Abstract
In yeast, two aminoacyl-tRNA synthetases, MetRS and GluRS, are associated with Arc1p. We have studied the mechanism of this complex formation and found that the non-catalytic N-terminally appended domains of MetRS and GluRS are necessary and sufficient for binding to Arc1p. Similarly, it is the N-terminal domain of Arc1p that contains distinct but overlapping binding sites for MetRS and GluRS. Localization of Arc1p, MetRS and GluRS in living cells using green fluorescent protein showed that these three proteins are cytoplasmic and largely excluded from the nucleus. However, when their assembly into a complex is inhibited, significant amounts of MetRS, GluRS and Arc1p can enter the nucleus. We suggest that the organization of aminoacyl-tRNA synthetases into a multimeric complex not only affects catalysis, but is also a means of segregating the tRNA- aminoacylation machinery mainly to the cytoplasmic compartment.
Keywords: aminoacyl-tRNA synthetase/ARC1/GluRS/MetRS/tRNA
Introduction
Aminoacyl-tRNA synthetases are required for the faithful translation of the genetic code, as they catalyse charging of tRNAs with their cognate amino acids (Martinis et al., 1999a,b; Ibba and Söll, 2000). The 20 different aminoacyl-tRNA synthetases, one for each amino acid, are divided into two classes (I and II) according to the sequence and structure of their catalytic domains, which are usually conserved in all kingdoms (Francklyn et al., 1997; Woese et al., 2000). They are also characterized by idiosyncratic tRNA-binding domains that are attached to, or inserted in, the class-defining catalytic core (Ibba et al., 1997). When compared with the prokaryotic synthetases, the corresponding enzymes from yeast or higher eukaryotes often contain extensions to the N- or C-termini, which are not essential for catalytic activity (Mirande, 1991; Kisselev and Wolfson, 1994; Reed and Yang, 1994; Weygand-Durasevic et al., 1996). Their function is not always clear. In some cases, they serve as non-specific tRNA-binding platforms that facilitate the association of the synthetases with their cognate tRNAs (Whelihan and Schimmel, 1997; Wang and Schimmel, 1999; Cahuzac et al., 2000; Frugier et al., 2000). In other cases, these eukaryote-specific appendices are required for assembly into multisynthetase complexes (Rho et al., 1996, 1999; Agou and Mirande, 1997; Kim et al., 2000a), although catalytic domains may also be responsible for the formation of these complexes (Kim et al., 2000a). Higher eukaryotes are indeed characterized by the presence of a supramolecular multienzyme complex containing nine aminoacyl-tRNA synthetases and three non-catalytic polypeptides, p43 (pro-EMAPII), p38 and p18 (Filonenko and Deutscher, 1994; Quevillon and Mirande, 1996; Norcum and Warrington, 1998; Quevillon et al., 1999). The function of this complex is still unknown. It is possible that p43 could modulate the activity of the enzymes, as is shown to be the case for human ArgRS (Park et al., 1999).
We previously identified in yeast a smaller complex of aminoacyl-tRNA synthetases consisting of methionyl- tRNA (MetRS) and glutamyl-tRNA (GluRS) synthetases, as well as the protein Arc1p, the yeast homologue of p43 (Simos et al., 1996; Quevillon et al., 1997). Arc1p associates with the two enzymes both in vivo and in vitro. Furthermore, the catalytic efficiency of the in vitro reconstituted complexes of MetRS or GluRS with Arc1p is substantially increased compared with the monomeric enzymes (Simos et al., 1998; Deinert et al., 2001). This stimulation is due to the increased affinity of the complexes for the cognate tRNAs and requires the C-terminal part of Arc1p. Indeed, this part of Arc1p harbours a tRNA-binding domain (TRBD), which is conserved in p43 as well as other proteins (Kleeman et al., 1997; Morales et al., 1999; Kaminska et al., 2000). Surprisingly, in mammals this domain can also function as a cytokine (Knies et al., 1998; Wakasugi and Schimmel, 1999; Behrensdorf et al., 2000), possibly linking the progression of apoptosis to the inhibition of protein translation (Weiner and Maizels, 1999). Recently, the structure of the TRBD of p43 has been solved, showing that part of it adopts the OB fold, an oligonucleotide-binding structural motif (Kim et al., 2000b; Renault et al., 2001). In yeast, Arc1p is required for optimal cell growth and is essential for viability in the absence of the tRNA nuclear export factor Los1p (Simos et al., 1996).
Apart from their established role in tRNA aminoacylation, aminoacyl-tRNA synthetases are also implicated in several other cellular processes such as mitochondrial RNA splicing or transcriptional, as well as translational, regulation (reviewed in Martinis et al., 1999a,b). Recently, aminoacyl-tRNA synthetases have also been implicated in nuclear tRNA export as inhibition of tRNA- aminoacylation causes accumulation of mature tRNAs inside the nucleus of Xenopus oocytes (Lund and Dahlberg, 1998; Simos and Hurt, 1999). The requirement of tRNA-aminoacylation for efficient nuclear tRNA export was also observed in yeast cells (Sarkar et al., 1999; Grosshans et al., 2000). These observations indicate that aminoacyl-tRNA synthetases can enter the nucleus. Cell fractionation and biochemical experiments have demonstrated the presence of tRNA-aminoacylation activities in the nuclear compartment of higher eukaryotic cells (Lund and Dahlberg, 1998; Nathanson and Deutscher, 2000). However, direct intranuclear localization by microscopic methods has been performed for only a few aminoacyl- tRNA synthetases in mammalian cells (Kisselev and Wolfson, 1994; Popenko et al., 1994; Ko et al., 2000), and for one enzyme in yeast cells (Azad et al., 2001).
In order to understand the molecular details of the assembly of the Arc1p–MetRS–GluRS complex, we have expressed and purified from yeast truncated versions of all three components. Our results show that the formation of the complex in vivo requires the N-terminally appended non-catalytic domains of the synthetases, both of which interact with overlapping sites on the N-terminal domain of Arc1p. Furthermore, localization of these three components demonstrated that all of them can be located in the nucleus only if they are not assembled in a complex. Therefore, association of eukaryotic aminoacyl-tRNA synthetases into a multimeric complex provides a means of regulating their subcellular distribution.
Results
A functional link between the N-terminally appended domain of GluRS and Arc1p
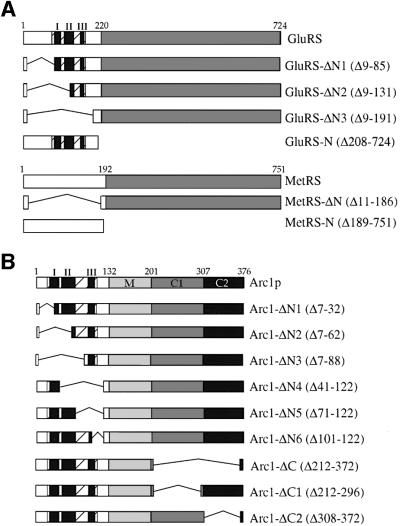
The structural genes for yeast cytoplasmic MetRS and GluRS, MES1 and YGL245W (from now on called GUS1), respectively, encode proteins that contain N- terminal extensions when compared with the corresponding Escherichia coli enzymes. In the case of MetRS, the 185 residue long N-terminal domain is not homologous to known proteins and can be removed without affecting the activity or stability of monomeric MetRS (Walter et al., 1989). In the case of GluRS, sequence alignment reveals an N-terminal extension of 210 amino acids, a part of which (residues 87–170) is homologous to several other proteins (Figure 1 and data not shown). These polypeptides include the N-terminal parts of GluRSs from other species as well as other aminoacyl-tRNA synthetases and proteins involved in translation, such as the β and γ subunits of the eukaryotic elongation factor EF1B and the C-terminal regions of p18 and p38, the two non-catalytic components of the mammalian multisynthetase complex (Koonin et al., 1994; Quevillon and Mirande, 1996; Quevillon et al., 1999). Strikingly, this domain is also present in the N-terminal region of Arc1p (amino acids 24–106) and its closely related homologue from Schizosaccharomyces pombe (S.pombe Arc1p).
