Abstract
According to the nucleated polymerization model, in vivo prion proliferation occurs via dissociation (shearing) of the huge prion polymers into smaller oligomeric ‘seeds’, initiating new rounds of prion replication. Here, we identify the deletion derivative of yeast prion protein Sup35 (Sup35-Δ22/69) that is specifically defective in aggregate shearing and ‘seed’ production. This derivative, [PSI+], previously thought to be unable to turn into a prion state, in fact retains the ability to form a prion ([PSI+]Δ22/69) that can be maintained in selective conditions and transmitted by cytoplasmic infection (cytoduction), but which is mitotically unstable in non-selective conditions. MorePSI+]Δ22/69 retains its mitotic stability defect. The [PSI+]Δ22/69 cells contain more Sup35 protein in the insoluble fraction and form larger Sup35 aggregates compared with the conventional [PSI+] cells. Moderate excess of Hsp104 disaggregase increases transmission of the [PSI+]Δ22/69 prion, while excess Hsp70-Ssa chaperone antagonizes it, opposite to their effects on conventional [PSI+]. Our results shed light on the mechanisms determining the differences between transmissible prions and non-transmissible protein aggregates.
Keywords: chaperone/oligopeptide repeats/prion/protein aggregation/Saccharomyces cerevisiae
Introduction
Prions are ‘infectious’ isoforms of normal proteins that are capable of reproducing themselves by converting a normal protein, encoded by the same gene, into a prion (Prusiner, 1982). First discovered as infectious agents causing neurodegenerative diseases in mammals and humans (for review see Prusiner, 1998), prions were then implicated in certain cytoplasmically inherited traits in yeast (Wickner, 1994). In this way, prions provide a new mechanism for genetic transmission of information, coded in structural rather than sequence templates (for review see Chernoff, 2001).
It has been proposed that prions are aggregated proteins propagated via nucleated polymerization, so that pre-existing multimeric prion ‘seeds’ initiate polymerization of the soluble protein (for reviews see Lansbury and Caughey, 1995; Kushnirov and Ter-Avanesyan, 1998). Indeed, the prion proteins characterized thus far form insoluble aggregates in vivo, when present in prion state, and in vitro (for reviews see Serio et al., 1999; Serio and Lindquist, 2000; Chernoff, 2001). In some cases, these aggregates possess characteristic features of amyloids (ordered fiber-like structures with high β-sheet content). This relates prion phenomena to amyloidoses and neural inclusion disorders, such as Alzheimer’s disease, Parkinson’s disease, Huntington’s disease, amyotrophic lateral sclerosis and others (for review see Koo et al., 1999).
Yeast prion [PSI+], a polymerized isoform of the translation termination factor Sup35 (eRF3), has been successfully used as a model to study cellular control of prion transmission (for reviews see Serio and Lindquist, 2000; Chernoff, 2001). The [PSI+] strains are defective in translation termination, which provides a convenient experimental assay for [PSI+]. Prion formation can be induced de novo in the [psi–] background by transient overproduction of the Sup35 protein (Chernoff et al., 1993; Derkatch et al., 1996), possibly due to increased aggregation at higher protein concentrations. Successful transmission of [PSI+] in cell generations requires an intermediate level of the chaperone protein Hsp104 (Chernoff et al., 1995), a disaggregase previously implicated in solubilizing temperature-induced protein aggregates (Parsell et al., 1994). Excess Hsp104 causes frequent loss of [PSI+] (Chernoff et al., 1995), apparently due to increased solubilization of prion aggregates (Patino et al., 1996; Paushkin et al., 1996). However, inactivation or loss of Hsp104 also results in the loss of [PSI+] (Chernoff et al., 1995) via blockage of aggregate partitioning or shearing (Wegrzyn et al., 2001). This leads to accumulation of the large agglomerates lacking prion replicating activity. It appears that the ability of prion polymers to dissociate into the small oligomeric ‘seeds’ (shearing) is crucial for prion transmission by infection or in cell generations, and provides the major distinction between prion aggregates and non-infectious protein inclusions. Yeast prions other than [PSI+], such as [PIN+] (Derkatch et al., 1997) and [URE3] (Moriyama et al., 2000), also appear to require Hsp104, implicating it as a universal trans-catalyst of prion transmission. However, the cis-factors that control aggregate shearing remain unknown.
Yeast Sup35 consists of three regions: Sup35C, essential and sufficient for the translation termination function; middle Sup35M; and the N-terminal prion-forming domain (Sup35N), responsible for prion formation and propagation (for reviews see Serio and Lindquist, 2000; Chernoff, 2001). Sup35N includes an N-terminal Gln- and Asn-rich stretch (QN-stretch) resembling huntingtin and other proteins associated with poly-Gln diseases (De Pace et al., 1998), and oligopeptide repeats (ORs) between amino acid (aa) positions 40 and 90, resembling those observed in the mammalian prion protein PrP (Cox, 1994). Deletion of SUP35N makes yeast cells unable to propagate [PSI+] (Ter-Avanesyan et al., 1994), while overproduction of Sup35N alone is sufficient to induce [PSI+] formation in the cells bearing an intact chromosomal copy of SUP35 (Derkatch et al., 1996). A number of sup35N mutations were identified that negatively affect prion propagation, leading to the ‘[PSI+] no more’ (PNM) phenotype. These include aa substitutions within the QN-stretch (De Pace et al., 1998; King, 2001) and substitutions or deletions within the OR region (Ter-Avanesyan et al., 1994; Liu and Lindquist, 1999; Parham et al., 2001). In several cases, it was observed that the mutant Sup35 derivatives are poorly recruited into the Sup35PSI+ aggregates and/or show markedly decreased rates of amyloid formation in vitro (De Pace et al., 1998; Kochneva-Pervukhova et al., 1998; King, 2001; Parham et al., 2001). The molecular effects of the other mutations remain unclear.
Among PNM mutations, the sup35-Δ22/69 (formerly called sup35-ΔBstEII) deletion, lacking a piece of Sup35N between aa positions 22 and 69, is of specific interest. It abolishes the interaction between Sup35N and some of its partners in vivo (Bailleul et al., 1999) and slightly decreases the rates of spontaneous Sup35 aggregation in vitro (Glover et al., 1997). This led to the suggestion that the defect in prion propagation is due to inability of the mutant protein to enter the Sup35PSI+ aggregates (Kochneva-Pervukhova et al., 1998; Serio and Lindquist, 2000). However, overproduction of Sup35-Δ22/69, and especially of Sup35N-Δ22/69 in cells bearing full-size Sup35 protein resulted in partial loss of the Sup35 termination function, detected as nonsense suppression, a phenotype similar to that of [PSI+] (Ter-Avanesyan et al., 1993; Derkatch et al., 1996). In contrast to [PSI+], suppression was co-retained and co-lost with the Sup35- Δ22/69 (or Sup35N-Δ22/69) encoding plasmid.
Here, we perform a detailed analysis of the Sup35-Δ22/69 prion-forming potential in vivo. Our data show that the Sup35-Δ22/69 protein can form prion aggregates. However, these aggregates are unusually large and are rapidly lost in non-selective conditions, apparently due to the inability to produce small ‘seeds’. These features and patterns of interaction with Hsp104 implicate sup35-Δ22/69 as the first prion protein mutation that is specifically defective in aggregate shearing and transmission.
