Abstract
Initiation of phage Mu DNA transposition requires assembly of higher order protein–DNA complexes called Mu transpososomes containing the two Mu DNA ends and MuA transposase tetramer. Mu transpososome assembly is highly regulated and involves multiple DNA sites for transposase binding, including a transpositional enhancer called the internal activation sequence (IAS). In addition, a number of protein cofactors participate, including the target DNA activator MuB ATPase. We investigated the impact of the assembly cofactors on the kinetics of transpososome assembly with the aim of deciphering the reaction steps that are influenced by the cofactors. The transpositional enhancer IAS appears to have little impact on the initial pairing of the two Mu end segments bound by MuA. Instead, it accelerates the post-synaptic conformational step(s) that converts the reversible complex to the stable transpososome. The transpososome assembly stimulation by MuB does not require its stable DNA binding activity, which appears critical for directing transposition to sites distant from the donor transposon.
Keywords: DNA–protein complex/DNA transposition/enhancer element/transposase
Introduction
Higher order protein–DNA complexes play important roles in the initiation of transcription, recombination and DNA replication in all organisms. The assembly of these complexes is often a target of regulation. The protein– DNA complexes involved in transpositional and site-specific recombination are among the best characterized and thus are excellent systems for elucidating general principles governing complex assembly, disassembly and organization. The phage Mu transposition system is one of the most thoroughly studied examples of transpositional recombination (Mizuuchi, 1992; Chaconas et al., 1996).
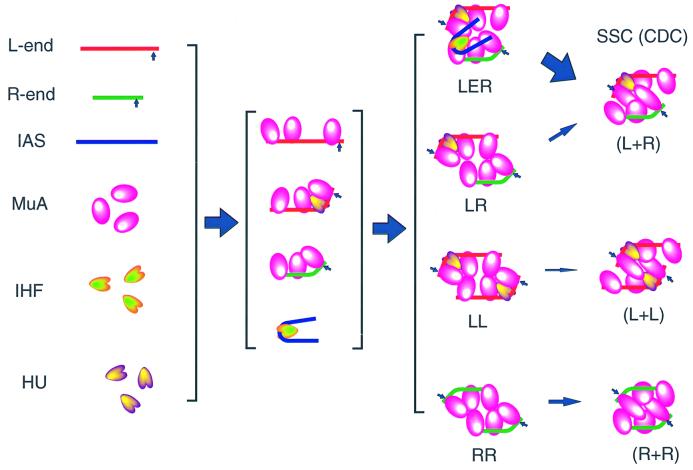
The Mu transposition reaction occurs through a series of higher order protein–DNA complexes called transpososomes (Craigie and Mizuuchi, 1987; Surette et al., 1987; Mizuuchi et al., 1992). Mu transpososomes contain the two ends of the Mu genome synapsed by a tetramer of Mu transposase (MuA) (Lavoie et al., 1991; Baker and Mizuuchi, 1992; Mizuuchi et al., 1992). The first of these complexes is the stable synaptic complex (SSC, or type 0 complex) in which the two Mu ends (L-end and R-end) have not been nicked. The basic structure of the complex is maintained through the subsequent chemical steps of donor cleavage and target DNA strand transfer. In the presence of Mg2+, MuA cleaves at the two Mu DNA ends to expose 3′ OH to generate the cleaved donor complex (CDC, or type 1 complex). Next, the 3′ ends of the Mu DNA attack a pair of phosphodiester bonds in the target DNA to generate the strand transfer complex (STC, or type 2 complex). Formation of the SSC is a prerequisite for the above two chemical reactions and its assembly is a critical control point in Mu transposition.
Each Mu end carries three MuA-binding sites with a 22 bp consensus sequence; the left end sites are designated L1, L2 and L3, and those on the right end are called R1, R2 and R3 (Craigie et al., 1984). The two Mu ends bound by MuA synapse early in the reaction prior to SSC assembly. However, this complex, called the LR complex, is unstable and has been observed only after protein cross-linking (Watson and Chaconas, 1996). While all six sites appear to participate in transpososome assembly (Allison and Chaconas, 1992), only three sites, L1, Rl and R2, are stably bound by the MuA tetramer within a transpososome (Lavoie et al., 1991; Mizuuchi et al., 1991, 1992).
Efficient SSC assembly involves several cofactors in addition to MuA and Mu DNA ends: a DNA cofactor, the IAS (internal activation sequence, or transpositional enhancer), and two DNA-bending proteins, HU and integration host factor (IHF), are required (reviewed in Haniford and Chaconas, 1992; Mizuuchi, 1992). HU binds to the spacer between L1 and L2 at the Mu L-end DNA to bend the DNA and is thought to juxtapose L1 and L2 (Lavoie et al., 1996). The IAS is ∼100 bp long and overlaps the Mu operator sequence (Leung et al., 1989; Mizuuchi and Mizuuchi, 1989; Surette et al., 1989). It consists of three components, two clusters of MuA-binding sequences that also bind the Mu repressor (O1 and O2) and a binding site for IHF in the middle. Mu repressor, which shares homology in its DNA-binding domain with the IAS-binding domain of MuA, binds the operator sites to block not only early transcription but also transposition directly by blocking use of the IAS by the transposase (Mizuuchi and Mizuuchi, 1989; Mizuuchi et al., 1992). MuA binds the IAS using a different domain from the one that binds to the Mu ends. The IAS is required for the assembly of the SSC, but not for donor cleavage and strand transfer (Mizuuchi et al., 1992; Surette and Chaconas, 1992). The IAS is not a stable component of the SSC after its formation. However, prior to SSC assembly, a complex that can be stabilized by protein cross-linking, called the LER, which contains the two ends of Mu and the IAS, has been observed (Watson and Chaconas, 1996).
A second phage-coded transposition protein, MuB, stimulates the target DNA strand transfer reaction and controls transposition target site selection. This ATP-dependent non-specific DNA-binding protein directs transposition to target DNA locations to which it is bound and, by using the energy of ATP hydrolysis, directs transposition away from the Mu genome (Adzuma and Mizuuchi, 1988, 1989). Through its interaction with MuA, MuB also stimulates the assembly of Mu transpososomes prior to the strand transfer step (Surette et al., 1991; Naigamwalla and Chaconas, 1997).
