Abstract
The MYC protooncogene is frequently deregulated in human cancers. Here, by screening a kinase-directed library of small inhibitory RNAs, we identify glycogen synthase kinase 3β (GSK3β) as a gene whose inactivation potentiates TNF-related apoptosis-inducing ligand death receptor-mediated apoptosis specifically in MYC-overexpressing cells. Small inhibitory RNA-induced silencing of GSK3β prevents phosphorylation of MYC on T58, thereby inhibiting recognition of MYC by the E3 ubiquitin ligase component FBW7. Attenuating the GSK3β–FBW7 axis results in stabilization of MYC, up-regulation of surface levels of the TNF-related apoptosis-inducing ligand death receptor 5, and potentiation of death receptor 5-induced apoptosis in vitro and in vivo. These results identify GSK3β and FBW7 as potential cancer therapeutic targets and MYC as a critical substrate in the GSK3β survival-signaling pathway. The results also demonstrate paradoxically that MYC-expressing tumors might be treatable by drug combinations that increase rather than decrease MYC oncoprotein function.
Keywords: FBW7, MYC, TNF-related apoptosis-inducing ligand
The MYC protooncogene encodes a basic helix–loop–helix leucine zipper transcription factor that plays a central role in promoting the development of many human cancers (1, 2). In addition to driving cancer cell growth and proliferation, MYC is also capable of sensitizing cells to apoptosis (3, 4), an activity that we recently proposed might be exploitable pharmacologically to selectively kill MYC-expressing tumors (5). We showed that MYC activation results in up-regulation of cell surface levels of death receptor 5 (DR5; also known as TRAIL-R2, TNFRSF10B), an apoptosis-inducing receptor for the cytokine TNF-related apoptosis-inducing ligand (TRAIL) (reviewed in refs. 6 and 7). MYC-overexpressing cells can therefore be killed preferentially over isogenic normal cells by agonists of DR5 apoptotic signaling. This MYC-induced apoptotic sensitivity may be a primary mechanism underlying TRAIL's unusual ability, unique among the TNF family of death ligands, to induce apoptosis in tumor cells preferentially over normal cells (5, 8–11).
Recombinant human TRAIL and agonistic antibodies against its two death-inducing receptors, DR4 and DR5, are currently undergoing development as cancer therapeutics. However, because many tumors, including MYC-expressing tumors, are resistant or only weakly sensitive to their effects (10), it would be desirable to identify agents that potentiate TRAIL-induced apoptosis. Here, to this end, we screened a library of small inhibitory RNAs (siRNAs) directed primarily against the protein kinase superfamily to identify genes whose inactivation potentiates DR5-mediated apoptosis specifically in MYC-expressing cells. This screen can be thought of as a sensitized synthetic lethal genetic screen (12, 13) in which the phenotypic output, lethality, is sensitized not only by a genetic alteration, MYC activation, but also by an environmental condition, i.e., by the presence of a suboptimal dose of DR5 agonistic antibody. Among the genes identified in this screen was glycogen synthase kinase 3β (GSK3β), which plays a central role in a wide variety of physiological and pathological processes (14, 15). We show that GSK3β's ability to phosphorylate MYC and target it for recognition by the E3 ubiquitin ligase component and tumor suppressor FBW7 (also known as FBXW7, hCDC4, AGO, and SEL10) underlies its ability to potentiate apoptosis selectively in MYC-expressing cells. These results identify the GSK3β–FBW7 axis as a potential MYC-specific cancer therapeutic target and demonstrate a counterintuitive approach toward cancer therapy, that of potentiating apoptosis by increasing rather than decreasing oncogene function.
Methods
Cell Lines. Genetically defined immortalized HA1E cells were obtained from Robert Weinberg (Whitehead Institute, Cambridge, MA), HCT116 and its FBW7+/- and FBW7-/- derivatives were obtained from Bert Vogelstein (The Johns Hopkins University, Baltimore), and HT115 cells were obtained from the European Collection of Cell Cultures (Salisbury, U.K.).
siRNA Library Screening. siRNAs were arrayed in 384-well microtiter plates in duplicate at 8 ng per well (8). For each 384-well plate the signal was normalized by dividing each well by the average of 24 wells on the same plate containing siRNAs against luciferase (siGL3) and multiplying by 100 to obtain normalized viability (percent viability).
Viability Assays. Cells (8,000–10,000 per well) were plated in 96-well plates in the appropriate cell culture medium, incubated with LiCl orKClfor8hortransfected with siRNAs, and treated with DR5-A 48 h after transfection for an additional 20 h. Cell viability was measured in triplicate by CellTiterGlo (Promega) according to the manufacturer's instructions.
Supporting Information. Further details are available in Supporting Methods, which is published as supporting information on the PNAS web site.
Results
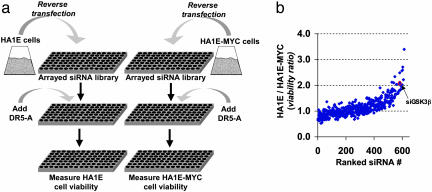
An siRNA Screen Identifies GSK3β as a MYC-Dependent Potentiator of DR5-Mediated Apoptosis. To identify genes that modulate cell survival in a MYC-dependent fashion, we screened an arrayed library of 624 largely kinase-directed siRNAs (8) for their ability to differentially potentiate DR5-mediated apoptosis in an isogenic pair of cell lines differing only in their level of MYC expression (Fig. 1a). The siRNA library was reverse-transfected into the immortalized but nontransformed kidney epithelial cell line HA1E or into its derivative, HA1E-MYC, which ectopically expresses MYC from an integrated retrovirus (5). The siRNA-transfected cells were assayed for sensitivity to DR5-A, an agonistic monoclonal antibody specific for DR5 (5, 16). Thirteen of the 624 siRNAs sensitized the MYC-expressing cells to DR5-A >2-fold over their non-MYC-expressing counterparts (Fig. 1b and Table 1, which is published as supporting information on the PNAS web site). We further characterized the function of one of the genes identified in this screen, GSK3β for several reasons: (i) A second siRNA targeting GSK3β present in the library also potentiated DR5-A sensitivity in MYC-expressing cells; (ii) GSK3β has a recently reported and testable relationship with MYC; and (iii) GSK3β was readily validated in these assays by various strategies as described below.