Results
Construction of yeast strains bearing the sup35-Δ22/69 allele
Isogenic haploid strains bearing the complete sup35::HIS3 deletion (Chernoff et al., 2000) on the chromosome and either sup35-Δ22/69 or the wild-type (SUP35+) allele on the single-copy (centromeric) plasmid were constructed as described in Materials and methods. We have confirmed that the sup35-Δ22/69 spore clones that originate from [PSI+] diploids have lost [PSI+], while [PIN+] prion, needed for [PSI+] induction by overproduced Sup35 (Derkatch et al., 1997, 2001), was not affected (data not shown). Most of the experiments described below were performed using strains GT247-1C, GT247-1D, GT249-21A (all sup35-Δ22/69), GT255-2D (SUP35+) and their derivatives (see Table I).
Table I. Genotypes and origins of the yeast strains.
| Strain | Genotype/description | Source |
|---|---|---|
| GT81 | MATa/MATα ade1-14/ade1-14 his3/his3 leu2/leu2 lys2/lys2 ura3/ura3 trp1/trp1 [PSI+ PIN+] (self-diploid) | Chernoff et al. (2000) |
| GT159 | the haploid MATa [psi– PIN+] derivative of GT81 | Chernoff et al. (1999) |
| GT109 | the SUP35/sup35::HIS3 derivative of GT81 | Chernoff et al. (2000) |
| GT111 | the [psi– PIN+] derivative of GT109 | Chernoff et al. (2000) |
| GT113 | the [psi– pin–] derivative of GT109 | Chernoff et al. (2000) |
| GT247-1C | MATa ade1-14 his3 leu2 lys2 ura3 trp1 sup35::HIS3 [LEU2 sup35-Δ22/69] [psi– PIN+] | this study |
| GT247-1D | the same as GT247-1C but MATα | this study |
| GT249-21A | the same as GT247-1D | this study |
| GT255-2D | MATa ade1-14 his3 lys2 ura3 leu2 trp1 sup35::HIS3 [LEU2 SUP35] [psi– PIN+] | this study |
| c10B-H49 [ρ–] (OT102) | MATα lys1-1 his3 leu1 ade2-1 SUQ5 kar1-1 cyhR [psi– rho–] | Kochneva-Pervukhova et al. (1998) |
| GT388 | MATα lys1-1 his3 leu1 ura3 ade2-1 SUQ5 kar1-1 cyhR [psi– rho–] | this study |
The Sup35-Δ22/69 protein can both induce the prion state and be turned into a prion
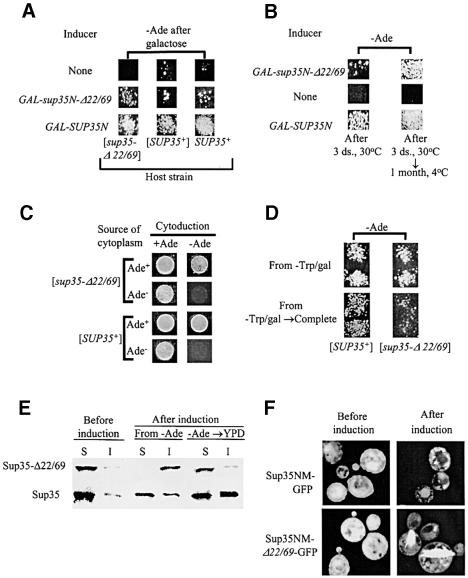
In contrast to the previously reported results (Derkatch et al., 1996, 1997), we have observed that the transient galactose-induced overexpression of the sup35N-Δ22/69 construct under the GAL promoter in the strains containing the UGA allele ade1-14 slightly but reproducibly increased the frequency of Ade+ colonies on glucose medium (i.e. after the GAL promoter was turned off; Figure 1A). This effect was detected in both GT255-2D bearing the SUP35+ allele on the CEN plasmid and the isogenic [psi– PIN+] strain GT159 bearing the SUP35+ allele on the chromosome. Apparently, such a low level of Ade+ induction (∼4-fold over background) was not detected in the previous experiments employing the multicopy (rather than galactose-inducible) sup35-Δ22/69 constructs (Derkatch et al., 1996, 1997) due to lower sensitivity of that experimental system.
Fig. 1. The Sup35-Δ22/69 protein can both induce the prion state and be turned into a prion. (A) Transient overproduction of Sup35N or Sup35N-Δ22/69 induces the appearance of Ade+ colonies in the [psi– PIN+] strains bearing sup35-Δ22/69 or SUP35+. Yeast strains: GT247-1C ([sup35-Δ22/69], left column); GT255-2D ([SUP35+], central column) and GT159 (SUP35+, right column). Plasmids: left and central columns, pRS316GAL control (none), pRS316GAL-SUP35N (GAL-SUP35N) and pGAL-SUP35NΔBst (GAL-sup35N-Δ22/69); right column, pFL39GAL control (none), pFL39GAL-SUP35N (GAL-SUP35N) and pFL39GAL-SUP35NΔBst (GAL-sup35N-Δ22/69). The –Ade/glucose plates were photographed after 10 days at 30°C. Similar results were obtained at 25°C (not shown). The strains GT247-1D and GT249-1A (not shown) produced the same result as GT247-1C. The plasmid pGAL-SUP35, bearing a complete SUP35 gene under the GAL promoter, induced Ade+ formation at about the same efficiency as pRS316GAL-SUP35N (not shown). (B) Induction of prion derivatives of Sup35-Δ22/69 is increased at low temperature. Yeast strain: GT249-21A ([sup35-Δ22/69]). Plasmid designations are as above (A, left and central columns). After induction on –Ura/galactose medium for 3 days at 30°C (left) or for 3 days at 30°C followed by 1 month at 4°C (right), cultures were velveteen replica plated onto –Ade/glucose medium. Plates were photographed after 10 days at 30°C. Strains GT247-1C and GT247-1D (not shown) produced the same result as GT249-21A. (C) Transmission of the newly induced prion derivatives of Sup35-Δ22/69 by cytoduction. Ade+ derivatives, induced in GT247-1C ([sup35-Δ22/69], line 1) or in GT255-2D ([SUP35+], line 3) by pFL39GAL-SUP35N, were mated to the lawn of the recipient strain c10B-H49 on YPD, and following 1 day of incubation, transferred to medium selective for cytoductants (+Ade; see Materials and methods) and the same medium lacking adenine (–Ade) to identify cytoductants able to suppress the ade2-1 mutation. The original uninduced strains (lines 2 and 4) were used as cytoplasm donors in the control experiment. Plates were photographed after 3 (left column) or 7 (right column) days of incubation. The same results were observed with the recipient strain GT388 (not shown). (D) Instability of the newly induced prion derivatives of Sup35-Δ22/69. Yeast strains are as in (A), left and central columns. Plasmid: pFL39GAL-SUP35N. After induction on –Trp/galactose medium for 3 days at 30°C, cultures were replica plated either directly onto –Ade/glucose medium (plate on the top) or first onto complete synthetic glucose-containing medium and then (after 1 day of incubation) onto –Ade/glucose medium. Plates were photographed after 10 days at 30°C. Post-induction incubation on complete medium decreases the frequency of Ade+ colonies in sup35-Δ22/69 but not in SUP35+. The same result was obtained with the inducer plasmid pGAL-SUP35 and also in the case when YPD was used instead of the synthetic complete medium (not shown). (E) Centrifugation analysis of aggregation of the Sup35 and Sup35-Δ22/69 prions. Yeast strains GT247-1C (Sup35-Δ22/69) and GT255-2D (Sup35), bearing the plasmid pRS316GAL-SUP35N, were incubated on –Ura/galactose medium for 3 days and velveteen replica plated onto –Ade/glucose medium. The Ade+ colonies, induced in each strain, were collected after 2 weeks of growth at 30°C, pooled together and either used directly for protein isolation (from –Ade) or incubated in liquid YPD on the shaker at 30°C and 200 r.p.m. (–Ade → YPD) for 24 h (Sup35-Δ22/69) or up to 72 h (Sup35) prior to protein isolation. Uninduced cultures of the respective strains (before induction) were used as [psi–] controls. Proteins were isolated and fractionated into the soluble (S) and insoluble (I) fractions by centrifugation at 12 000 g as described (Newnam et al., 1999), run on the SDS–PAGE gel, transferred onto the nitrocellulose membrane and reacted with the Sup35NM specific antibodies. The larger fraction of the Sup35-reacting material was insoluble in the Ade+ cultures bearing Sup35-Δ22/69, compared with those bearing full-size Sup35. However, insoluble Sup35-Δ22/69 disappeared after 24 h in YPD, while full-size Sup35 aggregates remained even after 72 h in YPD. (F) Fluorescence microscopy of the Sup35 and Sup35-Δ22/69 prion aggregates. Derivatives of the yeast strains GT255-2D ([SUP35+]) and GT247-1C ([sup35–22/69]), bearing the plasmids pRS316-NMGFP (Sup35NM–GFP) and pRS316-NMΔBstGFP (Sup35NM-Δ22/69–GFP), respectively, were transformed with the plasmid pFL39GAL-SUP35N, incubated on –Ura/galactose medium for 3 days and velveteen replica plated onto –Ade/glucose medium. Cells from Ade+ colonies, grown after 2 weeks of incubation at 30°C, were analyzed by fluorescence microscopy as described in Materials and methods. Uninduced cultures of the respective strains (before induction) were used as [psi–] controls. Prion aggregates are seen as fluorescent clumps.