We have developed an experimental system to study the roles of the cofactors for SSC assembly. With the previous in vitro transposition reactions that depended on a mini-Mu plasmid DNA containing two Mu ends and the IAS site on the same molecule as a substrate, the concentrations of each DNA site in a reaction could not be changed independently. We therefore devised a reaction system using three separate DNA fragments: Mu L-end, Mu R-end and the IAS. We studied the stimulation of SSC assembly by the cofactors as a function of the MuA-bound Mu end concentration. Here we try to determine whether the IAS stimulates the initial pairing of the two Mu end DNA segments, or if it accelerates post-synaptic conformational changes. Our results suggest that the IAS does not affect the initial pairing of two Mu ends bound by MuA, which results in the formation of the LR complex. Instead, the IAS appears to accelerate the conversion of the LR complex to the SSC. Using similar approaches, we also investigated how MuB stimulates transpososome assembly. The effects of MuB on the assembly process appeared more complex. However, stable DNA binding activity of MuB is not essential for stimulation of transpososome assembly.
Results
The IAS stimulates the assembly of transpososomes containing L- and R-end fragments
The IAS is known to stimulate the Mu transposition reaction at the step of transpososome assembly, but once the SSC is formed it does not have a significant effect on later reaction steps. In the past, the assembly process has been studied by using supercoiled mini-Mu plasmid as the substrate DNA, an experimental set-up that does not allow detailed kinetic dissection of the assembly process. Previously, it was shown that the IAS can be supplied as a DNA fragment separate from a supercoiled plasmid donor containing R- and L-ends (Surette and Chaconas, 1992). We tested whether the IAS stimulation of the assembly can be detected when all three substrate DNA sites are present on separate linear DNA fragments (Figure 1). To compensate for the absence of DNA supercoiling, a requirement for efficient assembly under normal reaction conditions, dimethylsulfoxide (DMSO), which alleviates this requirement, was added in the reaction.
Fig. 1. Mu transpososome assembly with short DNA fragments. Steps involved in assembly of the stable synaptic complex (SSC). First, each protein component binds to its cognate binding sites on the DNA substrate. These protein-bound DNA segments will associate with each other to form transient higher order complexes. In the absence of the IAS, the paired complex LR would form, but its conversion to the SSC is inefficient. RR complexes and LL complexes would also form. In the presence of the IAS, the LER complex in which both L- and R-ends and the IAS are held together by MuA is formed. This complex is considered to be an efficient precursor to the SSC. In the presence of Mg2+, both Mu ends are cleaved by MuA to generate the cleaved donor complex (CDC). In addition to MuA, HU and IHF are required. See the text for details.
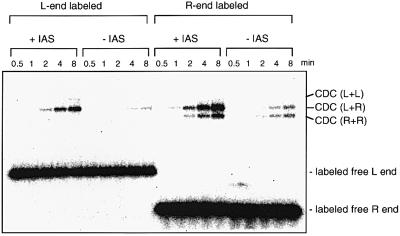
The L- and R-end DNA fragments were incubated with MuA, IHF and HU, in the presence or absence of a third DNA fragment that contained the IAS. The formation of stable MuA–DNA complexes was assayed by autoradiography after electrophoresis of the samples on a native agarose gel (Figure 2). The reaction was carried out in the presence of Mg2+, which allows it to proceed through the DNA cleavage step to make the CDC. The strand transfer step to make the STC was blocked by the addition of a low concentration of heparin. This simplified the identification of transpososomes formed with different combinations of Mu end fragments by avoiding the presence of the target DNA fragment of different sizes. While higher concentrations of heparin inhibited transpososome assembly, the lower concentrations used here did not inhibit the SSC assembly or the donor cleavage reaction. Instead, it stimulated transpososome assembly several fold, possibly due to inhibition of non-specific DNA binding by MuA (data not shown). Other than general stimulation of the SSC assembly and inhibition of strand transfer, inclusion of heparin did not significantly affect the Mu end partner preference or the IAS stimulation. The electrophoresis conditions used separate Mu transpososomes according to the sizes and number of DNA fragments they contain. Transpososomes containing L- and R-ends, as well as those with two copies of the R-end, were easily detected. Those with two copies of the L-end appeared to be significantly less abundant. The identities of the complexes in the gel were determined by labeling different DNA partners, or by omission of the partner DNA from the reaction (data not shown). This electrophoresis method does not allow detection of unstable complexes such as LR or LER. The SSC also cannot be quantitated reliably by this analysis. Previous studies indicated that SSC formation is a rate-limiting step in the reaction, and is followed rapidly within 1–2 min by donor DNA cleavage to form the CDC (Mizuuchi et al., 1992). Therefore, the linear phase of the rate after the short lag measured by this method reflects the initial rate of SSC formation.
Fig. 2. The IAS accelerates assembly of the transpososome composed of Mu R- and L-ends. Agarose gel analysis of the products of assembly reactions in the presence and absence of the IAS shows paired end complexes with both L- and R-ends, or L- or R-end alone. The assembly reactions were carried out as described in Materials and methods, except that the concentrations of MuA and DNA fragments were increased: 400 nM MuA, 25 nM each L- and R-end fragment, 100 nM IAS or non-IAS control fragment, which was included in reactions without the IAS fragment. The fastest migrating CDC species contained two copies of the R-end fragment held by a MuA tetramer. The intermediate CDC species contained one copy each of the L- and R-end fragments. The slowest migrating CDC species contained two copies of the L-end fragment. The reactions were incubated at 30°C for the times indicated. Only the L-end fragment was labeled with 32P in the reactions shown on the left half of the autoradiograph. Only the R-end was labeled in the reactions on the right half.
The IAS fragment clearly stimulated complex formation specifically between L- and R-end fragments (Figure 2). The stimulatory effect of the IAS was strongly dependent on the presence of IHF, as has been shown in reactions involving linear IAS fragment and plasmid Mu donor DNA (Surette and Chaconas, 1992). CDC formation with paired R-end or paired L-end fragments was not stimulated significantly. Likewise, in reactions that contained either the R-end or L-end fragment alone, no stimulation by the IAS was observed (data not shown).
