Abstract
With the euchromatic portion of several mammalian genomes now sequenced, emphasis has turned to ascertaining the functions of gene products. A method for targeting destruction of selected proteins in mammalian cells is described, based on the ubiquitin-independent mechanism by which ornithine decarboxylase (ODC) is degraded by the 26S proteasome in collaboration with antizyme (AZ). We show that expressing whole proteins, protein domains, or peptide ligands fused to the N terminus of ODC promotes proteasome-dependent degradation of these chimeric fusion proteins and their interacting cellular target proteins. Moreover, the degradation of the interacting (targeted) protein depends on coexpression of AZ in about half of cases, providing an inducible switch for triggering the degradation process. By using 12 pairs of interacting proteins for testing, direct comparisons with several alternative strategies for achieving targeted protein destruction based on the concept of induced ubiquitination revealed advantages of the ODC/AZ system, which does not require posttranslational attachment of ubiquitin to target proteins. As proof of concept, the ODC/AZ system was used to ablate expression of specific endogenous proteins (e.g., TRAF6; Rb), and was shown to create the expected lesions in cellular pathways that require these proteins. Altogether, these findings reveal a strategy for achieving targeted destruction of cellular proteins, thus providing an additional tool for revealing the cellular phenotypes of gene products.
Keywords: antizyme, ornithine decarboxylase, proteasome, protein degradation
Functions are known for fewer than half the genes identified in mammalian genomes to date, indicating a need for gene functionalization technologies that can reveal the phenotypes of genes and their encoded proteins in various cellular contexts. Methods targeting gene-expression pathways at the level of mRNA, such as antisense oligonucleotides and small interfering RNA, offer exquisite specificity for gene silencing through Watson–Crick-type hybridization of synthetic or vector-derived nucleotide sequences to target mRNAs. These gene-silencing methods, however, sometimes prove ineffective, either because the targeted mRNAs encode long-lived stable proteins or because of functional redundancy among members of multigene families. These deficiencies have prompted attempts to inducibly target degradation of the protein products of genes by tapping into components of the eukaryotic ubiquitination machinery, which induces proteasome-dependent degradation of ubiquitin-modified proteins (reviewed in ref. 1). Accordingly, expression systems have been described in which whole proteins or fragments constituting functional protein-interaction domains or peptide ligands are expressed as chimeric fusion proteins together with modified versions of ubiquitin (2, 3), ubiquitin ligases (e.g., HECT-domain-containing proteins) (4), or adapter proteins that associate with ubiquitin ligase complexes (e.g., F-box proteins of Skp1/cullin/F-box protein complexes) (5–8), with the intention of inducing polyubiquitination of cellular proteins that bind these chimeric proteins, thus creating holes in pathways that reveal cellular phenotypes.
Three-dimensional structures of E3 ligase complexes with their substrate proteins reveal a precise geometric relationship between substrate and ligase, which poises ubiquitin-conjugating enzymes (E2s) for catalytic addition of ubiquitin onto susceptible lysine residues in the target protein (9–13). These structural constraints suggest that attempts to generically induce ubiquitination of target proteins may prove difficult because of incompatibility between substrate and ligase and, thus, limit the utility of gene-functionalization strategies predicated by ubiquitin modification of target proteins. Direct methods for targeting protein complexes for proteasome-dependent degradation without the requirement for posttranslational modification with ubiquitin conceivably could overcome this limitation.
A naturally occurring alternative pathway for proteasome-dependent protein degradation has been identified in ornithine decarboxylase (ODC) and antizyme (AZ) (reviewed in ref. 14). ODC directly binds to and is degraded by the 26S proteasome through a mechanism that is catalyzed by AZ and is independent of ubiquitin (15). We therefore tested the hypothesis that expressing protein-interaction domains as chimeric fusions with ODC could confer AZ-dependent degradation of interacting target proteins.