Moreover, transient overexpression of SUP35, SUP35N or sup35N-Δ22/69 under the GAL promoter induced intense formation of Ade+ colonies in the sup35-Δ22/69 [psi– PIN+] strains (Figure 1A). Although GAL-sup35N- Δ22/69 remained the least efficient inducer, it induced more Ade+ colonies in the sup35-Δ22/69 background than in the SUP35+ background (Figure 1A). Like [PSI+] induction in the SUP35+ strains (for review see Chernoff, 2001), Ade+ induction in the sup35-Δ22/69 strains was strictly dependent on the presence of [PIN+] (data not shown), was higher at 25°C than at 30°C (data not shown) and was significantly increased by prolonged incubation of induced cultures at 4°C (Figure 1B).
As Ade+ phenotype is indicative of ade1-14UGA suppression due to a translation termination defect, one possible explanation of our results is that Sup35-Δ22/69 protein can be turned into a partially defective state in response to overproduction, as has been shown previously for the complete Sup35 protein. Alternatively, one could suggest that transient suppression of ade1-14 by the overproduced Sup35 derivative in the [PIN+] environment leads to accumulation of the Ade1 read-through product, which is retained in cell generations at a level sufficient for growth on –Ade medium. To distinguish between these possibilities, we studied a transfer of the suppressor state to the [psi–] strains c10B-H49[ρ–] and GT388 (Table I) bearing a different nonsense reporter (ade2-1UAA) by cytoplasmic infection (cytoduction). Ade+ derivatives induced in the sup35-Δ22/69 or SUP35+ strains were used as cytoplasm donors. In both cases, Ade+ cytoductants were obtained, although the efficiency of transfer was apparently lower with the sup35-Δ22/69 donor, as could be judged from heterogeneous growth on the selective medium (Figure 1C). As read-through Ade1 protein would be incapable of compensating for the ade2 defect, this result shows that nonsense suppression in the sup35-Δ22/69 background was caused by the transmissible isoform of the Sup35-Δ22/69 protein. As this isoform is maintained and reproduced for the number of cell generations sufficient to form a visible Ade+ colony, it certainly bears the major characteristics of the [PSI+] prion.
Mitotic instability of the Sup35-Δ22/69 prion isoform
To check whether prion derivatives of Sup35-Δ22/69 are capable of stable propagation in non-selective conditions, the galactose-induced cultures were incubated on complete glucose medium, non-selective for suppression, for 1–2 days before replica plating onto –Ade medium. While this step did not have any detectable effect on Ade+ induction by complete Sup35 or Sup35N in the SUP35+ strain, it greatly decreased the frequency of Ade+ colonies induced by Sup35, Sup35N or Sup35N-Δ22/69 in the sup35-Δ22/69 strain (Figure 1D), suggesting that most of the newly induced Sup35-Δ22/69 prion derivatives are mitotically unstable and can easily be lost in non-selective conditions. To confirm this, we picked the individual Ade+ colonies induced by Sup35N in the SUP35+ background and by Sup35N or Sup35N-Δ22/69 in the sup35-Δ22/69 background, streaked them out on non-selective (YPD) medium and analyzed four subclones of each colony. While the vast majority of the SUP35+ Ade+ colonies produced Ade+ subclones, most of the sup35-Δ22/69 Ade+ colonies produced Ade– subclones (Table II). Eight exceptional stable Ade+ derivatives of the sup35-Δ22/69 strain were found to contain nuclear suppressor mutations, apparently acquired during growth of the initially unstable Ade+ culture under the selective pressure (data not shown). These data confirm mitotic instability of the Sup35-Δ22/69 prion derivatives in non-selective conditions.
Table II. Individual analysis of Ade+ derivatives, obtained in the SUP35+ and sup35-Δ22/69 backgrounds.
| Inducee | Inducer | Cytoplasm recipient | Ade+ derivatives |
||
|---|---|---|---|---|---|
| Stable Ade+ | Unstable Ade+ | Total Ade+ analyzed | |||
| Originally induced Ade+ colonies | |||||
| SUP35+ | Sup35N | N/A | 24 | 0 | 24 |
| SUP35+ | Sup35N-Δ22/69 | N/A | 10 | 14 | 24 |
| sup35-Δ22/69 | Sup35N | N/A | 0 | 58 | 58 |
| sup35-Δ22/69 | Sup35N-Δ22/69 | N/A | 8a | 52 | 60 |
| Ade+ cytoductants | |||||
| SUP35+ | Sup35N | SUP35+ | 16 | 0 | 16 |
| sup35-Δ22/69 | Sup35N | SUP35+ | 0 | 40 | 40 |
aProven to be suppressor mutants.
Quite remarkably, a large fraction of the Ade+ colonies induced by the transient overproduction of Sup35N-Δ22/69 in the SUP35+ strain (Figure 1A) were also mitotically unstable (Table II). As the frequency of Ade+ induction in this experiment was low (see above), it is possible that most, if not all, of the remaining stable Ade+ colonies originated from spontaneous [PSI+] formation or from ade1-14 reversions, rather than from Sup35N-Δ22/69-mediated induction. Likewise, the Ade+ cytoductants that received cytoplasm from the sup35-Δ22/69 Ade+ donor (Figure 1D) were mitotically unstable, in contrast to cytoductants that received cytoplasm from the SUP35+ Ade+ donor (Table II), despite the fact that the recipient strain contained the wild-type SUP35+ allele in both cases. Cytoduction experiments with the ura3 recipient strain GT388 confirmed that the transfer of the Ade+ phenotype is not associated with the transfer of the URA3 plasmid bearing the sup35-Δ22/69 allele (data not shown). This indicates that the mitotic stability defect of the initial Sup35-Δ22/69 prion derivative can not be corrected by the addition of or substitution for the full-size Sup35 protein. A similar phenomenon was described recently for chimeric Candida–Saccharomyces Sup35 constructs (Chien and Weissman, 2001).
