants branch at U881 rather than at U880, which lies above a more perfect 4 bp stem. These data can be explained by considering the presence of a G–U wobble pair immediately above the selected branch-site in all mutants capable of branching; this is consistent with the significant phylogenetic conservation of this pairing. This was confirmed by the creation of mutants 3C and 3D, which selectively activated the shifted branch-site of mutant 3B by incorporating an upstream G–U wobble pair. The relative importance of this G–U pair as a recognition determinant is reflected in the unusually efficient branching of mutant 3C, which underscores the importance of local structure surrounding the branch site of group II introns.
Comparison with the spliceosome
There are major differences between group II introns and the spliceosome in terms of branch-site recognition and function. Unlike group II introns, a first step hydrolysis reaction has not been observed during spliceosomal RNA processing. Eukaryotic pre-mRNA splicing (i.e. yeast and metazoan) occurs exclusively through the branching reaction. The system that most resembles group II intron branching in terms of fidelity is spliceosomal processing by Saccharomyces cerevisiae. Within the introns of yeast genes, a highly conserved UACUAAC sequence contains the branch point (underlined), which is located at varying distances from the 3′-splice site (Spingola et al., 1999). Like group II introns, substituting adenosine with cytosine completely abolishes branching and splicing activity (Langford et al., 1984; Newman et al., 1985).
In contrast to yeast, branch-site selection in higher eukaryotes is more loosely constrained. The branch-site region (consensus YURAC) often contains two adjacent adenosines. Although branching usually occurs from the invariant downstream adenosine, the adjacent upstream purine has also been observed to serve as a branch site (Konarska et al., 1985; Ruskin et al., 1985; Hornig et al., 1986; Noble et al., 1987, 1988). Deletion or substitution of the branch-point adenosine to guanosine or uridine leads to a reduction in the rate of splicing, reduced efficiency of the second step reaction and the activation of cryptic branch sites (Ruskin et al., 1985; Hornig et al., 1986). In marked constrast with group II introns, substitution of the branch-point adenosine with cytidine has the smallest effects on splicing (Hornig et al., 1986). Furthermore, a bulge structure appears to be essential for presenting the reactive nucleophile to the catalytic core (Query et al., 1994, 1996). Taken together, almost every feature concerning selection of the lariat branch point (identity of branch-point nucleotide, requirement of a bulge and distance from the 3′ splice site) differs between group II introns and the mammalian spliceosome.
Functional implications of the branch-site recognition determinants
It is interesting to consider why group II introns have been selected to maintain such stringent fidelity for proper branch-site formation. High fidelity branching is puzzling in light of the fact that group II introns maintain a second pathway for self-splicing that does not even involve branching. This alternative hydrolysis pathway has been shown to be viable both in vitro and in vivo (Jarrell et al., 1988b; Daniels et al., 1996; Podar et al., 1998). One explanation for stringent branch-site fidelity might be that it is required for proper 5′- and 3′-splice site selection. However, cryptic mutants 4B and 4C were found to choose both the splice sites properly, thus leading to correctly spliced exons. Another explanation may have nothing to do with self-splicing and relate instead to the second reaction that attenuates the evolution of group II introns, i.e. intron mobility (Yang et al., 1996). It is possible (although untested) that cryptic lariat structures fail to perform the reverse-splicing reactions that are critical for group II intron retrotransposition.
Taken together, the mapping results, splicing kinetics and phylogenetic analysis all indicate that there are three major structural determinants for branch-point activation in group IIB introns: (i) a 4 bp stem beneath the branch site; (ii) a D56 linker of at least 3 nt; and (iii) a G–U wobble pair upstream of the branch-site. The data indicate that no single determinant is absolutely required, and that all three determinants function together to ensure proper branch-site selection. This redundancy may help explain the unusually high degree of fidelity observed for group II intron branch-site choice: multiple sufficient recognition determinants will ensure that the branch-site is always chosen correctly, even in the event of local structural disruptions.