Stimulation of transpososome assembly required proper arrangement of the enhancer-type MuA-binding sites on a continuous piece of DNA. A non-IAS DNA fragment with a length similar to the IAS fragment, or longer (883 and 929 bp), a linear pBR322 DNA and single-stranded φX174 DNA all failed to stimulate the reaction (data not shown). The IAS fragment used in these experiments contained the O1 and O2 Mu repressor (and MuA) binding sites and the intervening IHF-binding site. We digested the IAS fragment with the restriction enzyme MluI, which cuts in the middle of the IHF site, to test whether unlinked O1 and O2 fragments could substitute for the IAS. We observed no stimulatory effect with either the O1 or O2 fragment alone or with a mixture of the two (data not shown). At higher concentrations (ranging from 36 to 1076 nM), the mixture of the O1 and O2 fragments inhibited the assembly reaction rather than stimulating it (data not shown). Substitution of the O1 and O2 sites with non-IAS DNA sequence, while retaining the IHF-binding site, abolished the activity of the IAS. This fragment was added in all the reactions that did not include the IAS fragment in order to keep the concentration of the IHF-binding site constant, although omission of this DNA fragment did not impact the reaction noticeably when tested. Thus, the integrity of the tripartite structure of the IAS is essential for the stimulation of transpososome assembly.
The IAS accelerates a post-synaptic conformational step of transpososome assembly
We are interested in identifying which reaction step is accelerated by the IAS during assembly of the SSC. While the transpososome assembly process is almost certainly complex, with many elementary steps, one can divide the process conceptually into three stages. The first stage involves binding of MuA to its recognition sites at the ends of the Mu genome. With the Kd being of the order of 10–8 M, this process is usually rapid. Our past experience suggested that in the presence of a saturating concentration of MuA >100 nM, the association half-time should be less than a few seconds and the dissociation half-time should be less than a minute (data not shown). The next two stages are (i) pairing of two Mu ends bound by MuA to form the non-stable LR complex and (ii) the subsequent conformational rearrangements to form the SSC (see Figure 1).
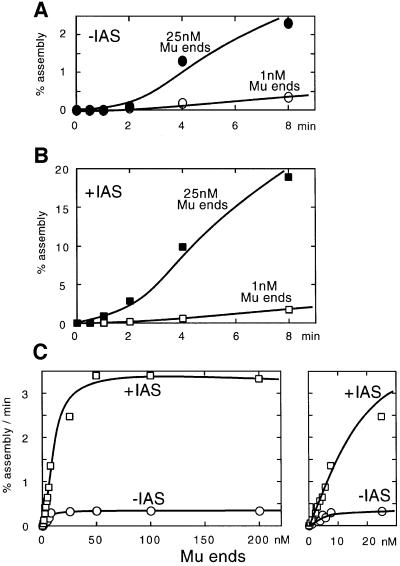
A prediction based on this simplified kinetic model is the following. At low Mu end concentrations, the overall rate of the reaction should exhibit a second-order response with respect to Mu end fragment concentration, while at higher Mu end fragment concentrations, the bimolecular pairing step becomes no longer rate limiting and the reaction kinetics should become first order with respect to the Mu end fragment concentration. To test this prediction, we measured the initial rate of assembly at various concentrations of Mu ends. The concentrations of the two Mu ends were varied in parallel from 0.5 to 200 nM. The concentrations of MuA and HU that gave the highest reaction rate were determined for each Mu end concentration. Broad optima were observed for the protein concentrations; for MuA the optimum centered around a few hundred nanomolar excess of the binding site concentration in the reaction (see Figure 3 legend for details). The time course of the reaction exhibited a linear phase after a short lag of a minute or two before the reaction slowed (Figure 3A). We attribute the initial lag mainly to the conversion of the SSC to CDC (Mizuuchi et al., 1992). We estimated the initial assembly rate from the steep part of the time course curve for each condition. In this experiment, only the L-end fragment was radiolabeled and only the complex containing the L- and R-ends was assayed.
Fig. 3. Mu end concentration dependency of transpososome assembly kinetics with or without the IAS. The assembly reactions were carried out as described in Figure 2, except the concentration of Mu end DNA was varied from 0.1 to 200 nM each of the L- and R-ends. Since higher concentrations of the Mu end fragments require elevated MuA concentrations, the optimal MuA concentration was determined for each Mu end concentration. The optimal IAS, IHF and HU concentrations were also determined for each Mu end concentration. For the concentration range of each Mu end fragment from 0.1 to 5 nM, 200 nM MuA, 50 nM the IAS, 150 nM IHF, 200 nM HU and 2.5 µg/ml heparin were added; for 50 and 100 nM each Mu end fragment, 900 nM MuA, 200 nM IAS, 250 nM IHF, 200 nM HU and 3.5 µg/ml heparin were added; and for 200 nM Mu ends, 1800 nM MuA, 300 nM IAS, 350 nM IHF, 200 nM HU and 3.5 µg/ml heparin were added. For reactions in the absence of the IAS, a control DNA fragment was included at the same concentration as for the IAS. The reactions were incubated at 30°C for the times indicated. Only the L-end was labeled with 32P. (A) Time course of assembly in the absence of the IAS. The ordinate is expressed as the percentage of the labeled Mu L-end fragment converted to the CDC containing both L- and R-end fragments. (B) Time course of assembly in the presence of the IAS. (C) Mu end concentration dependency of the assembly rate in the presence or absence of the IAS. The right panel shows an expansion of the lower concentration area.
A plot of the observed assembly rates as a function of the Mu end concentration revealed the predicted biphasic behavior, namely the second-order regime and the first-order regime (Figure 3C). Note that in these figures, the rate of assembly was expressed as the fraction of the substrate converted to the product per unit time. Thus, a linear slope intersecting the origin at the lower Mu end concentrations indicates a second-order response to the Mu end concentration changes, while the plateau at higher Mu end concentrations indicates a first-order response. Above ∼10–50 nM Mu end concentration, the relative reaction rate becomes independent of the Mu end fragment concentration. Presumably, at higher concentrations, essentially all the Mu ends are paired quickly. If we assume a relatively fast pre-equilibrium of the Mu end pairing prior to a slower conformational step, we obtain an apparent Kd of between 4 and 20 nM for the pairing equilibrium (note that the concentrations of the two ends were changed in parallel).
We were interested in directly assaying the LR complex formed at different Mu end concentrations. However, our repeated efforts to detect the LR complex by using different protein cross-linking reagents and different gel methods did not succeed, presumably due to the absence of the stabilizing effects of DNA supercoiling, which was present in the experiments involving plasmid DNA substrates (Watson and Chaconas, 1996).
