Abstract
Transcription initiation includes a phase in which short transcripts dissociate from the transcription complex and the polymerase appears not to move away from the promoter. During this process DNA may scrunch within the complex or the polymerase may transiently break promoter contacts to transcribe downstream DNA. Promoter release allowing extended downstream movement of the polymerase may be caused by RNA-mediated disruption of promoter contacts, or by limits on the amount of DNA that can be scrunched. Using exonuclease and KMnO4 footprinting of T7RNAP transcription complexes we show that the DNA scrunches during progression through initial transcription. To determine whether promoter release is determined by RNA length or by the amount of DNA scrunched, we compared release at promoters where the polymerase is forced to initiate at +2 with those where it initiates at +1. For RNAs of identical length, release is greater when more DNA is scrunched. Release is inhibited when a nick introduced into the template relieves the strain of scrunching. DNA scrunching therefore makes an important contribution to T7 promoter release.
Keywords: DNA scrunching/promoter release/transcription initiation/T7RNAP
Introduction
The process of transcription initiation is remarkably similar for polymerases as disparate as the single-subunit RNAPs homologous to the well-characterized T7RNAP, and the multi-subunit RNAPs of which Escherichia coli RNAP is representative. Following promoter opening, both types of RNAP go through a process of abortive transcription characterized by the synthesis and release of transcripts up to ∼9 nucleotides (nt) in length (although in some cases abortive transcripts up to ∼13 nt in length are seen) (Carpousis and Gralla, 1980; Hansen and McClure, 1980; Martin et al., 1988; Jia and Patel, 1997; Sen et al., 2000; Jiang et al., 2001). Footprints of T7 or E.coli RNAP do not move away from the promoter during this initial phase of transcription, although the downstream borders of the footprints do appear to extend downstream as the RNA is extended (Ikeda and Richardson, 1986; Gunderson et al., 1987; Krummel and Chamberlin, 1989; Place et al., 1999). The RNA length at which the upstream border of the footprint is first observed to move downstream is interpreted as reflecting promoter release and has been mapped to between 6 and 13 nt (for T7RNAP) (Ikeda and Richardson, 1986) and between 8 and 11 nt (for E.coli RNAP) (Straney and Crothers, 1987; Krummel and Chamberlin, 1989).
Three mechanisms have been proposed to explain why the transcription complex (TC) footprint does not move away from the promoter during initial transcription. The inchworming model suggests that the RNAP is flexible so that the domain containing the active site moves away from the promoter binding domain to allow transcription of downstream DNA while upstream promoter contacts remain in place (Krummel and Chamberlin, 1989). The DNA looping or scrunching mechanism proposes that the template strand is threaded through the active site and either loops out or packs (scrunches) into a pocket in the RNAP (Cheetham and Steitz, 1999). The transient excursion model explains the footprinting observations in terms of the dynamic nature of the initial transcription complex (ITC). The polymerase is proposed to spend most of its time bound to the promoter during initial transcription (accounting for the observation of a static upstream footprint boundary), but occasionally disengages the promoter and moves downstream to synthesize a short transcript. Release of the transcript may be followed by RNAP backsliding to re-establish promoter contacts. The crystal structure of T7RNAP seems incompatible with the conformational changes required for inchworming, and an ITC structure with a 3mer RNA reveals at least limited template scrunching (Cheetham and Steitz, 1999). How ever, the transient excursion model cannot be formally excluded, particularly as the RNA grows longer. Footprinting data do not fully resolve this issue. MPE-Fe2+ EDTA footprinting of T7RNAP initiation complexes, for example, does reveal downstream extension of the footprint during initial transcription (Ikeda and Richardson, 1986; Gunderson et al., 1987). However, this could reflect increased DNA opening in the downstream region, rather than downstream movement of the leading edge of the RNAP. Investigators using a non-intercalating reagent to footprint initiation complexes detected a significantly smaller extension of the footprint and suggested that this extended footprint might simply represent the time-average of a distribution of abortively cycling polymerase molecules (Muller et al., 1989).
The question of what triggers promoter release so as to allow extended downstream movement of the polymerase also remains unanswered. One class of mechanisms, which may be described as RNA length mediated, proposes that the growth of the RNA to a particular length allows it to contact a site on the RNAP through which it triggers release, for example by causing a change in polymerase conformation. The other type of mechanism may be described as DNA mediated. It suggests that strain is introduced into the promoter-bound complex as increasing amounts of DNA are looped out or scrunched within the TC. Eventually this strain accumulates to a point where it forces RNAP to release the promoter (Carpousis and Gralla, 1985; Straney and Crothers, 1985). Recent structural studies have refined each of these models. For example, the E.coli RNAP structure reveals a ‘flap’, which may close to form a tunnel through which the RNA emerges, and cross-linking studies reveal that RNA 9–12 nt away from the 3′-end contacts this flap (Zhang et al., 1999; Korsheva et al., 2000). The flap may also interact with the σ subunit, which confers promoter specificity on the polymerase. Extension of the RNA to ∼9 nt may therefore dislodge σ from its interaction with the flap, releasing the polymerase from the promoter (Mooney and Landick, 1999). A recent cross-linking study suggests an analogous mechanism for T7RNAP. In this case it was found that the RNA 8–10 nt from the 3′-end interacts with a T7RNAP element important for promoter binding, suggesting that release is mediated by the RNA reaching a length that allows it to compete with promoter for binding to this element (Temiakov et al., 2000). In contrast to these RNA-mediated release mechanisms, the structure of a T7RNAP ITC reveals a pocket into which the template strand appears to scrunch. This pocket can accommodate no more than 6–8 nt of DNA, suggesting that release occurs when the filling of this pocket forces the polymerase to move away from the promoter (Cheetham and Steitz, 1999).
In this study we address the following questions. At what point does promoter release occur? Are all of the TCs released at the same point (i.e. 100% release when the RNA reaches 8 nt) or is release distributed over a range of RNA lengths? Does the DNA scrunching occur during initial transcription or does the polymerase make transient excursions? Is release RNA length mediated, DNA mediated or some combination of both? Though we use T7RNAP to address these questions, the similarities in the abortive transcription and promoter release reactions of the single- and multi-subunit RNAPs suggest that our observations are generally relevant to the mechanisms of transcription initiation.