Fig. 1. Top: schematic representation of the S.cerevisiae GluRS and Arc1p protein sequences. In GluRS the grey box represents the catalytic domain, while in Arc1p it represents the different domains as indicated. In both proteins the hatched box represents a conserved motif. The parts of the motif corresponding to the homology blocks shown below are marked as black bars numbered I–III. The M, C1 and C2 domains of Arc1p correspond to amino acids 132–201, 202–307 and 308–376, respectively. Bottom: alignment of three homologous sequence blocks (I–III) from the β and γ subunits of the human and S.cerevisiae (Sc) EF-1B, mouse cysteinyl-tRNA synthetase (CysRS), human valyl-tRNA synthetase (ValRS), the non-catalytic components of the Chinese hamster ovary multisynthetase complex p18 and p38, human bifunctional glutamyl-prolyl-tRNA synthetase (GluProRS), S.cerevisiae glutamyl-tRNA synthetase (GluRS) and S.cerevisiae and S.pombe (Sp) Arc1p. Identical residues are shown white on a black background, while partially conserved residues are in grey boxes. The numbers of the first and last residue in each block are indicated. Alignment was performed using the ClustalW program.
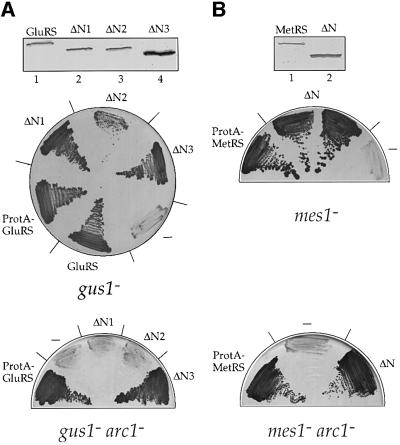
To study the function of the appended N-terminal regions of GluRS and MetRS in the formation of the Arc1p complex, we generated truncation mutants as shown schematically in Figure 2A. Three different N-terminal deletion constructs were made in the case of GluRS: GluRS-ΔN1 (lacking residues 9–85), GluRS-ΔN2 (lacking residues 9–131) and GluRS-ΔN3 (lacking residues 9–191). In the case of MetRS, one mutant (MetRS-ΔN) lacking the entire N-terminal part (residues 11–186) was constructed. Subsequently, full-length MetRS, GluRS and the N-terminal deletion constructs were tagged with protein A (ProtA) to facilitate their detection and purification. All the truncated constructs were stably expressed and functional, as shown by their ability to complement the corresponding gus1 and mes1 null strains (Figure 3). Only in the case of GluRS-ΔN2 did the cells grow more slowly, while all other mutants complemented to wild-type levels. This shows that the N-terminal appended domains of both GluRS and MetRS are not required for cell growth and, consequently, aminoacylation activity. However, since GluRS-ΔN3 and MetRS-ΔN are expressed in higher amounts than the corresponding full-length proteins (Figure 3), it is possible that the reduced functionality of the N-terminal truncated proteins is compensated for by overproduction.
Fig. 2. Schematic representation of deletion mutants used in this study. (A) Deletion mutants of GluRS and MetRS. White and grey boxes indicate the non-catalytic N-terminal domains and the catalytic domains of the enzymes, respectively. The hatched boxes represent the conserved motif in the N-terminal domain of GluRS and the vertical black bars the homology blocks shown in Figure 1. (B) Deletion mutants of Arc1p. The N-terminal (N), middle (M) and C-terminal (C1 and C2) domains of Arc1p are indicated by different shades of grey. The hatched box in the N-domain represents the conserved motif also found in GluRS and the vertical black bars the homology blocks shown in Figure 1. The M, C1 and C2 domains of Arc1p correspond to amino acids 132–201, 202–307 and 308–376, respectively. The position of each deletion is represented by a line. Numbers refer to amino acid residues. Numbers in parentheses indicate the deleted amino acids.
Fig. 3. Genetic analysis of the N-terminal truncation mutants of (A) GluRS and (B) MetRS. Upper panels: western blotting analysis of extracts from cells expressing ProtA-tagged full-length or N-terminally truncated forms of GluRS (A, lanes1–4) and MetRS (B, lanes 1 and 2) in the strains gus1– and mes1–, respectively, using an anti-ProtA antibody. Equal amounts of extracts were loaded in each lane. Middle panels: complementation of the knock-out strains gus1– (A) and mes1– (B) by constructs expressing full-length or N-terminally truncated ProtA-tagged GluRS or MetRS, respectively, as indicated. Lower panels: complementation of the double knock-out strains gus1– arc1– (A) and mes1– arc1– (B) by constructs expressing full-length or N-terminally truncated ProtA-tagged GluRS or MetRS, respectively, as indicated. Complementation is shown by the ability to lose the pURA3-GUS1 (A) or pURA3-MES1 (B) plasmids and grow in the presence of 5-FOA. – indicates transformation with an empty plasmid.
We then tested whether the deletions in the N-terminal domains of GluRS and MetRS affect cell growth when the expression of Arc1p is simultaneously abolished. For this reason we constructed haploid yeast strains in which the deletion of the chromosomal copies of either MES1 or GUS1 genes was combined with the disruption of the ARC1 gene. As shown in Figure 3A, GluRS-ΔN3 lacking the entire N-domain could still complement the double-disrupted gus1– arc1– strain. However, GluRS-ΔN1 or GluRS-ΔN2, which lacked parts of the N-domain, could not rescue these cells, exhibiting a synthetic lethal relationship with Arc1p. This demonstrates a functional link between the N-domain of GluRS and Arc1p. Since both GluRS-ΔN1 and GluRS-ΔN2 contain the catalytic domain of GluRS, which is sufficient for survival (as in GluRS-ΔN3), these results also indicate that the shortened N-domain exhibits a dominant-negative phenotype when Arc1p is missing (see Discussion). The double-disrupted mes1– arc1– strain was equally well complemented by full-length MetRS or MetRS-ΔN, showing that the complete removal of the N-domain of MetRS can be tolerated even in the absence of Arc1p (Figure 3B).
The N-terminally appended domains of MetRS and GluRS are both necessary and sufficient for binding to Arc1p
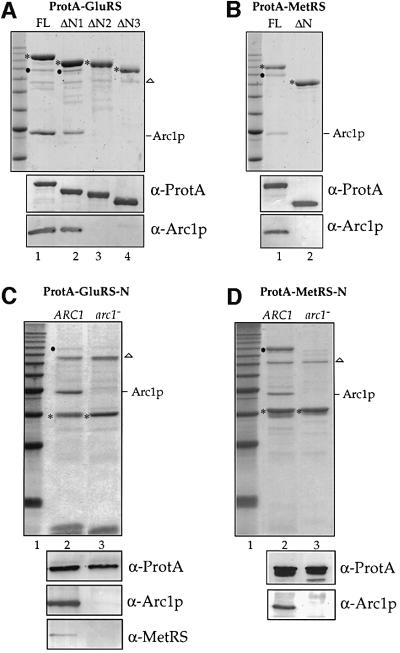
To find out whether the appended N-domains of GluRS and MetRS are involved in the formation of the Arc1p–MetRS–GluRS complex, the ProtA tagged truncation mutants were purified by IgG–Sepharose affinity chromatography from total cell extracts of yeast cells expressing the corresponding mutants. This analysis revealed that when the N-terminal domains of GluRS and MetRS are missing, binding to Arc1p and formation of the ternary complex are abolished (Figure 4A, lanes 3 and 4, and B, lane 2). However, in the case of ProtA–GluRS-ΔN1, association with Arc1p and MetRS still persists, showing that the non-conserved first 85 residues of GluRS do not participate in complex formation (Figure 4A, lane 2). The apparent stoichiometry of the complexes isolated from crude extracts deviates from the 1:1:1 ratio for GluRS, MetRS and Arc1p that is observed upon in vitro reconstitution of the Arc1p–GluRS–MetRS complex from purified components (Deinert et al., 2001). This difference is most likely due to the dilution of the extracts and to the limited proteolysis that preferentially removes the N-terminal appended domains of the enzymes.