Materials and Methods
Plasmids. The cDNAs encoding human AZ (70–228) and human ODC (full-length) were PCR-amplified from either human placenta or human fetal brain randomly primed cDNA libraries (Stratagene). To create the C-terminal fusion cassette vectors, cDNAs encoding ODC (full-length), Siah-interacting protein (SIP) (full-length), Siah1 (full-length), monoubiquitin (full-length), and S5a (full-length) were PCR-amplified with a forward primer containing BglII site on the 5′ end and a reverse primer containing EcoRI site on the 3′ end and subcloned into the BamHI and EcoRI sites of pcDNA3 plasmid (Invitrogen) with an N-terminal Myc epitope-tag (MEQKLISEEDL) and flexible linkers (FLs) consisting of either the Bcl-2 loop (residues of 31 to 94), GGS, or AGGGSGGGGSGGGGSGGGGS. To create N-terminal fusion cassette vectors, cDNAs encoding E7 (31–105) and Fbx7 (full-length) were PCR-amplified with a forward primer containing a XhoI site on the 5′ end and a reverse primer containing ApaI site on the 3′ end and subcloned into the XhoI and ApaI sites of the pcDNA3-myc vector. Ubiquitin mutant (G76V) was generated by two-step PCR-based mutagenesis using a full-length human monoubiquitin cDNA. Adaptors, TRAF6 (271–530), TRAF2 (ΔRING), E7 (2–34), p21waf-1 (full-length), Caspase-9 (1–92), Apaf-1 (1–87), FADD (1–78), IKKα (304–758), IKKβ (305–745), and BAG-1 (full-length), were PCR-amplified and subcloned into each of the cassette vectors. To create the luciferase reporter plasmid containing an E2F binding site, primers 5′-CTGCAATTTCGCGCCAAACTTGTGCAATTTCGCGCCAAACTTGC-3′ and 5′-TCGAGCAAGTTTGGCGCGAAATTGCACAAGTTTGGC GCGAAATTGCACTCGA-3′ were annealed and ligated into pGL3E vector cleaved with SacI and HindIII (16). The reporter gene plasmid containing four tandem HIV-NFκB response elements has been described in ref. 17.
Transfections and Cell Culture. HEK293T cells were maintained in high-glucose DMEM containing 10% FCS, 1 mM l-glutamine, and antibiotics. Cells (≈5 × 105) in six-well plates were transfected with plasmid DNAs by using Lipofectamine 2000 (Invitrogen). In some cases, 50 μg/ml cycloheximide was added to prevent protein synthesis.
Immunoblots. Cells were lysed in 1 ml of HKMEN solution containing 0.5% Nonidet P-40, 0.1 μM PMSF, 5 μg/ml leupeptin, 1 μg/ml aprotinin, 1 μg/ml pepstatin, and 20 μM MG132. Lysates were analyzed by SDS/PAGE/immunoblotting using various antibodies, followed by HRPase-conjugated goat anti-mouse or anti-rabbit Ig (Amersham Pharmacia) and detected by using enhanced chemiluminescence (Amersham Pharmacia).
Pulse–Chase Analysis. For pulse–chase analysis of ectopically expressed HA-tagged TRAF6 (271–530) or IKKβ (305–745), HEK239T cells were transiently transfected in six-well plates. After 24 h, cells were pulse-labeled for 1 h with 0.1 mCi(1 Ci = 37 GBq) of [35S]methionine and [35S]cysteine per well and then chased with cold media. Cells were lysed in RIPA buffer (0.05 M Tris·HCl, pH 7.2/0.15 M NaCl/1% Triton X-100/1% deoxycholate/0.1% SDS) supplemented with protease inhibitors. After preclearing with 20 μl of protein G-Sepharose for 1 h at 4°C, HA-TRAF6 or HA-IKKβ were immunoprecipitated by using rat anti-HA monoclonal antibody (3F10, Invitrogen) adsorbed to protein G-Sepharose beads at 4°C for 4 h. After three washes with RIPA buffer, immunoprecipitates were subjected to SDS/PAGE. Dried gels were analyzed with a PhosphorImager (Molecular Dynamics).
