Abstract
Myocardin and the myocardin-related transcription factors (MRTFs) A and B act as coactivators for serum response factor, which plays a key role in cardiovascular development. To determine the functions of MRTF-B in vivo, we generated MRTF-B mutant mice by targeted inactivation of the MRTF-B gene. We show that mice homozygous for an MRTF-B loss-of-function mutation die during mid-gestation from a spectrum of cardiovascular defects that includes abnormal patterning of the branchial arch arteries, double-outlet right ventricle, ventricular septal defects, and thin-walled myocardium. These abnormalities are accompanied by a failure in differentiation of smooth muscle cells within the branchial arch arteries, which are derived from the neural crest. The phenotype of MRTF-B mutant mice is distinct from that of mice lacking myocardin, revealing unique roles for these serum response factor coactivators in the development of different subsets of smooth muscle cells in vivo.
Keywords: cardiovascular development, congenital heart disease, heart, neural crest
Myocardin and the myocardin-related transcription factors (MRTFs) A and B comprise a family of coactivators that provides transcriptional activity to serum response factor (SRF) (refs. 1 and 2; reviewed in refs. 3 and 4). SRF binds a DNA sequence known as a CArG box and serves as a docking site for myocardin and MRTFs (1, 2). CArG boxes are required for expression of numerous smooth, cardiac, and skeletal muscle genes (5, 6). Tissue-specific deletion of the Srf gene in these muscle cell types results in severe abnormalities in muscle cell differentiation (7–10).
Members of the myocardin family share homology in a basic and glutamine-rich region that interacts with SRF (1, 2). An adjacent (Scaffold attachment factor, Acinus, PIAS) domain is involved in target gene specificity, such that its deletion prevents activation of a subset of SRF target genes (1). A coiled-coil domain resembling a leucine zipper mediates dimerization of myocardin and MRTFs (11–13) and has been proposed to allow cooperativity between CArG boxes (11). The C-terminal regions of myocardin and MRTFs function as transcription activation domains (1, 2).
Myocardin and the MRTFs enhance the transcriptional activity of SRF by forming a ternary complex with SRF on DNA and providing their powerful transcription activation domains (1, 2). MRTF-A and -B also convey stimulatory signals from the Rho GTPase and the actin cytoskeleton to SRF via their regulated translocation into the nucleus (12, 14–16).
Myocardin expression is restricted to cardiac and smooth muscle cells (1), whereas MRTF-A and MRTF-B are expressed in a broad range of cell types (2, 17). Consistent with the postulated role of myocardin as a regulator of smooth muscle gene expression, myocardin knockout mice die by embryonic day (E)10.5 with an absence of differentiated smooth muscle cells (SMCs) (18). Although a dominant negative myocardin mutant is sufficient to prevent cardiac gene expression in Xenopus embryos (1), myocardin null mouse embryos do not show cardiac defects (18). Mice homozygous for a lacZ enhancer trap allele of MRTF-B display perinatal lethality (19), which has been attributed to abnormalities in vascular development (20). However, this lacZ insertion has the potential to generate a dominant negative mutant of MRTF-B that would be predicted to perturb the function of other myocardin family members. Moreover, a fraction of transcripts generated from this allele encode the wild-type protein, thereby preventing determination of a true loss-of-function phenotype (19).
To unequivocally determine the function of MRTF-B in vivo, we created an MRTF-B mutant allele by homologous recombination in embryonic stem cells, which we used to generate MRTF-B null mice. These mice die between E13.5 and E14.5 and display severe defects in the anatomy of branchial arch arteries and cardiac outflow tract, accompanied by a defect in smooth muscle differentiation. We conclude that MRTF-B plays an essential early role in development of a specific subset of vascular SMCs.
Methods
Transfection Assays. Transfection assays were performed as described in ref. 11. Briefly, COS cells were transiently transfected with 100 ng of pcDNA expression plasmids encoding wild-type or mutant forms of myocardin, MRTF-A and MRTF-B, and an SM22-luciferase reporter. Forty-eight hours later, cell extracts were prepared, and luciferase activity was determined.