Fig. 1.
siRNA screen for genes synthetically lethal with MYC activation in the presence of suboptimal doses of DR5 agonists. (a) Screen schematic. Arrayed siRNAs were reverse-transfected into HA1E or HA1E-MYC cells and assayed for sensitivity to DR5-A (see Methods). (b) Genotype-dependent effects of siRNAs on cell viability. siRNAs were ranked in increasing order on the x axis by their ability to sensitize HA1E-MYC cells to DR5-A-induced apoptosis relative to the sensitization of HA1E cells (see Supporting Methods). The siRNA for GSK3β (siGSK3β) is indicated.
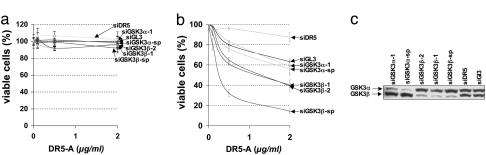
To confirm that depletion of GSK3β potentiates apoptosis specifically in MYC-expressing cells, we repeated the cell viability assays with multiple nonoverlapping siRNAs directed against GSK3β or against the related gene GSK3α across a range of DR5-A concentrations (Fig. 2 a and b). At all DR5-A concentrations tested, multiple siRNAs targeting GSK3β sensitized the MYC-overexpressing HA1E-MYC cells to the action of DR5-A (Fig. 2b) and had little or no effect on the isogenic parental cell line (Fig. 2a). siRNAs diminishing the GSK3α isoform failed to sensitize HA1E or HA1E-MYC cells, despite reducing target protein expression to a similar extent as the GSK3β siRNAs (Fig. 2c).
Fig. 2.
Depletion of GSK3β but not GSK3α potentiates DR5-mediated apoptosis specifically in MYC-overexpressing cells. (a and b) Multiple siRNAs targeting nonoverlapping sequences of GSK3β (siGSKβ) or GSK3α (siGSKα) were reverse-transfected into HA1E or HA1E-MYC cells and assayed for sensitivity to DR5-A. Numbers designate distinct siRNAs, and sp denotes a “smartpool” of four distinct siRNAs. Error bars indicate standard deviations of triplicate measurements. siGL3 were used as controls. (c) siRNA efficacy was determined by Western blot analysis using antibodies specific for each siRNA target.
The commonly used GSK3 inhibitor LiCl (15) also sensitized HA1E-MYC cells, but not HA1E cells, to DR5-A in a concentration-dependent manner (Fig. 8, which is published as supporting information on the PNAS web site). Substitution of KCl for LiCl to control for changes in osmolarity did not significantly influence DR5-A sensitivity in either cell line. The small-molecule GSK3 inhibitor 6-bromoindirubin-3-oxime (17) similarly sensitized HA1E-MYC cells (Fig. 9, which is published as supporting information on the PNAS web site) but not HA1E cells (data not shown). Although the chemical inhibitors do not distinguish between the α and β GSK3 isoforms, these data, when combined with the siRNA results presented above, provide strong evidence that reducing or inhibiting the GSK3β isoform potentiates DR5-mediated apoptosis specifically in MYC-overexpressing cells.
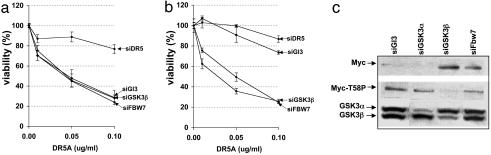
GSK3β Regulates Sensitivity to DR5-Mediated Apoptosis by Means of Phosphorylation of MYC T58. MYC is a known GSK3 substrate, being phosphorylated at threonine residue T58, which negatively regulates MYC protein stability (18, 19). To determine whether MYC T58 phosphorylation mediates GSK3β's prosurvival activity, we tested the effects of a MYC allele, MYCT58A, in which T58 was mutated to a nonphosphorylatable alanine residue. MYCT58A was significantly more potent than WT MYC in sensitizing HA1E cells to DR5-A-induced apoptosis, sensitizing to a similar extent as the combination of WT MYC plus siRNA against GSK3β (Fig. 3 a and b and Fig. 10 a and b, which is published as supporting information on the PNAS web site). Furthermore, GSK3β siRNAs were unable to further sensitize cells expressing the MYCT58A mutant to DR5-A (Fig. 3a), in striking contrast to similar experiments performed with WT MYC (Fig. 3b).
Fig. 3.