Results
Monitoring of ITC boundaries during progressive RNA extension
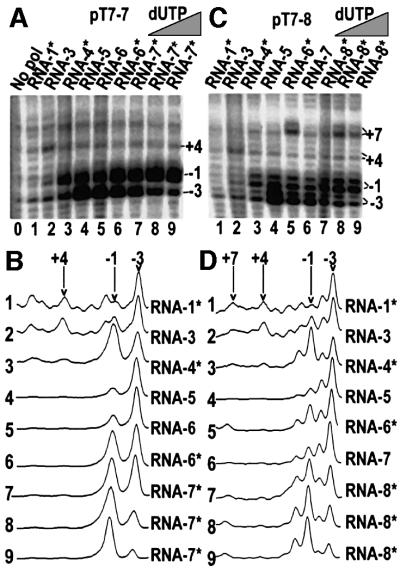
Previous studies have shown that the T7RNAP footprint and transcription bubble do not move away from the promoter as the RNA is extended to +6, but that both the transcription bubble and the footprint have left the promoter once the RNA reaches 13 nt (Ikeda and Richardson, 1986; Brieba and Sousa, 2001). To define the point at which release occurs we designed two promoters, pT7-7 and pT7-8, whose initially transcribed sequences (ITSs) are GGGAGCTT and GGGAGACTT, respectively (see Table I for a description of all promoter constructs used in this study). Inclusion of different sets of NTPs and 3′-dNTPs in transcription reactions with these promoters allows progressive extension of the RNA up to 7 or 8 nt in length. TC boundaries on the template strand in reactions allowing RNA extension to different lengths were then mapped with exonuclease III (exo III) and λ exonuclease (λ exo) (Figure 1). Exo III experiments reveal that the upstream boundary of the TC remains stationary (at approximately –19) even as the RNA is extended up to 7 nt (Figure 1A, lanes 5–11 and 13–18). In reactions containing 3′-dGTP only (lanes 5 and 13) a set of feint bands is also observed between –4 and –10. This probably reflects the instability of a T7RNAP–promoter complex stabilized only by binding of initiating NTPs. T7RNAP–promoter complexes on double-stranded, relaxed templates are unstable until they initiate transcription (Ikeda and Richardson 1986; Basu and Maitra 1986; Diaz et al., 1996; Place et al., 1999; Brieba and Sousa, 2001). We could not detect exonuclease-defined boundaries until NTPs were added to the reactions. The observation that limited exo III digestion into the promoter occurs in complexes stabilized only by binding of the initiating NTPs indicates that these complexes are less stable than those in which transcription has been initiated, so that the exonuclease either pushes the polymerase off the promoter or digests into the promoter when the polymerase transiently dissociates. When the RNA is extended to 8 nt a new boundary appears at approximately –4, accounting for ∼40% of the complexes (Figure 1A, lane 19). A similarly positioned boundary appears in lane 11 of Figure 1A (where the RNA has been extended to only 7 nt), although it accounts for only 5–10% of the TCs. However, this band is not seen in lane 18. Although the RNAs in both lanes 11 and 18 are extended to 7 nt and are terminated with a 3′-dNMP, the TCs in these lanes differ in that the incoming NTP is present in the reaction in lane 11, but not in lane 18. Previous studies have shown that binding NTP to the TC stabilizes it in the post-translocated position (Huang and Sousa, 2000). The presence of a forward-shifted boundary in lane 11, but not in lane 18, likely reflects increased promoter release due to the downstream pulling effect of NTP binding on the active site. This interpretation is supported by observations that addition of the incoming NTP to reactions like that in lane 18 leads to the appearance of a downstream-shifted boundary and that increasing the concentration of this NTP increases the percentage of the shifted boundary (Figure 2; unpublished observations).
Table I. Sequences of promoters used.
| Designation | +1 to +13 template strand sequencea |
|---|---|
| pT7-7 | CCCTCGAAGGCTG |
| pT7-8 | CCCTCTGAAGGCT |
| pT7 | CCCTCTGGCCTTA |
| pT7-A | ACCCTCTGGCCTT |
| pT72G | CCGCGCAACGCGT |
| pT72G-T | TCCGCGCAACGCG |
| pT73G | CCCGCGAACGCCT |
| pT73G-T | TCCCGCGAACGCC |
| pT7-cont | CCCTCCTGAGGCT |
| pT7-nickb | CCCTCCTGAGGCT |
aAll promoters share a –1 to –23 sequence identical to the T7 class III promoter consensus.
bpT7-nick differs from all other promoters in that its template strand is composed of two covalently discontinuous segments extending from –32 to –5 and –4 to +19.
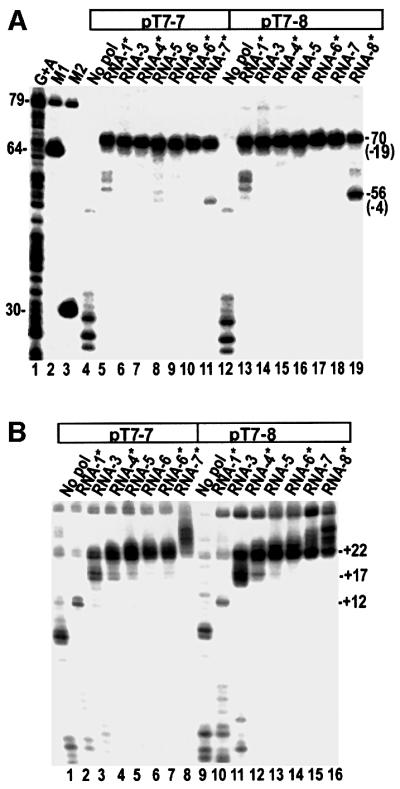
Fig. 1. (A) Exo III mapping of the upstream boundary of T7RNAP ITCs formed with either 5′-template strand-labeled pT7-7 (lanes 4–11) or pT7-8 (lanes 12–19). The length of the RNA that can be made, based on the NTPs present, is specified above each lane. Reactions in lanes 5 and 13 (‘RNA-1*’) contain 3′-dGTP only. In lanes marked with an asterisk, the RNA lacks a 3′-OH group and the incoming NTP is present in the reaction at 1 mM, so that complexes with bound NTP can form. Lane 1, Maxam–Gilbert (G+A) sequencing reaction with labeled pT7-7; lanes 2 and 3, restriction digests of pT7-7. DNA fragment lengths are as indicated, and the positions at which exo III digestion is halted (relative to the transcription start site) are in parentheses. (B) λ exo digestion of the downstream boundary of T7RNAP ITCs formed with either 3′-template strand-labeled pT7-7 (lanes 1–8) or pT7-8 (lanes 9–16).
Fig. 2. Permanganate probing of T7RNAP ITCs formed with either 3′-template strand-labeled pT7-7 (A) or pT7-8 (C). Lengths of the RNA in the complexes are as specified in Figure 1. Lanes 8, 9 and 10 contain, respectively, 0.01, 0.1 or 1 mM 3′-dUTP. Reactive Ts are labeled relative to the transcription start site (+1). (B) and (D) show scans of the reactions in (A) and (C) respectively. In (C) and (D) the doublets are due to incomplete fill-in by Klenow in the labeling reaction.
The downstream boundary detected by λ exo (Figure 1B) shows a different pattern of movement as the RNA is extended. The boundary of the complex stabilized by binding of the initiating NTPs (Figure 1B, lanes 2 and 10) is at +12/+13, though it is quantitatively weak and much of the exonuclease digests past this position (note the shorter DNAs in these lanes). Extension of the RNA to 3 nt shifts the boundary to between +16 and +22 (lanes 3 and 11). Extension of the RNA to 4 nt shifts more of the boundary to +22 (lanes 4 and 12); extension to 5 or 6 nt progressively reduces the boundary at +17 (lanes 5, 6, 13 and 14) and further extension to 7 or 8 nt shifts part of the boundary downstream of +22 (lanes 8, 15 and 16).