Next, we repeated the same experiment in the presence of the IAS fragment. Again, the concentrations of the IAS fragment and IHF were optimized for each Mu end concentration. If the IAS assists only the pairing of the two Mu ends, the IAS acceleration of the assembly rate will be observed only at subsaturating Mu end concentrations. On the other hand, if the IAS assists only the post-synaptic conformational step, the effect of the IAS will be independent of the Mu end concentration. The rate of the assembly reaction in the presence of the IAS was 4- to 12-fold higher than that in its absence (Figure 3B and C). The stimulation by the IAS was observed throughout the range of Mu end concentrations we examined. The fact that we do not observe a lowering of the apparent Kd in the presence of the IAS, and that at least a similar or slightly higher level of stimulation by the IAS is observed above the saturating Mu end concentration, indicates that the IAS does not promote Mu end pairing. Instead, it accelerates the rate of subsequent conformational steps. We did not observe a significant change in the optimal MuA concentration in the presence of the IAS, indicating that the IAS does not critically affect the binding of transposase to Mu ends.
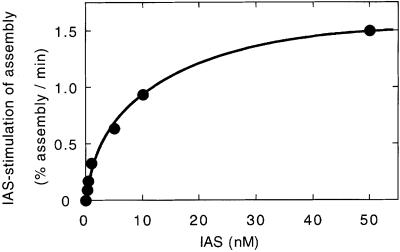
In order to estimate the apparent affinity of the IAS for the LR complex, reactions were carried out at a limiting concentration of Mu ends (0.5 nM) in the presence of a varying concentration of the IAS fragment (Figure 4). We observed the maximum stimulation of transpososome assembly by the IAS above 40 nM IAS, with a half-maximum stimulation at 5–10 nM. At present, we do not know whether the observed concentration for the half-maximum rate reflects the IAS-binding equilibrium or the rate of binding. At higher concentrations of the Mu ends, the same or a slightly higher concentration of the IAS as of the Mu end was required for maximum stimulation. For technical reasons, we have not tested whether a copy of the IAS can turn over to stimulate multiple transpososome assembly events. Thus, we do not know at present whether the IAS comes off the SSC that assembled.
Fig. 4. The IAS concentration dependency for assembly stimulation. The assembly reactions were carried out as in Figure 3 at a Mu end concentration of 0.5 nM with different concentrations of the IAS fragment. The IHF concentration was changed to match the IAS concentration. The assembly rate in the absence of the IAS (0.19%/min) was subtracted from the observed rate in the presence of the IAS.
MuB appears to promote the initial paring of two Mu ends bound by MuA
MuB protein enhances the efficiency of the Mu transposition reaction in multiple ways. MuB binds to DNA non-specifically in the presence of ATP, and DNA bound by MuB is a preferred target for Mu transposition, resulting in a stimulation of strand transfer (Adzuma and Mizuuchi, 1988; Baker et al., 1991). MuB also stimulates earlier steps in the transposition reaction pathway by assisting the assembly of MuA–Mu end DNA complexes (Surette et al., 1991; Naigamwalla and Chaconas, 1997). MuA interacts with MuB through its C-terminal domain (Harshey and Cuneo, 1986; Baker et al., 1991).
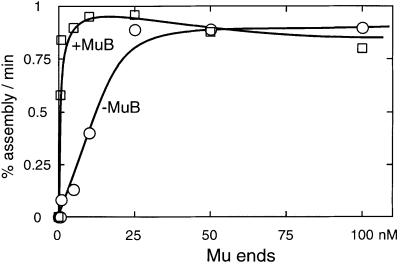
We examined the effect of MuB on the Mu transpososome assembly kinetics (Figure 5). The reaction conditions were essentially the same as in Figure 3, except for the addition of ATP and omission of the IAS and heparin, since MuB stimulation is inhibited by heparin. MuB was added together with a 50 bp DNA fragment that was intended to serve as the ‘target’ DNA at a constant ratio of one MuB molecule per 18 bp. The concentration of Mu ends was varied over a range of 0.1–100 nM and the optimal concentrations of MuA and the MuB/target were determined experimentally for each Mu end DNA concentration. For the purpose of quantitation, all stable complexes were summed together because, unlike the IAS stimulation, MuB stimulation was not limited to the complexes containing the L- and R-ends. We observed a broad optimum. For example, at 5 nM Mu ends and 100 nM MuA, anywhere between 56 and 560 nM MuB stimulated the overall rate of transpososome formation to approximately the same extent (data not shown).
Fig. 5. MuB stimulates transpososome assembly at low Mu end concentrations. The assembly reactions were carried out as described in Materials and methods, except that the concentration of Mu end DNA was varied from 0.5 to 100 nM. The concentrations of MuA and MuB–target complex were optimized for each Mu end concentration. The assembly time course was analyzed as described for Figure 3, except that the labeled L-end that was incorporated into all transpososomes was counted and the percentage assembly rate was plotted against the Mu end concentration.
When the observed percentage assembly rates were plotted as a function of Mu end concentration, the assembly process was again shown to be saturable with respect to the Mu end concentration (Figure 5). In the absence of MuB, the assembly reaction exhibited a first-order response to the Mu end concentration above 25 nM, indicating that the apparent Kd for Mu end pairing is ∼5–10 nM under these conditions. When MuB and target DNA were added to the reaction, transpososome assembly was greatly stimulated at lower concentrations of the Mu ends. However, at the saturating Mu end concentrations, no stimulation was observed. The apparent Kd for end pairing was below 0.5 nM in the presence of MuB (Figure 5). These data suggest that MuB assists the initial pairing of two Mu ends bound by MuA. Qualitatively similar results were observed when the stable MuB–target complex was pre-formed in the presence of ATPγS and then added to the reaction, instead of adding MuB, ATP and the target DNA to the assembly reaction without pre-incubation. However, ADP failed to support MuB stimulation of transpososome assembly (data not shown). Assembly of the complex containing two L-end copies, an inefficient reaction in the absence of MuB, became almost as efficient as other combinations of the ends in the presence of MuB (data not shown). The MuB-stimulated reaction exhibited less initial time lag, reflecting acceleration of the donor cleavage and strand transfer steps by MuB (see Figure 6).
