Abstract
Cell differentiation in Trypanosoma brucei involves highly regulated changes in morphology, proliferation and metabolism. However, the controls of these developmental processes are unknown. We have identified two novel proteins from the rare CCCH zinc finger family, each <140 amino acids in length and implicated in life cycle regulation. TbZFP1 is transiently enriched during differentiation from the bloodstream to procyclic form, whereas tbZFP2, when ablated in bloodstream forms by RNA interference, inhibits this developmental step. Moreover, expressing an ectopic copy of tbZFP2 results in a dramatic procyclic stage-specific remodelling of the trypanosome cytoskeleton similar to the morphogenic events of differentiation. This phenotype, we term ‘nozzle’, involves polar extension of microtubules at the posterior end of the cell and is dependent upon a motif hitherto restricted to E3 ubiquitin ligases. TbZFP1 and tbZFP2 represent the first molecules implicated in the control of trypanosome differentiation to the procyclic form.
Keywords: CCCH protein/differentiation/morphology/Trypanosoma brucei
Introduction
Differentiation is a central requirement for the adaptation of eukaryote cells to specialized functions or to a changing environment. In particular, microbial eukaryotes exploit programmed developmental changes to maximize their proliferation, transmission and survival. Trypanosoma brucei, the causative agent of human sleeping sickness and nagana in cattle, is transmitted between hosts by tsetse flies (Vickerman, 1985). These protozoan parasites undergo major developmental changes during the course of their life cycle, mediated by regulated changes in gene expression. Although the regulation of particular stage-specific molecules has been described (Vanhamme and Pays, 1995), molecules involved in controlling the events of differentiation have not yet been identified. These molecules may exert control over one or several of the cellular processes in differentiation between life cycle stages, which require changes in morphology, surface antigen expression and metabolism.
A fundamental component of differentiation events in both unicellular and multicellular organsims is the regulation of cell division and morphogenesis. In metazoans, differentiation frequently involves the elaboration of cellular processes or the modulation of shape to perform specialized functions, for example during neurite outgrowth of nerve cells (Valtorta and Leoni, 1999) or myoblast extension in the generation of muscle fibres (Perry and Rudnick, 2000). In unicellular organisms, morphological transitions may represent adaptation to specialized environments (Fulton, 1977), or the expression of shape characteristics specific for particular stages in the life cycle. Such processes have been used to identify and define events of differentiation, although the molecular processes that can elicit such changes are understood in only a few cases (Verde, 1998).
In T.brucei there is an intimate association between the regulated events of the cell cycle, cell morphogenesis and the ability of cells to undergo differentiation (Matthews and Gull, 1994). In particular, the position of its single copy organelles is strictly regulated during both the cell cycle and the life cycle. For example, the trypanosome’s unusual mitochondrial genome (kinetoplast) lies adjacent to the posterior of the cell in bloodstream forms but is repositioned to midway between the nucleus and the posterior during differentiation to the tsetse midgut (procyclic) form (Brown, 1973). This configuration is then strictly preserved during the procyclic form cell cycle (Robinson et al., 1995). This organelle repositioning represents one of the major events of differentiation between these two forms and involves outgrowth at the posterior end of the cell, a process mediated by microtubule extension (Matthews et al., 1995). This is co-ordinated with cell cycle progression and establishes the appropriate architecture for cell division of the differentiated parasite.
We have identified two small proteins, tbZFP1 and tbZFP2, implicated in regulated morphogenesis and differentiation in T.brucei. These molecules share the unusual CCCH zinc finger motif found in a diverse range of RNA-binding proteins involved in various aspects of the control of cell homeostasis and differentiation (Lai et al., 2000). Such proteins have an established role in RNA binding, localization and stability in a wide variety of organisms (Carballo et al., 1998; Tabara et al., 1999; Lai et al., 2000; Reese et al., 2000). We show that tbZFP1 is transiently enriched during differentiation from bloodstream to procyclic form parasites, the first description of this pattern of regulated expression in trypanosomes. Significantly, inducible ablation of tbZFP2 using RNA interference (RNAi) in bloodstream form cells compromises differentiation to the procyclic form. Ectopic expression of this molecule also generates a stage-specific, inducible and sequence-specific morphological phenotype in procyclic forms.
Using cytoskeletal modulation as an assay, we have dissected the functional domains within tbZFP2 and identify a motif shared with E3 ubiquitin ligases that is essential for tbZFP2-dependent morphological remodelling. This represents the first characterization of a novel family of proteins able to modulate fundamental events in the trypanosome life cycle.
Results
TbZFP1 and tbZFP2 are small CCCH zinc finger proteins
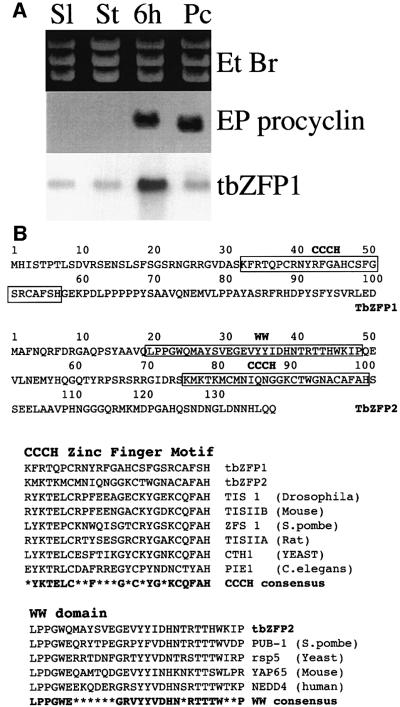
The transformation of T.brucei from its bloodstream to procyclic stage is synchronous if initiated with a population enriched for stumpy forms, a subtype of the bloodstream parasite population adapted for differentiation. This allows identification not only of transcripts that are elevated in one particular life cycle stage, but also those that are enriched during the events of differentiation itself. Using a simplified differential display procedure (Matthews and Gull, 1998), tbZFP1 was identified as a transcript transiently elevated early (2–8 h) in the differentiation process. Although tbZFP1 message was detectable at a low level in both bloodstream and procyclic stages of the life cycle, there was a clear elevation in mRNA 6 h into the differentiation process (Figure 1A).
Fig. 1. Identification and sequence analysis of tbZFP1 and tbZFP2. (A) The tbZFP1 transcript is elevated 6 h through synchronous differentiation. EP1 procyclin is also indicated as a stage-regulated transcript. The ethidium bromide-stained agarose gel indicates the integrity of the RNA used for analysis. Sl, slender form RNA; St, stumpy form RNA; Pc, procyclic form RNA. (B) Sequence analysis of tbZFP1 and tbZFP2. The CCCH and WW motifs are boxed and each domain is aligned with related sequences identified in the databases. A consensus for each domain is derived from the compared sequences; residues that are not conserved are shown as an asterisk.
tbZFP1 is an ∼3.5 kb mRNA predicted to encode a 101 amino acid protein containing a C(x)8C(x)5C(x)3H (CCCH) motif (Figure 1B). The CCCH motif is recognized to have a regulatory role in a diversity of proteins, principally through interaction with RNA (Lai et al., 2000). We searched the T.brucei genome database for sequences encoding further proteins bearing this unusual motif and from this identified tbZFP2. As with tbZFP1, the predicted protein encoded by this gene was small, comprising only 139 amino acids (Figure 1B). Although the structure and basic conservation of the predicted zinc finger region in tbZFP1 and 2 matched that of other CCCH proteins, each possessed only a single CCCH motif, in contrast to the reiterated motifs (at least two) common to this family (Lai et al., 2000). Upstream of the CCCH motif in tbZFP2 was a second highly conserved sequence, a WW domain (Figure 1B). This is an ∼20 amino acid sequence, which functions in protein–protein interactions in a large group of molecules (Pirozzi et al., 1997).