Materials and methods
Transcripts and plasmid templates
WT precursor RNA was transcribed from plasmid pJD20 (Jarrell et al., 1988a). Mutant transcripts 2A, 2C and 2B were transcribed from plasmids pJD20-J(56) 5, pJD20-J(56) 5ΔC and pJD20-J(56) 5ΔT (Boulanger et al., 1996). Mutant transcripts 1A, 1B, 3A, 3B, 3C, 3D, 4A, 4B, 4C, 4D and 4E were transcribed from plasmids pVC05, pVC06, pVC11, pVC10, pQL126, pQL127, pVC12, pVC13, pVC14, pVC15 and pVC16, which were constructed from pJD20 using the QuikChange mutagenesis kit (Stratagene). Plasmids were linearized with HindIII restriction enzyme and transcribed under standard conditions (Daniels et al., 1996) using T7 RNA polymerase (Davanloo et al., 1984). In each case, a trace amount of [α-32P]UTP was used to label the transcripts internally so that reaction products could be identified easily.
Branching kinetics
Branching kinetics for all the mutants was conducted and analyzed as described previously (Chu et al., 1998). Briefly, the transcripts were incubated in a splicing reaction buffer containing 40 mM 3-(N-morpholino)propane sulfonic acid (MOPS) (pH 7.5), 100 mM MgCl2, and 500 mM NH4(SO4)2. The reaction mixtures were incubated at 42°C and at specific time points, aliquots were removed and added to tubes containing quench buffer (70% formamide, 10 mM EDTA pH 8, 0.1% xylene cyanol and Bromophenol Blue dyes). Samples were then subjected to 6% denaturing PAGE, gels were dried and quantitated on a Hewlett Packard Instant Imager using methods described previously (Daniels et al., 1996). Data were plotted (Kaleidagraph Abelbeck software) and analyzed using a parallel kinetic model for simultaneous branching and hydrolysis reactions in order to extract branching rates (kbr). Equations and procedures for quantitation were described previously (Daniels et al., 1996; Liu et al., 1997; Chu et al., 1998).
Isolation of branched fragments using DNAzymes
Lariat introns were prepared and isolated as described previously (Chu et al., 1998). The branched region was then excised by simultaneous cleavage with two DNAzymes. The same two DNAzymes were used to map the branch-sites of all mutants studied to date (Chu et al., 1998). DNAzyme 1 was designed to cleave the phosphodiester linkage between A862 and C863. DNAzyme 2 was designed to cleave the phosphodiester linkage between A103 and U104 of the intron. Reactions were performed under single turnover conditions with both DNAzymes added simultaneously. Reactions contained 300 nM lariat RNA, 60 µM each of DNAzyme 1 and 2, 150 mM NaCl, 100 mM MgCl2 and 40 mM Tris, pH 8.0, and were incubated at 37°C for 5 h. After DNAzyme cleavage, the reaction was quenched and loaded on a 6% polyacrylamide gel. The Y-shaped RNA fragment generated from DNAzyme cleavage is 127 nt in length. This molecule was excised from the gel, eluted overnight at 4°C, precipitated and resuspended in a MOPS storage buffer (Chu et al., 1998).
The Y-shaped fragment was labeled at the 5′-terminus with [γ-32P]ATP and T4 polynucleotide kinase (NEB) or at the 3′-terminus with [32P]pCp and T4 RNA ligase (NEB) using standard reaction conditions (Chu, 2000). The labeling reaction was loaded on a 6% polyacrylamide gel and the labeled product excised, eluted, precipitated and resuspended in a MOPS storage buffer.
Mapping branched fragments by alkaline hydrolysis
In order to observe ∼2000 c.p.m. per band, 254 000 c.p.m. of labeled (either 5′ or 3′) Y-fragment was added to a reaction buffer containing 50 mM NaHCO3, pH 9.0 and 5 µg tRNA (Kuchino and Nishimura, 1989) in a total volume of 10 µl. The reaction was incubated at 90°C for 5 min, combined with an equal volume of quench buffer and then immediately placed on ice. The reaction mixture was loaded on a 20% denaturing polyacrylamide gel, which was dried and then analyzed on a PhosphorImager (Molecular Dynamics).