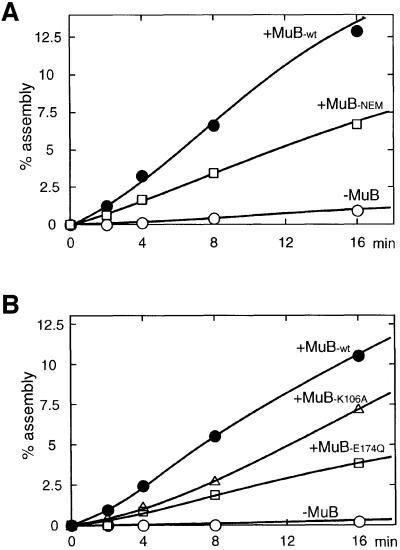
Fig. 6. N-ethylmaleimide-treated MuB and ATPase-deficient MuB can stimulate transpososome assembly. (A) The time course of the assembly reaction at 5 nM Mu end in the presence or absence of 280 nM untreated or NEM-treated MuB is shown. (B) Mutant MuB defective in the ATPase activity (K106A and E174Q) can stimulate transpososome assembly.
Modified MuB and mutant MuB also stimulate the assembly reaction
MuB variants that are partially defective in certain activ ities have been studied in the past. N-ethylmaleimide (NEM) modification of MuB results in much reduced DNA binding activity, but the modified protein retains activity to stimulate MuA to carry out the strand transfer reaction (Baker et al., 1991; Surette and Chaconas, 1991). We tested whether NEM-modified MuB can stimulate transpososome assembly. As shown in Figure 6A, NEM-treated MuB also stimulated complex formation with only a modest decrease in efficiency compared with untreated MuB.
We also tested MuB stimulation of assembly with mutant MuB proteins that have amino acid substitutions in the ATPase active site. These mutants (K106A and E174Q) lack ATPase activity (Yamauchi and Baker, 1998). MuB K106A has a 10-fold reduction in DNA binding, whereas E174Q has only a 2-fold defect, and both mutant proteins retain activity to stimulate intramolecular strand transfer (Yamauchi and Baker, 1998). As shown in Figure 6B, these mutants also stimulated the assembly reaction with an efficiency similar to NEM-treated MuB. Therefore, neither stable DNA binding nor ATP hydrolysis by MuB is essential for the stimulation of transpososome assembly.
Why does MuB not stimulate transpososome assembly at higher concentrations of Mu DNA ends?
The results described above suggest that MuB assists initial pairing of Mu end DNA segments bound by MuA, but not the conformational steps that follow pairing. This was unexpected. Previous observations pointed to a later role for MuB in transpososome assembly. For example, if the terminal CA sequence at one of the Mu donor ends is mutated, transpososome assembly is severely inhibited in the absence of MuB (Surette et al., 1991). However, the mutant Mu end forms the LR complex readily without MuB (Watson and Chaconas, 1996), indicating that the terminal CA sequence is not involved in the initial pairing. Thus, MuB appeared to assist the mutant Mu ends for transpososome assembly at later steps.
One possibility is that the conformational step with normal Mu ends does not need MuB stimulation, and only the assembly involving mutant Mu ends requires MuB stimulation for that step. To test this possibility, we mutated the cleavage sites (A–G) at the L- or R-end DNA. We tested the stimulatory effect of MuB on complex assembly with these mutated Mu end DNA fragments in the presence and absence of MuB. Irrespective of the presence or absence of MuB, the Mu end concentration dependency was similar to that with normal Mu end sequences, except that the assembly rate was ∼10-fold lower (data not shown). Most importantly, the mutation did not significantly influence the apparent Kd of end pairing. Therefore, even in the reactions involving mutant Mu ends, the results appear to suggest that MuB does not stimulate the conformational steps of transpososome assembly.
If MuB did indeed promote initial pairing of the two Mu ends, we expected a higher level of stimulation of assembly when both the IAS and MuB were included together than with the IAS or MuB alone. However, we have not been able to find conditions under which the two cofactors function co-operatively. We have not found a satisfactory explanation for this observation (see Discussion below). Further study is needed to clarify the relationship between the effects of the IAS and MuB on transpososome assembly.
Discussion
Assembly of Mu transpososomes under physiological conditions requires cis-acting DNA sequences, a certain donor DNA topology and the DNA-bending proteins IHF and HU (Mizuuchi, 1992; Chaconas et al., 1996). These multiple requirements make it difficult to study the basic protein–DNA interactions that influence assembly of the transpososomes. We showed previously that this obstacle can be overcome by using modified reaction conditions that allow assembly of active Mu transpososome without the requirement for donor DNA superhelicity, accessory proteins or additional DNA sequence cofactors (Craigie and Mizuuchi, 1986; Mizuuchi and Mizuuchi, 1989; Savilahti et al., 1995). In this study, we investigated how the accessory proteins and DNA sites stimulate the process of transpososome assembly.
The use of the short DNA fragments as the substrates was necessary in order to investigate the effects of the substrate concentration on the rate of the assembly reaction. On the other hand, this necessitated the use of reaction conditions that made the transpososome assembly less stringently dependent on cofactors such as the IAS. Nevertheless, we found that the assembly reaction still responds to the stimulatory effects of the IAS and MuB under these reaction conditions. However, because of this technical limitation, it is possible that we are investigating only limited aspects of the stimulatory effects that these cofactors have on the transpososome assembly reaction, as will be discussed below.
The IAS appears to stimulate a post-pairing conformational step(s) for SSC assembly
The IAS contains two clusters of MuA-binding sequences that coincide with the O1 and O2 operator sites to which Mu repressor binds. When the two halves of the IAS were separated by cleavage with a restriction enzyme, no stimulatory effect was observed, even at elevated concentrations of the resulting O1 and O2 DNA segments. Thus, the integrity of the entire IAS region is required for stimulation of transpososome assembly. Binding of IHF to the site between O1 and O2 introduces a sharp bend (Rice et al., 1996) in the middle of the IAS and this bending must be important for IAS function in addition to the continuity between O1 and O2. The IAS stimulatory effect was observed mainly for the assembly of complexes with the L- and R-end combination. Assembly of complexes with a pair of L- or R-ends was not strongly stimulated by the IAS. This indicates that the IAS helps to avoid incorrect pairing of Mu ends. The symmetry of R1 and L1 sites and the functional symmetry of the transposition reaction suggest a structural element of symmetry in the transpososome. However, this is not a true symmetry due to the asymmetry in the overall arrangement of the MuA-binding sites at the L- and R-ends. The end pair specificity of the IAS must reflect the corresponding asymmetry in the IAS structure. An interesting question would be which part of the IAS interacts with the MuA molecules bound to each of the two Mu ends. Allison and Chaconas (1992) proposed that the O1 and O2 sites of the IAS interact with the L3 site and the R3 site, respectively, via a MuA bridge, while Jiang et al. (1999) proposed O1–R1 bridging and O2–L1 bridging. While it is tempting to assume that O1 and O2 each interacts with either the R- or L-end-bound MuA molecules, this does not have to be the case. With the U-turn bend between O1 and O2 imposed by IHF, it is possible that two clusters of MuA-binding sites are on two nearly parallel duplexes in the functional conformation of the IAS, bringing the left half of O1 close to the right half of O2, and vice versa. If so, it may be equally possible that one half of both O1 and O2 interacts with the MuA molecules bound to each of the Mu ends.