Antibodies were raised to a peptide antigen from the C-terminal 15 amino acids of each predicted protein (anti-ZFP1 and anti-ZFP2). The resulting antibodies were used to detect each protein in samples derived from bloodstream forms of T.brucei undergoing synchronous differentiation to the procyclic form. In each case, proteins of the size predicted from the gene sequence were identified (12 and 16 kDa, respectively), confirming that tbZFP1 and 2 are translated into protein. Although only one intron has been identified in T.brucei to date, this analysis also confirmed that tbZFP1 and 2 did not represent exons of much larger genes.
The profile of tbZFP1 demonstrated the very low levels of this protein in bloodstream forms. However, 2 h after the initiation of differentiation, its expression was transiently elevated, being maximal at 4–6 h before dropping to a lower level at 8–10 h and then returning to higher levels in established procyclic forms (Figure 2A). This profile demonstrates that tbZFP1 is differentiation enriched and procyclic stage expressed. This regulated pattern of expression was also supported by earlier northern analysis (Figure 1A). In contrast, tbZFP2 was present at the same level in bloodstream and procyclic forms and was not enriched during differentiation (Figure 2B).
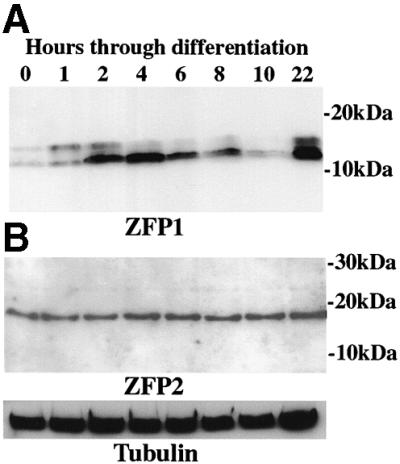
Fig. 2. TbZFP1 and tbZFP2 protein expression profile during differentiation to procyclic forms. Proteins were prepared from cells undergoing synchronous differentiation from bloodstream stumpy forms (0 h) and at time points through differentiation to early procyclic forms (22 h) and probed with the anti-ZFP1 (A) or anti-ZFP2 antibody (B). α-tubulin is shown as a loading control. A weak shadow band is detected by the anti-ZFP1 antibody, which is variable between experiments. Anti-ZFP1 and 2 pre-immune serum did not react with trypanosome proteins (not shown).
TbZFP2 RNA interference compromises differentiation
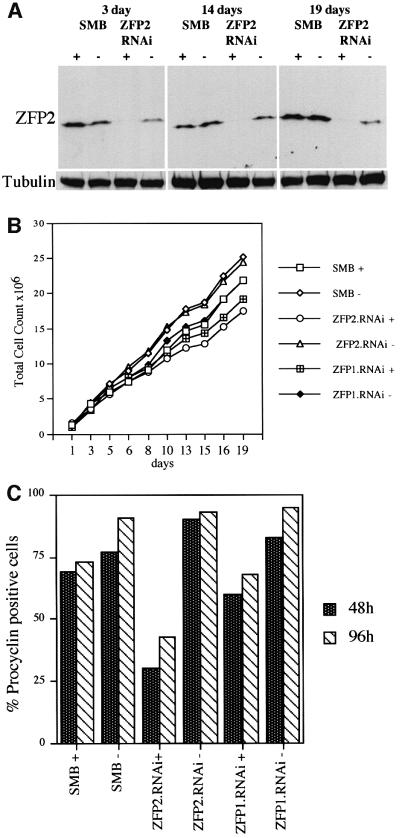
To investigate the function of tbZFP1 and 2 during differentiation to the procyclic form, transcript ablation by RNAi was employed. This technology uses the tetracycline-inducible in vivo expression of a double-stranded gene-specific RNA, resulting in gene silencing (Ngo et al., 1998). RNAi has only been described in procyclic forms to date, thus the ability to suppress tbZFP2 expression was assessed in s427 SMB bloodstream forms. These cells have been engineered to express the tetracycline repressor and T7 RNA polymerase (a kind gift of George Cross, Rockefeller University, New York). When transfected with the RNAi expression vector p2T7i (LaCount et al., 2000) containing the tbZFP2 coding region, a >90% reduction in the level of tbZFP2 protein was observed upon induction with tetracycline (Figure 3A). This confirmed that RNAi gene silencing was operating in an efficient and inducible manner in bloodstream forms. Over 19 days, during which the RNAi effect was stable (Figure 3A), there was no growth or morphological phenotype due to the reduction in tbZFP2. Although there was a slightly decreased growth in the presence of tetracycline, this effect was also observed in control lines (Figure 3B). Similarly, no phenotype was observed when ZFP1.RNAi was induced in bloodstream forms (Figure 3B).
Fig. 3. RNAi gene ablation of tbZFP2 generates a differentiation defect. (A) Western analysis of ZFP2.RNAi effect using the anti-ZFP2 antibody. SMB cells are the parental line expressing the tetracycline repressor and T7 polymerase, into which the ZFP2.RNAi construct was transfected. Induction with tetracycline shows a 90–95% reduction in tbZFP2 levels within 3 days and is maintained at this level for at least 19 days. (B) Growth of ZFP1/ZFP2.RNAi cells monitored in the presence and absence of tetracycline. No obvious growth phenotype is observable, although SMB, ZFP1.RNAi and ZFP2.RNAi cell lines all grew marginally more slowly in the presence of tetracycline. (C) The percentage of EP procyclin-positive cells after ZFP1/ZFP2.RNAi lines were stimulated to differentiate to procyclic forms. ZFP2.RNAi-induced cells show compromised ability to undergo differentiation into procyclic forms.
ZFP1.RNAi and ZFP2.RNAi cell lines were then examined for the effects of gene ablation during differentiation. It should be noted that the cultured transgenic parasites used for these experiments are not capable of generating stumpy forms and therefore differentiate asynchronously to the procyclic form. Suppression of tbZFP1 in ZFP1.RNAi cells showed no reproducible differentiation phenotype, although the expression of EP procyclin was slightly reduced (65% versus 75% for controls, Figure 3C). This observation was variable for several independent cell lines, and western analysis later confirmed that ZFP1.RNAi suppression of tbZFP1 was considerably leaky during differentiation (not shown). This meant that any tbZFP1 differentiation effects could not be assessed reliably in ZFP1.RNAi cells.
In contrast, ZFP2.RNAi cells showed a differentiation phenotype. Induced cells, expressing only ∼5% of wild-type levels of tbZFP2, showed a significantly reduced expression of EP procyclin as assessed by immunofluorescence (25% versus 75% in controls; Figure 3C). These induced ZFP2.RNAi cells also did not undergo the other differentiation-associated events such as morphological restructuring and kinetoplast repositioning. Importantly, uninduced cells differentiated as efficiently as control cells, as assessed by EP procyclin gain (95% at 48 h; Figure 3C) and the morphological events of differentiation. Thus, tbZFP2 ablation compromises differentiation in an inducible manner.