Reporter Gene Assays. NFκB and E2F transcriptional activity were measured in HEK293T cells by transient transfection reporter gene assays. Cells at ≈50% confluence in 24-well plates were cotransfected with 0.1 μg of reporter plasmids containing NFκB- or E2F-binding sites cloned upstream of a Luciferase gene, 0.01 μgof pCMVβ-LacZ control plasmid, and 0.1 μg of various additional plasmids, as indicated. After 24 h, cells were lysed, and the relative amount of luciferase activity was measured according to the manufacturer's instructions (Promega), normalizing all values relative to β-galactosidase activity.
Results
To explore technologies for inducing proteasome-dependent degradation of target proteins, we engineered plasmids to express in mammalian cells various chimeric proteins in which a protein involved in ubiquitination mechanisms was fused to proteins or protein domains known to interact with specific target proteins. Twelve pairs of interacting proteins were studied, chosen randomly from reagents available in our laboratory, including: (i) the C-terminal TRAF domain (residues 271–530) of the adapter protein TRAF6, which binds TRAF6 (18), (ii) a peptide (residues 341–349) from the cytosolic domain of the TNF receptor (TNFR)-family protein RANK, known to bind TRAF6 (19); (iii) the cytosolic domain of TNFR-family member CD40, known to bind TRAF2 (20), (iv) I-TRAF, a TRAF2-binding protein (21); (v and vi) the leucine zipper of IKKβ, which binds IKKα and IKKβ (22); (vii) a peptide from papillomavirus E7 protein, which binds Rb; (viii) the CARD domain of proCaspase-9, which binds Apaf1; (ix) The CARD domain of Apaf-1, which binds proCaspase-9 (23); (x) the DED domain of adapter protein FADD, which binds proCaspase-8; (xi) BAG1, a Hsp70-binding cochaperone (24), and (xii) p21Waf-1, a Cdk2 inhibitor (25). Protein or peptide ligands were expressed in HEK293T cells as fusion proteins with an N or C terminus appended protein known to participate in protein-ubiquitination reactions, including (i) SIP, a protein known to bind the E3 ligase Siah1 and the Skp1 protein of Skp1/cullin/F-box protein complexes (26); (ii) Siah-1, a RING-domain containing E3 ligase (27); (iii) E7, a papillomavirus protein with reported E3 ligase activity (28, 29); (iv) a fragment of the F-box protein Fbx7, in which the substrate-binding domain was substituted, leaving the Skp1-binding region; (v) ubiquitin G76V, a nonhydrolyzable variant of ubiquitin previously used to confer proteasome-sensitivity on proteins; (vi) a tandem 4′ oligomer of ubiquitin G76V (2); and (vii) S5a, a subunit of the proteasome involved in substrate recognition (30). In most cases, the protein ligand was separated from the ubiquitin-pathway domain by a FL consisting of the sequence AGGGS(GGGGS)3 (31). All constructs included an epitope tag, allowing verification of protein production by immunoblotting and confirmation of binding to cellular target proteins by coimmunoprecipitation assays (data not shown).
The ability of these fusion proteins to induce reductions in the steady-state levels of endogenous and plasmid-expressed interacting proteins was then explored by immunoblot analysis of HEK293T cells transfected to high efficiency (>90%) with plasmids encoding the fusion proteins. The target proteins were coexpressed with epitope tags by cotransfection by using a plasmid with a strong constitutive promoter (CMV immediate-early region promoter) to ensure continuous production of target proteins and avoid artifactual reductions that might be caused by unanticipated effects of the chimeric fusion proteins on pathways that affect endogenous gene expression and to avoid false-negatives due to nontransfected cells. Although not every possible combination was tested (Table 1), none of these fusion proteins successfully reduced levels of target proteins, based on 40 combinations tested.