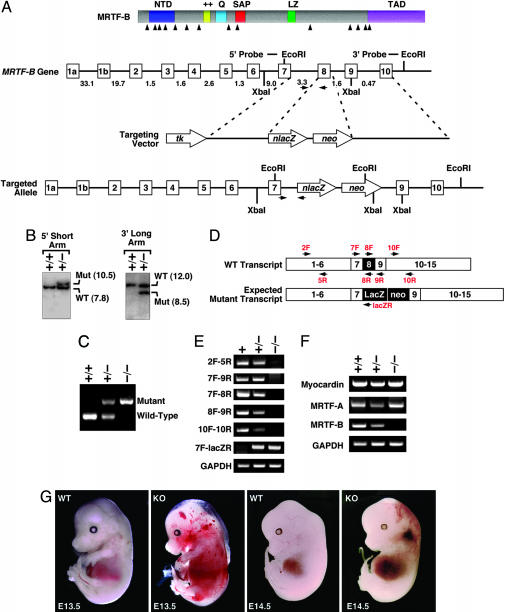
Generation of MRTF-B Mutant Mice. We created an MRTF-B targeting vector so as to delete exon 8 of the gene and introduce a lacZ and neomycin-resistance gene. The targeting vector was electroporated into 129 SvEv-derived ES cells, and selection was performed with G-418 and 1-(2′-deoxy-2′-fluoro-β-d-arabinofuranosyl)-5-iodouracil, respectively. Four hundred ES cell clones were isolated and analyzed for homologous recombination by Southern blotting with both 5′ and 3′ probes. Three clones with a targeted MRTF-B gene were used for blastocyst injection, and the resulting chimeric male mice were bred to C57BL/6 females to obtain germ-line transmission of the mutant allele. Genotyping was performed by PCR with genomic DNA prepared from tail biopsies or from embryo yolk sacs.
RT-PCR. Total RNA was purified from the hearts of E10.5 embryos with TRIzol reagent (Invitrogen) according to the manufacturer's instructions. For RT-PCR, total RNA was used as a template for reverse transcriptase and random hexamer primers. Primer sequences are available on request.
India Ink Injection. Embryos were collected at E11.5, and India ink was injected intracardially by using a finely drawn glass pipette. The embryos were then immediately fixed in 4% paraformaldehyde overnight, dehydrated, and cleared in benzyl alcohol:benzyl benzoate.
Histology and Immunohistochemistry. Histology and LacZ staining were performed as described in ref. 21. For immunohistochemistry, paraffin sections were deparaffinized, rehydrated, and stained with monoclonal α-smooth muscle (SM) actin antibody (clone 1A4, Sigma).
Results
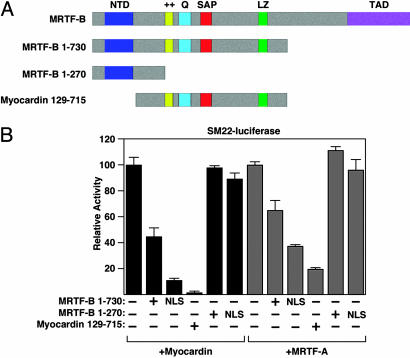
Potential Dominant Negative Function of an MRTF-B lacZ Insertion Allele. Our goal was to analyze the function of MRTF-B in vivo by creating MRTF-B null mice. We were aware of a mutation created by random insertion of a β-geo cassette between exons 10 and 11 of the MRTF-B gene (20). This lacZ insertion allele would generate an MRTF-B protein truncated at amino acid 730 (Fig. 1A). Based on prior studies, we anticipated that this truncated protein would act as a dominant negative mutant (1, 2, 13, 16). Before embarking on the creation of a new MRTF-B mutation, we examined the function of this mutant protein in transfected COS cells. The truncated MRTF-B protein (1–730) failed to activate an SRF-dependent reporter alone (data not shown); however, as anticipated, it interfered with the transcriptional activity of the wild-type myocardin and MRTF-A proteins when comparable amounts of each plasmid were transfected (Fig. 1B). The dominant negative activity of the MRTF-B 1–730 mutant was weaker than that of a previously described myocardin mutant protein (129–715) (1), presumably because the N-terminal domain of MRTF-B causes partial sequestration in the cytoplasm (12, 16). The 1–730 mutant protein, like wild-type MRTF-B, showed nuclear localization in ≈15% of transfected COS cells. Fusion of an SV40 nuclear localization sequence to the amino terminus of this truncated protein to more closely reflect the nuclear localization of the endogenous MRTF-B protein in SMCs enhanced its dominant negative activity (Fig. 1B).