Effects of progressive RNA extension and NTP binding on transcription bubble structure during initial transcription
To characterize the transcription bubble as the RNA was progressively extended we probed the template strand with KMnO4 (Figure 2). T7RNAP open complexes are unstable until transcription is initiated (Villemain et al., 1997; Bandwar and Patel, 2001), and reactivity with permanganate cannot be detected in polymerase–promoter binary complexes (Place et al., 1999; Brieba and Sousa, 2001). However, we detect weak reactivity at –3 upon addition of 3′-dGTP to polymerase–promoter complexes (Figure 2A and C, lanes 1). Addition of GTP, which allows RNA extension to +3, increases the reactivity at –3 and also reveals weak reactivity at –1 and +4 (Figure 2A and C, lanes 2). Extension of the RNA to 4 nt further increases reactivity at –1 and –3 but suppresses the weak reactivity at +4 (Figure 2A and C, lanes 3). Maximal reactivity with permanganate develops upon extension of the RNA to 5 nt (Figure 2A and C, lanes 4).
We tested the effects of NTP binding on transcription bubble structure or interactions with polymerase by adding templated NTPs to complexes with 3′-dNMP terminated transcripts. Unexpectedly, NTP binding markedly alters the relative permanganate reactivity of the –1 and –3 bases. This is seen most clearly in the lane scans shown in Figure 2B and D. For example, in, in scans 3 from (B) and (D), scan 5 from (D) and scans 7–9 from (B) and (D), reactivity at –1 is strong and is usually stronger than reactivity at –3. These scans are all of reactions in which the transcripts were 3′-dNMP terminated and the templated NTP was present in the reaction. In contrast to this pattern of reactivity, scans of reactions in which the templated NTP was absent from the reaction (Figure 2B and D, scans 2 and 4 of panels B and D, scan 5 of panel B and scan 6 of panel D) show reduced reactivity at –1 relative to –3. That this is not an RNA length effect or due to 3′-dNMP termination of the transcript can be seen by comparing scans 5 and 6 of Figure 2B. Both of these are of reactions in which the RNAs are 6 nt in length and are 3′-dCMP terminated. However, scan 6 shows a reaction to which the templated NTP (UTP) is added to 1 mM, and clearly reveals that addition of UTP enhances reactivity at –1 relative to –3. That this effect is a function of the concentration of the added NTP is shown in Figure 2B and D, scans 7–9, which are of reactions in which the concentration of the templated NTP was progressively increased. These scans show clearly that progressively increasing the concentrated of templated NTP progressively enhances reactivity at –1 relative to –3.
Effects of RNA length versus active site position on promoter release
The experiment shown in Figure 1 suggests that the polymerase first moves permanently away from the promoter when the RNA reaches ∼8 nt in length. This release of the promoter could therefore be triggered by the RNA reaching a specific length. Alternatively, it could be triggered by the active site reaching +8 on the template strand, leading to scrunching or looping out of the DNA between +8 and upstream promoter sequences (i.e. RNA- versus DNA-mediated release). To distinguish between these two mechanisms we took advantage of the fact that T7RNAP has a strong preference for initiating with a G, so that it will initiate from +2 (numbering relative to the –4 to –1 TATA element) if the template base at +1 is G, A or T and the base at +2 is C (Imburgio et al., 2000; Jiang et al., 2001). We therefore designed promoters that contained canonical or near canonical ITSs placed at either +1 or shifted 1 nt downstream by insertion of an A or T into the template at +1 (Table I). Figure 3A shows transcripts obtained with different NTP combinations on a promoter whose ITS is either GGGAGACCGGAAU (pT7; lanes 1–6) or UGGGAGACCGGAAU (pT7-A; lanes 7–12). On a promoter whose +1 to +3 sequence is ‘GGG’, T7RNAP will synthesize oligo(G) transcripts up to 14 or more bases in length if GTP is the only NTP present, as is seen in lane 1 of Figure 3A. On pT7-A, oligo(G) synthesis is also evident (though it is less processive than on pT7 and does not extend beyond approximately five bases; lane 7) showing that initiation on this promoter is directed to +2 when only GTP is present. Addition of GTP+3-′dATP, GTP+ATP, GTP+ATP+3-′dCTP, GTP+ATP+CTP+3′-dUTP or all four NTPs results in synthesis of transcripts up to 4, 6, 7, 13 and ∼59 nt in length, respectively, on both promoters (lanes 2–6 and 8–12). On pT7 we also detect smaller amounts of transcripts longer than the expected lengths in lanes 2–4. These probably reflect synthesis of transcripts with extra Gs at their 5′-ends (Huang et al., 2000). The absence of such transcripts in the pT7-A reactions (lanes 8–10) may reflect either the reduced efficiency of oligo(G) synthesis, or the general reduction in transcription efficiency on this promoter (largely due to an increase in abortion after dimer synthesis). In any case, the critical observation is that, with identical combinations of NTPs, the predominant transcripts are of identical length and sequence with both promoters.
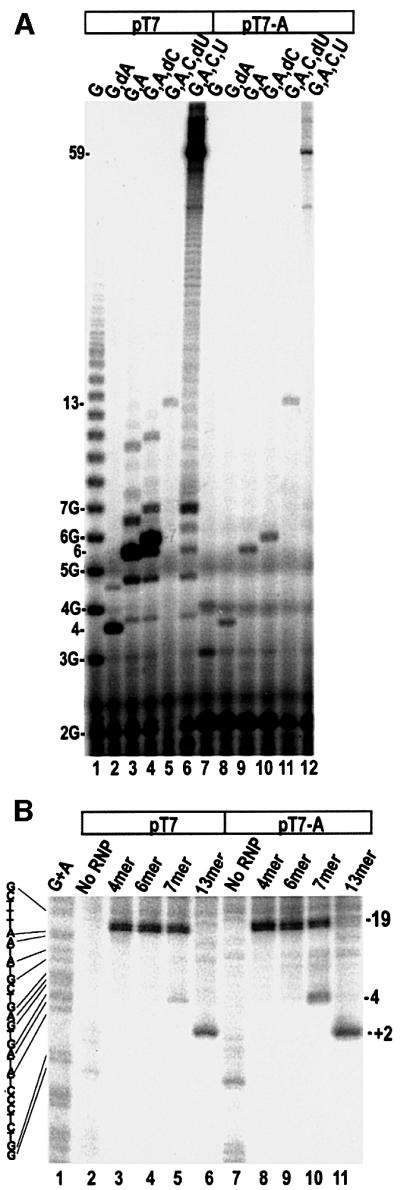
Fig. 3. (A) Transcription from pT7 (lanes 1–6) or pT7-A (lanes 7–12) in the presence of the NTPs indicated over each lane. Transcripts lengths are indicated, and oligo(G) transcript lengths are specified as 3G, 4G, etc. (B) Exo III digestion of ITCs formed with 5′-template strand-labeled pT7 (lanes 2–4) or pT7-A (lanes 7–11), with the RNA lengths indicated above each lane. The template strand sequence is given next to the Maxam–Gilbert (G+A) sequencing reaction shown in lane 1. Promoter positions at which exo III is halted by the TCs are given to the right of lane 11.