Fig. 4. The N-domains of MetRS and GluRS are both essential and sufficient for stable association with Arc1p. Affinity purification by IgG–Sepharose chromatography of (A) the full-length (FL) ProtA–GluRS and the three N-terminal truncated forms expressed in the gus1– strain; (B) the full-length (FL) ProtA–MetRS and the N-terminal truncated form expressed in the mes1– strain; (C) the N-terminal domain of GluRS (ProtA–GluRS-N) from wild-type (ARC1) and arc1– strains; (D) the N-domain of MetRS (ProtA–MetRS-N) from wild-type (ARC1) and arc1– strains. Upper panels: analysis of the column eluates by SDS–PAGE and Coomassie Blue staining. The first lane from the left contains the 10 kDa ladder molecular weight markers (the stronger band corresponds to 50 kDa). The asterisks indicate the positions of the ProtA–fusion proteins. The black dots indicate the position of co-purifying MetRS (A and C) or GluRS (B and D). The identity of the protein bands corresponding to GluRS and MetRS in (A) and (B) was confirmed by mass spectroscopic analysis. The position of co-purifying Arc1p is indicated on the right. The protein band (which is indicated by a triangle) at 75 kDa corresponds to an irrelevant contaminant. Lower panels: western blotting analysis of the eluates using anti-ProtA, anti-Arc1p or anti-MetRS antibodies as indicated. Only the relevant parts of the blots are shown.
To determine whether the N-domains of MetRS and GluRS (MetRS-N and GluRS-N, respectively, shown schematically in Figure 2A) are sufficient for binding to Arc1p, they were tagged with ProtA, expressed in wild-type and arc1– cells, and isolated by affinity chromatography. As shown in Figure 4C (lane 2), ProtA–GluRS-N co-purified with both Arc1p and MetRS. The specificity of this interaction is shown by the fact that both Arc1p and MetRS were absent when GluRS-N was purified from an arc1– strain (Figure 4C, lane 3). This also shows that no direct interaction takes place between the N-domain of GluRS and MetRS, i.e. these two proteins can only assemble into the complex via Arc1p. Similar results were also obtained for ProtA–MetRS-N: this domain also co-purified specifically with Arc1p and GluRS (Figure 4D). Furthermore, the stoichiometry of the co-purified complexes suggests that Arc1p binds more efficiently to the N-domain of GluRS than to that of MetRS. Taken together, our data demonstrate that the N-terminally appended domains of MetRS and GluRS are necessary and sufficient for binding to Arc1p. In the case of GluRS, the Arc1p binding site was mapped to the conserved part of the N-domain, between amino acids 86 and 207.
The N-domain of Arc1p harbours distinct but overlapping binding sites for MetRS and GluRS
We have previously demonstrated that the 132 amino acid long N-terminal domain of Arc1p (Arc1-N) can form a stable complex with both MetRS and GluRS in vivo as well as in vitro (Simos et al., 1998; Deinert et al., 2001). As revealed by the alignment shown in Figure 1, the central part of Arc1-N (residues 24–106) contains a motif also found in GluRS. To reveal the role of this motif in the formation of the Arc1p–GluRS–MetRS complex, we introduced serial deletions in the N-domain of Arc1p and constructed the mutants Arc1-ΔN1–Arc1-ΔN6, which are shown schematically in Figure 2B. These truncated Arc1p forms were tagged with ProtA, expressed in the arc1 null strain and purified by affinity chromatography from total cell extracts of yeast cells expressing the corresponding mutants. As expected, full-length ProtA-tagged Arc1p co-purified with GluRS and MetRS (Figure 5A, lane 2). Deletion of the first 32 residues of Arc1p caused dis sociation of MetRS but not of GluRS, which was still recovered with Arc1-ΔN1 (Figure 5A, lane 3). Western blot analysis using an antibody against MetRS confirmed that MetRS was indeed completely absent from the ProtA–Arc1-ΔN1 eluate (data not shown). In all other cases (Arc1-ΔN2–Arc1-ΔN6), the deletions caused dissociation of both GluRS and MetRS, as revealed by their absence from the corresponding eluates (Figure 5A, lanes 4–8). These data show that an intact N-terminal domain of Arc1p is required for binding to MetRS. However, the first part of this domain (residues 7–32) is dispensable for the Arc1p–GluRS interaction. Therefore, we conclude that the N-domain of Arc1p contains distinct but overlapping binding sites for MetRS and GluRS. Furthermore, the Arc1p–GluRS interaction is mediated by domains sharing the same conserved sequence motif, which may, accordingly, represent a dimerization element.
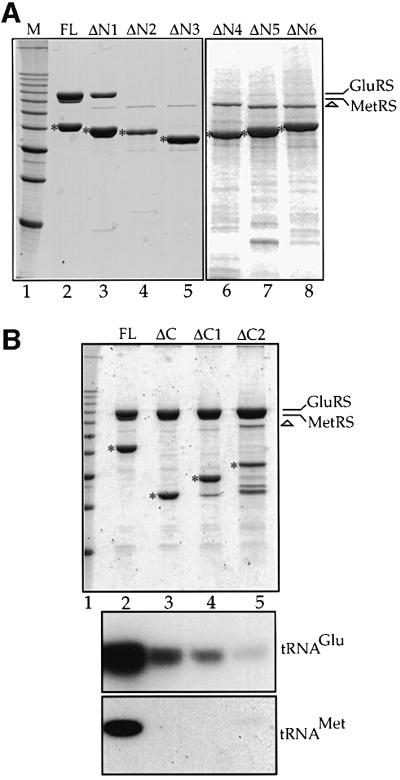
Fig. 5. The N-domain of Arc1p contains distinct but overlapping binding sites for MetRS and GluRS, while an intact C-domain is only required for tRNA binding. IgG–Sepharose affinity purification of (A) ProtA-tagged full-length (FL) Arc1p (lane 2) and six versions carrying truncations at the N-terminal domain (lanes 3–8) expressed in the arc1– strain and (B) ProtA-tagged full-length (FL) Arc1p (lane 2) and three versions carrying complete or partial deletions of the C-terminal domain (lanes 3–5) expressed in the arc1– strain. (A and B, upper panel): analysis of the eluates by SDS–PAGE and Coomassie Blue staining. The asterisks indicate the positions of the ProtA–fusion proteins. The positions of co-purifying GluRS and MetRS are indicated on the right. The apparent lack of stoichiometry in (B), lanes 3–5, is due to the co-migration of MetRS and GluRS. The protein band at 75 kDa (indicated by a triangle) corresponds to an irrelevant contaminant. Lane 1, molecular weight markers (10 kDa ladder; 30–120 kDa). (B, lower panels) Northern blot analysis of co-purifying RNAs using radioactive probes against tRNAGlu and tRNAMet. Only the relevant part of the autoradiographs is shown.
The biochemical evidence for the interaction sites on Arc1-N was also confirmed genetically using the previously isolated synthetic lethal yeast strains slk117 and slk88 (Simos et al., 1996). Strain slk117, which carries a mutation in the MES1 gene and lacks full-length Arc1p, could not be complemented by any of the Arc1p N-domain deletion mutants. On the other hand, strain slk88, which carries a defective GluRS and also lacks Arc1p, survived when transformed by Arc1-ΔN1 but not by any of Arc1-ΔN2–Arc1-ΔN6 (data not shown). Therefore, the lack of biochemical interaction correlates with the lack of complementation and vice versa.