Fig. 1.
Transcriptional activities of MRTF-B mutant proteins. (A) The structures of wild-type and mutant MRTF-B and myocardin proteins are shown. The lacZ gene trap line (20) is predicted to generate a truncated MRTF-B protein containing residues 1–730. The targeted mutation we introduced into the MRTF-B gene is predicted to generate a truncated MRTF-B protein containing residues 1–270. A dominant negative mutant of myocardin is also shown. NTD, N-terminal domain; ++, basic domain; SAP, Scaffold attachment factor. Q, glutamine-rich domain; LZ, leucine zipper; TAD, transcription activation domain. (B) Wild type and mutant proteins were transfected into COS cells and assayed for their ability to activate an SM22-luciferase reporter. Truncated MRTF-B proteins containing the SV40 nuclear localization sequence (NLS) at the amino terminus were also tested. The MRTF-B 1–730 mutant functions as a dominant negative, whereas the 1–270 mutant lacks inhibitory activity.
Based on these findings, we were concerned that the potential dominant negative activity of the lacZ insertion allele might cloud the interpretation of the potential role of MRTF-B in vivo. In addition, it has been reported that a minor fraction of wild-type transcripts are generated from this allele by splicing around the lacZ insertion (20). Thus, the allele does not function as a true null.
Generation of MRTF-B Knockout Mice. To create a loss-of-function allele of the mouse MRTF-B gene, we introduced a lacZ cassette after exon 7 and deleted exon 8 of the gene by homologous recombination in ES cells (Fig. 2 A–C). This mutation results in a truncated protein containing only the first 270 amino acids of MRTF-B and lacking the SRF-binding, dimerization, and transcription activation domains. In contrast to the dominant negative activity of the predicted product of the lacZ insertion allele, the 270-aa mutant protein is functionally inert (Fig. 1B).
Fig. 2.
Generation and analysis of MRTF-B knockout mice. (A) Gene targeting strategy. The mouse MRTF-B protein is schematized at the top. The targeting vector contained a 3.2-kb 5′ arm and a 4.7-kb 3′ arm and replaced a 0.5-kb region of the gene with a lacZ-neo cassette. Intron junctions within the coding region are shown by arrowheads beneath the schematized protein. Exons 1–10 are shown in boxes, and sizes of introns are indicated. Positions of 5′ and 3′ probes used for Southern analysis in B are indicated. Positions of PCR primers used for genotyping are shown by horizontal arrows. (B) Southern analysis. Genomic DNA from ES cell clones was isolated and analyzed by Southern blot with 5′ and 3′ probes after digestion with XbaI and EcoRI, respectively. (C) PCR analysis. Genomic DNA from E11.5 embryos was analyzed by PCR with primers shown in A.(D) Positions of primers used for RT-PCR. A schematic of the exons of the MRTF-B gene and positions of primers used for RT-PCR is shown. The expected mutation would contain a lacZ-neo cassette between exons 7 and 9 and would delete exon 8. (E) RT-PCR was performed by using RNA isolated from hearts of E10.5 embryos, and the primers shown in D. Genotypes of embryos are shown at the top. The truncated transcript generated from the mutant allele (2F-5R) was expressed at a much lower level than the WT MRTF-B transcript. (F) RT-PCR was performed by using RNA isolated from hearts of E10.5 embryos with primers specific for myocardin, MRTF-A, and MRTF-B. Transcripts for GAPDH were detected as a control for RNA loading and integrity. (G) Appearance of wild type and MRTF-B mutant embryos at E13.5 and E14.5.
Germ-line transmission of the mutant MRTF-B allele was achieved from three independent ES cell clones. Intercrosses of MRTF-B+/– mice in the isogenic C57BL/6 background or in mixed backgrounds failed to produce any MRTF-B null offspring among >100 offspring analyzed. Contrary to findings observed with the MRTF-B mutant mice generated by random insertion of a β-geo cassette (20), we conclude that our homozygous MRTF-B mutation results in embryonic lethality with complete penetrance.