Supplementary data
Supplementary data for this paper are available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
The authors would like to thank Phil Pang, Leven Wadley and Eckhard Jankowsky for guidance with alignments and distribution analysis. In addition, we thank Magda Konarska, Charles Query and Olga Fedorova for helpful discussions. This work was supported by grants GM50313 (to A.M.P.) and GM31480 (to P.S.P.) from the National Institutes of Health and grant I-1211 from the Robert A. Welch Foundation (to P.S.P.). A.M.P. is an Assistant Investigator of the Howard Hughes Medical Institute.
References
- Bonen L. and Vogel,J. (2001) The ins and outs of group II introns. Trends Genet., 17, 322–331. [DOI] [PubMed] [Google Scholar]
- Boudvillain M. and Pyle,A.M. (1998) Defining functional groups, core structural features and inter-domain tertiary contacts essential for group II intron self-splicing: a NAIM analysis. EMBO J., 17, 7091–7104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger S.C., Faix,P.H., Yang,H., Zhou,J., Franzen,J.S., Peebles,C.L. and Perlman,P.S. (1996) Length changes in the joining segment between domain 5 and 6 of a group II intron inhibit self-splicing and alter 3′ splice site selection. Mol. Cell. Biol., 16, 5896–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkard M.E., Kierzek,R. and Turner,D.H. (1999) Thermodynamics of unpaired terminal nucleotides on short RNA helices correlates with stacking at helix termini in larger RNAs. J. Mol. Biol., 290, 967–982. [DOI] [PubMed] [Google Scholar]
- Cech T.R. (1986) The generality of self-splicing RNA: relationship to nuclear mRNA splicing. Cell, 44, 207–210. [DOI] [PubMed] [Google Scholar]
- Chanfreau G. and Jacquier,A. (1996) An RNA conformational change between the two chemical steps of group II self-splicing. EMBO J., 15, 3466–3476. [PMC free article] [PubMed] [Google Scholar]
- Chin K. and Pyle,A.M. (1995) Branch-point attack in group II introns is a highly reversible transesterification, providing a possible proof-reading mechanism for 5′-splice site selection. RNA, 1, 391–406. [PMC free article] [PubMed] [Google Scholar]
- Chu V.T. (2000) Mechanism of branch-point selection in a catalytic group II intron. PhD thesis, Department of Biochemistry, Columbia University, New York, NY.
- Chu V.T., Liu,Q., Podar,M., Perlman,P.S. and Pyle,A.M. (1998) More than one way to splice an RNA: branching without a bulge and splicing without branching in group II introns. RNA, 4, 1186–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Deme,E., Jacquier,A. and Michel,F. (1997) Multiple tertiary interactions involving domain II of group II self-splicing introns. J. Mol. Biol., 267, 520–536. [DOI] [PubMed] [Google Scholar]
- Daniels D., Michels,W.J. and Pyle,A.M. (1996) Two competing pathways for self-splicing by group II introns: a quantitative analysis of in vitro reaction rates and products. J. Mol. Biol., 256, 31–49. [DOI] [PubMed] [Google Scholar]
- Davanloo P., Rosenburg,A.H., Dunn,J.J. and Studier,F.W. (1984) Cloning and expression of the gene for bacteriophage T7 DNA. Proc. Natl Acad. Sci. USA, 81, 2035–2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib-Hajj S.D., Boulanger,S.C., Hebbar,S.K., Peebles,C.L., Franzen,J.S. and Perlman,P.S. (1993) Domain 5 interacts with domain 6 and influences the second transesterification reaction of group II intron self-splicing. Nucleic Acids Res., 21, 1797–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehara M., Watanabe,K.I. and Ohama,T. (2000) Distribution of cognates of group II introns detected in mitochondrial cox1 genes of a diatom and a haptophyte. Gene, 256, 157–167. [DOI] [PubMed] [Google Scholar]
- Gaur R.K., Mclaughlin,L.W. and Green,M.R. (1997) Functional group substitutions of the branchpoint adenosine in a nuclear pre-mRNA and a group II intron. RNA, 3, 861–869. [PMC free article] [PubMed] [Google Scholar]