Previously, we have demonstrated that in order for the IAS to stimulate transpososome assembly, all four MuA monomers that constitute the complex must be able to interact with the IAS. From this observation, we proposed that the IAS functions as the assembly ‘platform’ for the transpososome (Mizuuchi et al., 1995). However, it has been unclear whether this ‘platform’ assists pairing of the two Mu DNA ends bound by MuA, or the post-pairing conformational step(s). Our results strongly support the notion that the IAS does not help the pairing of the two Mu ends. Instead, it accelerates post-pairing conformational step(s). This is consistent with the previous observation that the LR complex, presumed to be the initial pairing intermediate on the pathway for transpososome assembly, can be formed efficiently in the absence of the IAS (Watson and Chaconas, 1996). The apparent Kd for pairing of ∼10 nM we observed here shows that the effective intramolecular concentration of the two Mu ends on a small supercoiled plasmid would be high enough to saturate the end pairing step (Vologodskii and Cozzarelli, 1996). Then, how does the IAS with multivalent interactions with the MuA monomers within the LR complex accelerate the structural transition necessary for transpososome assembly? One scenario might be the following. With three MuA monomers bound at each Mu end, there may be many combinations of inter-MuA interactions and therefore many ways in which the ends may initially pair. In other words, the LR complex may be an ensemble of different complexes, only one of which is the true precursor for the transpososome. The IAS may be able selectively to stabilize the correct subspecies of the LR complex on the pathway to the transpososome. If the equilibrium among the subspecies of the LR complexes is relatively fast, this action of the IAS alone may be sufficient for the acceleration of transpososome assembly.
However, we suspect that the IAS does more than what is proposed above. If the stabilization of the right subspecies of the LR complexes is the only action of the IAS in transpososome assembly stimulation, it is difficult to explain why the assembly rate has a steep temperature dependency (Baker and Mizuuchi, 1992) while the LR complex forms quickly at room temperature (Watson and Chaconas, 1996). In addition, careful examination of the results presented in Figure 3 suggests that the IAS may slightly destabilize rather than stabilize the LR complex on the way to the transpososome. This suggestion derives from the fact that the apparent Kd for end pairing may, if anything, be slightly higher in the presence of the IAS than in its absence; thus the extent of stimulation by the IAS is higher at saturating end concentrations. This observation can be the result of many technical complications associated with the experiment. However, it argues against ground-state stabilization. Combined with the observation that the IAS is not a stable component of the transpososome (Mizuuchi et al., 1992; Surete and Chaconas, 1992) and the IAS is more readily cross-linkable in the form of the LER complex than to the transpososome (Watson and Chaconas, 1996), the above consideration suggests that the IAS preferentially stabilizes the transition state between the LR complex and transpososome. In other words, the IAS acts like an ‘enzyme’ that catalyzes the transition from the LR complex to transpososome. This model nicely explains why the IAS becomes dispensable once the transpososome is formed (Mizuuchi et al., 1992; Surette and Chaconas, 1992).
The role of MuB protein in the transpososome assembly reaction
MuB stimulated the transpososome assembly at lower concentrations of Mu ends. This stimulation required ATP or ATPγS as a cofactor. ADP was inefficient as the cofactor. Assembly stimulation by MuB was not dependent on stable DNA binding by MuB, ATPase activity of MuB or the ability of MuA to stimulate the ATPase activity of MuB. These conditions parallel closely, but not precisely, the ability of MuB to stimulate intramolecular strand transfer (Baker et al., 1991; Yamauchi and Baker, 1998). The exception is that intramolecular strand transfer can be stimulated reasonably efficiently in the presence of ADP (Yamauchi and Baker, 1998). The physical state of MuB under a variety of conditions is currently under investigation.
The results of the kinetic experiments presented here indicated that on the surface, unlike the IAS, MuB specifically stimulated the initial pairing of two Mu ends bound by MuA. However, we are not satisfied with this interpretation for two reasons. First, this model cannot explain earlier observations that suggested MuB stimulation of the post-pairing assembly steps in reactions with supercoiled plasmid DNA in the absence of DMSO. Secondly, we failed to observe synergistic stimulation of transpososome assembly by MuB and the IAS. One possibility might be that MuB stimulates both the Mu end pairing step and the post-pairing conformational step, but the latter effect is masked by some unknown effect due to the presence of DMSO in the reaction and/or by the absence of DNA superhelicity. However, if MuB does stimulate the pairing step, why did we fail to detect a synergistic effect of MuB and the IAS at lower Mu end concentrations?
An alternative possibility might be that like the IAS, MuB also stimulates only the post-pairing conformational steps, and the effects of the IAS and MuB are redundant. Our failure to observe assembly stimulation by MuB when the Mu end concentration is high may reflect an unknown inhibitory effect of the higher concentration(s) of the reaction component(s). Inhibitory side reactions that are insignificant at lower concentrations of the reactants may become significant at higher concentrations. Indeed, when the Mu end or IAS fragment concentration was raised higher to 200–500 nM, MuB significantly inhibited transpososome assembly (data not shown). We suspect that the MuA–MuB interaction may produce a form of MuA or MuB that is inhibitory for transpososome assembly stimulation, in some ways related to transposition target immunity. We found that like the Mu end sequence, the IAS sequence confers weaker, but significant target immunity to nearby DNA regions (data not shown). MuA promotes dissociation of MuB from DNA and, at higher MuA concentrations, MuB unbound to DNA accumulates (Baker et al., 1991). While these forms of MuB appear to stimulate intramolecular strand transfer by the assembled CDC, we still do not know the detailed physical states and functional activities of these MuB forms. It is possible that the high steady-state concentration of this MuB state includes a form that is detrimental to assembly stimulation. However, we have failed so far to obtain evidence that supports or refutes this possibility.