The gene-targeting event was confirmed by RT-PCR analysis of mRNA from the hearts of E10.5 embryos by using primers representing exon sequences within and surrounding the deleted region of the gene (Fig. 2D). These assays showed that exon 7 was spliced to the lacZ allele as predicted (primers 7F and lacZR in Fig. 2 D and E). Sequencing of this PCR product confirmed that the mutant allele generated a protein in which amino acid 270 of MRTF-B was fused inframe to lacZ (data not shown). Portions of the MRTF-B transcript 3′ of the lacZ insertion site could not be detected in homozygous mutant embryos. There was no change in expression of myocardin or MRTF-A in mutant hearts (Fig. 2F), indicating that these genes were not up-regulated to compensate for the lack of MRTF-B.
Early Embryonic Lethality of MRTF-B–/– Mice. Genotyping of litters from timed pregnancies revealed no homozygous mutants after E14.5 (Table 1). At E14.5, we observed only 2 homozygous mutants of 35 embryos analyzed, a number substantially lower than expected from Mendelian inheritance. Homozygous mutant embryos at E13.5 and E14.5 showed pericardial edema and widespread hemorrhaging, suggesting that vascular abnormalities contributed to embryonic demise (Fig. 2G and data not shown). At E12.5 and earlier, homozygous mutants appeared outwardly normal and were present approximately at Mendelian ratios. We conclude that the targeted mutation in the MRTF-B gene results in complete lethality between E13.5 and 14.5.
Table 1. Genotypes of offspring from MRTF-B+/- intercrosses.
| Genotype | +/+ | +/- | -/- | Total |
|---|---|---|---|---|
| P10 | 33 | 61 | 0 | 94 |
| E15.5-16.5 | 17 | 36 | 0 | 53 |
| E14.5 | 14 | 19 | 2 (6) | 35 |
| E13.5 | 17 | 30 | 12 (20) | 59 |
| E12.5 | 14 | 41 | 14 (20) | 69 |
| E9.5≈11.5 | 24 | 50 | 25 (25) | 99 |
The number of offspring of each genotype from intercrosses of MRTF-B+/- is shown. Numbers in parentheses indicate % of total offspring at each age.
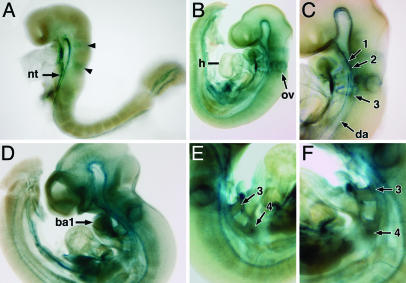
Expression of lacZ from the Targeted MRTF-B Allele. The lacZ reporter integrated into the targeted MRTF-B allele was expressed in a pattern corresponding to that of the wild-type gene. At E8.5, strong expression of lacZ was observed along the ventral neural tube and in two zones in the developing hindbrain, previously reported to correspond to rhombomeres 3 and 5 (Fig. 3A) (20). At E9.5, lacZ expression was also detected in the otic vesicle, heart, dorsal aorta, and branchial arch arteries 1, 2, and 3 (Fig. 3 B and C). At E10.5, lacZ continued to be expressed in the above structures, as well as the first branchial arch and the third and fourth branchial arch arteries of homozygous mutant embryos (Fig. 3 D–F).
Fig. 3.
LacZ staining of MRTF-B mutant embryos. Embryos were stained for lacZ expression. At E8.5, staining was apparent in the ventral neural tube and two distinct stripes (arrowheads) in the developing hindbrain (A). LacZ staining is observed in the otic vesicle, heart, dorsal aorta, and branchial arch arteries 1, 2, and 3 at E9.5 (B and C). At E10.5, lacZ expression becomes more widespread, with high level of expression in the first branchial arch (D). LacZ expression is also detected in the third and fourth arch arteries of MRTF-B mutant embryos as shown in the right and left lateral view (E and F). Branchial arch arteries are numbered. ba1, first branchial arch; nt, neural tube; ov, otic vesicle, h, heart; da, dorsal aorta.