- Gautheret D., Konings,D. and Gutell,R.R. (1994) A major family of motifs involving G–A mismatches in ribosomal RNA. J. Mol. Biol., 242, 1–8. [DOI] [PubMed] [Google Scholar]
- Hornig H., Aebi,M. and Weissman,C. (1986) Effect of mutations at the lariat branch acceptor site on β-globin pre-mRNA splicing in vitro. Nature, 324, 589–591. [DOI] [PubMed] [Google Scholar]
- Jarrell K.A., Dietrich,R.C. and Perlman,P.S. (1988a) Group II intron domain 5 facilitates a trans-splicing reaction. Mol. Cell. Biol., 8, 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrell K.A., Peebles,C.L., Dietrich,R.C., Romiti,S.L. and Perlman,P.S. (1988b) Group II intron self-splicing: alternative reaction conditions yield novel products. J. Biol. Chem., 263, 3432–3439. [PubMed] [Google Scholar]
- Konarska M.M., Grabowski,P.J., Padgett,R.A. and Sharp,P.S. (1985) Characterization of the branch site in lariat RNAs produced by splicing of mRNA precursors. Nature, 313, 552–557. [DOI] [PubMed] [Google Scholar]
- Kuchino Y. and Nishimura,S. (1989) Enzymatic RNA sequencing. Methods Enzymol., 180, 154–163. [DOI] [PubMed] [Google Scholar]
- Langford D.J., Klinz,F.J., Donath,C. and Gallwitz,D. (1984) Point mutations identify the conserved, intron-contained TACTAAC box as an essential splicing signal sequence in yeast. Cell, 36, 645–653. [DOI] [PubMed] [Google Scholar]
- Liu Q., Green,J.B., Khodadadi,A., Haeberli,P., Beigelman,L. and Pyle,A.M. (1997) Branch-site selection in a group II intron mediated by active recognition of the adenine amino group and steric exclusion of non-adenine functionalities. J. Mol. Biol., 267, 163–171. [DOI] [PubMed] [Google Scholar]
- Lund J., Tange,T.O., Dyhr-Mikkelsen,J., Hansen,J. and Kjems,J. (2000) Characterization of human RNA splice signals by interative functional selection of splice sites. RNA, 6, 528–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madhani H.D. and Guthrie,C. (1992) A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell, 71, 803–817. [DOI] [PubMed] [Google Scholar]
- Martinez-Abarca F. and Toro,N. (2000) Group II introns in the bacterial world. Mol. Microbiol., 38, 917–926. [DOI] [PubMed] [Google Scholar]
- Mathews D.H., Andre,T.C., Kim,J., Turner,D.H. and Zuker,M. (1998) An updated recursive algorithm for RNA secondary structure prediction with improved thermodynamic parameters. In Leontis,N.B. and Santa Lucia,J. (eds), Molecular Modeling of Nucleic Acids. American Chemical Society, New York, NY, pp. 246–257.
- Michel F. and Ferat,J.-L. (1995) Structure and activities of group II introns. Annu. Rev. Biochem., 64, 435–461. [DOI] [PubMed] [Google Scholar]
- Michel F., Umesono,K. and Ozeki,H. (1989) Comparative and functional anatomy of group II catalytic introns—a review. Gene, 82, 5–30. [DOI] [PubMed] [Google Scholar]
- Newman A.J., Lin,R.-J., Cheng,S.-C. and Abelson,J. (1985) Molecular consequences of specific intron mutations on yeast mRNA splicing in vivo and in vitro. Cell, 42, 335–344. [DOI] [PubMed] [Google Scholar]
- Noble J.C.S., Pan,Z.-Q., Prives,C. and Manley,J.L. (1987) Splicing of SV40 early pre-mRNA to large T and small t mRNAs utilizes different patterns of lariat branch sites. Cell, 50, 227–236. [DOI] [PubMed] [Google Scholar]
- Noble J.C.S., Prives,C. and Manley,J.L. (1988) Alternative splicing of SV40 pre-mRNA is determined by branch-site selction. Genes Dev., 2, 1460–1475. [DOI] [PubMed] [Google Scholar]
- Padgett R.A., Konarska,M.M., Grabowski,P.J., Hardy,S.F. and Sharp,P.A. (1984) Lariat RNAs as intermediates and products in the splicing of messenger RNA precursors. Science, 225, 898–903. [DOI] [PubMed] [Google Scholar]
- Peebles C.L., Perlman,P.S., Mecklenburg,K.L., Petrillo,M.L., Tabor,J.H., Jarrell,K.A. and Cheng,H.-L. (1986) A self-splicing RNA excises an intron lariat. Cell, 44, 213–223. [DOI] [PubMed] [Google Scholar]
- Podar M. (1997) Biochemical, molecular and genetic investigations of the structure and mechanism of a group II intron catalytic RNA. Genetics and Development Program, University of Texas Southwestern Medical Center, Dallas, TX.