While the N-domain of Arc1p is involved in the formation of the Arc1p–GluRS–MetRS ternary complex, its C-terminal domain (residues 201–376) has been shown to mediate broad-specificity binding of free Arc1p to several tRNA species and high-specificity binding of the Arc1p–MetRS–GluRS complex to the cognate tRNAs (Simos et al., 1998; Deinert et al., 2001). According to the homology between Arc1p and the mammalian protein EMAPII, part of the C-domain of Arc1p (residues 203–303; marked C1 in Figure 2B) contains a distinct structural motif called the OB fold. The OB fold is also part of the anticodon binding domain of three aminoacyl-tRNA synthetases and, interestingly, represents the core structure of the prokaryotic structure-specific tRNA-binding protein Trbp111 (Swairjo et al., 2000). To find out whether the tRNA-binding properties of Arc1p are due to the presence of this OB fold, we prepared Arc1-ΔC1 and Arc1-ΔC2, two constructs in which either the region corresponding to the OB-fold (C1) or the rest of the Arc1p C-domain (residues 308–372; C2) was deleted, respectively (see Figure 2B). These ProtA-tagged mutant forms as well as full-length Arc1p and Arc1-ΔC (which lacks the entire C-domain) were purified by affinity chromatography. As shown in Figure 5B, in all four cases similar amounts of MetRS and GluRS were co-purified. However, analysis of the co-purifying tRNAs showed that not only removal of the whole C-domain but also deletion of either C1 or C2 caused a dramatic reduction in the amounts of cognate tRNAGlu and tRNAMet that bound to the Arc1p–MetRS–GluRS complex. This shows that the OB fold subdomain (C1) is not sufficient for stable tRNA binding, but that the presence of the C2 subdomain is also required. Furthermore, the inability of Arc1p to bind tRNA does not influence at all the protein–protein inter actions between Arc1p and the two tRNA-aminoacylation enzymes.
Formation of the Arc1p–MetRS–GluRS complex causes nuclear exclusion of its components
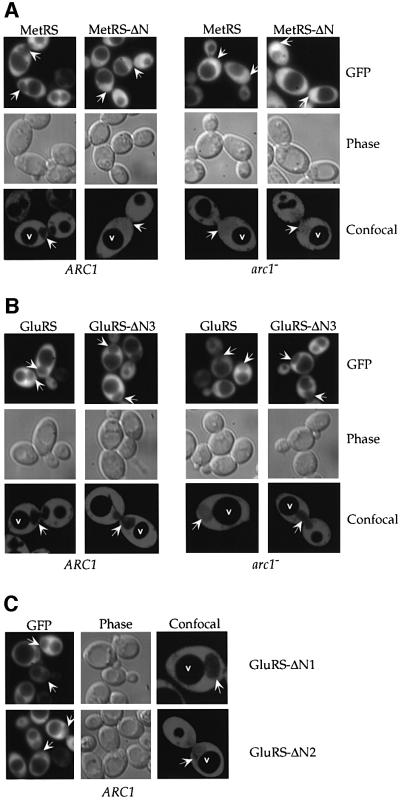
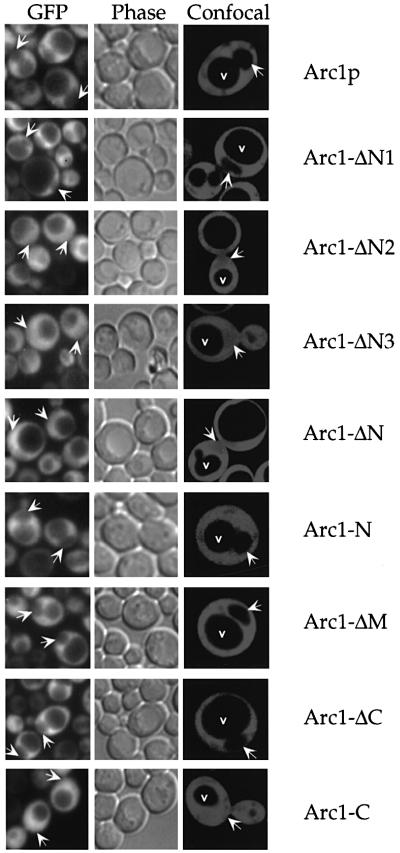
Aminoacyl-tRNA synthetases are normally found in the cytoplasm. However, recent results have suggested the existence of minor nuclear pools of these enzymes, which may be required for the charging of the newly synthesized tRNAs before nuclear export (Lund and Dahlberg, 1998; Nathanson and Deutscher, 2000; Azad et al., 2001). To study the intracellular distribution of the Arc1p– MetRS–GluRS complex and its individual components, we tagged MetRS, GluRS, Arc1p and their described truncation mutants with green fluorescent protein (GFP) and expressed them in yeast. All the GFP-tagged constructs were expressed normally when checked by western blotting analysis and were functional because they could complement the corresponding mutants (data not shown). Furthermore, the N-terminal GFP tag did not influence the integrity of the Arc1p ternary complex because the GFP-tagged enzymes co-purified with Arc1p as efficiently as the untagged native ones (data not shown). The cells expressing the GFP-tagged proteins were examined by conventional fluorescence microscopy as well as by confocal laser scanning microscopy in order to eliminate the contribution of cytoplasmic fluorescence to the nuclear signals. As shown in Figure 6 and summarized in Table I, GFP–GluRS and GFP–MetRS were detected exclusively in the cytoplasm of ARC1 cells, with distinct nuclear and vacuolar exclusion. The same was also true for GFP–Arc1p expressed in arc1– cells (Figure 7; Table I). We have observed that those mutant proteins that retained the ability to associate into a complex (GFP–GluRS- ΔN1, GFP–Arc1-ΔN1, GFP–Arc1-N, GFP–Arc1-ΔM and GFP–Arc1-ΔC) exhibited similar localization to the corresponding full-length proteins, with very little or no nuclear signal (Figures 6C and 7; Table I). Strikingly, however, when the GFP-tagged enzymes were unable to assemble into the Arc1p–MetRS–GluRS complex, either because of deletions in their N-domains or due to the absence of Arc1p, they could be detected inside the nucleus. In the case of GFP–MetRS-ΔN or when GFP–MetRS was expressed in arc1– cells, the nuclear signal was identical to the cytoplasmic signal, suggesting the presence of a significant nuclear pool of free enzyme (Figure 6A). In the case of GluRS, the effects of the deletions were variable. GFP–GluRS, when expressed in the absence of Arc1p, as well as GFP–GluRS-ΔN2 could be clearly detected inside the nucleus, although their nuclear signals were not as strong as in the cytoplasm (Figure 6B and C). GFP–GluRS-ΔN3, on the other hand, despite the fact that it is 26 amino acid residues shorter than GFP–GluRS-ΔN2, exhibited only very weak nuclear staining (Figure 6B; also see Discussion). Concerning the Arc1p mutants, GFP–Arc1-ΔN2, GFP–Arc1-ΔN3, GFP–Arc1-ΔN and GFP–Arc1-C, all of which lack the binding sites for MetRS and GluRS, displayed nuclear signals (Figure 7). In summary, there was a strong correlation between the inability of the various synthetase or Arc1p mutants to form a complex and their presence inside the nucleus (Table I). These data suggest that since most of the fusion proteins used in these experiments exceed in size the diffusion limit of the nuclear pore complex (∼50 kDa), Arc1p, GluRS and MetRS are capable of actively entering the nucleus. However, when organized into the Arc1p–MetRS–GluRS complex, they are excluded from the nucleus. The formation of this complex, which is mediated by the N-terminal domains of its individual components, may therefore trigger cytoplasmic reten tion or, alternatively, active nuclear export (also see Discussion).