Differences in cytoductant stability also confirm that cytoplasmic transmission of the Ade+ state is not due to the presence of residual inducer protein in the donor cytoplasm, as both sup35-Δ22/69 and SUP35+ Ade+ derivatives used for cytoduction were induced by one and the same inducer, Sup35N. Indeed, the control experiment using the donor cultures taken directly from galactose medium and containing high levels of the Sup35N inducer generated stable Ade+ cytoductants independently of whether the donor strain was SUP35+ or sup35-Δ22/69 (data not shown).
Aggregation of the Sup35-Δ22/69 protein in the cells containing mitotically unstable prions
Sup35 aggregation is associated with the [PSI+] state (Patino et al., 1996; Paushkin et al., 1996). To check whether the Sup35-Δ22/69 prion isoform is also aggregated, we isolated and analyzed proteins from the Ade+ colonies, freshly induced by GAL-SUP35N in the sup35-Δ22/69 and SUP35+ backgrounds. In contrast to the control (before induction) cultures containing soluble Sup35-reacting material, the Ade+ derivatives contained a significant fraction of insoluble Sup35. Moreover, the sup35-Δ22/69 Ade+ derivatives grown in the selective conditions contained almost all of the Sup35-reacting material in the low-speed precipitated fraction corresponding to large-size aggregates, while the SUP35+-based Ade+ derivatives grown in the same conditions showed less than half of the total Sup35 protein in this fraction (Figure 1E). Further growth of the sup35-Δ22/69 Ade+ derivatives in non-selective (YPD) medium resulted in the shift of all Sup35-Δ22/69 protein back to the soluble fraction, while the SUP35+ Ade+ derivatives grown in non-selective medium retained insoluble Sup35 (Figure 1E). This correlated with the rapid loss of the Sup35-Δ22/69 prion in non-selective conditions (Figure 1D; Table II).
To visualize prion aggregates, we transformed the isogenic [psi– PIN+] sup35-Δ22/69 and [psi– PIN+] SUP35+ yeast strains with centromeric plasmid bearing the SUP35NM-Δ22/69 or SUP35NM fragment, respectively, fused in-frame to green fluorescent protein (GFP) and expressed under the control of the endogenous SUP35 promoter. Prion induction by GAL-SUP35N was performed as described previously. Cells from the Ade+ colonies of both strains contained fluorescent clumps, which were not present in the ‘before induction’ samples and appear to have resulted from aggregation of the prion isoform of Sup35NM (or Sup35NM-Δ22/69)–GFP (Figure 1F). In agreement with differential centrifugation results, 10–30% of cells in the sup35-Δ22/69 Ade+ colonies contained huge rod-like structures, which were not observed in the cells from the SUP35+ Ade+ colonies (Figure 1F). Further incubation in non-selective conditions resulted in a sharp decrease in frequency of the aggregate-containing cells in the sup35-Δ22/69 but not in the SUP35+ population (data not shown).
Effects of the chaperone proteins on the Sup35-Δ22/69 prion
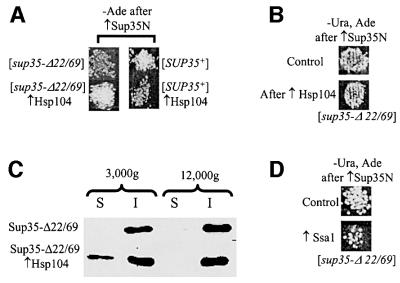
To check whether low stability of the Sup35-Δ22/69 prion is related to the large aggregate size, we performed the Ade+ induction experiments in the SUP35+ and sup35-Δ22/69 strains bearing an extra copy of the HSP104 gene on the centromeric plasmid. Normal levels of the Hsp104 disaggregase promote shearing and propagation of the [PSI+] aggregates, while excess Hsp104 leads to aggregate solubilization that results in prion loss (Chernoff et al., 1995; Patino et al., 1996; Paushkin et al., 1996; Wegrzyn et al., 2001). As expected, an extra copy of HSP104, expressed from its own promoter, decreased the frequency of Ade+ colonies induced in the SUP35+ background. However, it increased the frequency of Ade+ colonies induced in the sup35-Δ22/69 background (Figure 2A). This stimulatory effect was observed only at moderate levels of excess Hsp104, as high-level Hsp104 overexpression from the copper-induced CUP1 promoter antagonized formation of detectable Ade+ colonies in both strains (data not shown). Transient overexpression of HSP104 (simultaneously with the SUP35N inducer) from the GAL promoter did not influence the frequency of Ade+ colonies in the sup35-Δ22/69 background (Figure 2B), confirming that the continuous presence of excess Hsp104 is needed for its effect on the Sup35-Δ22/69 prion.
Fig. 2. Effects of the chaperone proteins on the Sup35-Δ22/69 prion. (A) The continuous presence of an extra copy of HSP104 increases the frequency of the detectable Ade+ derivatives in the sup35-Δ22/69 strain but decreases it in the SUP35+ strain. The yeast strains GT247-1C ([sup35-Δ22/69]) and GT255-2D ([SUP35+]) were co-transformed with the TRP1-based plasmid pFL39GAL-SUP35N and either URA3-based plasmid pYS104, containing the HSP104 gene (↑Hsp104), or matching control plasmid pRS316GAL. Co-transformants were incubated on –Trp, Ura/galactose medium for 3 days and velveteen replica plated onto –Ade/glucose medium. Plates were photographed after 10 days at 30°C. (B) Transient overproduction of Hsp104 does not influence Ade+ induction in the sup35-Δ22/69 background. The yeast strain GT247-1C ([sup35Δ22/69]) was co-transformed with the TRP1-based plasmid pFL39GAL-SUP35N and either URA3-based plasmid pGAL104-URA3, containing the HSP104 gene under the GAL promoter (↑Hsp104), or matching control plasmid pRS316GAL. Co-transformants were incubated on –Trp, Ura/galactose medium for 3 days and velveteen replica plated onto –Ade, Ura/glucose medium. Plates were photographed after 10 days at 30°C. (C) Moderate excess of Hsp104 promotes partial dissociation of the Sup35-Δ22/69 aggregates. Ade+ colonies induced in GT247-1C (Sup35-Δ22/69) by pFL39GAL-SUP35N in the presence of plasmid pYS104, containing the HSP104 gene (↑Hsp104), or matching control plasmid pRS316GAL (control), were collected after 2 weeks of growth on –Ade, Ura at 30°C, pooled together and used for protein isolation and centrifugation analysis of the Sup35-Δ22/69 aggregation, as described above (see the legend to Figure 1E). Centrifugation was performed either at 12 000 or 3000 g. The presence of excess Hsp104 increases the relative amount of the Sup35-Δ22/69 protein remaining in the soluble fraction at 3000 g, which appears to correspond to the smaller aggregates. (D) The presence of an extra copy of SSA1 decreases the frequency of Ade+ colonies in the sup35-Δ22/69 strain. The yeast strain GT247-1C ([sup35-Δ22/69]) was co-transformed with the TRP1-based plasmid pFL39GAL-SUP35N and either URA3-based plasmid pRS316K-SSA1-K10, containing the SSA1 gene (↑Ssa1), or matching control plasmid pRS316GAL. Co-transformants were incubated on –Trp, Ura/galactose medium for 3 days and velveteen replica plated onto –Ade, Ura/glucose medium. Plates were photographed after 14 days at 30°C.