We used exo III digestion to measure promoter release as a function of RNA length on these two promoters (Figure 3B). When RNA extension is limited to 4 or 6 nt (lanes 3, 4, 8 and 9), the upstream boundary of the complex is at –19 on both promoters, and extension of the RNA to 13 nt shifts most of the complex to +2 on both promoters (lanes 6 and 11). A difference between the two promoters is, however, seen in the reactions allowing transcript extension to 7 nt. On pT7 most of the complexes remain at –19, but a small fraction (5–10%) shift to –4 (lane 5). However, on pT7-A 20–30% of the complexes move to –4 (lane 10). Note that, for identical RNA lengths, release on pT7-A should result in downstream movement of the complex by 1 nt more than on pT7. The positions of the exonuclease-defined boundaries in these experiments are ±1 nt, so for simplicity we map the released complexes on both pT7 and pT7-A to approximately –4. However, the difference in electrophoretic migration between the bound (–19) and released (–4) complex boundaries is detectably ∼5% larger with pT7-A than on pT7, consistent with scrunching of more DNA and downstream movement by an additional nucleotide upon release with pT7-A.
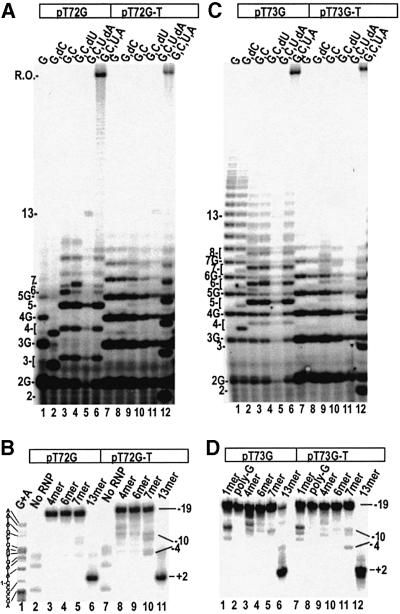
Similar experiments were carried out with two other sets of promoters (Table I). The ITS of pT72G is GGCGCG UUGCGCA. The ITS of pT72G-T contains an additional A at +1. Figure 4A shows the transcripts obtained with different NTPs on either pT72G (lanes 1–6) or pT72G-T (lanes 7–12). Relative to the transcript pattern seen with pT7 (Figure 3A), transcription on pT72G is altered in that: (i) oligo(G) synthesis is less processive (lane 1), and (ii) there is a high level of abortion at the 5mer (lanes 3 and 4). The major transcripts are of the expected length but, as with pT7, we observe small amounts of transcripts that are longer than expected (lanes 2–4). The transcript pattern obtained with pT72G-T is very different. In all reactions (except in lane 12 where UTP is present so that initiation at +1 is possible), oligo(G) synthesis predominates, though the oligo(G) synthesis reaction is less processive than on a canonical ITS. [In the presence of GTP only, oligo(G) synthesis on pT72G-T extends to ∼8 nt while on a canonical ITS it extends almost indefinitely as in lane 1 of Figure 4C.] The synthesis of oligo(G) transcripts even in the presence of multiple NTPs indicates that normal transcript extension is inefficient on this promoter, especially when transcription starts at +2. However, low levels of heterogeneous sequence transcripts that co-migrate with the predominant pT72G transcripts in the reactions with identical NTP mixes are visible against the background of oligo(G) transcripts in the pT72G-T reactions (Figure 4, compare lanes 8–10 with lanes 2–4), and with both pT72G and pT72G-T a 13mer is made in reactions containing G, C, U and dA (Figure 4A, lanes 5 and 11), indicating that productive initiation occurs even when transcription initiates at +2.
Fig. 4. (A) Transcription of pT72G (lanes 1–6) or pT72G-T (lanes 7–12) in the presence of the NTPs indicated. Normal and poly(G) transcript lengths are indicated. (B) Exo III digestion of ITCs formed with 5′-template strand-labeled pT72G (lanes 2–6) or pT72G-T (lanes 7–11), with the RNA lengths indicated over each lane. The template strand sequence is given next to the Maxam–Gilbert (G+A) sequencing reaction shown in lane 1. Promoter positions at which exo III is halted by the TCs are given to the right of lane 11. (C) Transcription of pT73G (lanes 1–6) or pT73G-T (lanes 7–12) in the presence of the NTPs indicated. Normal and poly(G) transcript lengths are as indicated. (D) Exo III digestion of ITCs formed with 5′-template strand-labeled pT73G (lanes 1–6) or pT73G-T (lanes 7–12), with the RNA lengths indicated above each lane. Promoter positions at which exo III is halted by the TCs are given to the right of lane 12.
Promoter release as a function of RNA length on these two promoters is shown in Figure 4B. No release is detected on either promoter for RNAs 4 or 6 nt long (lanes 3, 4, 8 and 9) and complete release is seen for RNAs 13 nt long (lanes 6 and 11). In the reactions with the 7mer RNAs (lanes 5 and 10) two new digestion products appear: a doublet centered at approximately –10 and a band at approximately –4. The band at –4 is probably due to released complexes since its distance from the 3′-end of the RNA is identical to that seen in an elongation complex (EC) (i.e. the upstream boundary of the released complex with the 13mer RNA in lanes 6 and 11 is at approximately +2), and it is at the same position as the released complex boundary in the experiments with pT7-7, pT7-8, pT7 and pT7-A. Interpretation of the doublet at approximately –10 is ambiguous, but it could represent released complexes that have backtracked on the DNA. Together, the bands at approximately –10 and –4 account for ∼5% of the complexes in lane 5 and ∼20% in lane 10. Figure 4C and D shows experiments performed with pT73G, whose ITS is GGGCGCUUGCGCAA, and with pT73G-T, which has a similar ITS with an A inserted at +1 (Table I). As with pT72G and pT72G-T, normal transcript extension is inefficient on these promoters, especially when initiating from +2 on pT73G-T, so that oligo(G) synthesis predominates, even in reactions where multiple NTPs are present (Figure 4C, lanes 8–11). Nevertheless, synthesis of a 13mer is observed in lanes 5 and 11, indicating that promoter release can occur on both promoters. This is confirmed by the experiment in Figure 4D, which shows complete release on both promoters in reactions that allow synthesis of a 13mer (lanes 6 and 12). No release is detected in the 1mer, poly(G), 4mer or 6mer reactions with either promoter (lanes 1–4 and 7–10), but ∼10% release is detected in the 7mer reaction with pT73G-T (lane 11; 10% = sum of products at –10 and –4), while no (<5%) release is seen with pT73G (lane 5).