Fig. 6. Nuclear entry of MetRS and GluRS through deletions in their N-terminal domains or by disruption of the arc1 gene. Localization by fluorescence and confocal laser scanning microscopy of (A) GFP–MetRS and GFP–MetRS-ΔN in mes1– (left panels) and mes1– arc1– cells (right panels); (B) GFP–GluRS and GFP–GluRS-ΔN3 in gus1– (left panels) and gus1– arc1– cells (right panels); (C) GFP–GluRS-ΔN1 and GFP–GluRS-ΔN2 in gus1– cells. Note that GFP–GluRS-ΔN1 and GRP–GluRS-ΔN2 cannot be localized in the double-disrupted gus1– arc1– cells because the partial deletions of the N-domain of GluRS are lethal in the absence of Arc1p. In all cases, arrows indicate the position of the nucleus in representative cells, as inferred from the phase-contrast fields. In the confocal images, which show sections through the middle of the cells, v indicates the vacuoles and arrows point to the nuclei.
Table I. Localization of GFP-tagged Arc1p, GluRS and MetRS constructs.
| Construct | Strain | Formation of complex | Localization |
|---|---|---|---|
| Arc1p | arc1– | + | C |
| Arc1-N | arc1– | + | C |
| Arc1-ΔN | arc1– | – | C + N |
| Arc1-ΔN1 | arc1– | + | C |
| Arc1-ΔN2 | arc1– | – | C + N |
| Arc1-ΔN3 | arc1– | – | C + N |
| Arc1-C | arc1– | – | C + N |
| Arc1-ΔC | arc1– | + | C |
| Arc1-ΔM | arc1– | + | C |
| MetRS | mes1– ARC1 | + | C |
| MetRS | mes1– arc1– | – | C + N |
| MetRS-ΔN | mes1– ARC1 | – | C + N |
| MetRS-ΔN | mes1– arc1– | – | C + N |
| GluRS | gus1– ARC1 | + | C |
| GluRS | gus1– arc1– | – | C + n |
| GluRS-ΔN1 | gus1– ARC1 | + | C |
| GluRS-ΔN2 | gus1– ARC1 | – | C + n |
| GluRS-ΔN3 | gus1– ARC1 | – | C |
| GluRS-ΔN3 | gus1– arc1– | – | C |
C, cytoplasmic signal and very little or no nuclear signal; C + N, cytoplasmic signal plus nuclear signal of similar intensity; C + n, cytoplasmic signal plus weaker nuclear signal.
Fig. 7. Deletions in the N-terminal domain of Arc1p allow entry into the nucleus. Localization by fluorescence microscopy and confocal laser scanning microscopy of GFP-tagged Arc1p or its various deletion mutants in arc1– cells as indicated. Arc1-ΔN1, Arc1-ΔN2, Arc1-ΔN3 and Arc1-ΔC are described in Figure 2B. Arc1-ΔN (Δ7–122), Arc1-N (Δ134–372), Arc1-ΔM (Δ134–198) and Arc1-C (Δ7–198) are described in Simos et al. (1998). The arrows indicate the position of the nucleus in representative cells, as inferred from the phase-contrast fields. The confocal images show sections through the middle of the cells; v indicates the vacuoles and arrows point to the nuclei.
Discussion
A stable complex is formed in yeast between Arc1p and two aminoacyl-tRNA synthetases, GluRS and MetRS. The results presented in this work, together with our previous findings (Simos et al., 1998; Deinert et al., 2001), suggest the following model for the organization of this complex (Figure 8). Both enzymes interact directly with Arc1p. These protein–protein interactions are mediated, on the one hand, by the N-terminally appended domains of the enzymes and, on the other hand, by the N-domain of Arc1p, which contains distinct but overlapping binding sites for the two enzymes. The catalytic domains of the two enzymes are therefore free to associate with, and aminoacylate, the cognate tRNAs. Selection and binding of these tRNAs are facilitated by the TRBD of Arc1p, which would recognize the conserved core ‘elbow’ structure of the tRNA and direct them to the active sites of the enzymes. This requires both the OB-fold subdomain (C1) as well as the eukaryote-specific C-terminal part (C2) of Arc1p. However, it remains unknown whether both cognate tRNAs can bind to the complex at the same time or one at a time. In either case, both enzymes obtain a catalytic advantage when associated with Arc1p, as shown by the in vitro reconstitution and analysis of the corresponding complexes (Simos et al., 1998; Deinert et al., 2001).
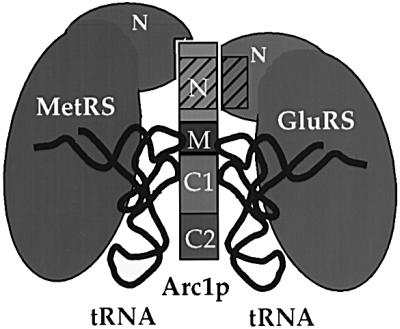
Fig. 8. A schematic hypothetical model for the organization of the yeast Arc1p–MetRS–GluRS complex. The domains of Arc1p (N, M, C1 and C2) are indicated by different shades of grey. The N-terminal domains of MetRS and GluRS are labelled. The hatched areas represent the conserved motifs present in the N-terminal domains of both Arc1p and GluRS. For details see text.
This interaction model may serve as a simple ‘prototype’ for the mammalian multisynthetase complex. Apart from nine synthetases, this complex contains three non-catalytic components (p43, p38 and p18), all of which share homologies with Arc1p. The TRBD of Arc1p is conserved in p43, which has also been shown to be able to bind to tRNA and stimulate the activity of at least one enzyme of the complex (Quevillon et al., 1997; Park et al., 1999). The N-domain of Arc1p contains a motif that is present in both p18 and p38 as well as in the N-terminal appended domain of the bifunctional synthetase GluProRS, another component of the multisynthetase complex (Quevillon and Mirande, 1996; Quevillon et al., 1999). This domain, originally identified in the glutathione S-transferase superfamily, also occurs within human valyl-tRNA synthetase (ValRS), mouse cysteinyl-tRNA synthetase (CysRS), the putative GluRSs from S.pombe (ID: O13775) and Arabidopsis thaliana (Day et al., 1998), and the β and γ subunits of the eukaryotic elongation factor EF-1B (Koonin et al., 1994; our unpublished results). We have shown here that this motif represents a protein– protein interaction domain responsible for the Arc1p–GluRS association in yeast. This motif appears to function only in heterodimerization as neither GluRS nor the N-domain of Arc1p forms homo-oligomers in solution, as shown previously by their migration in gel-filtration columns (Deinert et al., 2001). Therefore, we also predict that in mammals this motif may play a role in the formation and stabilization of higher order aminoacyl-tRNA synthetase complexes. Indeed, this conserved motif most likely represents the protein interface between human ValRS and the δ subunit of the eukaryotic elongation factor 1 complex (Bec et al., 1994; Janssen et al., 1994). The occurrence of this motif in both tRNA-aminoacylation and protein translation factors may indicate the presence of a network of interactions that brings together and coordinates these two machineries. In several cases, interaction of an aminoacyl-tRNA synthetase with the translation elongation machinery has indeed been shown to offer a catalytic advantage (Reed et al., 1994; Negrutskii et al., 1996, 1999). However, it may also provide the means for subcellular spatial organization, i.e. co-localization of the tRNA- aminoacylation machinery with the sites of ongoing protein translation (Kisselev and Wolfson, 1994; Stapulionis and Deutscher, 1995; Negrutskii and El’skaya, 1998).
Aminoacylation of the tRNAs that come off the translating ribosomes in the cytoplasm is the major function of aminoacyl-tRNA synthetases. On the other hand, and according to recent data (Lund and Dahlberg, 1998; Nathanson and Deutscher, 2000), at least a minor pool of synthetases has to enter the nucleus in order to aminoacylate the newly synthesized tRNAs as a pre-condition for their nuclear export. Data from the yeast system have also pointed to a link between nuclear tRNA-aminoacylation and tRNA nuclear export. Inhibition of aminoacylation leads to accumulation of mature tRNAs inside the yeast nucleus (Sarkar et al., 1999; Grosshans et al., 2000). Furthermore, there is a synthetic lethal relationship between Arc1p and the yeast nuclear tRNA export receptor Los1p (Hellmuth et al., 1998), and mutations that reduce the nuclear pool of tyrosyl-tRNA synthetase (TyrRS) have recently been shown to impair tRNA nuclear export (Azad et al., 2001). This demonstration of minor intranuclear pools of aminoacyl-tRNA synthetases in both yeast and vertebrates raises the following question: how is this relative distribution of aminoacyl-tRNA synthetases between nucleus and cytoplasm controlled? As demonstrated in several cases of regulated nucleocytoplasmic transport (reviewed in Kaffman and O’Shea, 1999), the answer to this can be the formation of a complex that is actively excluded from the nucleus due to cytoplasmic retention, enhanced nuclear export or masking of nuclear localization signals.