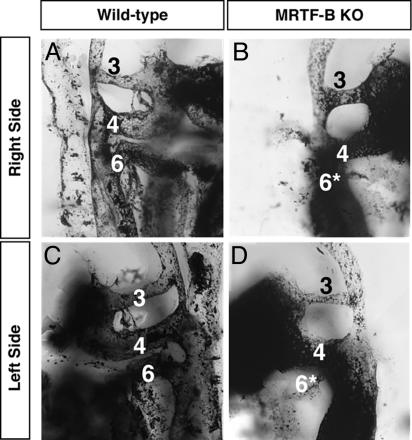
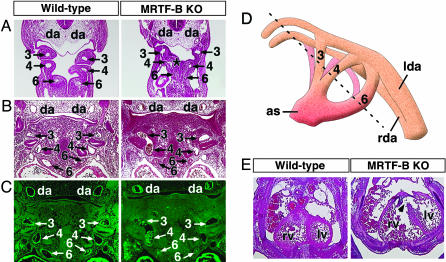
Cardiovascular Abnormalities in MRTF-B Mutant Embryos. Because of the expression of MRTF-B in the developing cardiovascular system and the mid-gestation lethality with obvious cardiovascular demise, we examined the cardiovascular system of mutant embryos by injecting India ink into the beating hearts of E11.5 embryos. In contrast to wild-type embryos, in which the third, fourth, and sixth branchial arch arteries were clearly intact bilaterally, in the mutant embryo both sixth branchial arch arteries were hypoplastic and narrow, whereas the third and fourth branchial arch arteries were normal (Fig. 4).
Fig. 4.
Visualization of vasculature by India ink injection. India ink was injected into the beating hearts of wild-type (A and C) and MRTF-B–/– embryos (B and D) at E11.5. The right (A and B) and left (C and D) branchial arch arteries for each embryo are shown in lateral view and numbered. Note that both the right and left sixth arch arteries of mutant embryo are hypoplastic and prematurely regressed (*).
Histological sections through the branchial arch arteries at E10.5 and E11.5 consistently confirmed that the sixth branchial arch arteries of mutant embryos were abnormally narrow and prematurely regressed compared with those of wild type. Furthermore, in serial histological sections, fusion between the fourth and sixth arteries was detected in E10.5 mutant embryos (Fig. 5A).
Fig. 5.
Branchial arch artery defects in MRTF-B mutant embryos. (A–C) Wild-type and MRTF-B mutant embryos at E10.5 (A) and E11.5 (B and C). Hematoxylin/eosin sections show hypoplastic sixth arch arteries in mutant embryos. Abnormal communication between the fourth and sixth arteries at E10.5 is indicated (*). Histological sections stained for SM α-actin show smooth muscle differentiation within the wall of branchial arch arteries and dorsal aortae in the wild-type embryo. In contrast, SM α-actin expression is not detected in the branchial arch arteries of the mutant embryo, but expression is seen in the dorsal aorta and heart. Branchial arch arteries are numbered as 3, 4, and 6. da, dorsal aorta. (D) Schematic of branchial arch arteries. The plane of section in the embryos shown in A–C is indicated by a dashed line. as, aortic sac; lda, left dorsal aorta; rda, right dorsal aorta. (E) Transverse section through the heart shows incomplete ventricular septation (arrowhead) and thin myocardial wall in the mutant embryo at E13.5. lv, left ventricle; rv, right ventricle.
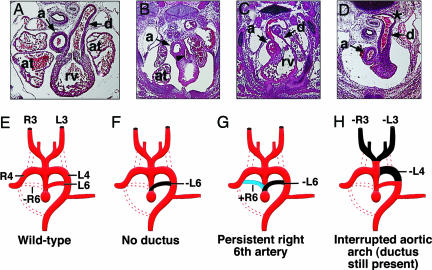
Serial histological sections revealed numerous branchial arch artery defects in mutant embryos at E13.5. In several mutant embryos, the ductus arteriosus, normally derived from the left sixth arch artery, abnormally regressed and did not connect to the aortic arch (Fig. 6 B and F). The pulmonary trunk was hypoplastic, possibly secondary to the lack of the ductus, but did give rise to the pulmonary arteries that projected to the primitive lung buds. In other E13.5 mutant embryos, the ductus arteriosus developed on the right side and passed behind the trachea before joining the descending aorta, reflecting abnormal regression of the left sixth arch artery accompanied by abnormal persistence of the right sixth arch artery (Fig. 6 C and G). We also observed interruption of the aortic arch because of abnormal regression of the left fourth arch artery. In such embryos, the ductus arteriosus connected with the descending aorta distal to the defect (Fig. 6 D and H). Interestingly, this abnormality was associated with an absence of carotid arteries bilaterally due to abnormal patterning of the third arch arteries.
Fig. 6.