Inhibition of promoter release on nicked templates
If the steric strain associated with DNA scrunching contributes to promoter release, then we would predict that relief of this strain would inhibit release. It has recently been reported that T7RNAP will transcribe promoters in which a nick or gap is introduced in the template strand between –5 and +1, though the amount of run-off transcript obtained on such promoters is markedly reduced relative to promoters with covalently continuous template strands (Jiang et al., 2001). Changes in the abortive transcript patterns with these promoters led to the conclusion that the introduction of a nick or gap in the template facilitated the threading of the template strand through the active site during initial transcription, presumably because the 3′-end of the template can emerge from the TC and relieve the strain of scrunching. We therefore evaluated release on two promoters that were identical except that the template strand of one promoter comprised two oligonucleotides extending from –32 to –5 and –4 to +19 (Table I). Transcription of the promoter with the continuous template strand (pT7-cont) in the presence of different NTPs gives rise to transcripts of the expected length (Figure 5A, lanes 1–6). Notably, there is a severe drop in total transcription products upon extension of the RNA from 8 to 9 nt (lanes 4 and 5), similar to the drop seen in experiments with other promoters upon addition of NTPs, allowing synthesis of 13 nt RNAs (i.e. Figures 3 and 4). Quantification showed that the amount of 9 nt RNA made in a 10 min reaction corresponds to approximately one transcript per template molecule (template is stoichiometrically limiting in these experiments). These observations indicate that upon synthesis of a 9 nt RNA, a stable TC that turns over slowly is formed. This was confirmed by time-course experiments (not shown), which revealed that the lifetime of the TC with the 9 nt RNA was >15 min. Thus 9 nt appears to be the minimal RNA length required to form a stable TC. On the promoter with a discontinuous template strand (pT7-nick) high levels of oligo(G) synthesis (lane 7) and 4mer synthesis (lane 8) are evident, though, in agreement with previous observations (Jiang et al., 2001), oligo(G) synthesis is less processive and transcription initiation is more heterogeneous. Though less efficient than with pT7-cont, extension of the RNA to 7 and 8 nt also occurs on pT7-nick (lanes 9 and 10), and quantification reveals that 7- and 8mers are made in molar excess of the amount of template (i.e. the polymerase cycles abortively under these conditions). Upon extension of the RNA to 9 nt we again observe a severe drop in total transcription products (lane 11), and turnover rate measurements confirm formation of a stable TC with a 9 nt RNA. Thus, on both pT7-cont and pT7-nick, the RNA can be extended to 9 nt, leading to formation of a stable TC. Further extension of the RNA, however, appears to be severely impeded on pT7-nick as evidenced by the extreme drop in 19 nt run-off transcript on pT7-nick versus pT7-cont (compare lanes 6 and 12). When we use exo III digestion to measure promoter release on pT7-cont, we observe no release for RNAs up to four bases in length (Figure 5B, lanes 2–4). Extension of the RNA to 7 or 8 nt results in ∼5 and ∼20% release, respectively (Figure 5B, lanes 5 and 6). Addition of the templated NTP to the complex with the 8mer RNA or extension of the RNA to 9 nt causes, respectively, ∼80 or ∼90% of the complexes to release the promoter. However, when we try to measure release on pT7-nick in the same way we obtain a very different result: no promoter release is observed on pT7-nick even upon addition of NTPs allowing extension of the RNA to 9 nt (Figure 5C).
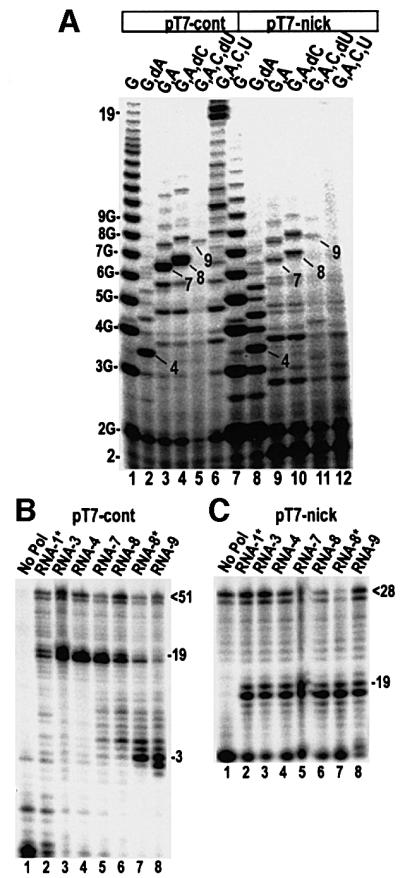
Fig. 5. (A) Transcription reactions with either pT7-cont (lanes 1–6) or pT7-nick (lanes 7–12) carried out with the NTPs indicated. Both promoters are synthetic 51 bp duplexes with a –23 to –1 sequence corresponding to the T7 class III consensus. The +1 to +19 non-template strand sequence is GGGAGGACTCCGACAGAGC. pT7-nick differs from pT7-cont in that its template strand is covalently discontinuous and is composed of two oligonucleotides extending from –32 to –5 and –4 to +19. (B and C) Exo III digestion of ITCs formed on either pT7-cont (B) or pT7-nick (C). Reactions allow RNA extension to the lengths indicated above each gel lane. Asterisked RNA lengths indicate that the incoming NTP is present but cannot be incorporated. Positions of the bound and released complexes are indicated as is the length of the undigested template strand at the top of the gel.
Discussion
Promoter release occurs when the RNA reaches 8 nt
If the point at which promoter release occurs is defined as the RNA length at which ∼50% of all complexes have left the promoter then, as detected by exo III digestion, we find that on a canonical T7 promoter release occurs when the RNA reaches 8 nt (Figures 1A and 5A: 20–80% of the complexes released at the 8 nt point). Release occurs sharply as a function of RNA length but not all complexes release at the same point. In reactions in which the RNA can be extended to 7 nt between 0 and 10% release is detected (Figures 1A and 5A). Synthesis of a 9mer results in ∼90% release (Figure 5A), and by the time a 13mer has been made essentially 100% of the complexes have left the promoter (Figures 4 and 5). A caveat to these conclusions is that the processive action of the exonuclease may be capable of pushing the polymerase along the template. If so, this would still indicate a change in the nature of the initiation complex when the RNA reaches ∼8 nt since the exonuclease is unable to push the polymerase off the promoter when the RNA is shorter than this. Therefore, either the polymerase is releasing the promoter and moving downstream when the RNA reaches ∼8 nt, or the promoter–polymerase interaction first becomes weak enough to be displaced by a processive exonuclease at this point.