TbZFP2 ectopic expression generates a differentiation phenotype in bloodstream forms
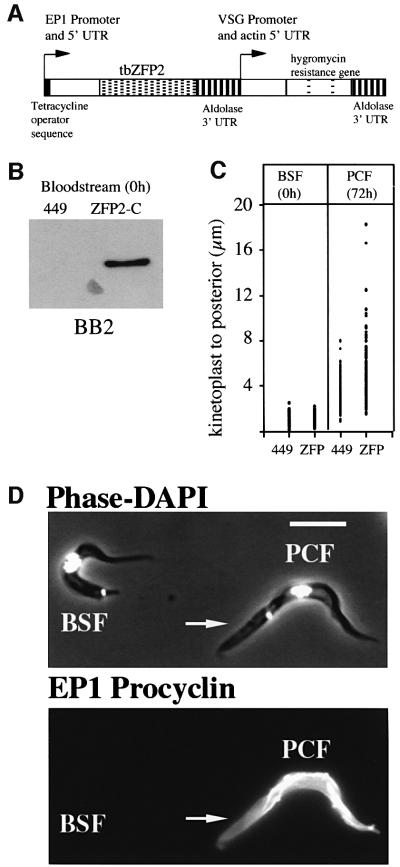
Due to the effect of tbZFP2 ablation on differentiation, further experiments were concentrated on this molecule. Initially, the consequence of expressing an ectopic copy of tbZFP2 in bloodstream forms and during differentiation to the procyclic form was investigated. Specifically, tbZFP2 was modified by the incorporation of a TY1 epitope tag (Bastin et al., 1996) sequence at the 3′ end and cloned into the trypanosome expression vector pHD451. This contains a modified EP1 promoter, allowing the expression of the transgene to be regulated by tetracycline (Figure 4A). Figure 4B shows the expression of tbZFP2 in bloodstream parasites, detected with an antibody recognizing the epitope tag sequence, BB2. TbZFP2 could be clearly detected in the transgenic parasites, confirming the efficient function of the expression vector at this life cycle stage. Bloodstream forms induced to express ectopic tbZFP2 grew at an identical rate and were morphologically indistinguishable from both uninduced lines and the parental line, 449.
Fig. 4. Ectopic tbZFP2 expression generates a morphological phenotype in differentiating cells. (A) pHD451 engineered to express the tbZFP2 gene. The TY1 epitope tag was incorporated into the C-terminus to generate ZFP2-C. (B) Western blot of the expression of ZFP2-C in bloodstream cells. (C) Analysis of the kinetoplast–posterior dimension in bloodstream forms transfected with the 449 construct alone, or with ZFP2-C, or in the same cell lines induced to undergo differentiation to procyclic forms for 72 h. (D) ZFP2-C cells undergoing differentiation to the procyclic form. The cells have been stained with the procyclic form marker, EP1 procyclin. Note that the undifferentiated cell (procyclin-negative) has a normal bloodstream form (BSF) morphology, with a small kinetoplast–posterior dimension. In contrast, the procyclin-positive, differentiated cell (PCF) has undergone cell elongation (arrow). Bar = 10 µm.
Under differentiation conditions, the parental line differentiated well and expressed EP1 procyclin at a level of >60% after 72 h. These cells had also repositioned their kinetoplast, the major morphological event of this differentiation step (the mean kinetoplast–posterior distance had increased from 1.1 to 3.4 µm; Figure 4C). Induced ZFP2-C cell lines gained EP1 procyclin at similar levels to the 449 line. However, many differentiated ZFP2-C cells had grossly extended the kinetoplast–posterior dimension, reaching in some cells up to 18 µm, with the average population dimension being increased to 5.4 µm (Figure 4C and D). Thus, tbZFP2 ablation by RNAi inhibits differentiation to procyclic forms, whereas ectopic expression results in gross morphological restructuring during this developmental step.
Transgenic tbZFP2 expression induces a specific morphological phenotype in procyclic forms
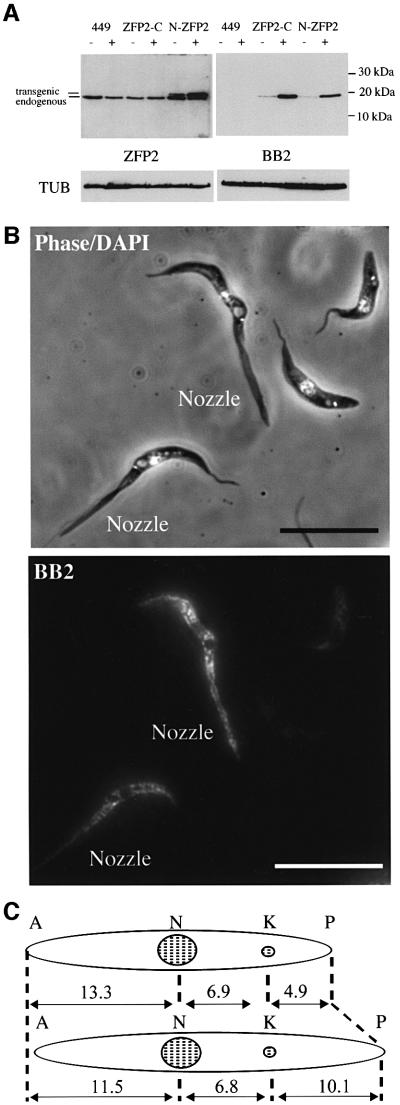
In order to define the morphological phenotype associated with ectopic tbZFP2 expression during differentiation, transgenic procyclic cells were generated expressing either ZFP2-C (C-terminal tag) or N-ZFP2 (N-terminal tag). In each case, inducible expression was verified using the anti-TY1 epitope tag antibody, BB2 (Figure 5A). Probing the same samples with the antibody raised to tbZFP2 detected both the endogenous and the transgenic protein and demonstrated that the transgenic protein was not grossly overexpressed with respect to physiological levels, being elevated ∼2-fold. Interestingly, ZFP2-C could not be detected with the anti-ZFP2 antibody despite this protein being clearly detected with the anti-tag antibody, BB2 (Figure 5A). Sequencing of genomic DNA from these parasites confirmed that the sequence of the integrated transgene was completely correct and that there was no change in the region of the predicted protein sequence recognized by the anti-peptide antibody (data not shown). Apparently, recognition by the anti-ZFP2 antibody is prevented when in the context of a C-terminal TY1 epitope tag.
Fig. 5. TbZFP2 induces morphological change (‘nozzle’ formation) in procyclic form trypanosome cells. (A) Expression of ZFP2-C and N-ZFP2 in procyclic trypanosomes under the control of the tetracycline-inducible promoter. Western analysis of either strain 449 (tetracycline repressor alone), or 449 cells transfected with ZFP2-C or N-ZFP2 in the presence (+) and absence (–) of tetracycline. The left panel shows the blot probed with the antipeptide antibody specific for tbZFP2. The right panel shows the blot probed with the anti-tag antibody, BB2. α-tubulin antibody (Sherwin et al., 1987) indicates loading. (B) Top panel: phase contrast images of procyclic ZFP2-C cells grown in the presence of tetracycline and counterstained with DAPI. Cells displaying the characteristic phenotype of ZFP2-C overexpression are labelled ‘Nozzle’. Bottom panel: immuno fluorescence of the same cells using the anti-tag antibody BB2. Cells displaying the elongate phenotype show detectable expression levels of ZFP2-C. Bar = 20 µm. (C) Representation of cellular landmarks defining the dimensions of procyclic cells. The respective dimensions of a wild-type procyclic trypanosome population (upper panel) and a procyclic population overexpressing ZFP2-C (lower panel) are shown. A, anterior; N, nucleus; K, kinetoplast; P, posterior. Measurements are in micrometres and represent the mean length taken from 200 cells.
The transgenic parasites were examined for phenotype associated with tbZFP2 expression. Interestingly, induced ZFP2-C procyclic cells generated a morphological phenotype identical to that observed during differentiation of ZFP2-C bloodstream forms. Whilst the population comprised cells with apparently normal morphology, many parasites demonstrated an elongate form extending up to twice the normal cell length (Figure 5B). Importantly, this phenotype was inducible, being observed only in those parasite populations induced to express ZFP2-C with tetracycline. Although dependent upon induction, increasing tetracycline levels did not increase the proportion of elongate cells due to the exquisite sensitivity of the expression system, which is maximally induced above a threshold of only 0.001 µg/ml tetracycline (not shown). Immunofluorescence microscopy showed that all cells with the elongate morphology expressed the transgenic protein (Figure 5B), although some cells in the population with normal morphology also showed expression. Perhaps these cells were in the early stages of morphological change, displayed only a minor phenotype or for some reason were unable to change morphologically. We could define no cellular location for tbZFP2 by immunofluorescence, although cell fractionation experiments indicate that the endogenous and transgenic tbZFP2 are cytosolic (not shown).