GSK3β regulates sensitivity to DR5-mediated apoptosis via phosphorylation of MYC T58. (a and b) Mutation of the GSK3β MYC T58 phosphorylation site mimics GSK3β or FBW7 loss of function. Derivatives of HA1E cells expressing retrovirally transduced WT MYC (b) or the T58 phosphorylation site mutant MYCT58A (a) were transfected with the indicated siRNAs and treated with DR5-A and assayed for viability (see Fig. 10 a and b for comparison with higher DR5-A concentrations). (c) In HA1E-MYC cells transfected with siRNAs directed against GSK3α (siGSK3α), GSK3β (siGSK3β), or FBW7 (siFBW7), MYC protein levels and T58 phosphorylation status were determined by Western blot analysis using MYC or MYCT58-phospho-specific antibodies. By densitometry, siRNAs directed against GSK3α, GSK3β, and FBW7 reduced MYC T58 phosphorylation by 1.7-fold, >100-fold, and 1.5-fold, respectively, relative to siGl3.
Importantly, whereas siRNA against GSK3β strongly reduced MYC T58 phosphorylation and increased the amount of total cellular MYC protein in HA1E-MYC cells, siRNA-mediated knockdown of GSK3α had only a modest influence on MYC phosphorylation and did not result in MYC accumulation (Fig. 3c). Thus, the ability of siRNAs against the two GSK3 isoforms to prevent MYC T58 phosphorylation and stabilize MYC correlates with their ability to potentiate MYC-dependent apoptosis. Collectively, these data identify MYC as a substrate specifically for the GSK3β isoform, and strongly argue for a model in which inhibition of GSK3β enhances sensitivity to DR5 agonists by preventing phosphorylation of MYC at T58.
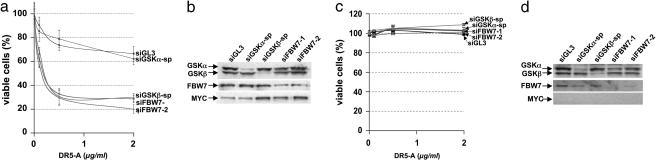
The GSK3β–FBW7 MYC Degradation Pathway Determines Sensitivity to DR5-A-Induced Apoptosis. Phosphorylation of MYC at T58 by GSK3β initiates a series of posttranslational modifications that allows it to be recognized and targeted for degradation by FBW7, a tumor-suppressor protein and F-box-containing component of the SCF (Skp–Cullin–F-box) ubiquitin ligase complex (20–22). To determine whether FBW7-dependent degradation of MYC plays a role in the control of cell survival mediated by GSK3β, we tested whether suppression of FBW7 function would mimic depletion of GSK3β. siRNAs targeting FBW7 and GSK3β increased MYC protein levels and enhanced sensitivity to DR5-A to a similar extent in HA1E-MYC cells (Fig. 4 a and b) but had little effect in HA1E cells (Fig. 4 c and d), suggesting that GSK3β and FBW7 act in a common pathway regulating MYC stability and apoptosis. Consistent with this idea, a functional siRNA against FBW7, like those against GSK3β, failed to further sensitize HA1E cells expressing stabilized MYC (HA1E-MYCT58A) (Fig. 3a).
Fig. 4.
The GSK3β–FBW7 MYC degradation pathway determines sensitivity to DR5-A-induced apoptosis. (a and c) HA1E-MYC (a) and HA1E (c) cells were transfected with one of two nonoverlapping siRNAs targeting the MYC ubiquitination component FBW7 (siFBW7-1 or siFBW7-2) and assayed for sensitivity to DR5-A. (b and d) siRNA efficacy in HA1E-MYC (b) and HA1E (d) cells was determined by Western blot analysis. Note that depletion of FBW7 or GSK3β, but not GSK3α, results in increased MYC protein levels in only HA1E-MYC cells.
Notably, both GSK3β and FBW7 siRNAs enhanced MYC protein levels in HA1E-MYC cells to a similar extent as the chemical GSK3 inhibitors (compare Fig. 4b with Fig. 9b). Whereas siRNAs targeting GSK3β prevented MYC T58 phosphorylation, FBW7 specific siRNAs did not (Fig. 3c), in accordance with observations that FBW7 functions downstream of GSK3β-dependent MYC T58 phosphorylation (21, 22). Combined with our previous observations that MYC overexpression sensitizes cells to DR5-mediated apoptosis (5), our results suggest that stabilization and accumulation of MYC protein resulting from the inactivation of the GSK3β/FBW7 axis is responsible for the hypersensitivity of MYC-expressing cells to DR5-A-induced apoptosis.
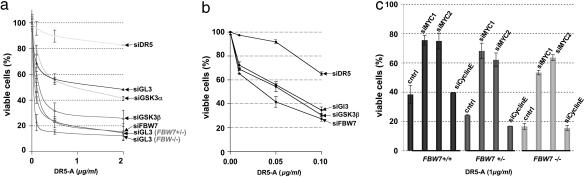
Mutation of the FBW7 Tumor Suppressor Enhances Apoptotic Sensitivity in MYC-Expressing Tumor Cells. The experiments presented thus far were performed in derivatives of normal primary cells engineered to overexpress MYC. Importantly, these results could also be recapitulated in human tumor-derived cell lines that endogenously express MYC. Like HA1E-MYC cells, the human colon carcinoma cell line HCT116 could be sensitized to DR5-A by siRNAs against GSK3β and FBW7, but not GSK3α (Fig. 5a and Fig. 11a, which is published as supporting information on the PNAS web site). Furthermore, derivatives of HCT116, in which one or both copies of FBW7 were disrupted by homologous recombination (FBW7+/- and FBW7-/-, respectively) (Fig. 11b) (23), were hypersensitive to DR5-A when compared with the parental HCT116 line (Fig. 5a). In FBW7-/- cells, GSK3β siRNA was unable to further increase apoptotic sensitivity (Fig. 5b and Fig. 12, which is published as supporting information on the PNAS web site), confirming that, in these cells, GSK3β and FBW7 function in a linear pathway to control the cellular response to DR5 signaling. Interestingly, both heterozygous and homozygous disruption of FBW7 strongly enhanced apoptosis by DR5-A (Fig. 5a), consistent with the observation that FBW7 can act as a haploinsufficient tumor-suppressor gene (24).