Progression through initial transcription is accompanied by DNA scrunching
Since crystal structures of T7 RNAP do not easily accommodate the type of conformational changes required for inchworming, our experiments were directed at distinguishing between the transient excursion and scrunching models. Our results support DNA scrunching and argue against the transient excursion model. First, we note that exo III does not detect downstream-shifted complexes in reactions making RNAs 3–7 nt in length (Figure 1A, lanes 13–18). One of the advantages of exonuclease digestion over area footprinting reagents is that detection of a complex does not require high occupancy of a particular site. Thus, in lane 11 of Figure 1A, we can easily detect a complex at –4 that represents only 5–10% occupancy of this site. If even a small (5–10%) percentage of the complexes were transiently releasing the promoter and moving downstream at any given moment, we should still detect them, but we fail to do so until the RNA reaches 7–8 nt in length. Furthermore, when release does occur exo III detects only two boundaries in these reactions, presumably corresponding to promoter-bound and released complexes. Additional boundaries that might correspond to RNAPs shuttling between bound and released positions are not seen. Digestion with λ exo reveals further that the downstream boundary of the TC moves downstream as the RNA is extended even while the upstream boundary remains in place (Figure 1B). However, the downstream boundaries of these TCs are expected to be dynamic since the RNAP is constantly cycling between initiation, transcript extension, abortive transcript release and re- initiation. Since TCs in reactions that allow RNA extension to three or more nucleotides show downstream-shifted boundaries (Figure 1B, lanes 4–8 and 12–16), it is likely that these cycling complexes spend most of their time paused at the point where NTP limitation (or 3′-dNMP incorporation) halts transcript extension. This is consistent with kinetic studies showing that turnover during abortive cycling is limited by the rate of transcript release (Jia and Patel, 1997; Huang et al., 1999). These observations argue against the transient excursion mechanism, since if the RNAP moved downstream only occasionally, it would be unlikely to present a sufficient barrier to the processive action of λ exo to cause a shift in the downstream boundary of the TC. It is also useful to compare the distance between the exonuclease-defined boundaries of an EC (where the DNA is not expected to scrunch) with those measured for ITCs. If the DNA is actually scrunching during initial transcription while promoter contacts are maintained then we expect that the distance between the RNA 3′-end and the λ exo-defined downstream boundary will be similar in an ITC and an EC. However, the distance between the upstream exo III-defined boundary and the RNA 3′-end should be greater in a scrunched ITC than in an EC, and should increase as the RNA is extended. In an EC with bound NTP, λ exo halts ∼15 nt downstream of the 3′-end of the RNA, while exo III halts ∼12 nt upstream (Huang and Sousa, 2000). In the ITCs studied here λ exo halts 14–19 nt downstream of the RNA 3′-end, while exo III halts 20–27 nt upstream. The distance between the downstream boundary and the RNA 3′-end in the ITC is therefore similar to that seen in an EC, while the distance between the upstream boundary and the RNA 3′-end is much greater in the ITC than in an EC (and grows larger as the RNA grows). This is consistent with the DNA scrunching during initial transcription.
NTP binding to the initiation complex alters transcription bubble structure or interactions
Strong evidence for scrunching also comes from the effects of NTP binding on the structure of the transcription bubble in ITCs. Addition of templated NTPs to reactions in which TCs contain 3′-dNMP-terminated RNAs increases permanganate reactivity at –1 and decreases reactivity at –3 (Figure 2). This argues strongly that the RNAP active site is positioned at the templating base or else the templated NTP could not bind to the complex. Since exo III digestion reveals that, simultaneously, upstream promoter contacts remain in place, this implies that the intervening DNA must be looping out or scrunching within the complex.
It is unclear how NTP binding causes a change in transcription bubble structure or interactions during initial transcription. It is possible that conformational changes that accompany NTP binding alter RNAP–DNA interactions in the melted region of the promoter. Alternatively, NTP binding may hold the polymerase in a forward translocated position. The decreased reactivity at –3 is what would be expected as the polymerase begins to leave the promoter and the –3 base pair closes. The strongest suppression of –3 reactivity is seen in the reactions with 7- or 8mer RNAs with templated NTP present (Figure 2B and D, scans 7–9). Based on exo III digestion, these reactions contain mixed populations of released and promoter-bound complexes, so it is possible that the decreased reactivity at –3 in these reactions reflects an increasing fraction of released complexes. However, exo III digestion detects only a small fraction of released complexes in reactions making 7mer with pT7-7 and dUTP (Figure 1A, lane 11), but suppression of permanganate reactivity at –3 under similar conditions is ∼70% (Figure 2B, scans 7–9). Furthermore, the NTP-binding-driven changes in reactivity in reactions making shorter RNAs (Figure 2B and D, scans 3–6), cannot be due to release, since exo III detects no released complexes in such reactions. We therefore conclude that the NTP-binding-driven changes in KMnO4 reactivity reflect changes in RNAP conformation.
One other aspect of the changes in KMnO4 reactivity seen during initial transcription deserves comment. In reactions that allow RNA extension to +3, the template T at +4 is KMnO4 reactive (Figure 2A and C, lanes 3 and 2, respectively; B and D, scans 2). Reactivity is weak, but is comparable to that seen at +7 in reactions with pT7-8, which allow extension to +6 (Figure 2C, lane 5), or to the reactivity at the downstream edge of the RNA–DNA hybrid in an EC. Reactivity at +4 diminishes upon further extension of the RNA, as expected if this base becomes paired with RNA. However, based on the crystal structure of an ITC, it has been proposed that steric constraints limit the RNA–DNA hybrid in the complex to 2 or 3 bp at most (Cheetham and Steitz, 1999). If so, reactivity at +4 should be detected upon extension of the RNA to ∼7 nt. However, in our experiments reactivity at +4 is not seen in complexes making RNAs 7 or 8 nt in length (Figure 2B, lanes 8–10 and C, lanes 6–9). It is therefore likely that, even during initial transcription, the RNA–DNA hybrid reaches at least 4 or even 5 bp in length.
Scrunching contributes to promoter release
The clearest evidence that DNA scrunching contributes to promoter release was obtained with pT7 and pT7-A (Figure 3), where there was a reproducible ∼3-fold increase in the percentage of release at the 7mer RNA length with pT7-A versus pT7. One concern is that heterogeneity in initiation may result in synthesis of transcripts longer than the expected length. However, analysis of the transcripts made on pT7-A reveals few such transcripts (Figure 3A). In fact, such transcripts are more abundant in the reactions with pT7 than with pT7-A. We therefore conclude that the increased release at the 7mer length with pT7 versus pT7-A is not due to heterogeneous initiation resulting in synthesis of longer transcripts with pT7-A, but to the increased amount of DNA that has to be scrunched to make a 7mer with pT7-A versus pT7.
The percentage release at the 7mer length with pT7-A is, however, less than that seen with pT7-8 at the 8mer length (compare lane 19, Figure 1A with lane 10, Figure 3B), although in both cases an identical number of nucleotides would be scrunched. This may indicate that release is mediated by both RNA length and DNA scrunching, so that synthesis of a 7mer from initiation at +2 results in a level of release intermediate between that seen when a 7mer versus an 8mer is made from initiation at +1. Alternatively, since these reactions involve populations of cycling complexes, it is possible that a reduced fraction of the complexes are actually engaged with longer RNAs in the pT7-A reactions. Progression through initial transcription on pT7-A is less efficient than on pT7, primarily due to an increase in abortion after dimer synthesis. If, at any one time, a substantial fraction of the complexes on pT7-A are re-initiating or halted at dimer synthesis, then the percentage of complexes engaged with a 7mer RNA will be reduced, as will be the percentage release observed relative to a situation where close to 100% of the complexes are engaged with 7mer. This may also account for the low levels of release seen at the 7mer length with the pT72G-T and pT73G-T promoters, since progression through initial transcription is very inefficient with these promoters. Nevertheless, with either pT72G-T or pT73G-T, we observe increased release, relative to pT72G or pT73G, in reactions allowing 7mer synthesis (Figure 4B and D, lane 5 versus lane 10 and lane 5 versus lane 11, respectively). We conclude that on all three sets of promoters, increasing the DNA that has to be scrunched by 1 nt increases the amount of release seen when the RNA reaches 7 nt. The lack of quantitative agreement between the amount of release observed in the case where an 8mer is made starting at +1 versus a 7mer starting at +2, is due either to an RNA length-mediated contribution to release or to a decrease in the efficiency of progression through initial transcription when the polymerase is forced to start at +2 from a non-canonical ITS.