In contrast to ZFP2-C, cells that had an N-terminal tag fused to tbZFP2 did not demonstrate an elongate morphology, despite equivalent expression of this transgenic protein (Figure 5A). Therefore, to confirm that the elongate morphology was caused by tbZFP2, rather than incorporation of the tag, a cell line that expressed tbZFP2 lacking an epitope tag was generated (ZFP2-No-tag). These cells, which expressed tbZFP2 at the same levels as ZFP2-C, exhibited an elongate phenotype identical to that of ZFP2-C (data shown in Figure 8B). This established that the observed phenotype was specific for the expression of tbZFP2, rather than a pleotropic effect generated by transgenic protein overexpression in T.brucei.
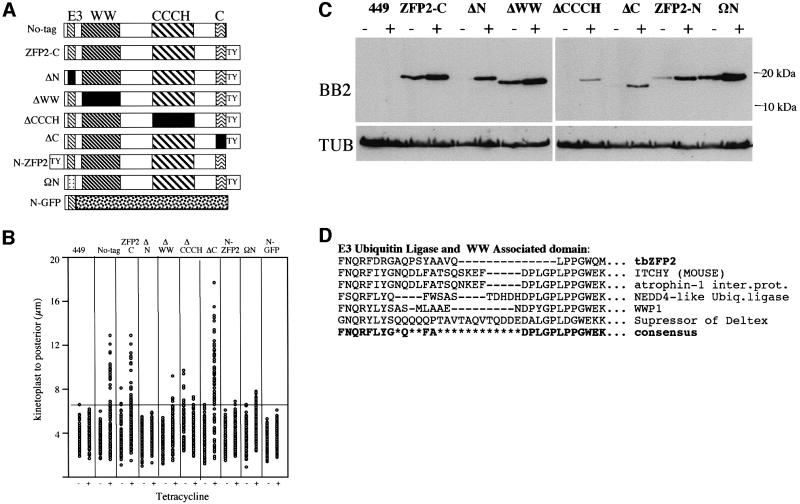
Fig. 8. (A) Schematic representation of the expression constructs used to dissect the role of each domain in tbZFP2. Those constructs and cell lines in which a specific domain was deleted have that domain shown in black. (B) Morphometric assay of the kinetoplast–posterior dimension for mutant forms of ZFP2 grown in the presence (+) and absence (–) of tetracycline. 449 is the procyclic cell line expressing the tetracycline repressor. Open circles show the length of the kinetoplast–posterior end dimension of the cell for 100 random individuals within each population. The horizontal line shows the maximum kinetoplast–posterior dimension seen in the 449 population and marks the length beyond which cells are considered to have nozzled. (C) Expression levels of the mutant forms of ZFP2 in procyclic trypanosomes. Western analysis of each ZFP2 mutant cell line grown in the presence (+) or absence (–) of tetracycline. The upper panels show proteins probed with the anti-tag antibody BB2. The lower panel shows the same samples probed with an antibody recognizing α-tubulin as a loading control. Proteins were prepared from the same cell samples used for morphometric analysis in (B). (D) Sequence alignment of the identified E3 ubiquitin ligase domain associated with the WW domain in tbZFP2.
TbZFP2 causes polar extension of the trypanosome cytoskeleton
The morphological events of differentiation involve an increase in only the kinetoplast–posterior dimension. To establish whether the elongate form of procyclic cells induced to overexpress ZFP2-C was caused by an overall increase in all cellular dimensions or whether this was restricted to only the kinetoplast–posterior dimension, a detailed morphometric analysis was carried out. The linear distance between the cell posterior and kinetoplast, between the kinetoplast and nucleus, and between the nucleus and the cell anterior was measured (Figure 5C). This clearly demonstrated that ZFP2-C induced an increase in only one cellular parameter, with the mean kinetoplast–posterior distance increasing from 4.9 to 10.1 µm in the presence of tetracycline. In addition to measuring the population as a whole, we also measured the cellular dimensions of only those parasites showing an obvious morphological phenotype. Although the kinetoplast–posterior dimension (14 µm) was elevated massively over the wild-type cell dimension in this subset, all other cellular dimensions were preserved (data not shown). This established that ZFP2-C induction results in polar extension of one specific morphological parameter posterior to the kinetoplast, a phenotype we have termed ‘nozzle’ formation.
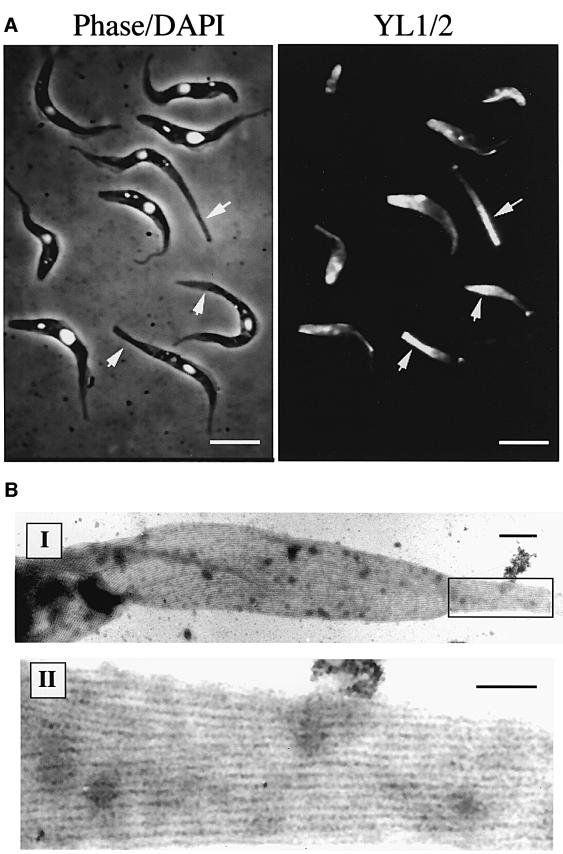
The shape of the trypanosome cell is defined by its microtubule cytoskeleton. To assess whether nozzle formation involved cytoskeletal microtubules, we stained cells overexpressing ZFP2-C with YL1/2, an antibody specific for tyrosinated α-tubulin. This marker for newly assembled microtubules (Kilmartin et al., 1982) has provided a useful indicator of the cytoskeleton during trypanosome cell division (Sherwin and Gull, 1989) and kinetoplast repositioning during differentiation to procyclic forms, a process which involves posterior microtubule extension (Matthews et al., 1995). Figure 6A shows cells undergoing nozzle formation and stained with YL1/2. In those cells with normal morphology, the staining pattern was characteristic for procyclic forms, with basal bodies, newly growing flagella and the cell posterior being labelled. In those cells with a clearly developed nozzle, however, the cell body staining remained rather weak but there was an unusually bright labelling of the cell coincident with the nozzle tip (arrowed in Figure 6A). This indicated that the cell nozzle contained tyrosinated microtubules. To distinguish whether this involved the extension of existing microtubules within the posterior end of the cell, or the interdigitation or assembly of short additional microtubules, we examined the microtubule architecture of the nozzle by electron microscopy. Figure 6B shows detergent-extracted cytoskeletons of cells expressing ZFP2-C visualized by negative staining. In these cells, it is clear that the nozzle was comprised of microtubules, confirming our YL1/2 analysis. Furthermore, close examination demonstrated that there were no short interdigitating microtubules, but rather the nozzle comprised microtubules that had undergone extreme elongation at the posterior end of the cell. This is where the ‘plus ends’ of the microtubule cytoskeleton congregate in T.brucei (Robinson et al., 1995). Significantly, however, unregulated microtubule extension was not observed for all microtubules because the flagellum of the transgenic parasites appeared completely normal and in proportion to the remainder of the cell body. We conclude that ZFP2-C-induced nozzle formation involves polarized extension of microtubules only at the posterior end of the cell.