Fig. 5.
Depletion or mutation of the FBW7 tumor suppressor enhances DR5-A sensitivity in tumor-derived cell lines in a MYC-dependent manner. (a) The colon carcinoma-derived cell line, HCT116, was transfected with the indicated siRNAs as previously described, and sensitivity to DR5-A was compared with genetically engineered HCT116 cells (see Methods) heterozygous (+/-) or homozygous (-/-) for FBW7. Confirmation of the efficacy of siRNA-mediated knockdown and of FBW7 genotypic status is shown in Fig. 9. (b) HCT116 FBW7-/- cells were transfected with siRNAs and analyzed as in a with lower DR5-A concentrations (see Fig. 12 for comparison with higher DR5-A concentrations). (c) HCT116 FBW7+/+, FBW7+/-, and FBW7-/- cells were transfected with control siRNAs (siGL3), two distinct nonoverlapping siRNAs targeting MYC (siMYC1 and siMYC2), and an siRNA specific for cyclin E (siCyclinE). After 48 h, transfected cells were treated with DR5-A for 20 h, and cell viability was determined.
Similarly, the colon carcinoma cell line HT115, which carries a naturally occurring heterozygous mutation in FBW7 was highly sensitive to DR5-A (Fig. 13, which is published as supporting information on the PNAS web site). The mutation present in HT115, C1153T, is among the most frequently occurring FBW7 mutations found in colon cancer (23), resulting in a mutated arginine residue (R465) critical for substrate recognition (23, 25). Notably, sensitivity to DR5-A could be reversed by stable overexpression of a WT FBW7 cDNA in HT115 cells (Fig. 13).
In HCT116 and HT115 cells, as in HA1E-MYC cells, the increased sensitivity to DR5-A caused by FBW7 mutation critically depended on MYC levels, given that knockdown of MYC by siRNA could suppress DR5-A-induced apoptosis in these cell lines (Fig. 5c and Fig. 14, which is published as supporting information on the PNAS web site). Qualitatively similar rescue in the HCT116 series of cell lines was also obtained if MYC function was reduced by stable retroviral expression of a dominant negative MYC allele (26) (Fig. 15, which is published as supporting information on the PNAS web site). In contrast, an siRNA that reduced levels of cyclin E, another substrate of GSK3 (27) and FBW7 (28), had no significant effects on cell viability (Fig. 5c), despite strongly suppressing micronuclei formation (a proxy for chromosomal instability) induced by FBW7 disruption (Fig. 16, which is published as supporting information on the PNAS web site) as reported in ref. 23. Because MYC siRNAs had no influence on micronuclei formation in FBW7-depleted cells (Fig. 16), our data delineate the function of two critical FBW7 targets, MYC and cyclin E: In FBW7-depleted cells, the resulting increase in DR5-A sensitivity depends on MYC stabilization, whereas the increase in chromosomal instability depends on cyclin E stabilization.
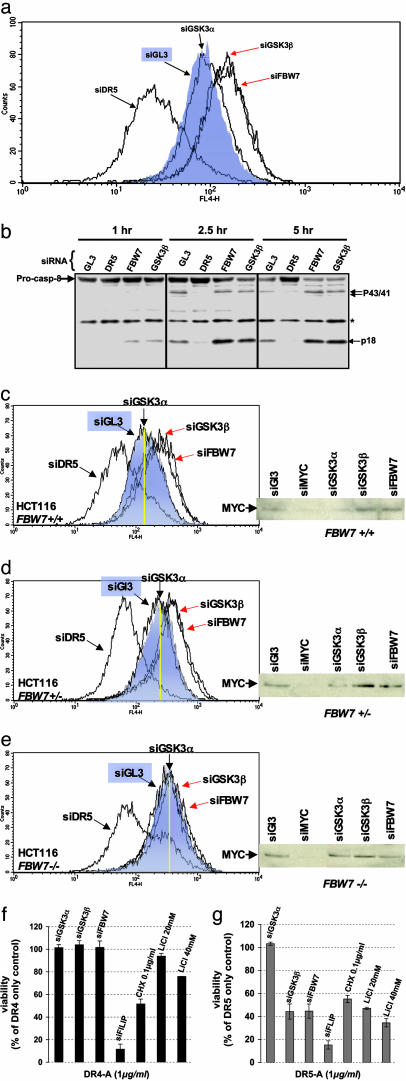
Inactivation of the GSK3β–FBW7 Axis Increases DR5 Receptor Levels and Stimulates DR5-Dependent Caspase-8 Processing. Previously, we showed that ectopic expression of MYC induces the up-regulation of DR5 receptor on the cell surface and subsequent sensitization to apoptosis induced by DR5 agonists (5). Thus, we asked whether the additional stabilization of MYC caused by FBW7 or GSK3β depletion could further influence surface DR5 receptor levels. Transfection of HA1E-MYC cells with siRNAs against GSK3β and FBW7 resulted in increased levels of DR5 receptor relative to cells transfected with control or GSK3α siRNAs (Fig. 6a).
Fig. 6.