Individual analysis of the Ade+ colonies that were induced in the sup35-Δ22/69 background at moderately increased Hsp104 levels confirmed that excess Hsp104 increases longevity of the Sup35-Δ22/69 prion derivatives. Among 11 independent Ade+ derivatives analyzed, seven produced Ade+ colonies in non-selective conditions. Moreover, the Ade+ phenotype was retained more frequently in the progeny retaining the HSP104 plasmid (Ura+) than in those losing the HSP104 plasmid (Ura–; Table III). However, excess Hsp104 was not able to compensate completely for the mitotic transmission defect. All the colonies, even those retaining the HSP104 plasmid, had lost the Ade+ phenotype upon the next round of incubation in non-selective conditions (data not shown).
Table III. Analysis of colonies produced by Ade+ derivatives of the sup35-Δ22/69 [URA3 HSP104] strain in non-selective conditions.
| Class | Phenotype |
Total number | |
|---|---|---|---|
| Ade+ | Ade– | ||
| Retained [URA3 HSP104] |
13 |
32 |
45 |
| Lost [URA3 HSP104] | 3 | 36 | 39 |
| Total | 16 | 68 | 84 |
To check whether a moderate excess of Hsp104 influences Sup35-Δ22/69 prion aggregation, we isolated proteins from the fresh sup35-Δ22/69 Ade+ colonies, induced in the presence or absence of the HSP104 CEN plasmid, and analyzed them by differential centrifugation. In both cases, most Sup35-Δ22/69 was precipitated at 12 000 g. However, the presence of excess Hsp104 led to the appearance of a Sup35-Δ22/69 fraction remaining in solution at lower centrifugation speed, 3000 g (Figure 2C). This shows that an increase in the mitotic stability of the Sup35-Δ22/69 prion correlates with the appearance of the smaller aggregates, apparently generated by the disaggregating activity of Hsp104.
The Ssa protein of the Hsp70 family was previously shown to counteract the [PSI+]-antagonizing effect of excess Hsp104 (Newnam et al., 1999), to increase [PSI+]-mediated suppression (Newnam et al., 1999) and to stimulate de novo [PSI+] induction by excess Sup35 or Sup35N (K.Allen and Y.Chernoff, unpublished observations). In contrast, we have observed that excess Ssa protein decreases the frequency of detectable Ade+ colonies induced in the sup35-Δ22/69 background (Figure 2D). As we have previously proposed that Ssa is a factor promoting aggregate growth (Chernoff, 2001; Wegrzyn et al., 2001), its antagonizing effect on the Sup35-Δ22/69 prion agrees with the notion that the low mitotic stability of this prion derivative is related to its abnormally large aggregate size.
Discussion
Our data show that the deletion derivative of the Sup35 protein, Sup35-Δ22/69, formerly described as ‘[PSI+] no more’ (Ter-Avanesyan et al., 1994), retains both aggregate-forming and -inducing activities and is able to enter protein aggregates in vivo (Figure 1A, E and F). The overproduction-induced aggregation-prone isoform of Sup35-Δ22/69 possesses the major characteristic features of the [PSI+] prion. When selection for the partial loss of the translation termination function is applied (i.e. in the ade1-14UGA strain incubated on –Ade medium), the Sup35-Δ22/69 aggregation-prone isoform can be maintained and propagated in cell divisions long enough to form Ade+ colonies (Figure 1A). No low-molecular-weight protein fragments corresponding to the Sup35N inducer protein were detected in the Ade+ cultures (data not shown), confirming that inducer is not retained by the time the Ade+ colony is formed. Moreover, the suppressor state could be transmitted by cytoduction to a recipient strain bearing a different nonsense reporter, ade2-1UAA (Figure 1C), confirming that suppression is not caused by the residual read-through Ade1 protein accumulated during the inducer’s overproduction. Thus, it is clear that partial loss of termination activity becomes an inherent property of the Sup35-Δ22/69 protein itself. Therefore, we recognize the Sup35-Δ22/69 aggregation-prone isoform as a prion and designate it [PSI+]Δ22/69.
However, in contrast to the conventional [PSI+] prions formed by the full-size Sup35 protein, the [PSI+]Δ22/69 prion is rapidly lost once cells resume growth in non-selective conditions. This loss was detected both by recovery of Sup35-Δ22/69 translation termination function (Figure 1D; Table II) and shift of the Sup35-Δ22/69 protein from the insoluble to soluble fraction (Figure 1E). Moreover, this mitotic instability pattern is retained by the [PSI+]Δ22/69-derived prion that was transmitted by cytoduction to the strain containing full-size Sup35 protein (Table II). This shows that Sup35 lacking the 22/69 region forms a prion that is inherently defective in mitotic transmission in the absence of selective pressure. This defect appears to be determined by the structural patterns of the initial prion ‘nucleus’, as it can not be corrected by the following addition of the full-size Sup35 molecules.
The physical properties of the Sup35-Δ22/69 aggregates also differ from the conventional Sup35PSI+ prions by a larger proportion of protein in the low speed precipitating fraction, and a larger size and unusual morphology of GFP-tagged clumps (Figure 1E and F). Our previous data show that [PSI+] propagation in vivo probably occurs via small oligomers, while large cytologically visible aggregates represent the ‘dead ends’ of the prion replication cycle (Wegrzyn et al., 2001). This agrees with the recent in vitro results suggesting that the process of Sup35NM amyloid formation proceeds via oligomeric intermediates (Serio et al., 2000). Thus, accumulation of the huge aggregates in the [PSI+]Δ22/69-containing cells suggests a defect in aggregate shearing and production of the small oligomeric ‘seeds’.
In strong support of this model, we have observed that a moderate excess of Hsp104 disaggregase exhibits opposite effects on the newly formed [PSI+] and [PSI+]Δ22/69 prions: it is antagonizing conventional [PSI+] but increases the frequency of detectable [PSI+]Δ22/69 derivatives by improving their mitotic transmission (Figure 2A). As Hsp104 is responsible for aggregate shearing and required for production of new oligomeric ‘seeds’ (Wegrzyn et al., 2001), a moderate excess of Hsp104 most likely assists [PSI+]Δ22/69 by promoting partial dissociation of large aggregates, so that a larger number of ‘seeds’ is generated. This is confirmed by differential centrifugation results (Figure 2C). However, a further increase in Hsp104 levels becomes counterproductive as it leads to aggregate loss via solubilization. Completely stable [PSI+]Δ22/69 derivatives were not recovered even in the extra-HSP104-containing strain, indicating that excess Hsp104 is not able either to fully compensate for the transmission defect caused by the Δ22/69 deletion or irreversibly alter the inherent structural parameters of aggregates that are responsible for the shearing defect.
Another chaperone (Ssa of the Hsp70 family) that assists in propagation of the conventional [PSI+] prion in normal conditions (Newnam et al., 1999) exhibits an antagonistic effect on [PSI+]Δ22/69 (Figure 2D). This agrees with the previously observed inhibitory effect of excess Ssa on transmission of the large [PSI+] aggregates generated at low levels of Hsp104 activity (Wegrzyn et al., 2001) and is most likely explained by the previously hypothesized aggregation-promoting role of Ssa leading to a further increase of aggregate size (Chernoff, 2001; Wegrzyn et al., 2001).