Fig. 6. Analysis of the tubulin cytoskeleton in procyclic cells expressing ZFP2-C. (A) The left panel shows a phase contrast image of procyclic ZFP2-C cells grown in the presence of tetracycline, counterstained with DAPI to reveal the nucleus and kinetoplast. The right panel shows the same cells stained with the antibody YL1/2, specific for tyrosinated tubulin (Kilmartin et al., 1982). Note that in nozzled cells (arrowed), the posterior tip is brightly labelled, indicating the presence of dynamic microtubules (bar = 10 µm). (B) Electron micrographs of the cytoskeleton of a cell with a nozzle morphology (I) (bar = 0.75 µm). An enlarged section is also shown, revealing the microtubule architecture within the nozzle structure (II) (bar = 0.4 µm).
ZFP2-C-induced morphology is cell cycle position dependent
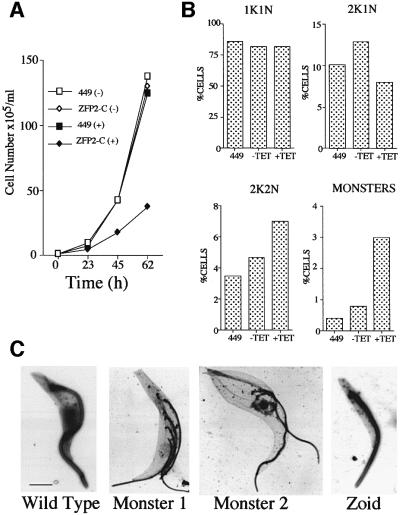
In addition to morphological restructuring, procyclic cells induced to express ZFP2-C also exhibited a reduced growth rate. Figure 7A shows the growth of procyclic form parasites transfected with either ZFP2-C or with the tetracycline repressor construct alone (cell line 449). Whereas all cell lines grew at similar rates in the absence of tetracycline, only those with ZFP2-C showed reduced growth when induced. To determine whether this represented accumulation in one specific cell cycle phase, we scored the percentage of parasites in different stages of the cell cycle by 4′,6-diamidino-2-phenylindole (DAPI) staining (Figure 7B). Initially, this analysis was performed on the population as a whole, without regard to the presence or extent of a nozzle on individual cells. Figure 7B demonstrates that ZFP2-C induction did not result in an accumulation in one particular cell cycle phase, although some increase in the proportion of cells in later stages of the cell cycle was observed (i.e. cells with two kinetoplasts and two nuclei; Figure 7B). Interestingly, the induced population was found to contain a significant proportion of cells with karyotypic defects, or possessing unusual numbers of flagella or kinetoplasts, and there was elevation in the proportion of anucleate cytoplasts (‘zoids’; Figure 7C). Such cells arise as a consequence of microtubule disruption (Ploubidou et al., 1999).
Fig. 7. Growth characteristics of procyclic form cells overexpressing ZFP2-C. (A) Growth of the strain 449 (the tetracycline repressor-expressing cell line; squares) and strain ZFP2-C (diamonds), in the presence (+; filled symbols) and absence (–; open symbols) of tetracycline, monitored for 62 h. (B) Karyotype analysis of strain 449 and strain ZFP2-C, in the presence (+TET) and absence (–TET) of tetracycline. Karyotypes represented are one kinetoplast and one nucleus (1K1N), two kinetoplasts and one nucleus (2K1N), two kinetoplasts and two nuclei (2K2N), and ‘Monsters’ representing the collective sum of cells with aberrant karyotypes (multiple nuclei or kinetoplasts; anucleate cytoplasts). The percentage of each karyotype from 200 cells is shown. (C) Electron micrographs of cells expressing ZFP2-C and with aberrant cytoskeletal architecture. The cell population had been grown for 48 h in the presence of tetracycline, cytoskeletons prepared by detergent extraction and these visualized by negative staining. Bar = 2 µm.
We also scored the cell cycle status of the subset of cells exhibiting an obvious nozzle phenotype. In this case, all nozzled cells were restricted to the early phase of the cell cycle (i.e. with a single kinetoplast and nucleus, or two very closely placed kinetoplasts). Indeed, from an analysis of >5000 nozzled cells, none were in later stages of the cell cycle, and none showed an unusual number or configuration of kinetoplasts or nuclei. This demonstrates that ZFP2-C expression generates a cell cycle-dependent growth and morphological phenotype, with cells early in the cell cycle undergoing nozzle formation, whereas those later in the cell cycle have normal morphology or karyotypic defects.
A novel motif found in E3 ubiquitin ligases is essential for nozzle formation
By exploiting the nozzle phenotype, the signals in tbZFP2 responsible for morphological restructuring were dissected. Thus, constructs were generated specifically deleted for the C-terminal 19 amino acids (ΔC), the WW domain (ΔWW) or the CCCH motif (ΔCCCH), or in which the first 10 amino acids of tbZFP2 were either deleted (ΔN) or exchanged for 10 unrelated amino acids (ΩN) (Figure 8A). These latter constructs were designed to investigate the inability of ZFP2 tagged at its N-terminus to induce nozzle formation. Once stable cell lines were generated, the expression of each mutant was induced and the morphology of the cells analysed, using nozzle formation as a phenotypic assay. Figure 8B shows morphometric analysis of the kinetoplast–posterior dimension in 100 cells from each cell line, either induced or not. Consistent with our earlier analysis, cells expressing either native tbZFP2 or ZFP2-C generated nozzled cells with the kinetoplast–posterior dimension extending up to 14 µm. In uninduced cells, there was a limited extension in a small proportion of cells (1%), presumably due to leaky expression. Deletion of either the WW or the CCCH domain in tbZFP2 reduced nozzle formation, with only a few cells demonstrating very limited posterior extension (Figure 8B). This indicated that these domains are important but not completely essential for nozzle formation. Interestingly, deletion of the C-terminal 19 amino acids of tbZFP2 resulted in an increase in the proportion and extent of nozzles in the population.
When the TY tag was positioned at the N-terminus of tbZFP2 or when the N-terminal 10 amino acids of tbZFP2 were disrupted, nozzle formation was completely ablated. Thus, removal of the N-terminal 10 amino acids of tbZFP2 prevented posterior extension and resulted in a cell population with a mean kinetoplast–posterior dimension indistinguishable from that of wild-type cells (Figure 8B). Significantly, this did not represent a steric consequence of shortening the overall length of the molecule, since substitution of these amino acids with unrelated sequence also compromised nozzle formation (ΩN, Figure 8B), nor did this reflect the abundance of the mutant protein (Figure 8C). Thus the 10 amino acids at the N-terminus of tbZFP2 were essential for nozzle formation. To establish whether this sequence was sufficient for nozzle formation, the 10 amino acids of tbZFP2 alone were fused to the N-terminus of green fluorescent protein (GFP) and procyclic form transfectants were assayed for morphological restructuring. In this case, the parasites demonstrated both normal morphology (Figure 8B) and growth rate, establishing that the integrity of the N-terminal 10 amino acids of tbZFP2 is necessary but not sufficient to induce the phenotype in procyclic form trypanosomes. Interestingly, database analysis of this essential region of the molecule demonstrated significant homology to a short motif found directly upstream of WW domains, specifically in the E3 ubiquitin ligase family of molecules (Figure 8D). These molecules are involved in the targeted degradation of proteins via the proteasome.
Discussion
The transformation of bloodstream to procyclic form trypanosomes requires the control and co-ordination of many cellular events. One of the major steps in dissecting this process is to isolate and define known regulatory proteins or to identify novel molecules whose expression pattern or induced phenotype implicates them in life cycle progression. We originally isolated and characterized a small protein, tbZFP1, which is transiently enriched during the differentiation programme. Interestingly, this protein contains a known regulatory motif, the unusual CCCH zinc finger. We used the CCCH domain of tbZFP1 as a search motif to identify tbZFP2, a second small T.brucei protein containing this motif. Inducible RNAi gene silencing of tbZFP2 compromised the ability of bloodstream parasites to differentiate to the procyclic form. Ectopic expression of tbZFP2 promoted extreme morphological restructuring during differentiation and in established procyclic forms.