Branchial arch artery defects in MRTF-B mutant embryos. (A–D) Defects of the great vessels in E13.5 mutant embryos. In the transverse section of the wild-type embryo, the pulmonary trunk communicates with the descending aorta via the ductus arteriosus (A). Mutant embryos display hypoplastic pulmonary trunk (arrowhead) and no ductus arteriosus (B), persistent right-sided ductus arteriosus (C), and interrupted aortic arch (D). Note the abnormal shape and position of the descending aorta where the ductus arterious joins (*). a, aorta; at, atrium; d, ductus arteriosus; rv, right rentricle. (E–H) Schematic diagrams of aortic arch defects seen in A–D. Dotted lines represent normal regression. Black areas depict abnormal regression, whereas blue areas indicate abnormal persistence. (E) Patterning of the branchial arch arteries in the wild-type embryo with normal regression of the right sixth artery (-R6). (F) Absence of the ductus arteriosus resulting from abnormal regression of the left sixth artery (-L6). (G) Persistent right-sided ductus caused by abnormal regression of the left sixth artery (-L6) and abnormal persistence of the right sixth artery (+R6). (H) Interrupted aortic arch and loss of both carotid arteries, due to abnormal regression of the left fourth (-L4) and both right and left third arteries (-R3, -L3). R3, R4, and R6, right arch arteries 3, 4, and 6, respectively. L3, L4, and L6, left aortic arch arteries, respectively.
In addition to the abnormal development of the branchial arch arteries, MRTF-B mutant embryos displayed abnormalities in the origin of the great vessels at E12.5 and E13.5. Double outlet right ventricle was observed in all mutant embryos (data not shown). High ventricular septal defects and thin-walled myocardium were observed as well (Fig. 5E).
Lack of Differentiation of Branchial Arch Artery SMCs in MRTF-B Null Embryos. Given the ability of myocardin and MRTFs to activate SMC gene expression in vitro (11, 17, 22–24), we anticipated that the vascular abnormalities of MRTF-B mutant embryos might be accompanied by defects in SMC differentiation. Indeed, immunostaining of histologic sections for SM-α-actin expression showed a clear reduction in smooth muscle differentiation within the walls of the third, fourth, and sixth branchial arch arteries of mutant embryos at E11.5 (Fig. 5C). In contrast, SM-α-actin expression was unaffected in the dorsal aorta or heart of mutant embryos. Thus, the loss of MRTF-B appears to specifically disrupt SMC differentiation within the branchial arch arteries where morphological abnormalities are also observed.
Discussion
The phenotype of MRTF-B null mice demonstrates that MRTF-B is an obligate regulator of early cardiovascular development. The absence of MRTF-B results in lethal malformations of the branchial arch arteries and cardiac outflow tract, as well as cardiac anomalies, accompanied by a failure in differentiation of a specific subset of SMCs.
Cardiovascular Abnormalities in MRTF-B Mutant Embryos. The vascular structures affected by the MRTF-B null mutation are derived from the neural crest, which migrates into the branchial arches giving rise to vascular SMCs (25, 26). Based on the lack of α-SM actin staining in the affected vascular structures of MRTF-B mutant embryos, we conclude that MRTF-B is required for activation of the SMC differentiation program in these cells, analogous to the function of myocardin in other SMCs (18). Whether the block to differentiation of these cells is causally related to or independent of the profound remodeling defects in the branchial arch arteries is unclear.
In principle, the failure in differentiation of neural crest-derived SMCs could reflect an abnormality in neural crest cell migration, a block in differentiation of smooth muscle precursors after they reach their destination, or an increase in apoptosis of these cells. Expression of Plexin A, a marker of neural crest cells, showed no difference in wild-type and MRTF-B mutant embryos, nor was there an increase in apoptosis within the branchial arches of mutant embryos (data not shown). Thus, we conclude that MRTF-B acts specifically to control differentiation of SMCs in the branchial arch arteries.
Numerous mouse mutants display defects in patterning of the branchial arch arteries (26), but these defects do not result in lethality until birth. We are uncertain why the homozygous MRTF-B mutation results in embryonic lethality. This early lethality may result from the lack of the ductus arteriosus, which is necessary for fetal life after ventricular and outflow tract septation. Thin myocardium may also contribute to embryonic lethality. Alternatively, MRTF-B may have functions elsewhere in the embryo, such as the liver, that are required for embryonic viability. Tissue-specific deletion of MRTF-B will be required to resolve these issues.