Inactivation of the GSK3β–FBW7 axis increases DR5 receptor levels and stimulates DR5-dependent caspase-8 processing in MYC-expressing cells. (a) DR5 cell surface expression was measured by flow cytometry in HA1E-MYC cells 48 h after siRNA transfection with the denoted siRNAs. (b) Caspase-8 activation was monitored by proteolytic processing of the caspase-8 zymogen [pro-form, arrow labeled Pro-casp-8 (p55/53); activated form, arrow labeled p18; transient p43/41 form, arrow labeled p43/41]. *, nonspecific band. (c–e) HCT116 FBW7+/+ (c), FBW7+/- (d), and FBW7-/- (e) cells were examined for cell surface DR5 levels by FACS analysis. For comparison, each cell line was transfected with siRNAs specific for DR5, GL3 (control shaded in blue), GSK3α, GSK3β, and FBW7.(Right) Correlation to MYC protein levels. (Left) The yellow line in each FACS panel denotes the control levels of DR5 receptor, which increases with decreasing FBW7 gene dosage. (f and g) FBW7 or GSK3β depletion specifically enhances apoptosis induced through DR5 (f) but not DR4 (g). Apoptosis was induced in siRNA-transfected or compound-treated HA1E-MYC cells of an agonistic antibody against DR4 (f) or DR5 (g). Note that treatments disrupting the GSK3β–FBW7 axis sensitize specifically to DR5-A, whereas depletion of FLIP or treatment with the translation inhibitor cycloheximide (CHX) sensitize to DR4- and DR5-mediated apoptosis. Data were normalized to antibody-treated cells transfected with a control siRNA (siGL3). Treatment with the DR4 and DR5 antibodies alone resulted in ≈5% and ≈30% reduction in viability under the conditions used for these assays.
In contrast, an siRNA targeting DR5 decreased cell surface DR5 expression, confirming the specificity of the reagents used in these assays. Furthermore, increased DR5 receptor levels in GSK3β- or FBW7-depleted cells were associated with increased activation and proteolytic processing of caspase-8 upon DR5 stimulation (Fig. 6b), thus correlating increased DR5 receptor levels to increased functional apoptotic signaling. GSK3β and FBW7 siRNAs had no detectable influence on DR5 receptors in HA1E cells (data not shown), suggesting that disrupting the GSK3β/FBW7-dependent MYC degradation process in cells with low MYC expression is not sufficient to enhance DR5 cell surface expression and subsequent sensitization to DR5 agonists.
Similar increases in DR5 surface levels were observed in HCT116 cancer cells upon transfection with GSK3β and FBW7 siRNAs (Fig. 6c). Furthermore, incrementally reducing FBW7 gene dosage by heterozygous or homozygous mutation in HCT116 resulted in increasing DR5 cell surface levels with decreasing FBW7 gene dosage (Fig. 6 d and e). Notably, in HCT116 FBW7+/+ and FBW7+/- cells, DR5 cell surface expression could be further increased by transfection of siRNAs against GSK3β or FBW7 (Fig. 6 c and d). However, no further increases could be observed after introduction of these siRNAs into HCT116 FBW7-/- cells devoid of functional FBW7 (Fig. 6e). These results demonstrate that GSK3β regulates DR5 receptor levels only in the presence of FBW7 and, thus, that GSK3β acts in a linear pathway with FBW7 to regulate DR5 receptor levels.
Although treatment with the GSK3 inhibitor LiCl or with siRNAs against GSK3β or FBW7 sensitized HCT116 cells to DR5-A, they had little effect on sensitivity to an agonistic antibody against the DR4 TRAIL receptor (Fig. 6, compare f and g). In contrast, treatment with the general enhancer of death receptor-induced apoptosis, cycloheximide, or with an siRNA against FLIP (a gene that inhibits caspase-8 activity and, thus, attenuates the DR4 and DR5 signaling pathways) sensitized cells similarly to DR4 or DR5 agonistic antibodies. These data suggest that the sensitization to apoptosis provoked by inactivation of the GSK3β–FBW7 axis is highly specific for DR5 apoptotic signaling and likely occurs at least in part at the level of DR5 itself, although other mechanisms may also play a role [e.g., see Ricci et al. (10)].
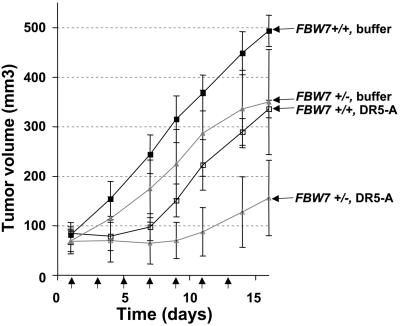
The GSK3β–FBW7 Axis as a Potential Therapeutic Target. Our data suggest that targeting the GSK3–FBW7 axis in MYC-expressing cells might be an effective therapeutic strategy in combination with DR5 agonists. To test this idea in vivo, we grew HCT116 FBW7+/+ and FBW7+/- cells as s.c. xenografted tumors in nude mice. HCT116 FBW7-/- cells grew poorly in vivo and were excluded from the study. When tumors reached a mean volume of ≈80 mm3, tumor-bearing mice were treated with 100 μg of DR5-A every other day for a total of seven treatments or with buffer only as a control. Upon completion of the treatment regimen, the FBW7+/- tumors exhibited a significantly greater response to DR5-A (Fig. 7), with a mean tumor volume of 88 mm3 versus 223 mm3 for the HCT116 FBW7+/+ tumors (P = 0.0087), suggesting that targeting GSK3 or FBW7 may be effective as a combination therapy with DR5 agonists.
Fig. 7.
Response of FBW7+/+ and FBW7+/- tumor xenografts to DR5-A therapy in vivo. HCT116 FBW7+/+ or FBW7+/- cells (n = 3 × 106 each) were implanted in the flank of BALB/c nude mice (see Methods). At a tumor volume of 80 mm3, four animals per group were i.p. injected with 100 μg of DR5-A or buffer every other day for a total of seven treatments. Tumor sizes were measured with calipers in three dimensions. Arrows indicate the days of treatment. Error bars represent standard deviations.