Along these lines, our results show that the formation of the Arc1p–synthetase complex does indeed dictate the predominant subcellular distribution of its components. We have shown that Arc1p, MetRS or GluRS can enter the nucleus only upon the condition that the Arc1p–MetRS–GluRS complex has been disrupted, i.e. only as free proteins. Disruption of the complex was achieved either by removing the N-terminal domains of the enzymes or Arc1p in various combinations. In all cases except one (GFP–GluRS-ΔN3), a correlation was observed between the inability to form the complex and the occurrence of a significant nuclear pool (Table I). The most likely explanation of our results is that Arc1p, MetRS and GluRS contain nuclear localization signals that can drive at least a certain amount of these proteins inside the nucleus. Such signals have indeed been predicted in the case of Arc1p and GluRS (Schimmel and Wang, 1999). However, when the proteins are organized into a complex, as is the case under wild-type steady-state conditions, they are found only or predominantly in the cytoplasm. The observation that only very small amounts of GFP–GluRS-ΔN3 enter the nucleus, despite the lack of interaction with Arc1p, may be due to the fact that the longer truncation in this mutant removed a putative nuclear localization signal together with the Arc1p interaction domain. An alternative explanation is that the structural integrity of this mutant is affected, although this seems unlikely as this mutant is both stably expressed and functional in vivo. In the same vein, GluRS-ΔN2 may be less functional than GluRS-ΔN3 (Figure 3A) due to the former’s increased nuclear import and corresponding depletion of its cytoplasmic pool (Figure 6C). However, it should be mentioned that most of the conditions that disrupt the Arc1p ternary complex and subsequently lead to a change in the subcellular distribution of its components do not affect significantly the levels of in vivo aminoacylation or the rate of nuclear export of the cognate tRNAs (our unpublished data). This is not unexpected, as despite the increased nuclear pools of GluRS and MetRS, which should not disturb nuclear aminoacylation and efficient tRNA export, the major parts of GluRS and MetRS remain in the cytoplasm and can therefore aminoacylate efficiently the bulk of tRNA.
The formation of a ternary complex that is excluded from the nucleus represents a simple way of controlling the relative subcellular distribution of Arc1p, GluRS and MetRS. At equilibrium, and depending on the relative affinities between the components of the complex and their cellular abundance, a certain pool will be non-associated and therefore free to enter the nucleus, while the rest would be found in the cytoplasm as part of the ternary complex. Interference with complex formation would then disrupt the equilibrium in favour of dissociation and subsequent nuclear entry, as we have observed experimentally. The identification of the nuclear localization sequences of Arc1p, MetRS and GluRS, as well as the nature of the nuclear exclusion mechanism of this and other related tRNA-aminoacylation complexes, are certainly interesting subjects for future research.
Materials and methods
Yeast strains, media and plasmids
The yeast strains RS453 and arc1– have been described previously (Simos et al., 1996). The following previously described plasmids were used: pUN100 (CEN/ARS, LEU2) (Elledge and Davis, 1988); pRS314 (CEN/ARS, TRP1), pRS315 (CEN/ARS, LEU2), pRS316 (CEN/ARS, URA3) (Sikorski and Hieter, 1989); pASZ11 (CEN/ARS, ADE2) (Stotz and Linder, 1990); pRS315-PNOP1-GFP, pRS315-PNOP1-ProtA (Hellmuth et al., 1998); pUN100-MES1, pPP1-ProtA-GluRS (Simos et al., 1996); pEMBLyex4-His8-MetRS (Deinert et al., 2001). Plasmid pASZ11-PNOP1-GFP was constructed by subcloning the NOP1::GFP cassette from pRS315-PNOP1-GFP. Yeast cells were grown in rich YPD medium or synthetic SDC medium containing the necessary amino acids and nutrients. For counterselection of cells containing URA3 plasmids, 5-fluoroorotic acid (FOA; Toronto Research Chem.) was used at 1 µg/ml.
Construction of strains carrying deletions of the chromosomal genes coding for MetRS and GluRS
Deletion of the chromosomal genes coding for GluRS and MetRS (GUS1 and MES1, respectively) was achieved by PCR-mediated single-step targeted integration. The selectable marker was the S.pombe his5+ gene, which can substitute the S.cerevisiae HIS3 gene and allow growth of the RS453 strain on media lacking histidine (Longtine et al., 1998). This gene was amplified by PCR from plasmid pFA6a-His3MX6 using 70 nucleotide long primers that had 5′-ends (50 nt) annealing to the upstream or downstream regions of the GluRS or MetRS open reading frames and 3′-ends (20 nt) annealing to the marker gene. The products of the PCR reactions were used to transform the diploid strain RS453 and select for His+ transformants. The integration of the his5+ gene in the correct loci and generation of gus1::his5+ or mes1::his5+ heterozygotes were confirmed by PCR. Sporulation and tetrad dissection of the heterozygous diploid cells carrying the null alleles of MES1 and GUS1 demonstrated 2:2 segregation for viability and histidine auxotrophy, showing as expected that the products of MES1 and GUS1 are essential. To construct haploid cells harbouring a gus1 or mes1 knockout allele and rescued by plasmid-encoded GluRS or MetRS (shuffle strains), the heterozygous gus1::his5+ or mes1::his5+ strains were transformed with pRS316-URA3-GluRS or pRS316-URA3-MetRS, respectively, sporulated and tetrads dissected. Haploid gus1::his5+ or mes1::his5+ cells complemented by the corresponding wild-type genes on the URA3-containing plasmid exhibited no growth defect but died on 5-FOA-containing plates unless co-transformed with a plasmid encoding functional parts of GluRS or MetRS, respectively.
Miscellaneous procedures
Expression of the fusion proteins transformed in yeast cells was checked by western blotting of whole-cell extracts using polyclonal antibodies against the GFP moiety (Clontech), rabbit PAP (Dako) to detect the ProtA tag or antibodies against Arc1p (Simos et al., 1996). Affinity purification of ProtA fusion proteins was performed essentially as described previously (Simos et al., 1996, 1998). The localization of the GFP-tagged proteins in living yeast cells was examined in the fluorescein channel of a Zeiss Axioskop fluorescence microscope. Images were obtained with a Xillix Microimager CCD camera and processed with Improvison Openlab 1.7 software.
Confocal laser scanning microscopy was performed using a Leica TCS MP confocal microscope. Series of 0.5 µm sections were recorded. The images shown in Figures 6 and 7 correspond to the sections approximately in the middle of the cells.
Protein concentrations were determined using the Protein Assay reagent from Bio-Rad. SDS–PAGE, DNA manipulations (restriction digests, PCR amplifications, etc.), RNA extraction and northern analysis were performed according to standard protocols. Detection of tRNAs on northern blots was conducted using the following end-labelled synthetic DNA oligonucleotides complementary to tRNA specific sequences: tRNAGlu(UUC), 5′-CGGTCTCCACGGTGAAAGC-3′; elongator tRNAMet(CAU), 5′-TCGACCTTCAGAT-3′.
Supplementary data
Owing to space limitations, cloning and tagging of GluRS and MetRS by ProtA and GFP as well as the construction of plasmids expressing ProtA- and GFP-tagged truncated forms of MetRS, GluRS and Arc1p are described in detail in the Supplementary data, available at The EMBO Journal Online.