Despite technical limitations of our current approach, we are encouraged by the fact that one can observe the assembly rate enhancements effected by the cofactors in the reaction based on short DNA substrates. This allows us to make use of a variety of substrate analogs for the study of transpososome assembly/disassembly rates in the presence of the cofactors. One of the technical limitations of our current approach has been that it depends on electrophoretic detection of the stable complexes. Less stable complexes could not be detected or quantitated reliably. We are currently developing assay systems that do not rely on electrophoretic methods for complex detection. It is hoped that these developments will open additional experimental paths to look into the mechanism of transpososome assembly. Controlled assembly of higher order macromolecular complexes is involved in many important biological processes. Further understanding of the regulatory mechanism involved in individual systems will benefit our understanding of the roles of macromolecular interactions in other reactions as well.
Materials and methods
Proteins
Wild-type MuA protein was purified essentially as described (Baker et al., 1993). Wild-type MuB was purified as described by Chaconas et al. (1985) with the additional step described by Adzuma and Mizuuchi (1991). Mutant MuB proteins with an N-terminal His tag were a gift from Yamauchi and Baker (1998) and were centrifuged to remove precipitates after dialysis against MuB diluent buffer containing 30 mM HEPES– NaOH pH 7.6, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol (DTT) and 30% glycerol. MuB protein modified with NEM was made as described (Baker et al., 1991). HU protein was a gift from A.Segal (SU, San Diego, CA). IHF protein was a gift from S.-W.Yang.
DNA and reagents
The Mu L-end fragment contained 184 bp of Mu L-end sequence and 9 bp of flanking DNA beyond the cleavage site. The Mu R-end fragment contained 90 bp of Mu R-end sequences plus 9 bp of flanking DNA beyond the cleavage site. The IAS fragment contained the 170 bp Mu repressor binding sequence, O1 and O2 (position 876–1039; Goosen and van de Putte, 1987), including the intervening IHF-binding site. A control non-IAS fragment (176 bp) retained the IHF-binding site in the middle, but the O1 and O2 sequences were replaced by pBR322 sequences from positions 2789 to 2843 and 2834 to 2900, respectively. For the above fragments, a CTG/CAG sequence was added to both ends. The above fragments were amplified from the mini-Mu donor plasmid DNA pMK586 (Mizuuchi et al., 1992) or pMK543 (Mizuuchi and Mizuuchi, 1989) using appropriate primers and purified with anion-exchange HPLC on a Gen-Pak FAX column (Millipore, Waters), with a 0–1 M NaCl linear gradient in 25 mM Tris–HCl pH 8.0 and 1 mM EDTA. A 50 bp fragment with the sequence derived from pBR322 (position 3946–3995) was synthesized as a target DNA (HHMI/Keck Oligonucleotide Synthesis Facility, Yale University). The 5′ ends of the Mu end fragments were radiolabeled using [32P]ATP and T4 polynucleotide kinase (New England Biolabs). [32P]ATP (6000 Ci/mmol) was from New England Nuclear. NuSieve GTG– agarose was from FMC Bioproducts.
Transposition reactions
Standard reactions to test the effects of the IAS contained 25 mM Tris–HCl, 120 mM NaCl, 10 mM MgCl2, 15% glycerol, 1 mM DTT, 100 µg/ml bovine serum albumin (BSA), 0.005% Triton X-100, 2.5 µg/ml heparin, 15% DMSO, 5 nM Mu L- and R-end DNA fragments, <0.05 nM 32P-labeled L-end fragment, 50 nM IAS fragment or control fragment, 150 nM IHF and 200 nM HU. Reactions were started by the addition of 200 nM MuA protein and were incubated at 30°C for the time indicated. To investigate the Mu end concentration effects on the reaction rate, the concentrations of the two Mu ends were changed in parallel and the concentrations of the IAS fragment and protein components were optimized for each Mu end concentration as described in the legend to Figure 3. Reactions were stopped by addition of 1 µl of 0.3 M EDTA (final 12 mM) and cooling in an ice bath. Standard reactions to test the effects of MuB were essentially the same as described above except that the IAS, IHF and heparin were omitted and 100 nM 55 bp target fragment, 10 mM ATP and 280 nM MuB were included, and the concentration of NaCl was increased to 156 mM.
Formation of Mu transpososomes was assayed by non-denaturing gel electrophoresis of the protein–DNA complexes using a 4% NuSieve agarose gel in 1× TAE buffer (40 mM Tris–acetate pH 7.8, 8 mM sodium acetate, 1 mM EDTA) containing 100 µg/ml BSA and 100 µg/ml heparin essentially as described (Mizuuchi et al., 1995). The labeled protein– DNA complexes were quantitated by autoradiography of the dried gels using Fuji imaging plates and a Fuji BAS 2000 scanner (Fuji Medical Systems).
Acknowledgments
Acknowledgements
We are grateful to Kenji Adzuma, Eric Greene and Tania Baker for discussion and careful reading of the manuscript. We thank Anka Segall for the supply of HU protein, Michael Yamauchi and Tania Baker for mutant MuB proteins, and Shu-Wei Yang for IHF protein. This work was supported in part by the NIH Intramural AIDS Targeted Antiviral Program.