Phenotypes Resulting from Different MRTF-B Mutant Alleles. As this work was being completed, another report (20) described the phenotype resulting from a lacZ insertion allele of MRTF-B. In contrast to our results, suggesting that the truncated MRTF-B protein predicted to arise from that lacZ insertion allele behaves as a dominant negative mutant, that study concluded that the mutant protein failed to act as a dominant negative mutant. We cannot explain the discrepancy between the results of the two studies. However, it should be pointed out that the mutant protein produced by the lacZ insertion allele contains the dimerization and SRF-interaction domains and would therefore be expected to possess dominant negative activity, based on our prior analyses of myocardin and MRTF functions (1, 2). The low level of the wild-type protein generated by that allele also further complicates the interpretation of phenotypes resulting from it (20). Although both studies conclude that MRTF-B is required for differentiation of SMCs in the branchial arch arteries, the timing of lethality, as well as the severity and extent of cardiovascular defects, are clearly different with the two mutations. It is difficult to know whether the different phenotypes resulting from the two mutant alleles reflect expression of the dominant negative protein or leaky expression of wild-type protein from the lacZ insertion allele or other variables.
Potential Involvement of Myocardin Family Members in Human Congenital Heart Disease. The requisite roles of myocardin and MRTF-B in vascular development raises the possibility that mutations or polymorphisms in these genes may contribute to outflow tract abnormalities in humans, which represent one of the most prevalent forms of congenital heart disease (25–27). Although mice heterozygous for loss-of-function mutations in these genes do not display obvious phenotypes, it is not uncommon for heterozygous mutations in cardiovascular developmental control genes to cause congenital heart disease in humans (28). Thus, myocardin and MRTFs warrant careful consideration as culprits in forms of congenital heart disease characterized by vascular abnormalities.
Acknowledgments
We thank Da-Zhi Wang for advice and assistance during the initial phase of this project; Shelby Hacker and Hiromi Yanagisawa for India ink injection; Alisha Tizenor for graphics; Jennifer Page for editorial assistance; and Vidu Garg and Rhonda Bassel-Duby for insightful comments on the work. This work was supported by grants from the National Institutes of Health, the Donald W. Reynolds Center for Clinical Cardiovascular Research, the Muscular Dystrophy Association, and the Robert A Welch Foundation (E.N.O.).
Author contributions: J.O., J.A.R., and E.N.O. designed research; J.O. and J.A.R. performed research; J.O., J.A.R., and E.N.O. analyzed data; and J.O., J.A.R., and E.N.O. wrote the paper.
Abbreviations: MRTF, myocardin-related transcription factor; SRF, serum response factor; En, embryonic day n; SMC, smooth muscle cell.
References
- 1.Wang, D., Chang, P. S., Wang, Z., Sutherland, L., Richardson, J. A., Small, E., Krieg, P. A. & Olson, E. N. (2001) Cell 105, 851–862. [DOI] [PubMed] [Google Scholar]
- 2.Wang, D.-Z., Li, S., Hockemeyer, D., Sutherland, L., Wang, Z., Schratt, G., Richardson, J. A., Nordheim, A. & Olson, E. N. (2002) Proc. Natl. Acad. Sci. USA 99, 14855–14860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang, D.-Z. & Olson, E. N. (2004) Curr. Opin. Genet. Dev. 14, 558–566. [DOI] [PubMed] [Google Scholar]
- 4.Cen, B., Selvaraj, A. & Prywes, R. (2004) J. Cell. Biochem. 93, 74–82. [DOI] [PubMed] [Google Scholar]