Acknowledgments
Acknowledgements
We would like to thank Dr H.D.Fahimi for providing access to the confocal laser scanning microscopy facility, Dr F.Fasiolo for providing the anti-MetRS antibody and Dr J.Lechner for performing the mass spectroscopic analysis. This work was supported by Deutsche Forschungsgemeinschaft Research Grant SI 661/2-1 (to G.S.).
References
- Agou F. and Mirande,M. (1997) Aspartyl-tRNA synthetase from rat: in vitro functional analysis of its assembly into the multisynthetase complex. Eur. J. Biochem., 243, 259–267. [DOI] [PubMed] [Google Scholar]
- Azad A.K., Stanford,D.R., Sarkar,S. and Hopper,A.K. (2001) Role of nuclear pools of aminoacyl-tRNA synthetases in tRNA nuclear export. Mol. Biol. Cell, 12, 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bec G., Kerjan,P. and Waller,J.P. (1994) Reconstitution in vitro of the valyl-tRNA synthetase-elongation factor (EF) 1βγδ complex. Essential roles of the NH2-terminal extension of valyl-tRNA synthetase and of the EF-1δ subunit in complex formation. J. Biol. Chem., 269, 2086–2092. [PubMed] [Google Scholar]
- Behrensdorf H.A., van de Craen,M., Knies,U.E., Vandenabeele,P. and Clauss,M. (2000) The endothelial monocyte-activating polypeptide II (EMAP II) is a substrate for caspase-7. FEBS Lett., 466, 143–147. [DOI] [PubMed] [Google Scholar]
- Cahuzac B., Berthonneau,E., Birlirakis,N., Guittet,E. and Mirande,M. (2000) A recurrent RNA-binding domain is appended to eukaryotic aminoacyl-tRNA synthetases. EMBO J., 19, 445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day I.S., Golovkin,M. and Reddy,A.S. (1998) Cloning of the cDNA for glutamyl-tRNA synthetase from Arabidopsis thaliana. Biochim. Biophys. Acta, 1399, 219–224. [DOI] [PubMed] [Google Scholar]
- Deinert K., Fasiolo,F., Hurt,E.C. and Simos,G. (2001) Arc1p organizes the yeast aminoacyl-tRNA synthetase complex and stabilizes its interaction with the cognate tRNAs. J. Biol. Chem., 276, 6000–6008. [DOI] [PubMed] [Google Scholar]
- Elledge S.J. and Davis,R.W. (1988) A family of versatile centromeric vectors designed for use in the sectoring-shuffle mutagenesis assay in Saccharomyces cerevisiae. Gene, 70, 303–312. [DOI] [PubMed] [Google Scholar]
- Filonenko V.V. and Deutscher,M.P. (1994) Evidence for similar structural organization of the multienzyme aminoacyl-tRNA synthetase complex in vivo and in vitro. J. Biol. Chem., 269, 17375–17378. [PubMed] [Google Scholar]
- Francklyn C., Musier-Forsyth,K. and Martinis,S.A. (1997) Aminoacyl-tRNA synthetases in biology and disease: new evidence for structural and functional diversity in an ancient family of enzymes. RNA, 3, 954–960. [PMC free article] [PubMed] [Google Scholar]
- Frugier M., Moulinier,L. and Giege,R. (2000) A domain in the N-terminal extension of class IIb eukaryotic aminoacyl-tRNA synthetases is important for tRNA binding. EMBO J., 19, 2371–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans H., Hurt,E. and Simos,G. (2000) An aminoacylation-dependent nuclear tRNA export pathway in yeast. Genes Dev., 14, 830–840. [PMC free article] [PubMed] [Google Scholar]
- Hellmuth K., Lau,D.M., Bischoff,F.R., Künzler,M., Hurt,E.C. and Simos,G. (1998) Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol., 18, 6374–6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibba M. and Soll,D. (2000) Aminoacyl-tRNA synthesis. Annu. Rev. Biochem., 69, 617–650. [DOI] [PubMed] [Google Scholar]
- Ibba M., Curnow,A.W. and Soll,D. (1997) Aminoacyl-tRNA synthesis: divergent routes to a common goal. Trends Biochem. Sci., 22, 39–42. [DOI] [PubMed] [Google Scholar]
- Janssen G.M., van Damme,H.T., Kriek,J., Amons,R. and Moller,W. (1994) The subunit structure of elongation factor 1 from Artemia. Why two α-chains in this complex? J. Biol. Chem., 269, 31410–31417. [PubMed] [Google Scholar]
- Kaffman A. and O’Shea,E.K. (1999) Regulation of nuclear localization: a key to a door. Annu. Rev. Cell Dev. Biol., 15, 291–339. [DOI] [PubMed] [Google Scholar]
- Kaminska M., Deniziak,M., Kerjan,P., Barciszewski,J. and Mirande,M. (2000) A recurrent general RNA binding domain appended to plant methionyl-tRNA synthetase acts as a cis-acting cofactor for aminoacylation. EMBO J., 19, 6908–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T., Park,S.G., Kim,J.E., Seol,W., Ko,Y.G. and Kim,S. (2000a) Catalytic peptide of human glutaminyl-tRNA synthetase is essential for its assembly to the aminoacyl-tRNA synthetase complex. J. Biol. Chem., 275, 21768–21772. [DOI] [PubMed] [Google Scholar]
- Kim Y., Shin,J., Li,R., Cheong,C., Kim,K. and Kim,S. (2000b) A novel anti-tumor cytokine contains a RNA-binding motif present in aminoacyl-tRNA synthetases. J. Biol. Chem., 275, 27062–27068. [DOI] [PubMed] [Google Scholar]
- Kisselev L. and Wolfson,A.D. (1994) Aminoacyl-tRNA synthetases from higher eukaryotes. Prog. Nucleic Acid Res. Mol. Biol., 48, 83–141. [DOI] [PubMed] [Google Scholar]
- Kleeman T.A., Wei,D., Simpson,K.L. and First,E.A. (1997) Human tyrosyl-tRNA synthetase shares amino acid sequence homology with a putative cytokine. J. Biol. Chem., 272, 14420–14425. [DOI] [PubMed] [Google Scholar]
- Knies U.E., Behrensdorf,H.A., Mitchell,C.A., Deutsch,U., Risau,W., Drexler,H.C. and Clauss,M. (1998) Regulation of endothelial monocyte-activating polypeptide II release by apoptosis. Proc. Natl Acad. Sci. USA, 95, 12322–12327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko Y.G., Kang,Y.S., Kim,E.K., Park,S.G. and Kim,S. (2000) Nucleolar localization of human methionyl-tRNA synthetase and its role in ribosomal RNA synthesis. J. Cell Biol., 149, 567–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin E.V., Mushegian,A.R., Tatusov,R.L., Altschul,S.F., Bryant,S.H., Bork,P. and Valencia,A. (1994) Eukaryotic translation elongation factor 1γ contains a glutathione transferase domain—study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci., 3, 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie,A.,III, Demarini,D.J., Shah,N.G., Wach,A., Brachat,A., Philippsen,P. and Pringle,J.R. (1998) Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast, 14, 953–961. [DOI] [PubMed] [Google Scholar]
- Lund E. and Dahlberg,S.G. (1998) Proofreading and aminoacylation of tRNAs before export from the nucleus. Science, 282, 2082–2085. [DOI] [PubMed] [Google Scholar]
- Martinis S.A., Plateau,P., Cavarelli,J. and Florentz,C. (1999a) Aminoacyl-tRNA synthetases: a family of expanding functions. EMBO J., 18, 4591–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinis S.A., Plateau,P., Cavarelli,J. and Florentz,C. (1999b) Aminoacyl-tRNA synthetases: a new image for a classical family. Biochimie, 81, 683–700. [DOI] [PubMed] [Google Scholar]
- Mirande M. (1991) Aminoacyl-tRNA synthetase family from prokaryotes and eukaryotes: structural domains and their implications. Prog. Nucleic Acid Res. Mol. Biol., 40, 95–142. [DOI] [PubMed] [Google Scholar]