Although we were not able to completely eradicate the FBW7+/- tumors, it should be noted that the increased treatment efficacy observed for the FBW7+/- tumors was a result of only a 50% reduction in FBW7 gene dosage. Furthermore, the HCT116 cell line is known to be defective in mismatch repair and to rapidly evolve resistance to TRAIL pathway agonists in vivo because of the acquisition of frameshift mutations in the BAX gene [LeBlanc et al. (29) and our unpublished data]. Nevertheless, depletion of FBW7, in vitro and in vivo, enhanced sensitivity to DR5 agonists. Because restoration of FBW7 tumor suppressor function may not be therapeutically feasible, our data may highlight an alternative strategy of exploiting enhanced sensitivity to DR5 agonists for targeting cancer cells with defective FBW7 function.
Discussion
The development of large-scale libraries of siRNAs directed against significant subsets of the genome (30–32) or against biologically interesting and therapeutically relevant gene families (8, 33) has ushered in an era in which the functions of large numbers of genes can be systematically interrogated based on their loss-of-function phenotypes. We and others (34–37) have proposed that one particularly rewarding application of this technology may be in identifying synthetic lethal interactions with genes known to play central roles in cancer cell proliferation or survival. Here we demonstrate the potential of such synthetic lethal screens to identify cancer drug targets, such as GSK3β and FBW7, a pair of genes mediating the ubiquitin-dependent degradation of MYC, as genes whose inactivation potentiates apoptosis specifically in MYC-overexpressing cells.
Implications for Cancer Therapy. The sensitization to DR5-mediated apoptosis resulting from suppression of the GSK3β–FBW7 axis has important implications for how DR5 agonists might be used in the clinic. Mutations at the T58 GSK3β phosphorylation site on MYC are frequently observed in Burkitt's lymphoma (38) and have been shown to inhibit its degradation by the proteasome (39), whereas inactivating FBW7 mutations have been reported in a variety of solid tumor types, including breast, ovarian, endometrial, pancreatic, and colorectal carcinomas (23, 40, 41). Such mutations can clearly act as determinants of the response to DR5-A treatment when experimentally introduced into cells (Figs. 3a and 5a). Thus, it will be of interest to determine whether their presence can be used to prospectively identify those patients who might benefit most from DR5 agonist treatment.
Small-molecule GSK3 inhibitors are currently under development for several indications, including diabetes, neurodegenerative diseases, and bipolar disorder (15). By indirectly blocking MYC degradation, such inhibitors may be safe (see below) and effective against MYC-overexpressing cancers, particularly when combined with DR5 agonists or other therapeutics that may act at least in part by harnessing TRAIL signaling pathways, such as histone deacetylase inhibitors, interferons, retinoids, and arsenic trioxide (42–46). Similarly, direct chemical inhibition of FBW7 functional activity may also be effective, although, to date, small-molecule antagonists of SCF ubiquitin ligase complexes, although currently under investigation, have yet to be clinically tested.
Employing Synthetic Lethality to Elucidate Tumor-Biased Cell Death Pathways. A synthetic lethal relationship between two genes frequently reflects an underlying biochemical interaction between the proteins encoded by the genes (12). Thus, in uncovering a genetic relationship between GSK3β and MYC, we identified MYC as a critical substrate mediating GSK3β's poorly understood cell survival function. Combining this knowledge with previous work from several laboratories, including our own (5, 18, 19, 21, 22), we propose a model for how GSK3β might regulate DR5-mediated apoptosis under conditions in which DR5 agonist levels are limiting or in weakly sensitive MYC-positive tumor cells. By phosphorylating MYC at T58, GSK3β targets MYC for FBW7-dependent ubiquitination and degradation, thereby keeping MYC levels low and cell surface DR5 levels below a threshold necessary for activation. In this model, the GSK3β–FBW7 axis can be thought of as a buffer that restrains the phenotypic consequences of MYC activation (12). Disruption of this buffering system in MYC-expressing cells, either by mutation or pharmacological intervention, thus unleashes MYC's effects, which in the presence of DR5 agonists, results in the catastrophic induction of apoptosis.
The idea that oncogenes might be exploited to suppress tumorigenesis, although unusual, is not without precedent. Like MYC, the oncogene E2F1 is capable of strongly promoting proliferation and apoptosis (47, 48). Surprisingly, E2F1-null mice develop a wide range of tumors not seen in similarly aged WT mice (49), suggesting that, under some circumstances, E2F1's apoptotic functions become critical for restraining tumor growth (50). This finding has led to the proposal that E2F1 can act either as a tumor-suppressor gene or as an oncogene, depending on context. Because of its essential role in proliferation, MYC does not act as a classical tumor-suppressor gene; its inactivation results in hypoproliferation rather than predisposition to cancer (51). Nonetheless, our results show that MYC's apoptotic function can be manipulated, e.g., by inhibiting GSK3β/FBW7 function in the presence of DR5 agonists, such that its proapoptotic functions outweigh its proliferative ones, resulting in tumor suppression rather than promotion.
Supplementary Material
Acknowledgments
We thank Dr. Bert Vogelstein for the HCT116 FBW7 deletion cell lines and Dr. Robert Weinberg for the HA1E derived cell lines.