- Podar M. and Perlman,P.S. (1999) Photocrosslinking of 4-thio uracil-containing RNAs supports a side-by-side arrangement of domains 5 and 6 of a group II intron. RNA, 5, 318–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podar M., Chu,V.T., Pyle,A.M. and Perlman,P.S. (1998) Group II intron splicing in vivo by first-step hydrolysis. Nature, 391, 915–918. [DOI] [PubMed] [Google Scholar]
- Pyle A.M., Chu,V.T., Jankowsky,E. and Boudvillain,M. (2000) Using DNAzymes to cut, process and map RNA molecules for structural studies or modification. Methods Enzymol., 317, 140–146. [DOI] [PubMed] [Google Scholar]
- Qin P.Z. and Pyle,A.M. (1998) The architectural organization and mechanistic function of group II intron structural elements. Curr. Opin. Struct. Biol., 8, 301–308. [DOI] [PubMed] [Google Scholar]
- Query C.C., Moore,M.M. and Sharp,P.A. (1994) Branch nucleophile selection in pre-mRNA splicing: evidence for the bulged duplex model. Genes Dev., 8, 587–597. [DOI] [PubMed] [Google Scholar]
- Query C.C., Strobel,S.A. and Sharp,P.A. (1996) Three recognition events at the branch-site adenine. EMBO J., 15, 1392–1402. [PMC free article] [PubMed] [Google Scholar]
- Ruskin B., Greene,J.M. and Green,M.R. (1985) Cryptic branch point activation allows accurate in vitro splicing of human β-globin intron mutants. Cell, 41, 833–844. [DOI] [PubMed] [Google Scholar]
- Spingola M., Grate,L., Haussler,D. and Ares,M. (1999) Genome-wide bioinformatic and molecular analysis of introns in Saccharomyces cerevisiae.RNA, 5, 221–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J.S. and Manley,J.L. (1995) A novel U2–U6 snRNA structure is necessary for splicing. Genes Dev., 9, 843–854. [DOI] [PubMed] [Google Scholar]
- van der Veen R., Arnberg,A.C., van der Horst,G., Bonen,L., Tabak,H.F. and Grivell,L.A. (1986) Excised group II introns in yeast mitochondria are lariats and can be formed by self-splicing in vitro. Cell, 44, 225–234. [DOI] [PubMed] [Google Scholar]
- van der Veen R., Kwakman,J.H.J.M. and Grivell,L.A. (1987) Mutations at the lariat acceptor site allow self-splicing of a group II intron without lariat formation. EMBO J., 6, 3827–3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Zimmerly,S., Perlman,P.S. and Lambowitz,A.M. (1996) Efficient integration of an intron RNA into double-stranded DNA by reverse-splicing. Nature, 381, 332–335. [DOI] [PubMed] [Google Scholar]
- Yu Y.-T., Maroney,P.A., Darzynkiewicz,E. and Nilsen,T.W. (1995) U6 snRNA function in nuclear pre-mRNA splicing: a phosphorothioate interference analysis of the U6 phosphate backbone. RNA, 1, 46–54. [PMC free article] [PubMed] [Google Scholar]