References
- Adzuma K. and Mizuuchi,K. (1988) Target immunity of Mu trans position reflects a differential distribution of Mu B protein. Cell, 53, 257–266. [DOI] [PubMed] [Google Scholar]
- Adzuma K. and Mizuuchi,K. (1989) Interaction of proteins located at a distance along DNA: mechanism of target immunity in the Mu DNA strand-transfer reaction. Cell, 57, 41–47. [DOI] [PubMed] [Google Scholar]
- Adzuma K. and Mizuuchi,K. (1991) Steady-state kinetic analysis of ATP hydrolysis by the B protein of bacteriophage Mu. Involvement of protein oligomerization in the ATPase cycle. J. Biol. Chem., 266, 6159–6167. [PubMed] [Google Scholar]
- Allison R.G. and Chaconas,G. (1992) Role of the A protein-binding sites in the in vitro transposition of Mu DNA: a complex circuit of interactions involving the Mu ends and the transpositional enhancer. J. Biol. Chem., 267, 19963–19970. [PubMed] [Google Scholar]
- Baker T.A. and Mizuuchi,K. (1992) DNA-promoted assembly of the active tetramer of the Mu transposase. Genes Dev., 6, 2221–2232. [DOI] [PubMed] [Google Scholar]
- Baker T.A., Mizuuchi,M. and Mizuuchi,K. (1991) MuB protein allosterically activates strand transfer by the transposase of phage Mu. Cell, 65, 1003–1013. [DOI] [PubMed] [Google Scholar]
- Baker T.A., Mizuuchi,M., Savilahti,H. and Mizuuchi,K. (1993) Division of labor among monomers within the Mu transposase tetramer. Cell, 74, 723–733. [DOI] [PubMed] [Google Scholar]
- Chaconas G., Gloor,G. and Miller,J.L. (1985) Amplification and purification of the bacteriophage Mu encoded B transposition protein. J. Biol. Chem., 260, 2662–2669. [PubMed] [Google Scholar]
- Chaconas G., Lavoie,B.D. and Watson,M.A. (1996) DNA transposition: jumping gene machine, some assembly required. Curr. Biol., 6, 817–820. [DOI] [PubMed] [Google Scholar]
- Craigie R. and Mizuuchi,K. (1986) Role of DNA topology in Mu transposition: mechanism of sensing the relative orientation of the two DNA segments. Cell, 45, 793–800. [DOI] [PubMed] [Google Scholar]
- Craigie R. and Mizuuchi,K. (1987) Transposition of Mu DNA: joining of Mu to target DNA can be uncoupled from cleavage at the ends of Mu. Cell, 51, 493–501. [DOI] [PubMed] [Google Scholar]
- Craigie R., Mizuuchi,M. and Mizuuchi,K. (1984) Site-specific recognition of the bacteriophage Mu ends by the Mu A protein. Cell, 39, 387–394. [DOI] [PubMed] [Google Scholar]
- Goosen N. and van de Putte,P. (1987) Regulation of transcription. In Symonds,N., Toussaint,A., van de Putte,P. and Howe,M.M. (eds), Phage Mu. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 41–52.
- Haniford D.B. and Chaconas,G. (1992) Mechanistic aspects of DNA transposition. Curr. Opin. Genet. Dev., 2, 698–704. [DOI] [PubMed] [Google Scholar]
- Harshey R.M. and Cuneo,S.D. (1986) Carboxyl-terminal mutants of phage-Mu transposase. J. Genet., 65, 159–174. [Google Scholar]
- Jiang H., Yang,J.Y. and Harshey,R. (1999) Criss-crossed interactions between the enhancer and the att sites of phage Mu during DNA transposition. EMBO J., 18, 3845–3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B.D., Chan,B.S., Allison,R.G. and Chaconas,G. (1991) Structural aspects of a higher order nucleoprotein complex: induction of an altered DNA structure at the Mu–host junction of the Mu type 1 transpososome. EMBO J., 10, 3051–3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavoie B.D., Shaw,G.S., Millner,A. and Chaconas,G. (1996) Anatomy of a flexer–DNA complex inside a higher-order transposition intermediate. Cell, 85, 761–771. [DOI] [PubMed] [Google Scholar]
- Leung P.C., Teplow,D.B. and Harshey,R.M. (1989) Interaction of distinct domains in Mu transposase with Mu DNA ends and an internal transpositional enhancer. Nature, 338, 656–658. [DOI] [PubMed] [Google Scholar]
- Mizuuchi K. (1992) Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu. Rev. Biochem., 61, 1011–1051. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M. and Mizuuchi,K. (1989) Efficient Mu transposition requires interaction of transposase with a DNA sequence at the Mu operator: implications for regulation. Cell, 58, 399–408. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Baker,T.A. and Mizuuchi,K. (1991) DNase protection analysis of the stable synaptic complexes involved in Mu transposition. Proc. Natl Acad. Sci. USA, 88, 9031–9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi M., Baker,T.A. and Mizuuchi,K. (1992) Assembly of the active form of the transposase–Mu DNA complex: a critical control point in Mu transposition. Cell, 70, 303–311. [DOI] [PubMed] [Google Scholar]
- Mizuuchi M., Baker,T.A. and Mizuuchi,K. (1995) Assembly of phage Mu transpososomes: cooperative transitions assisted by protein and DNA scaffolds. Cell, 83, 375–385. [DOI] [PubMed] [Google Scholar]
- Naigamwalla D.Z. and Chaconas,G. (1997) A new set of Mu DNA transposition intermediates: alternate pathways of target capture preceding strand transfer. EMBO J., 16, 5227–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P.A., Yang,S.W., Mizuuchi,K. and Nash,H.A. (1996) Crystal structure of an IHF–DNA complex: a protein-induced DNA U-turn. Cell, 87, 1295–1306. [DOI] [PubMed] [Google Scholar]
- Savilahti H., Rice,P.A. and Mizuuchi,K. (1995) The phage Mu transpososome core: DNA requirements for assembly and function. EMBO J., 14, 4893–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M.G. and Chaconas,G. (1991) Stimulation of the Mu DNA strand cleavage and intramolecular strand transfer reactions by the Mu B protein is independent of stable binding of the Mu B protein to DNA. J. Biol. Chem., 266, 17306–17313. [PubMed] [Google Scholar]
- Surette M.G. and Chaconas,G. (1992) The Mu transpositional enhancer can function in trans: requirement of the enhancer for synapses but not strand cleavage. Cell, 68, 1101–1108. [DOI] [PubMed] [Google Scholar]
- Surette M.G., Buch,S.J. and Chaconas,G. (1987) Transpososomes: stable protein–DNA complexes involved in the in vitro transposition of bacteriophage Mu DNA. Cell, 49, 253–262. [DOI] [PubMed] [Google Scholar]
- Surette M.G., Lavoie,B.D. and Chaconas,G. (1989) Action at a distance in Mu DNA transposition: an enhancer-like element is the site of action of supercoiling relief activity by integration host factor (IHF). EMBO J., 8, 3483–3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surette M.G., Harkness,T. and Chaconas,G. (1991) Stimulation of the Mu A protein-mediated strand cleavage reaction by the Mu B protein and the requirement of DNA nicking for stable type 1 transpososome formation. In vitro transposition characteristics of mini-Mu plasmids carrying terminal base pair mutations. J. Biol. Chem., 266, 3118–3124. [PubMed] [Google Scholar]
- Vologodskii A. and Cozzarelli,N.R. (1996) Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys. J., 70, 2548–2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M.A. and Chaconas,G. (1996) Three-site synapses during Mu DNA transposition: a critical intermediate preceding engagement of the active site. Cell, 85, 435–445. [DOI] [PubMed] [Google Scholar]
- Yamauchi M. and Baker,T.A. (1998) An ATP–ADP switch in MuB controls progression of the Mu transposition pathway. EMBO J., 17, 5509–5518. [DOI] [PMC free article] [PubMed] [Google Scholar]