Notably, while oligo(G) synthesis and heterogeneity in transcription initiation lead to synthesis of transcripts of longer than the expected length, these longer transcripts do not cause promoter release. For example, in lanes 3 of Figures 3A and 4A, where the predominant transcript is a 6mer or a 5mer, we also detect transcripts up to 10 nt in length, yet no promoter release is detected in these reactions. Furthermore, in reactions to which only GTP is added, oligo(G) transcripts ≥14 nucleotides in length are made (Figure 4C, lane 1), yet no release is detected. Initiation on pT72G-T, pT73G and pT73G-T is also characterized by extensive synthesis of oligo(G) transcripts and extended transcripts together with transcripts of the expected length, yet no release is observed until NTPs allowing synthesis of properly initiated transcripts ≥7 nt in length are added (Figure 4B and D). Such observations argue against a mechanism in which RNA length alone is the determinant of release. If RNA length does contribute to release, then it appears to be critical that the RNA be fully complementary to the template.
The final piece of evidence which argues that RNA length cannot be the sole determinant of release, and that the strain due to DNA scrunching is important, comes from experiments with a promoter containing a break in the template strand at –4. Abortive cycling is observed on such a promoter, as well as on a promoter of normal structure, if transcript extension is limited to 8 nt or less. However, on both types of promoter synthesis of a 9mer results in a drastic reduction in transcript production, indicating that 9 nt is the minimal length required for formation of a stable TC that turns over slowly (Figure 5A). Remarkably, while synthesis of a 9mer results in formation of a stable TC, it does not result in any detectable release on a promoter with a discontinuous template strand (Figure 5C), although 9mer synthesis results in ∼90% release on a promoter of normal structure (Figure 5B). It has been proposed that an interruption in the template strand could facilitate threading of the template through the active site without requiring scrunching, because it would allow the 3′-end of the template to emerge from the TC (Jiang et al., 2001). The structure of an ITC (Cheetham and Steitz, 1999) is consistent with this possibility. Thus, an appropriately placed break in the template strand apparently relieves the strain of scrunching and leads to formation of a highly unusual TC that has the stability of an EC, but remains promoter bound. The severe reduction in run-off transcript synthesis on pT7-nick (Figure 5A, lane 12) may reflect the polymerase’s inability to disengage the promoter.
While our results show that the steric strain generated by the scrunching of the template strand plays an important role in promoter release, they do not rule out a role for the RNA in this process. In general, promoter release might not be mediated by any one determinant (RNA length, amount of DNA scrunched), but by a competition between the strength of the RNAP–promoter interactions and forces favoring rupture of those interactions. Those forces could include disruption of promoter or polymerase interactions with elements important for promoter binding (the promoter specificity loop in the case of T7RNAP; σ subunit in the case of E.coli RNAP) by interactions with RNA, as well as DNA compression due to the scrunching of increas ing amounts of template within the TC. Evidence that RNA length, for example, cannot be the sole determinant of release during E.coli RNAP initiation emerges from observations that release on σ54 promoters occurs when the RNA is between 3 and 7 nt in length (Tintut et al., 1995), while on σ70 promoters it occurs between 8 and 11 nt. Though it is possible that this reflects a different disposition of σ54 compared with σ70 (so that a shorter RNA can displace σ54, but not σ70), Gralla and colleagues (Tintut et al., 1995) suggest that the earlier release on σ54 promoters is due to weaker interactions between σ54 and core RNAP. Conversely, some promoters that bind E.coli RNAP very strongly inhibit transcription initiation at the release step (Ellinger et al., 1994). A release mechanism mediated by multiple determinants may allow multiple mechanisms of regulation of release during transcription initiation.
Materials and methods
Promoters
pT7-A was constructed with the QuikChange site-directed mutagenesis kit (Stratagene) using the plasmid pT7-5 (Tabor and Richardson, 1985) as a template. Promoters pT7-7, pT7-8, pT73G, pT73G-T, pT72G and pTand2G-T were constructed by cloning a synthetic promoter containing the –29 to –1 sequence of the pT7-5 promoter and a +1 to + 13 ITS as specified in Table I, into BamHI/EcoRI-cut pUC19 (Vieira and Messing, 1982). To incorporate only one radioactive nucleotide at the 3′ end of the template strand, the EcoRI restriction site was changed to an NcoI restriction site using QuikChange. Promoter sequences were confirmed by sequencing. DNAs were purified using the Qiagen MaxiPrep kit according to the manufacturer’s instructions. Synthetic promoters (Figure 5) were assembled by mixing single-stranded DNA at 50 µM in 10 mM Tris–HCl pH 8, 1 mM EDTA and 50 mM NaCl, heating to 85°C for 10 min and slow cooling for 30 min.
Exonuclease digestion
T7 RNAP–promoter complexes (20 µl final reaction volume) were formed in 10 mM Tris–HCl pH 7.4, 10 mM NaCl, 4 mM MgCl2 and 8% glycerol with labeled promoter at 2 × 10–8 M and polymerase at 6 × 10–7 M. After a 10 min 37°C incubation, NTPs, as indicated in individual figures, were added at a final concentration of 5 mM. Following an additional 10 min, 37°C incubation digestion was initiated by addition of either 2.5 U of exo III or 1 U of λ exo and stopped after 4 min. Reactions were resolved by electrophoresis on 8% polyacrylamide/0.4% bis-acrylamide–8 M urea gels in 1 × TBE and analyzed by phosphoimaging.
KMnO4 treatment
Promoter–polymerase complexes were formed as described for exonuclease digestion, followed by reaction with 1.5 mM KMnO4 for 1 min before the reaction was quenched with 1/10 vol of 50% β-mercaptoethanol.
Transcription reactions
Transcription reactions were carried out with the NTPs indicated in individual figures at 5 mM and promoters at 1 × 10–7 M and polymerase at 3 × 10–7 M (for reactions with synthetic promoters) and with promoter at 2 × 10–8 M and polymerase at 1 × 10–7 M for plasmid templates. Transcripts were resolved by electrophoresis on 20% polyacrylamide/1% bis-acrylamide–6 M urea gels in 1 × TBE and analyzed by phosphoimaging.
Acknowledgments
Acknowledgements
This study was supported by NIH Grant GM 52522 (to R.S.) and a Fulbright-CONACYT fellowship (to L.G.B.).