Our data explain the previously observed nonsense-suppressor effect of overproduced Sup35- (or Sup35N-) Δ22/69 in the cells containing full-size Sup35 (Ter-Avanesyan et al., 1993; Derkatch et al., 1996). Apparently, overproduction induces unstable Sup35-Δ22/69 prions, which convert full-size Sup35 into a prion. However, the resulting prion remains unstable and is rapidly lost, as demonstrated in the cytoduction experiments. In support of this model, we have observed low-efficient induction of mitotically unstable Ade+ derivatives by the transient Sup35N-Δ22/69 overproduction in the strain bearing full-size Sup35 (Figure 1A; Table II). A low frequency of such ‘heterologous’ induction could be due to the previously reported anti-suppressor effect of the sup35-Δ22/69 allele on conventional [PSI+] (Ter-Avanesyan et al., 1994). It is possible that inclusion of the Sup35-Δ22/69 protein into wild-type [PSI+] aggregates inhibits shearing and reproduction. Indeed, continuous overproduction of Sup35N-Δ22/69 in the weak conventional [PSI+] strain results in an increased loss of the prion state (Y.O.Chernoff, unpublished observations).
Previous observations suggested that newly emerged isolates of [PSI+] and [PIN+] frequently display mitotic instability, but can be stabilized upon further propagation (Derkatch et al., 2000; I.Derkatch, Y.O.Chernoff and S.G.Inge-Vechtomov, unpublished observations). It is possible that prion generation occurs via a two-step process including initial formation of the unstable aggregate and its conversion into the ‘shearing-prone’ state that ensures stable transmission. Interestingly, the heterologous [PSI+] derivatives, formed by a chimeric Sup35 protein bearing the highly divergent Sup35N (or NM) region of the distantly related yeast Pichia methanolica, are characterized by lower mitotic stability (Chernoff et al., 2000; Kushnirov et al., 2000; Santoso et al., 2000), a larger proportion of protein in the insoluble fraction, lower sensitivity to excess Hsp104 (Kushnirov et al., 2000) and larger aggregate size (Y.O.Chernoff and E.Lewitin, unpublished observations), compared with the conventional endogenous Saccharomyces cerevisiae [PSI+] prions. This indicates that the Sup35 sequence divergence between Saccharomyces and Pichia might affect the shearing step.
The sup35-Δ22/69 deletion is the first documented sup35 allele that specifically affects aggregate shearing and ‘seed’ production. The Δ22/69 deletion overlaps two subregions of Sup35N previously shown to influence prion propagation: it removes the last several aa residues of the N-terminal QN-stretch (De Pace et al., 1998) and the first 2.5 ORs (Cox, 1994), as well as the region between the QN-stretch and ORs. It has yet to be determined which alteration contributes most to the shearing defect observed. Previous data indicate that point mutations (Kochneva-Pervukhova et al., 1998) or deletions (Parham et al., 2001) within ORs decrease the ability of Sup35 to enter prion aggregates. However, the shearing defect has not been reported for these mutants, nor was it reported for the aa substitutions within the QN-stretch (De Pace et al., 1998). It is not yet known whether correct spacing between the QN and OR regions is important for aggregate shearing. It is also possible that interactions between Sup35 and other proteins (Hsp104 and/or its co-factors) involved in aggregate shearing are impaired in the Sup35-Δ22/69 aggregates. Indeed, Δ22/69 decreases or abolishes in vivo interaction between Sup35N and some other proteins, including Sla1, which influences [PSI+] formation and propagation by an unknown mechanism (Bailleul et al., 1999). However, if the defect of prion shearing is indeed due to a lack of protein–protein interactions, this should mean that protein interaction patterns of prion aggregates are determined completely by the protein that initiated prion formation, as full-size Sup35 aggregates ‘seeded’ by Sup35-Δ22/69 appear to retain the same pattern (Table II).
Previously, it was reported that Sup35-Δ22/69 protein exhibits slow (relative to full-size Sup35) kinetics of aggregate formation in vitro (Glover et al., 1997). Part of that could be due to the shearing defect, although the complex nature of the Sup35-Δ22/69 deletion makes it possible that initial aggregation of Sup35-Δ22/69 is also impaired. Indeed, overproduced Sup35-Δ22/69 induced its own prionization less efficiently than overproduced Sup35 or Sup35N (Figure 1A). Further experiments are needed to uncover the specific molecular mechanisms of the shearing defect.
Our data provide a framework for detailed analysis of the mechanisms differentiating between the transmissible prions and non-transmissible protein aggregates. Our results also agree with the recent observations made in mammalian systems, indicating that huge deposits of aggregated proteins may represent ‘dead ends’ of amyloid propagation, while toxic effects are determined by the small proliferating intermediates (for review see Koo et al., 1999). In this case, aggregate shearing rather than aggregate formation per se could become the most prospective target of anti-prion treatments.
Materials and methods
Yeast plasmids
All the yeast plasmids used were single-copy (centromeric) shuttle vectors. The plasmids pRS315-SUP35ΔBst (LEU2) and pRS316-SUP35ΔBst (URA3) were constructed by inserting the XhoI–XbaI 3.5 kb fragment of pEMBL-SUP35-ΔBstEII (Ter-Avanesyan et al., 1993), which bears the sup35-Δ22/69 allele under the endogenous SUP35 promoter, into SalI–XbaI-cut pRS315 or XhoI–XbaI-cut pRS316 (Sikorski and Hieter, 1989), respectively. The plasmids bearing SUP35 and its derivatives under the galactose-inducible (GAL) promoter were constructed as follows. The TRP1 plasmid pFL39GAL-SUP35N and the control matching plasmid pFL39GAL were obtained by inserting, respectively, the 1.5 kb PvuII fragment of pRS316GAL-SUP35N (Bailleul et al., 1999) and the 1.2 kb PvuII fragment of pRS316GAL (Liu et al., 1992) into PvuII-cut pFL39 (Bonneaud et al., 1991). The URA3 plasmid pGAL-SUP35ΔBst and TRP1 plasmid pFL39GAL-SUP35NΔBst were constructed from pGAL-SUP35 (Derkatch et al., 1996) and pFL39GAL-SUP35N (see above), respectively, by deleting the 144 bp BstEII–BstEII region, corresponding to the Sup35 aa positions 22–69. The URA3 plasmid pGAL-SUP35NΔBst was constructed from pGAL-SUP35ΔBst by deleting the 1.8 kb region between HindIII sites, which removes SUP35C and part of SUP35M. The URA3 plasmids pRS316-SpGFP and pRS316-NMGFP were constructed by substituting the BamHI–SacI fragment of pRS315-Sp-SUP35HA (De Pace et al., 1998), bearing the SUP35 ORF, with either the 0.7 kb BamHI–SacI fragment of p316CG (Serio et al., 1999), containing the GFP ORF, or the 1.6 kb BamHI–SacI fragment of pmCNMG (Serio et al., 1999), containing the SUP35NM–GFP ORF, respectively. The resulting constructs express either superglow GFP or chimeric Sup35NM–GFP protein under the control of the SUP35 promoter. The plasmid pRS316-NMΔBstGFP has been obtained from pRS316-NMGFP by removing the 144 bp BstEII fragment of pRS316-NMGFP, coding for Sup35 aa positions 22–69. The URA3 plasmid pRS316K-SSA1-K10, containing the SSA1 gene under its own promoter, was kindly provided by E.Craig. The URA3 plasmid pCUP-HSP104 bearing HSP104 under CUP1 promoter has been constructed by K.Bapat in Y.O.Chernoff’s laboratory. The URA3 plasmids pYS104, which bears the HSP104 gene under its own promoter (Sanchez et al., 1992), pGAL104-URA3, which bears HSP104 under the GAL promoter (Chernoff et al., 1995), and the LEU2 plasmid pASB2, bearing the complete SUP35 gene under its own promoter (Borchsenius et al., 2000), were described earlier.