A number of lines of evidence indicate that the differentiation and morphological phenotypes associated with perturbed tbZFP2 expression are physiologically relevant. First, the differentiation phenotype observed with ablation of tbZFP2 in bloodstream forms was completely inducible, demonstrating that this was not a cell line-specific effect. Indeed the same phenotype was observed in several independent cell lines, whereas RNAi ablation of other unrelated genes in bloodstream forms has no detectable effect on these events (our unpublished observations). Secondly, the morphological restructuring we observe upon tbZFP2 ectopic expression involved modulation of only one defined parameter in the trypanosome cell (the kinetoplast–posterior dimension) rather than a general disruption of the trypanosome cell form. This contrasts with the few cases reported where abnormal morphologies result from gross alterations in vital cytoskeletal molecules (Ngo et al., 1998) or by more indirect perturbations of molecules involved in energy metabolism (Lamb et al., 2001). Thirdly, western blotting demonstrated that the nozzle phenotype was inducible and was seen when transgenic tbZFP2 was expressed at ∼2-fold endogenous levels. This is far less than the overexpression of unrelated proteins in procyclic T.brucei where phenotype is not observed (e.g. see Tasker et al., 2001), demonstrating that nozzle is not a pleotropic result of protein abundance. Supporting this, the phenotype was only seen in procyclic forms and not bloodstream forms. Finally, the mutation of only 10 amino acids in tbZFP2 completely ablated the nozzle phenotype without a reduction in the abundance of the transgenic protein, whereas deletion of the C-terminal 19 amino acids resulted in an exaggerated nozzle phenotype even at low expression levels. This establishes that the nozzle phenotype is stage and tbZFP2 sequence specific.
The tbZFP2-induced nozzle phenotype involved extension of microtubules only at the posterior end of the cell, where the plus ends of all but a specialized subset of cytoskeletal microtubules congregate (Robinson et al., 1995). Interestingly, those cells that exhibited this phenotype were positioned in the early stages of their cell cycle, while those at later stages either demonstrated no clear phenotype or exhibited a range of diverse morphological or cytoskeletal perturbations. Similar aberrant cell types have been generated in previous analyses of the trypanosome cytoskeleton using the microtubule inhibitor rhizoxin and it has been proposed that these reflect the timing during the cell cycle at which microtubule perturbation occurred (Ploubidou et al., 1999). Although the kinetics of nozzle formation (24–48 h) do not allow direct correlation with the action of rhizoxin (2–10 h), a similar phenomenon may explain the observed phenotypes in cells expressing ZFP2-C. In this case, microtubule extension would generate a nozzle phenotype in cells at the early stages of the cell cycle, these being the available plus ends at this phase of cell division. In contrast, later in the cell cycle, more gross morphological perturbation would result, as new microtubules interdigitate throughout the cytoskeleton in preparation for cytokinesis. This scenario would be compatible with microtubule outgrowth during differentiation, which, like nozzle formation, is co-ordinated with cell cycle progression. A contribution of the actin cytoskeleton to the observed phenotypes is unlikely since actin filaments have never been detected successfully in these organisms and ablation of actin transcripts by RNAi in T.brucei does not reveal an associated phenotype (Shi et al., 2000).
The unusual zinc finger motif shared by tbZFP 1 and 2 is of the characteristic structure CCCH. Although only a few cases have been examined in detail, proteins containing this recently characterized motif have been found to be associated with RNA. The RNA binding of the TTP and PIE-1 proteins is the best characterized. TTP associates with the 3′-untranslated region (3′-UTR) of tumour necrosis factor-α mRNA by its interaction with AU-rich elements and destabilizes this transcript in a regulated manner (Lai et al., 1999). In the case of PIE-1, two CCCH domains co-operate to recognize RNA and to position PIE-1 correctly with ribonuclear complexes (P granules) in the developing embryo of Caenorhabditis elegans (Reese et al., 2000). Other proteins bearing this motif are intimately associated with RNA stability, distribution or translation and operate at all stages of the gene expression pathway (Carballo et al., 1998). It is likely, therefore, that tbZFP1 and tbZFP2 interact with RNA or ribonuclear complexes in the cell via the CCCH domain, raising the intriguing possibility of the involvement of these proteins in regulated mRNA stability or translation. This is highly relevant in trypansomes where the organization of the genome into polycistronic transcription units necessitates that most genes be regulated at the post-transcriptional level (Vanhamme and Pays, 1995).
In addition to a CCCH motif, tbZFP2 possessed a conserved WW protein–protein interaction domain. Significantly, we found that sequences at the N-terminus of tbZFP2, immediately upstream of this domain, were essential though not sufficient for nozzle formation. Database analysis of this short sequence revealed that a related motif is found upstream of a WW core motif present in members of the E3 ubiquitin ligase (itchy) family. Although these molecules contain several WW domains positioned upstream of the HECT region responsible for ubiquitylation of target molecules, the short motif found in tbZFP2 is found only in association with one of these WW domains. The functional requirement for this motif in tbZFP2 has therefore defined this sequence as a novel WW-associated motif previously found exclusively in a specific subset of E3 ubiquitin ligases. Interestingly, a role for the proteasome has been uncovered recently for the regulated destruction of RNAs possessing an AU-rich element in their 3′-UTR (Laroia et al., 1999). This E3 ubiquitin ligase-related motif may, therefore, contribute to the involvement of tbZFP2 in RNA metabolism or translation.
TbZFP1 represents the first molecule characterized whose expression is enriched during trypanosome differentiation, whereas tbZFP2 is the first molecule with a differentiation phenotype when ablated by RNAi. Furthermore, the ectopic expression of tbZFP2 induced a morphological phenotype during differentiation and in procyclic cells, which mimics the ultrastructural events that define bloodstream to procyclic form differentiation. As in differentiating cells, this process involved microtubule outgrowth. These observations implicate tbZFP2 as a molecule able to modulate at least two differentiation-associated events (morphological restructuring and gain of EP procyclin). TbZFP1 and 2 are characterized by their unusually small size and their potential for interaction with partner molecules through their CCCH domain and, for tbZFP2, WW and E3 ubiquitin ligase domains. In this respect, it is important to note that tbZFP1, despite a very unusual pattern of regulated expression, showed no clear differentiation phenotype when suppressed. Although this may reflect an inability to suppress its expression completely using RNAi, this molecule may instead exert a controlling influence via an interacting partner. Interestingly, one potential partner of tbZFP1 may be tbZFP2, since WW domains specifically interact with proline-rich regions (Pirozzi et al., 1997). Such a motif (PPPPPY) is seen in the C-terminal region of tbZFP1. Experiments that elucidate the interactions of both tbZFP1 and 2 will prove invaluable in further dissecting their contribution to differentiation events in T.brucei.
Materials and methods
Trypanosome cultures, differentiation and transfection
Monomorphic bloodstream forms of T.brucei EATRO 2340 were cultured in HMI-9 at 37°C, 5% CO2 (Hirumi and Hirumi, 1989). Pleomorphic bloodstream trypanosomes were harvested from a rat 6 days post-infection when the population was predominantly stumpy in morphology (Matthews et al., 1995). For RNAi analysis, cultured T.brucei s427 ‘single-marker T7 RNAP/TETR’ bloodstream form cells (Wirtz et al., 1999) (‘SMB’) were used. These have been engineered to express the T7 polymerase and a tetracycline repressor. Procyclic forms were cultured in SDM-79 at 27°C (Brun and Schönenburger, 1979).
Transfection of bloodstream and procyclic form parasites was as described previously (Tasker et al., 2001). Drug concentrations used for selection were: hygromycin (20 µg/ml for procyclic forms, 2 µg/ml for bloodstream forms), neomycin G418 (2.5 µg/ml for bloodstream forms) or phleomycin (5 µg/ml for procyclic forms, 0.5 µg/ml for bloodstream forms). Selected cells were cloned by limiting dilution under drug selection.