Author contributions: S.R., Q.L.D., and K.C.Q. designed research; S.R. and Y.W. performed research; M.N. contributed new reagents/analytic tools; S.R., Q.L.D., and K.C.Q. analyzed data; and Q.L.D. and K.C.Q. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: siRNA, small inhibitory RNA; siGL3, siRNA directed against luciferase; DR5, death receptor 5; TRAIL, TNF-related apoptosis-inducing ligand.
References
- 1.Luscher, B. (2001) Gene 277, 1-14. [DOI] [PubMed] [Google Scholar]
- 2.Oster, S. K., Ho, C. S., Soucie, E. L. & Penn, L. Z. (2002) Adv. Cancer Res. 84, 81-154. [DOI] [PubMed] [Google Scholar]
- 3.Evan, G. I., Wyllie, A. H., Gilbert, C. S., Littlewood, T. D., Land, H., Brooks, M., Waters, C. M., Penn, L. Z. & Hancock, D. C. (1992) Cell 119-128. [DOI] [PubMed]
- 4.Pelengaris, S., Khan, M. & Evan, G. (2002) Nat. Rev. Cancer 2, 764-776. [DOI] [PubMed] [Google Scholar]
- 5.Wang, Y., Engels, I. H., Knee, D. A., Nasoff, M., Deveraux, Q. L. & Quon, K. C. (2004) Cancer Cell. 5, 501-512. [DOI] [PubMed] [Google Scholar]
- 6.Ozoren, N. & El-Deiry, W. S. (2002) Neoplasia 4, 551-557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.LeBlanc, H. N. & Ashkenazi, A. (2003) Cell Death Differ. 10, 66-75. [DOI] [PubMed] [Google Scholar]
- 8.Aza-Blanc, P., Cooper, C. L., Wagner, K., Batalov, S., Deveraux, Q. L. & Cooke, M. P. (2003) Mol. Cell 12, 627-637. [DOI] [PubMed] [Google Scholar]
- 9.Wang, Y., Quon, K. C., Knee, D. A., Nesterov, A. & Kraft, A. S. (2005) Cancer Res. 65, 1615-1616. [DOI] [PubMed] [Google Scholar]
- 10.Ricci, M. S., Jin, Z., Dews, M., Yu, D., Thomas-Tikhonenko, A., Dicker, D. T. & El-Deiry, W. S. (2004) Mol. Cell. Biol. 24, 8541-8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cui, H., Li, T. & Ding, H. F. (2005) J. Biol. Chem. 280, 9474-9481. [DOI] [PubMed] [Google Scholar]
- 12.Hartman, J. L., Garvik, B. & Hartwell, L. (2001) Science 291, 1001-1004. [DOI] [PubMed] [Google Scholar]
- 13.Reddy, A. & Kaelin, W. G. (2002) Curr. Opin. Pharmacol. 2, 366-373. [DOI] [PubMed] [Google Scholar]
- 14.Woodgett, J. R. (2001) Sci. STKE 100, re12. [DOI] [PubMed] [Google Scholar]
- 15.Cohen, P. & Goedert, M. (2004) Nat. Rev. Drug Discovery 3, 479-487. [DOI] [PubMed] [Google Scholar]
- 16.Ren, Y. G., Wagner, K. W., Knee, D. A., Aza-Blanc, P., Nasoff, M. & Deveraux, Q. L. (2004) Mol. Biol. Cell. 15, 5064-5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meijer, L., Skaltsounis, A. L., Magiatis, P., Polychronopoulos, P., Knockaert, M., Leost, M., Ryan, X. P., Vonica, C. A., Brivanlou, A., Dajani, R., et al. (2003) Chem. Biol. 10, 1255-1266. [DOI] [PubMed] [Google Scholar]
- 18.Sears, R., Nuckolls, F., Haura, E., Taya, Y., Tamai, K. & Nevins, J. R. (2000) Genes Dev. 14, 2501-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gregory, M. A., Qi, Y. & Hann, S. R. (2003) J. Biol. Chem. 278, 51606-51612. [DOI] [PubMed] [Google Scholar]
- 20.Yeh, E., Cunningham, M., Arnold, H., Chasse, D., Monteith, T., Ivaldi, G., Hahn, W. C., Stukenberg, P. T., Shenolikar, S., Uchida, T., et al. (2004) Nat. Cell. Biol. 6, 308-318. [DOI] [PubMed] [Google Scholar]
- 21.Welcker, M., Orian, A., Jin, J., Grim, J. A., Harper, J. W., Eisenman, R. N. & Clurman, B. E. (2004) Proc. Natl. Acad. Sci. USA 101, 9085-9090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yada, M., Hatakeyama, S., Kamura, T., Nishiyama, M., Tsunematsu, R., Imaki, H., Ishida, N., Okumura, F., Nakayama, K. & Nakayama, K. I. (2004) EMBO J. 23, 2116-2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rajagopalan, H., Jallepalli, P. V., Rago, C., Velculescu, V. E., Kinzler, K. W., Vogelstein, B. & Lengauer, C. (2004) Nature 428, 77-81. [DOI] [PubMed] [Google Scholar]
- 24.Mao, J. H., Perez-Losada, J., Wu, D., Delrosario, R., Tsunematsu, R., Nakayama, K. I., Brown, K., Bryson, S. & Balmain, A. (2004) Nature 432, 775-779. [DOI] [PubMed] [Google Scholar]
- 25.Orlicky, S., Tang, X., Willems, A., Tyers, M. & Sicheri, F. (2003) Cell 112, 243-256. [DOI] [PubMed] [Google Scholar]
- 26.Berns, K., Hijmans, E. M. & Bernards, R. (1997) Oncogene 15, 1347-1356. [DOI] [PubMed] [Google Scholar]