References
- Bandwar R.P. and Patel,S.S. (2001) Peculiar 2-aminoapurine fluores cence monitors the dynamics of open complex formation by bacteriophage T7 RNA polymerase. J. Biol. Chem., 276, 14075–14082. [DOI] [PubMed] [Google Scholar]
- Basu S. and Maitra,U. (1986) Specific binding of monomeric bacteriophage T3 and T7 RNA polymerase to their respective cognate promoters requires the initiating ribonucleoside triphosphate (GTP). J. Mol. Biol., 190, 425–437. [DOI] [PubMed] [Google Scholar]
- Brieba L.G. and Sousa,R. (2001) The T7 RNA polymerases intercalating hairpin is important for promoter opening during initiation but not for RNA displacement of transcription bubble stability during elongation. Biochemistry, 40, 3882–3890. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J. and Gralla,J.D. (1980) Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry, 19, 3245–3253. [DOI] [PubMed] [Google Scholar]
- Carpousis A.J. and Gralla,J.D. (1985) Interaction of RNA polymerase with lac UV5 promoter DNA during mRNA initiation and elongation. Footprinting, methylation and rifampicin-sensitivity changes accompanying transcription initiation. J. Mol. Biol., 183, 165–177. [DOI] [PubMed] [Google Scholar]
- Cheetham G.M. and Steitz,T.A. (1999) Structure of a transcribing T7 RNA polymerase initiation complex. Science, 286, 2305–2309. [DOI] [PubMed] [Google Scholar]
- Diaz G.A., Rong,M., McAllister,W.T. and Durbin,R.K. (1996) The stability of abortively cycling T7 RNA polymerase complexes depends upon template conformation. Biochemistry, 35, 10837–10843. [DOI] [PubMed] [Google Scholar]
- Ellinger T., Behnke,D., Bujard,H. and Gralla,J.D. (1994) Stalling of Escherichia coli RNA polymerase in the +6 to +12 region in vivo is associated with tight binding to consensus promoter elements. J. Mol. Biol., 239, 455–465. [DOI] [PubMed] [Google Scholar]
- Gunderson S.I., Chapman,K.A. and Burgess,R.R. (1987) Interactions of T7 RNA polymerase with T7 late promoters measured by footprinting with methidiumpropyl-EDTA-iron (II). Biochemistry, 26, 1539–1546. [DOI] [PubMed] [Google Scholar]
- Hansen U.M. and McClure,W.R. (1980) Role of sigma subunit of Escherichia coli RNA polymerase initiation. II. Release of sigma from ternary complexes. J. Biol. Chem., 255, 9564–9570. [PubMed] [Google Scholar]
- Huang J. and Sousa,R. (2000) T7 RNA polymerase elongation complex structure and movement. J. Mol. Biol., 303, 347–358. [DOI] [PubMed] [Google Scholar]
- Huang J., Villemain,J., Padilla,R. and Sousa,R. (1999) Mechanisms by which T7 lysozyme specifically regulates T7 RNA polymerase during different phases of transcription. J. Mol. Biol., 293, 457–475. [DOI] [PubMed] [Google Scholar]
- Huang J., Brieba,L.G. and Sousa,R. (2000) Misincorporation by wild-type and mutant T7 RNA polymerase: identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry, 39, 11571–11580. [DOI] [PubMed] [Google Scholar]
- Ikeda R.A. and Richardson,C.C. (1986) Interactions of the RNA polymerase of bacteriophage T7 with its promoter during binding and initiation of transcription. Proc. Natl Acad. Sci. USA, 83, 3614–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imburgio D., Rong,M., Ma,K. and McAllister,W.T. (2000) Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry, 39, 10419–10430. [DOI] [PubMed] [Google Scholar]
- Jia Y. and Patel,S.S. (1997) Kinetic mechanism of transcription initiation by bacteriophage T7 RNA polymerase. Biochemistry, 36, 4223–4232. [DOI] [PubMed] [Google Scholar]
- Jiang M., Rong,M., Martin,C. and McAllister,W.T. (2001) Interrupting the template strand of the T7 promoter facilitates translocation of the DNA during initiation, reducing transcript slippage and the release of abortive products. J. Mol. Biol., 310, 509–522. [DOI] [PubMed] [Google Scholar]
- Korsheva N., Mustaev,A., Kozlov,M., Malhotra,A., Nikiforov,V. and Darst,S.A. (2000) A structural model of transcription elongation. Science, 289, 619–625. [DOI] [PubMed] [Google Scholar]
- Krummel B. and Chamberlin,M. (1989) RNA chain initiation by Escherichia coli RNA polymerase. Structural transitions of the enzyme in early ternary complexes. Biochemistry, 28, 7829–7842. [DOI] [PubMed] [Google Scholar]
- Martin C.T., Muller,D.K. and Coleman,J.E. (1988) Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry, 27, 3966–3974. [DOI] [PubMed] [Google Scholar]
- Mooney R.A. and Landick,R. (1999) RNA polymerases unveiled. Cell, 98, 687–690. [DOI] [PubMed] [Google Scholar]
- Muller D.K., Martin,C.T. and Coleman,J.E. (1989) T7 RNA polymerase interacts with its promoter from one side of the DNA helix. Biochemistry, 28, 3306–3313. [DOI] [PubMed] [Google Scholar]
- Place C., Oddos,J., Buc,H., McAllister,W.T. and Buckle,M. (1999) Studies of contacts between T7 RNA polymerase and its promoter reveals features in common with multisubunit RNA polymerases. Biochemistry, 38, 4948–4957. [DOI] [PubMed] [Google Scholar]
- Sen R., Nagai,H. and Shimamoto,N. (2000) Polymerase arrest at the λP (R) promoter during transcription initiation. J. Biol. Chem., 275, 10899–10904. [DOI] [PubMed] [Google Scholar]
- Straney D.C. and Crothers,D.M. (1985) Intermediates in transcription initiation from the E.coli lac UV5 promoter. Cell, 43, 449–459. [DOI] [PubMed] [Google Scholar]
- Straney D.C. and Crothers,D.M. (1987) A stressed intermediate in the formation of stability initiated RNA chains at the Escherichia coli lac UV5 promoter. J. Mol. Biol., 193, 267–278. [DOI] [PubMed] [Google Scholar]
- Tabor S. and Richardson,C.C. (1985) A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl Acad. Sci. USA, 82, 1074–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temiakov D., Mentesanas,P.E., Ma,K., Mustaev,A., Borukhov,S. and McAllister,W.T. (2000) The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc. Natl Acad. Sci. USA, 97, 14109–14114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tintut Y., Wang,J.T. and Gralla,J.D. (1995) Abortive cycling and the release of polymerase for elongation at the sigma 54-dependent glnAp2 promoter. J. Biol. Chem., 270, 24392–24398 [DOI] [PubMed] [Google Scholar]
- Vieira J. and Messing,J. (1982) The pUC plasmids and M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene, 19, 259. [DOI] [PubMed] [Google Scholar]
- Villemain J., Guajardo,R. and Sousa,R. (1997) Role of complex instability in kinetic promoter selection by bacteriophage T7 RNA polymerase. J. Mol. Biol., 273, 958–977. [DOI] [PubMed] [Google Scholar]
- Zhang G., Campbell,E.A., Minakhin,L., Richter,C., Severinov,K. and Darst,S.A. (1999) Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell, 98, 811–824. [DOI] [PubMed] [Google Scholar]