Monomorphic and pleomorphic bloodstream trypanosomes were differentiated to procyclic forms by diluting cells to 2 × 106/ml with HMI-9 or SDM-79, respectively, and incubation at 27°C with 6 mM cis-aconitate. Differentiation was monitored by immunofluorescence for the expression of EP1 procyclin (Richardson et al., 1988).
Plasmid constructs
The trypanosome expression vector pHD 451 was used to direct transgene expression in bloodstream and procyclic forms previously engineered to express the tetracycline repressor by stable transfection with pHD 449 (Biebinger et al., 1997). To integrate the TY1 epitope tag sequence (Bastin et al., 1996) into the 3′ end of each tbZFP gene, PCR amplification was performed using a primer comprising the 3′ end sequence for each gene, the epitope tag followed by a stop codon and a BglII site (ZFP1 3′ C-TAG, ZFP2 3′ C-TAG; see Table I). This was used in combination with a primer comprising a HindIII site, a start codon and followed by the sequence encoding the N-terminus of either tbZFP gene (ZFP1 5′ C-TAG, ZFP2 5′ C-TAG). The PCR product cloned into pHD451 was designated ZFP1-C or ZFP2-C. To integrate a TY epitope tag into the N-terminus of tbZFP2, a similar strategy was followed using primer ZFP2 5′ N-TAG with primer ZFP2 3′ N-TAG. This was inserted into pHD451 and designated N-ZFP2. Modified forms of tbZFP2 were generated using PCR with the following primer sets: ZFP2 ΔN (ZFP2 ΔN with ZFP2 3′ C-TAG); ZFP2 ΔC (ZFP2 5′ C-TAG with ZFP2ΔC); and ZFP2 ΩN (ZFP2 ΩN5′ with ZFP2 3′ C-TAG).
Table I. Primer sequences used to generate tbZFP1 and tbZFP2 constructs.
| Oligonucleotide | Sequence |
|---|---|
| ZFP1 5′ C-TAG | 5′-CCCAAGCTTATGCACATTTCAACACCC-3′ |
| ZFP1 3′ C-TAG | 5′-CCCAGATCTTCAGTCAAGTGGATCCTGGTTAGTATGGACCTCATCCTCCAACCTTACGCT-3′ |
| ZFP2 3′ C-TAG | 5′-CCCAGATCTCTAGTCAAGTGGATCCTGGTTAGTATGGACCTCCTGCTGCAGATGGTTATT-3′ |
| ZFP2 5′ C-TAG: | 5′-CCCAAGCTTATGGCCTTCAACCAACGC-3′ |
| ZFP2 5′ N-TAG | 5′-CCCAAGCTATGGAGGTCCATACTAACCAGGATCCACTTGACGCCTTCAACCAACGCTTT-3′ |
| ZFP2 3′ N-TAG: | CCCAGATCTCTACTGCTGCAGATGGTT |
| ZFP2 ΔN | 5′-GGAAGCTTATGGCTCAGCCCTCTTATGCA-3′ |
| ZFP2 ΔC | 5′-CCCAGATCTCTAGTCAAGTGGATCCTGGTTAGTATGGACCTCCATCTTCATGCGCTGGC-3′ |
| ZFP2 ΩN5′ | 5′-GGAAGCTTATGCACATTTCAACACCCACTCTTTCCGACGCTCAGCCCTCTTATGCA-3′ |
| ZFP2 ΔWW 1F: | 5′-GCTCAAGCTTAGAGTGAAAACAAGG-3′ |
| ZFP2 ΔWW 1R: | 5′-GGGAATTCCACGGCTGCATAAG-3′ |
| ZFP2 ΔWW 2F: | 5′-CCTGAATTCGTGCTTAATGAGATG-3′ |
| ZFP2 ΔWW 2R: | 5′-TTTGGATCCCTTCTTCCATTCCTTCG-3′ |
| ZFP2 ΔWW 1F: | 5′-GCTCAAGCTTAGAGTGAAAACAAGG-3′ |
| ZFP2 ΔCCCH 2F: | 5′-CTCAGAATTCGGAGGAACTTGCG-3′ |
| ZFP2 ΔCCCH 2R | 5′-CCCATCTAGAATTTGGATCTCTTC-3′ |
| ZFP2 ΔCCCH 1R: | 5′-GCACGAATTCGTCTTCATTTTGG-3′ |
The deletion of the WW domain in tbZFP2 was achieved in a two-step process. First, a clone was generated in pBluescript which contained a 5′ end fragment of tbZFP2 fused to a 3′ end fragment via an engineered EcoRI site. The fused fragments excluded the WW domain and provided a template for PCR using primers ZFP2 5′ C-TAG and ZFP2 3′ C-TAG and insertion into pHD451. The primers used to generate the tbZFP2 5′ and 3′ fragments excluding the WW domain were ZFP2 ΔWW 1F; ZFP2 ΔWW 1R; ZFP2 ΔWW 2F and ZFP2 ΔWW 2R. Similarly, a deletion of the CCCH domain was generated with the primers ZFP2 ΔCCCH 1F, ZFP2 ΔCCCH 1R, ZFP2 ΔCCCH 2F and ZFP2 ΔCCCH 2R. This generated pHD451 ZFP2 ΔWW and pHD451 ZFP2 ΔCCCH. All constructs were verified by DNA sequencing.
RNAi constructs were made using the vector p2T7i (LaCount et al., 2000), which allows for tetracycline-inducible expression of double-stranded RNA from a T7 promoter in T.brucei s427 SMB. Essentially, the tbZFP1 or 2 coding regions were amplified using PCR primers ZFP1/2 5′ C-TAG and ZFP1/2 3′ N-TAG and cloned into the HindIII and BamHI sites of p2T7i. These constructs was designated ZFP1.RNAi and ZFP2.RNAi.
Protein isolation and western blotting
Proteins were prepared as described in Tasker et al. (2000). Western blots were incubated with either the anti-TY 1 epitope tag monoclonal antibody BB2 (diluted 1/20), or rabbit anti-ZFP1 or anti-ZFP2 antibody diluted 1/100. Horseradish peroxidase-conjugated secondary antibodies were used in each case. For antipeptide antibody production, a peptide containing the C-terminal 15 amino acids of either tbZFP1 (FRHDPYSFYSVRLED) or tbZFP2 (HQGNDNGLDNNHLQQ) was synthesized, coupled to keyhole limpet haemocyanin (KLH) and immunized into rabbits (Eurogentec, Belgium). Pre-immune serum and immune serum were screened against whole-cell trypanosome proteins.
Immunofluorescence and electron microscopy
Immunofluorescence was carried out as described in Sherwin et al. (1987) using parasites induced to express the tbZFP2 transgene by growth in tetracycline (10 µg/ml) for 24–48 h. Slides were examined on a Zeiss Axioscop 2 and images captured using NIH image 1.55. Figures were processed using Adobe Photoshop 5.0. Morphometric assays were performed using NIH image 1.55 (Matthews et al., 1995). For analysis of cytoskeletons by electron microscopy, trypanosomes were processed exactly as described in Sherwin et al. (1987).
DDBJ/EMBL/GenBank accession numbers
The sequences reported in this manuscript have been submitted to DDBJ/EMBL/GenBank with the accession Nos AAK39107 (tbZFP2) and AY049059 (tbZFP1).
Acknowledgments
Acknowledgements
We thank Professors I.Hagan and K.Gull for useful comments on the manuscript. We thank members of the Matthews lab for useful discussions and criticism. D.R.R. is a Wellcome Trust Career Development Fellow. K.R.M. is a recipient of a Wellcome Trust University Award. This work was funded by the Wellcome Trust.