- 27.Welcker, M., Singer, J., Loeb, K. R., Grim, J., Bloecher, A., Gurien-West, M., Clurman, B. E. & Roberts, J. M. (2003) Mol. Cell 12, 381-392. [DOI] [PubMed] [Google Scholar]
- 28.Koepp, D. M., Schaefer, L. K., Ye, X., Keyomarsi, K., Chu, C., Harper, J. W. & Elledge, S. J. (2001) Science 294, 173-177. [DOI] [PubMed] [Google Scholar]
- 29.LeBlanc, H., Lawrence, D., Varfolomeev, E., Totpal, K., Morlan, J., Schow, P., Fong, S., Schwall, R., Sinicropi, D. & Ashkenazi, A. (2002) Nat. Med. 8, 274-281. [DOI] [PubMed] [Google Scholar]
- 30.Zheng, L., Liu, J., Batalov, S., Zhou, D., Orth, A., Ding, S. & Schultz, P. G. (2004) Proc. Natl. Acad. Sci. USA 101, 135-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paddison, P. J., Silva, J. M., Conklin, D. S., Schlabach, M., Li, M., Aruleba, S., Balija, V., O'Shaughnessy, A., Gnoj, L., Scobie, K., et al. (2004) Nature 428, 427-431. [DOI] [PubMed] [Google Scholar]
- 32.Berns, K., Hijmans, E. M., Mullenders, J., Brummelkamp, T. R., Velds, A., Heimerikx, M., Kerkhoven, R. M., Madiredjo, M., Nijkamp, W., Weigelt, B., et al. (2004) Nature 428, 431-437. [DOI] [PubMed] [Google Scholar]
- 33.Brummelkamp, T. R., Nijman, S. M., Dirac, A. M. & Bernards, R. (2003) Nature 424, 797-801. [DOI] [PubMed] [Google Scholar]
- 34.Deveraux, Q. L., Aza-Blanc, P., Wagner, K. W., Bauerschlag, D., Cooke, M. P. & Hampton, G. M. (2003) Semin. Cancer Biol. 13, 293-300. [DOI] [PubMed] [Google Scholar]
- 35.Brummelkamp, T. R. & Bernards, R. (2003) Nat. Rev. Cancer 3, 781-789. [DOI] [PubMed] [Google Scholar]
- 36.Paddison, P. J. & Hannon, G. J. (2002) Cancer Cell 2, 17-23. [DOI] [PubMed] [Google Scholar]
- 37.Willingham, A. T., Deveraux, Q. L., Hampton, G. M. & Aza-Blanc, P. (2004) Oncogene 23, 8392-8400. [DOI] [PubMed] [Google Scholar]
- 38.Bhatia, K., Huppi, K., Spangler, G., Siwarski, D., Iyer, R. & Magrath, I. (1993) Nat. Genet. 5, 56-61. [DOI] [PubMed] [Google Scholar]
- 39.Gregory, M. A. & Hann, S. R. (2000) Mol. Cell. Biol. 20, 2423-2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calhoun, E. S., Jones, J. B., Ashfaq, R., Adsay, V., Baker, S. J., Valentine, V., Hempen, P. M., Hilgers, W., Yeo, C. J., Hruban, R. H. & Kern, S. E. (2003) Am. J. Pathol. 163, 1255-1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spruck, C. H., Strohmaier, H., Sangfelt, O., Muller, H. M., Hubalek, M., Muller-Holzner, E., Marth, C., Widschwendter, M. & Reed, S. I. (2002) Cancer Res. 62, 4535-4539. [PubMed] [Google Scholar]
- 42.Insinga, A., Monestiroli, S., Ronzoni, S., Gelmetti, V., Marchesi, F., Viale, A., Altucci, L., Nervi, C., Minucci, S. & Pelicci, P. G. (2005) Nat. Med. 11, 71-76. [DOI] [PubMed] [Google Scholar]
- 43.Nebbioso, A., Clarke, N., Voltz, E., Germain, E., Ambrosino, C., Bontempo, P., Alvarez, R., Schiavone, E. M., Ferrara, F., Bresciani, F., et al. (2005) Nat. Med. 11, 77-84. [DOI] [PubMed] [Google Scholar]
- 44.Clarke, N., Jimenez-Lara, A. M., Voltz, E. & Gronemeyer, H. (2004) EMBO J. 23, 3051-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Altucci, L., Rossin, A., Raffelsberger, W., Reitmair, A., Chomienne, C. & Gronemeyer, H. (2001) Nat. Med. 7, 680-686. [DOI] [PubMed] [Google Scholar]
- 46.Akay, C. & Gazitt, Y. (2003) Cell Cycle 2, 358-368. [PubMed] [Google Scholar]
- 47.Qin, X. Q., Livingston, D. M., Kaelin, W. G. & Adams, P. D. (1994) Proc. Natl. Acad. Sci. USA 91, 10918-10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shan, B. & Lee, W. H. (1994) Mol. Cell. Biol. 14, 8166-8173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yamasaki, L., Jacks, T., Bronson, R., Goillot, E., Harlow, E. & Dyson, N. J. (1996) Cell 85, 537-548. [DOI] [PubMed] [Google Scholar]
- 50.Pierce, A. M., Schneider-Broussard, R., Gimenez-Conti, I. B., Russell, J. L., Conti, C. J. & Johnson, D. G. (1999) Mol. Cell. Biol. 19, 6408-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Trumpp, A., Refaeli, Y., Oskarsson, T., Gasser, S., Murphy, M., Martin, G. R. & Bishop, J. M. (2001) Nature 414, 768-773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.