Yeast strains
Strains used in this study are listed in Table I. Isogenic diploid strains GT81, GT109, GT111 and GT113 were described previously (Chernoff et al., 2000). The original [PSI+] self-diploid GT81 is heterozygous by MAT locus and homozygous by all other genes. GT109 is a derivative of GT81 that is heterozygous by the sup35::HIS3 disruption. The strains GT111 and GT113 are [psi– PIN+] and [psi– pin–] derivatives of GT109, respectively. The haploid strain GT159 (Chernoff et al., 1999) is a [psi– PIN+] derivative of GT81-1C, a meiotic spore clone of GT81. Haploid strains bearing the sup35::HIS3 allele on the chromosome and either the sup35-Δ22/69 or SUP35+ allele on the single-copy (centromeric) plasmid were obtained by sporulating and dissecting GT111 (GT247-1C and GT247-1D) or GT109 (GT249-21A), transformed with the plasmid pRS315-sup35ΔBst. GT255-2D was obtained by sporulating and dissecting GT111 transformed with pASB2. If necessary, the plasmid shuffle was performed to substitute the LEU2 plasmids for the URA3 plasmids. The respiratory deficient kar1 strain c10B-H49 [ρ–] (Kochneva-Pervukhova et al., 1998) was used as a recipient in cytoduction experiments. This strain contains the tRNA suppressor SUQ5, able to suppress ade2-1 only in the presence of [PSI+] (Cox, 1994). The spontaneous ura3 derivative of this strain, GT388, was selected on the 5-FOA medium (see Kaiser et al., 1994).
Genetic and microbiological techniques
Standard yeast genetic techniques, media and cultivation conditions were used (Kaiser et al., 1994). Yeast cultures were incubated at 30°C, unless stated otherwise. The presence of [PSI+] was tested for by suppression of ade2-1UAA mutation in the presence of SUQ5 (strains c10B-H49 [ρ–] and GT388) or ade1-14UGA mutation (all the other strains), resulting in growth on the medium lacking adenine (–Ade) as described previously (Cox, 1994; Chernoff et al., 1995). Dissection was performed on the micromanipulator Ergaval Series 10 from Carl Zeiss (Jena, Germany). To induce [PSI+], yeast transformants were usually incubated on galactose medium, selective for the corresponding plasmid, for 3–4 days and velveteen replica plated onto –Ade/glucose medium, as described previously (Derkatch et al., 1996). Growth on –Ade was indicative of prion induction, leading to suppression of the ade1-14UGA allele. For cytoduction experiments, the donor strains GT247-1C, GT255-2D, GT159 or their freshly induced Ade+ derivatives, selected on –Ade medium, were mated to the karyogamy-defective kar1 (Conde and Fink, 1976) recipient strains c10B-H49 ([ρ–]) or GT388 (Table I). The recipients lack functional mitochondrial DNA and bear the recessive cyhr mutation. After 1 day mating on YPD medium, cell mixtures were velveteen replica plated to synthetic medium containing glycerol as the sole carbon source and cycloheximide (5 mg/l) to select for cytoductants receiving the mitochondria from the donor strain. In parallel, cell mixtures were replica plated onto the same medium lacking adenine to identify cytoductants able to suppress the ade2-1 mutation. At least eight independent repeats of each experiment demonstrated in Figures 1A–D, 2A, B and D were performed per strain/plasmid combination, with the same result.
DNA and protein analysis
Standard procedures were used for DNA isolation and plasmid constructions. Yeast proteins were isolated and fractionated by centrifugation according to Patino et al. (1996) with some modifications (Newnam et al., 1999; see the legend to Figure 2C). Proteins were run on SDS–PAGE gels and reacted with the Sup35NM-specific antibodies described earlier (Wegrzyn et al., 2001). Reaction to the secondary anti-rabbit antibodies and chemiluminescent detection were performed by using the ECL detection kit from Amersham.
Fluorescence microscopy
GFP images shown in Figure 1F were scanned using a Zeiss LSM510 UV confocal laser scanning microscope (Carl Zeiss Inc., New York, NY) at wavelength 488 nm and image analysis was conducted using a Zeiss LSM image browser (Carl Zeiss). Alternatively, the samples were analyzed using the microscope Lumam-I2 (LOMO, St Petersburg, Russia) and the video–computer interface Video-test FISH (ISTA, St Petersburg, Russia) at wavelength 400–460 nm (data not shown).
Acknowledgments
Acknowledgements
We thank K.Bapat for constructing the plasmid pCUP-HSP104, E.Craig for the gift of pRS316-SSA1-K10, A.Galkin for the guidance in fluorescence microscopy experiments and G.Zhouravleva for the helpful discussion. This work was supported by grants R01GM58763 from the National Institutes of Health (NIH) to Y.O.C., 99-04-49601 from the Russian Foundation for Basic Research (RFBR) to A.S.B. and RB1-2037 from the US Civilian Research and Development Foundation for the Independent States of the Former Soviet Union (CRDF). Any opinions, findings and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect those of the NIH, RFBR or CRDF.
References
- Bailleul P.A., Newnam,G.P., Steenbergen,J.N. and Chernoff,Y.O. (1999) Genetic study of interactions between the cytoskeletal assembly protein Sla1 and prion-forming domain of the release factor Sup35 (eRF3) in Saccharomyces cerevisiae. Genetics, 153, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneaud N., Ozier-Kalogeropoulos,O., Li,G.Y., Labouesse,M., Minvielle-Sebastia,L. and Lacroute,F. (1991) A family of low and high copy replicative, integrative and single-stranded S. cerevisiae/E.coli shuttle vectors. Yeast, 7, 609–615. [DOI] [PubMed] [Google Scholar]
- Borchsenius A.S., Tchourikova,A.A. and Inge-Vechtomov,S.G. (2000) Recessive mutations in SUP35 and SUP45 genes coding for translation release factors affect chromosome stability in Saccharomyces cerevisiae. Curr. Genet., 37, 285–291. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O. (2001) Mutation processes at the protein level: is Lamarck back? Mutat. Res., 488, 39–64. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Derkach,I.L. and Inge-Vechtomov,S.G. (1993) Multicopy SUP35 gene induces de novo appearance of psi-like factors in the yeast Saccharomyces cerevisiae. Curr. Genet., 24, 268–270. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Lindquist,S.L., Ono,B., Inge-Vechtomov,S.G. and Liebman,S.W. (1995) Role of the chaperone protein Hsp104 in propagation of the yeast prion-like factor [psi+]. Science, 268, 880–884. [DOI] [PubMed] [Google Scholar]
- Chernoff Y.O., Newnam,G.P., Kumar,J., Allen,K. and Zink,A. (1999) Evidence for a ‘protein mutator’ in yeast: role of the Hsp70-related chaperone Ssb in formation, stability and toxicity of the [PSI] prion. Mol. Cell. Biol., 19, 8103–8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chernoff Y.O., Galkin,A.P., Lewitin,E., Chernova,T.A., Newnam,G.P. and Belenkiy,S.M. (2000) Evolutionary conservation of prion-forming abilities of the yeast Sup35 protein. Mol. Microbiol., 35, 865–876. [DOI] [PubMed] [Google Scholar]
- Chien P. and Weissman,J.S. (2001) Conformational diversity in a yeast prion dictates its seeding specificity. Nature, 410, 223–227. [DOI] [PubMed] [Google Scholar]