- Morales A.J., Swairjo,M.A. and Schimmel,P. (1999) Structure-specific tRNA-binding protein from the extreme thermophile Aquifex aeolicus. EMBO J., 18, 3475–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson L. and Deutscher,M.P. (2000) Active aminoacyl-tRNA synthetases are present in nuclei as a high molecular weight multienzyme complex. J. Biol. Chem., 275, 31559–31562. [DOI] [PubMed] [Google Scholar]
- Negrutskii B.S. and El’skaya,A.V. (1998) Eukaryotic translation elongation factor 1α: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog. Nucleic Acid Res. Mol. Biol., 60, 47–78. [DOI] [PubMed] [Google Scholar]
- Negrutskii B.S., Budkevich,T.V., Shalak,V.F., Turkovskaya,G.V. and El’Skaya,A.V. (1996) Rabbit translation elongation factor 1α stimulates the activity of homologous aminoacyl-tRNA synthetase. FEBS Lett., 382, 18–20. [DOI] [PubMed] [Google Scholar]
- Negrutskii B.S., Shalak,V.F., Kerjan,P., El’skaya,A.V. and Mirande,M. (1999) Functional interaction of mammalian valyl-tRNA synthetase with elongation factor EF-1α in the complex with EF-1H. J. Biol. Chem., 274, 4545–4550. [DOI] [PubMed] [Google Scholar]
- Norcum M.T. and Warrington,J.A. (1998) Structural analysis of the multienzyme aminoacyl-tRNA synthetase complex: a three-domain model based on reversible chemical crosslinking. Protein Sci., 7, 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.G., Jung,K.H., Lee,J.S., Jo,Y.J., Motegi,H., Kim,S. and Shiba,K. (1999) Precursor of pro-apoptotic cytokine modulates aminoacylation activity of tRNA synthetase. J. Biol. Chem., 274, 16673–16676. [DOI] [PubMed] [Google Scholar]
- Popenko V.I., Ivanova,J.L., Cherny,N.E., Filonenko,V.V., Beresten,S.F., Wolfson,A.D. and Kisselev,L.L. (1994) Compartmentalization of certain components of the protein synthesis apparatus in mammalian cells. Eur. J. Cell Biol., 65, 60–69. [PubMed] [Google Scholar]
- Quevillon S. and Mirande,M. (1996) The p18 component of the multisynthetase complex shares a protein motif with the β and γ subunits of eukaryotic elongation factor 1. FEBS Lett., 395, 63–67. [DOI] [PubMed] [Google Scholar]
- Quevillon S., Agou,F., Robinson,J.C. and Mirande,M. (1997) The p43 component of the mammalian multi-synthetase complex is likely to be the precursor of the endothelial monocyte-activating polypeptide II cytokine. J. Biol. Chem., 272, 32573–32579. [DOI] [PubMed] [Google Scholar]
- Quevillon S., Robinson,J.C., Berthonneau,E., Siatecka,M. and Mirande, M. (1999) Macromolecular assemblage of aminoacyl-tRNA synthetases: identification of protein–protein interactions and characterization of a core protein. J. Mol. Biol., 285, 183–195. [DOI] [PubMed] [Google Scholar]
- Reed V.S. and Yang,D.C. (1994) Characterization of a novel N-terminal peptide in human aspartyl-tRNA synthetase. Roles in the transfer of aminoacyl-tRNA from aminoacyl-tRNA synthetase to the elongation factor 1α. J. Biol. Chem., 269, 32937–32941. [PubMed] [Google Scholar]
- Reed V.S., Wastney,M.E. and Yang,D.C.H. (1994) Mechanisms of the transfer of aminoacyl-tRNA from aminoacyl-tRNA synthetase to the elongation factor 1α. J. Biol. Chem., 269, 32932–32936. [PubMed] [Google Scholar]
- Renault L. et al. (2001) Structure of the EMAPII domain of human aminoacyl-tRNA synthetase complex reveals evolutionary dimer mimicry. EMBO J., 20, 570–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho S.B., Lee,K.H., Kim,J.W., Shiba,K., Jo,Y.J. and Kim,S. (1996) Interaction between human tRNA synthetases involves repeated sequence elements. Proc. Natl Acad. Sci. USA, 93, 10128–10133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rho S.B., Kim,M.J., Lee,J.S., Seol,W., Motegi,H., Kim,S. and Shiba,K. (1999) Genetic dissection of protein–protein interactions in multi-tRNA synthetase complex. Proc. Natl Acad. Sci. USA, 96, 4488–4493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Azad,A.K. and Hopper,A.K. (1999) Nuclear tRNA aminoacylation and its role in nuclear export of endogenous tRNAs in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 96, 14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schimmel P. and Wang,C.C. (1999) Getting tRNA synthetases into the nucleus. Trends Biochem. Sci., 24, 127–128. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,R. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simos G. and Hurt,E. (1999) Transfer RNA biogenesis: a visa to leave the nucleus. Curr. Biol., 9, R238–R241. [DOI] [PubMed] [Google Scholar]
- Simos G., Segref,A., Fasiolo,F., Hellmuth,K., Shevchenko,A., Mann,M. and Hurt,E.C. (1996) The yeast protein Arc1p binds to tRNA and functions as a cofactor for methionyl- and glutamyl-tRNA synthetases. EMBO J., 15, 5437–5448. [PMC free article] [PubMed] [Google Scholar]
- Simos G., Sauer,A., Fasiolo,F. and Hurt,E.C. (1998) A conserved domain within Arc1p delivers tRNA to aminoacyl-tRNA synthetases. Mol. Cell, 1, 235–242. [DOI] [PubMed] [Google Scholar]
- Stapulionis R. and Deutscher,M.P. (1995) A channeled tRNA cycle during mammalian protein synthesis. Proc. Natl Acad. Sci. USA, 92, 7158–7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz A. and Linder,P. (1990) The ADE2 gene from Saccharomyces cerevisiae: sequence and new vectors. Gene, 95, 91–98. [DOI] [PubMed] [Google Scholar]
- Swairjo M.A., Morales,A.J., Wang,C.C., Ortiz,A.R. and Schimmel,P. (2000) Crystal structure of trbp111: a structure-specific tRNA-binding protein. EMBO J., 19, 6287–6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K. and Schimmel,P. (1999) Two distinct cytokines released from a human aminoacyl-tRNA synthetase. Science, 284, 147–151. [DOI] [PubMed] [Google Scholar]
- Walter P., Weygand-Durasevic,I., Sanni,A., Ebel,J.-P. and Fasiolo,F. (1989) Deletion analysis in the amino-terminal extension of methionyl-tRNA synthetase from Saccharomyces cerevisiae shows that a small region is important for the activity and stability of the enzyme. J. Biol. Chem., 264, 17126–17130. [PubMed] [Google Scholar]
- Wang C.C. and Schimmel,P. (1999) Species barrier to RNA recognition overcome with nonspecific RNA binding domains. J. Biol. Chem., 274, 16508–16512. [DOI] [PubMed] [Google Scholar]
- Weiner A.M. and Maizels,N. (1999) A deadly double life. Science, 284, 63–64. [DOI] [PubMed] [Google Scholar]
- Weygand-Durasevic I., Lenhard,D., Filipic,S. and Söll,D. (1996) The C-terminal extension of yeast seryl-tRNA synthetase affects stability of the enzyme and its substrate affinity. J. Biol. Chem., 271, 2455–2461. [DOI] [PubMed] [Google Scholar]
- Whelihan E.F. and Schimmel,P. (1997) Rescuing an essential enzyme–RNA complex with a non-essential appended domain. EMBO J., 16, 2968–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R., Olsen,G.J., Ibba,M. and Soll,D. (2000) Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev., 64, 202–236. [DOI] [PMC free article] [PubMed] [Google Scholar]