References
- Bastin P., Bagherzadeh,Z., Matthews,K.R. and Gull,K. (1996) A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol. Biochem. Parasitol., 77, 235–239. [DOI] [PubMed] [Google Scholar]
- Biebinger S., Wirtz,L.E., Lorenz,P. and Clayton,C. (1997) Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol. Biochem. Parasitol., 85, 99–112. [DOI] [PubMed] [Google Scholar]
- Brown R.C., Evans,D.A. and Vickerman,K. (1973) Changes in oxidative metabolism and ultrastructure accompanying differentiation of the mitochondrion in Trypanosoma brucei. Int. J. Parasitol., 3, 691–704. [DOI] [PubMed] [Google Scholar]
- Brun R. and Schönenburger,M. (1979) Cultivation and in vitro cloning of procyclic forms of Trypanosoma brucei in a semi-defined medium. Acta Trop., 36, 289–292. [PubMed] [Google Scholar]
- Carballo E., Lai,W.S. and Blackshear,P.J. (1998) Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science, 281, 1001–1005. [DOI] [PubMed] [Google Scholar]
- Fulton C. (1977) Cell differentiation in Naegleria gruberi. Annu. Rev. Microbiol., 31, 597–629. [DOI] [PubMed] [Google Scholar]
- Hirumi H. and Hirumi,K. (1989) Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol., 75, 985–989. [PubMed] [Google Scholar]
- Kilmartin J.V., Wright,B. and Milstein,C. (1982) Rat monoclonal antitubulin antibodies derived by using a new nonsecreting rat cell line. J. Cell Biol., 93, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount D.J., Bruse,S., Hill,K.L. and Donelson,J.E. (2000) Double-stranded RNA interference in Trypanosoma brucei using head-to-head promoters. Mol. Biochem. Parasitol., 111, 67–76. [DOI] [PubMed] [Google Scholar]
- Lai W.S., Carballo,E., Strum,J.R., Kennington,E.A., Phillips,R.S. and Blackshear,P.J. (1999) Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor α mRNA. Mol. Cell. Biol., 19, 4311–4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai W.S., Carballo,E., Thorn,J.M., Kennington,E.A. and Blackshear,P.J. (2000) Interactions of CCCH zinc finger proteins with mRNA. Binding of tristetraprolin-related zinc finger proteins to AU-rich elements and destabilization of mRNA. J. Biol. Chem., 275, 17827–17837. [DOI] [PubMed] [Google Scholar]
- Lamb J.R., Fu,V., Wirtz,E. and Bangs,J.D. (2001) Functional analysis of the trypanosomal AAA protein TbVCP with trans-dominant ATP hydrolysis mutants. J. Biol. Chem., 276, 21512–21520. [DOI] [PubMed] [Google Scholar]
- Laroia G., Cuesta,R., Brewer,G. and Schneider,R. (1999) Control of mRNA decay by heat shock–ubiquitin–proteasome pathway. Science, 284, 499–502. [DOI] [PubMed] [Google Scholar]
- Matthews K.R. and Gull,K. (1994) Evidence for an interplay between cell cycle progression and the initiation of differentiation between life cycle forms of African trypanosome. J. Cell Biol., 125, 1147–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K. and Gull,K. (1998) Identification of stage specific and differentiation enriched transcripts during transformation of the African trypanosome from its bloodstream to procyclic form. Mol. Biochem. Parasitol., 95, 81–95. [DOI] [PubMed] [Google Scholar]
- Matthews K.R., Sherwin,T. and Gull,K. (1995) Mitochondrial genome repositioning during the differentiation of the African trypanosome between life cycle forms is microtubule mediated. J. Cell Sci., 108, 2231–2239. [DOI] [PubMed] [Google Scholar]
- Ngo H., Tschudi,C., Gull,K. and Ullu,E. (1998) Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl Acad. Sci. USA, 95, 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.L. and Rudnick,M.A. (2000) Molecular mechanisms regulating myogenic determination and differentiation. Front. Biosci., 5, D750–D767. [DOI] [PubMed] [Google Scholar]
- Pirozzi G., McConnell,S.J., Uveges,A.J., Carter,J.M., Sparks,A.B., Kay,B.K. and Fowlkes,D.M. (1997) Identification of novel human WW domain-containing proteins by cloning of ligand targets. J. Biol. Chem., 272, 14611–14616. [DOI] [PubMed] [Google Scholar]
- Ploubidou A., Robinson,D.R., Docherty,R.C., Ogbadoyi,E.O. and Gull,K. (1999) Evidence for novel cell cycle checkpoints in trypanosomes: kinetoplast segregation and cytokinesis in the absence of mitosis. J. Cell Sci., 112, 4641–4650. [DOI] [PubMed] [Google Scholar]
- Reese K.J., Dunn,M.A., Waddle,J.A. and Seydoux,G. (2000) Asymmetric segregation of PIE-1 in C.elegans is mediated by two complementary mechanisms that act through separate PIE-1 protein domains. Mol. Cell, 6, 445–455. [DOI] [PubMed] [Google Scholar]
- Richardson J.P., Beecroft,R.P., Tolson,D.L., Liu,M.K. and Pearson,T.W. (1988) Procyclin: an unusual immunodominant glycoprotein surface antigen from the procyclic stage of African trypanosomes. Mol. Biochem. Parasitol., 31, 203–216. [DOI] [PubMed] [Google Scholar]
- Robinson D.R., Sherwin,T., Ploubidou,A., Byard,E.H. and Gull,K. (1995) Microtubule polarity and dynamics in the control of organelle positioning, segregation and cytokinesis in the trypanosome cell cycle. J. Cell Biol., 128, 1163–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwin T. and Gull,K. (1989) Visualization of detyrosination along single microtubules reveals novel mechanisms of assembly during cytoskeletal duplication in trypanosomes. Cell, 57, 211–221. [DOI] [PubMed] [Google Scholar]
- Sherwin T., Schneider,A., Sasse,R., Seebeck,T. and Gull,K. (1987) Distinct localization and cell cycle dependence of COOH terminally tyrosinolated α-tubulin in the microtubules of Trypanosoma brucei brucei. J. Cell Biol., 104, 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Djikeng,A., Mark,T., Wirtz,E., Tschudi,C. and Ullu,E. (2000) Genetic interference in Trypanosoma brucei by heritable and inducible double-stranded RNA. RNA, 6, 1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabara H., Hill,R.J., Mello,C.C., Priess,J.R. and Kohara,Y. (1999) pos-1 encodes a cytoplasmic zinc-finger protein essential for germline specification in C.elegans. Development, 126, 1–11. [DOI] [PubMed] [Google Scholar]
- Tasker M., Wilson,J., Sarkar,M., Hendriks,E. and Matthews,K. (2000) A novel selection regime for differentiation defects demonstrates an essential role for the stumpy form in the life cycle of the African trypanosome. Mol. Biol. Cell, 11, 1905–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasker M., Timms,M., Hendriks,E. and Matthews,K. (2001) Cytochrome oxidase subunit VI of Trypanosoma brucei is imported without a cleaved presequence and is developmentally regulated at both RNA and protein levels. Mol. Microbiol., 39, 272–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valtorta F. and Leoni,C. (1999) Molecular mechanisms of neurite extension. Philos. Trans. R. Soc. Lond. B Biol. Sci., 354, 387–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanhamme L. and Pays,E. (1995) Control of gene expression in trypanosomes. Microbiol. Rev., 59, 223–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verde F. (1998) On growth and form: control of cell morphogenesis in fission yeast. Curr. Opin. Microbiol., 1, 712–718. [DOI] [PubMed] [Google Scholar]
- Vickerman K. (1985) Developmental cycles and biology of pathogenic trypanosomes. Br. Med. Bull., 41, 105–114. [DOI] [PubMed] [Google Scholar]
- Wirtz E., Leal,S., Ochatt,C. and Cross,G.A.M. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol., 99, 89–101. [DOI] [PubMed] [Google Scholar]