- Conde J. and Fink,G.R. (1976) A mutant of Saccharomyces cerevisiae defective for nuclear fusion. Proc. Natl Acad. Sci. USA, 73, 3651–3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox B.S. (1994) Prion-like factors in yeast. Curr. Biol., 4, 744–748. [DOI] [PubMed] [Google Scholar]
- De Pace A.H., Santoso,A., Hillner,P. and Weissman,J.S. (1998) A critical role for amino-terminal glutamine/asparagine repeats in the formation and propagation of a yeast prion. Cell, 93, 1241–1252. [DOI] [PubMed] [Google Scholar]
- Derkatch I.L., Chernoff,Y.O., Kushnirov,V.V., Inge-Vechtomov,S.G. and Liebman,S.W. (1996) Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics, 144, 1375–1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley,M., Zhou,P., Chernoff,Y.O. and Liebman,S.W. (1997) Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics, 147, 507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley,M.E., Masse,S.V., Zadorsky,S.P., Polozkov,G.V., Inge-Vechtomov,S.G. and Liebman,S.W. (2000) Dependence and independence of [PSI+] and [PIN+]: a two-prion system in yeast? EMBO J., 19, 1942–1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkatch I.L., Bradley,M.E., Hong,J.Y. and Liebman,S.W. (2001) Prions affect the appearance of other prions: the story of [PIN+]. Cell, 106, 171–182. [DOI] [PubMed] [Google Scholar]
- Glover J.R., Kowal,A.S., Schirmer,E.C., Patino,M.M., Liu,J.J. and Lindquist,S. (1997) Self-seeded fibers formed by Sup35, the protein determinant of [PSI+], a heritable prion-like factor of Saccharomyces cerevisiae. Cell, 89, 811–819. [DOI] [PubMed] [Google Scholar]
- Kaiser C., Michaelis,S. and Mitchell,A. (1994). Methods in Yeast Genetics: A Cold Spring Harbor Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- King C.Y. (2001) Supporting the structural basis of prion strains: induction and identification of [PSI] variants. J. Mol. Biol., 307, 1247–1260. [DOI] [PubMed] [Google Scholar]
- Kochneva-Pervukhova N.V., Paushkin,S.V., Kushnirov,V.V., Cox,B.S., Tuite,M.F. and Ter-Avanesyan,M.D. (1998) Mechanism of inhibition of Ψ+ prion determinant propagation by a mutation of the N-terminus of the yeast Sup35 protein. EMBO J., 17, 5805–5810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo E.H., Lansbury,P.T. and Kelly,J.W. (1999) Amyloid diseases: abnormal protein aggregation in neurodegeneration. Proc. Natl Acad. Sci. USA, 96, 9989–9990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnirov V.V. and Ter-Avanesyan,M.D. (1998) Structure and replica tion of yeast prions. Cell, 94, 13–16. [DOI] [PubMed] [Google Scholar]
- Kushnirov V.V., Kochneva-Pervukhova,N.V., Chechenova,M.B., Frolova,N.S. and Ter-Avanesyan,M.D. (2000) Prion properties of the Sup35 protein of yeast Pichia methanolica. EMBO J., 19, 324–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lansbury P.T. and Caughey,B. (1995) The chemistry of scrapie reaction: the ‘ice 9’ metaphore. Chem. Biol., 2, 1–5. [DOI] [PubMed] [Google Scholar]
- Liu H., Krizek,J. and Bretscher,A. (1992) Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics, 132, 665–673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.J. and Lindquist,S. (1999) Oligopeptide-repeat expansions modulate ‘protein-only’ inheritance in yeast. Nature, 400, 573–576. [DOI] [PubMed] [Google Scholar]
- Moriyama H., Edskes,H.K. and Wickner,R.B. (2000) [URE3] prion propagation in Saccharomyces cerevisiae: requirement for chaperone Hsp104 and curing by overexpressed chaperone Ydj1p. Mol. Cell. Biol., 20, 8916–8922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newnam G.P., Wegrzyn,R.D., Lindquist,S.L. and Chernoff,Y.O. (1999) Antagonistic interactions between yeast chaperones Hsp104 and Hsp70 in prion curing. Mol. Cell. Biol., 19, 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham S.N., Resende,C.G. and Tuite,M.F. (2001) Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J., 20, 2111–2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell D.A., Kowal,A.S., Singer,M.A. and Lindquist,S. (1994) Protein disaggregation mediated by heat-shock protein Hsp104. Nature, 372, 475–478. [DOI] [PubMed] [Google Scholar]
- Patino M.M., Liu,J.J., Glover,J.R. and Lindquist,S. (1996) Support for the prion hypothesis for inheritance of a phenotypic trait in yeast. Science, 273, 622–626. [DOI] [PubMed] [Google Scholar]
- Paushkin S.V., Kushnirov,V.V., Smirnov,V.N. and Ter-Avanesyan,M.D. (1996) Propagation of the yeast prion-like [psi+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J., 15, 3127–3134. [PMC free article] [PubMed] [Google Scholar]
- Prusiner S.B. (1982) Novel proteinaceous infectious particles cause scrapie. Science, 216, 136–144. [DOI] [PubMed] [Google Scholar]
- Prusiner S.B. (1998) Prions. Proc. Natl Acad. Sci. USA, 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y., Taulien,J., Borkovich,K.A. and Lindquist,S. (1992) Hsp104 is required for tolerance to many forms of stress. EMBO J., 11, 2357–2364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoso A., Chien,P., Osherovich,L.Z. and Weissman,J.S. (2000) Molecular basis of a yeast prion species barrier. Cell, 100, 277–288. [DOI] [PubMed] [Google Scholar]
- Serio T.R. and Lindquist,S.L. (2000) Protein-only inheritance in yeast: something to get [PSI+]-ched about. Trends Cell Biol., 10, 98–105. [DOI] [PubMed] [Google Scholar]
- Serio T.R., Cashikar,A.J., Moslehi,J.J., Kowal,A.S. and Lindquist,S.L. (1999) Yeast prion [Ψ+] and its determinant, Sup35p. Methods Enzymol., 309, 649–673. [DOI] [PubMed] [Google Scholar]
- Serio T.R., Cashikar,A.J., Kowal,A.S., Sawicki,G.J., Moslehi,J.J., Serpell,L., Arnsdorf,M.F. and Lindquist,S.L. (2000) Nucleated conformational conversion and the replication of conformational information by a prion determinant. Science, 289, 1317–1321. [DOI] [PubMed] [Google Scholar]
- Sikorski R.S. and Hieter,P. (1989) A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics, 122, 19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ter-Avanesyan M.D., Kushnirov,V.V., Dagkesamanskaya,A.R., Didichenko,S.A., Chernoff,Y.O., Inge-Vechtomov,S.G. and Smirnov,V.N. (1993) Deletion analysis of the SUP35 gene of the yeast Saccharomyces cerevisiae reveals two non-overlapping functional regions in the encoded protein. Mol. Microbiol., 7, 683–692. [DOI] [PubMed] [Google Scholar]
- Ter-Avanesyan M.D., Dagkesamanskaya,A.R., Kushnirov,V.V. and Smirnov,V.N (1994) The SUP35 omnipotent suppressor gene is involved in the maintenance of the non-Mendelian determinant [psi+] in the yeast Saccharomyces cerevisiae. Genetics, 137, 671–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegrzyn R.D., Bapat,K., Newnam,G.P., Zink,A.D. and Chernoff,Y.O. (2001) Mechanism of prion loss after Hsp104 inactivation in yeast. Mol. Cell. Biol., 21, 4656–4669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R.B. (1994) [URE3] as an altered URE2 protein: evidence for a prion analog in Saccharomyces cerevisiae. Science, 264, 566–569. [DOI] [PubMed] [Google Scholar]