- 5.Miano, J. M. (2003) J. Mol. Cell Cardiol. 35, 577–593. [DOI] [PubMed] [Google Scholar]
- 6.Owens, G. K., Kumar, M. S. & Wamhoff, B. R. (2004) Physiol. Rev. 84, 767–801. [DOI] [PubMed] [Google Scholar]
- 7.Parlakian, A., Tuil, D., Hamard, G., Tavernier, G., Hentzen, D., Concordet, J. P., Paulin, D., Li, Z. & Daegelen, D. (2004) Mol. Cell. Biol. 24, 5281–5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miano, J. M., Ramanan, N., Georger, M. A., de Mesy Bentley, K. L., Emerson, R. L., Balza, R. O., Jr., Xiao, Q., Weiler, H., Ginty, D. D. & Misra, R. P. (2004) Proc. Natl. Acad. Sci. USA 101, 17132–17137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Niu, Z., Yu, W., Zhang, S. X., Barron, M., Belaguli, N. S., Schneider, M. D., Parmacek, M., Nordheim, A. & Schwartz, R. J. (2005) J. Biol. Chem. 280, 32531–32538. [DOI] [PubMed] [Google Scholar]
- 10.Li, S., Czubryt, M. P., McAnally, J., Bassel-Duby, R., Richardson, J. A., Wiebel, F. F., Nordheim, A. & Olson, E. N. (2005) Proc. Natl. Acad. Sci. USA 102, 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang, Z., Wang, D.-Z., Teg Pipes, G. C. & Olson, E. N. (2003) Proc. Natl. Acad. Sci. USA 100, 7129–7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miralles, F., Posern, G., Zaromytidou, A. I. & Treisman, R. (2003) Cell 113, 329–342. [DOI] [PubMed] [Google Scholar]
- 13.Selvaraj, A. & Prywes, R. (2003) J. Biol. Chem. 278, 41977–41987. [DOI] [PubMed] [Google Scholar]
- 14.Cen, B., Selvaraj, A., Burgess, R. C., Hitzler, J. K., Ma, Z., Morris, S. W. & Prywes, R. (2003) Mol. Cell. Biol. 23, 6597–6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du, K. L., Chen, M., Li, J., Lepore, J. J., Mericko, P. & Parmacek, M. S. (2004) J. Biol. Chem. 279, 17578–17586. [DOI] [PubMed] [Google Scholar]
- 16.Kuwahara, K., Barrientos, T., Pipes, G. C., Li, S. & Olson, E. N. (2005) Mol. Cell. Biol. 25, 3173–3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Du, K. L., Ip, H. S., Li, J., Chen, M., Dandre, F., Yu, W., Lu, M. M., Owens, G. K. & Parmacek, M. S. (2003) Mol. Cell. Biol. 23, 2425–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li, S., Wang, D.-Z., Wang, Z., Richardson, J. A. & Olson, E. N. (2003) Proc. Natl. Acad. Sci. USA 100, 9366–9370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Skarnes, W. C., Auerbach, B. A. & Joyner, A. L. (1992) Genes Dev. 6, 903–918. [DOI] [PubMed] [Google Scholar]
- 20.Li, J., Zhu, X., Chen, M., Cheng, L., Zhou, D., Lu, M. M., Du, K., Epstein, J.A. & Parmacek, M.S. (2005) Proc. Natl. Acad. Sci. USA 102, 8916–8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, L., Miano, J. M., Mercer, B. & Olson, E. N. (1996) J. Cell Biol. 132, 849–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen, J., Kitchen, C. M., Streb, J. W. & Miano, J. M. (2002) J. Mol. Cell. Cardiol. 34, 1345–1356. [DOI] [PubMed] [Google Scholar]
- 23.Yoshida, T., Sinha, S., Dandre, F., Wamhoff, B. R., Hoofnagle, M. H., Kremer, B. E., Wang, D.-Z., Olson, E. N. & Owens, G. K. (2003) Circ. Res. 92, 856–864. [DOI] [PubMed] [Google Scholar]
- 24.Wang, Z., Wang, D.-Z., Hockemeyer, D., McAnally, J., Nordheim, A. & Olson, E. N. (2004) Nature 428, 185–189. [DOI] [PubMed] [Google Scholar]
- 25.Creazzo, T. L., Godt, R. E., Leatherbury, L., Conway, S. J. & Kirby, M. L. (1998) Ann. Rev. Physiol. 60, 267–286. [DOI] [PubMed] [Google Scholar]
- 26.Stoller, J. Z. & Epstein, J. A. (July 26, 2005) Sem. Cell Dev. Biol., 10.1016/j.semcdb.2005.06.004. [DOI]
- 27.Hoffman, J. I. & Kaplan, S. (2002) J. Am. Coll. Cardiol. 39, 1890–1900. [DOI] [PubMed] [Google Scholar]
- 28.Srivastava, D. & Olson, E. N. (2000) Nature 404, 221–226. [DOI] [PubMed] [Google Scholar]