Table 1. Summary of tested fusion proteins.
| Ligand | Target | ODC (N) | ODC plus AZ | SIP (N) | Siah (N) | E7 (C) | Fbx7 (C) | Ub1 (N) | Ub4 (N) | S5a (N) |
|---|---|---|---|---|---|---|---|---|---|---|
| TRAF6-C | TRAF6 | ↓ | ↓ | — | — | — | — | — | — | — |
| RANK-pep. | TRAF6 | — | ↓ | — | — | — | — | — | — | — |
| CD40CT | TRAF2 | — | — | nd | nd | nd | nd | nd | nd | — |
| I-TRAF | TRAF2 | — | — | — | nd | nd | nd | nd | nd | — |
| IKKα(LZ) | IKKα | — | — | — | nd | nd | nd | nd | nd | — |
| IKKβ(LZ) | IKKβ | ↓ | ↓ | — | nd | nd | nd | nd | nd | — |
| E7 | Rb | — | ↓ | — | — | nd | — | — | — | — |
| Caspase9(CARD) | Apaf1 | — | — | — | — | — | — | — | — | nd |
| Apaf1(CARD) | Caspase9 | nd | — | — | — | — | — | — | — | nd |
| FADD(DED) | Caspase8 | nd | — | — | — | — | — | — | — | nd |
| BAG-1 | HSP70 | — | — | nd | nd | nd | nd | nd | nd | nd |
| p21 | Cdk2 | ↓ | ↓ | nd | — | — | nd | nd | nd | nd |
| Success ratio | All targets | 3/12 | 5/12 | 0/9 | 0/7 | 0/6 | 0/6 | 0/6 | 0/6 | 0/7 |
N, N-terminal fusion; C, C-terminal fusion; nd, not done; ↓, decrease; —, no change.
Testing of an AZ-Based System for Targeted Protein Degradation. Given the lack of efficacy of fusion proteins based on known components of the ubiquitination machinery to effectively induce degradation of interacting proteins, we turn to an alternative strategy based on knowledge of the mechanism by which ODC is degraded by the proteasome through ubiquitin-independent mechanisms. For this purpose, protein ligands were expressed as fusion proteins with ODC at their C termini, thus exposing the C terminus of ODC, which is known to bind the proteasome independently of ubiquitin.
A FL sequence was also inserted between the protein ligands and ODC. Three types of linkers were tested, including a 63-aa segment from the Bcl-2 protein (residues 31–94), which is known from structural studies to constitute a nonstructured, flexible peptide rich in prolines and glycines (32), a Gly-Gly-Pro tripeptide, and the sequence [Gly-Gly-Gly-Gly-Ser]3. These ODC fusion proteins were then expressed in HEK293T cells alone or in combination with AZ, which binds ODC and catalyzes its degradation by the 26S proteasome (33) (Fig. 1). Again, protein targets were coexpressed from plasmids with epitope tags for initial experiments, and their levels of expression were evaluated by SDS/PAGE/immunoblotting.
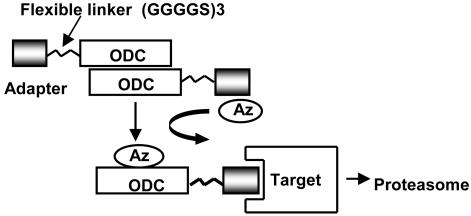
Fig. 1.
Model for AZ-assisted degradation of target proteins by ODC chimeric fusion proteins. A protein-interaction domain (adapter) is expressed with ODC attached to its C terminus with a FL sequence AGGGS(GGGGS)3. ODC is known to form homodimers. The ODC-adapter fusion binds target proteins and docks on the 26S proteasome. AZ catalyzes degradation of the ODC-adapter protein by the proteasome along with the interacting target protein.
Of the 12 pairs of protein interactions tested by using the ODC/AZ system, 5 resulted in specific reductions of the target protein (Table 1). For two of these five successful knock-downs, reductions of target protein were entirely dependent on coexpression of AZ with the ODC chimeric fusion protein, whereas, in another case, AZ enhanced the reduction caused by the ODC chimeric fusion alone. In the other two successful cases, the ODC chimeric fusion protein was sufficient by itself, suggesting that fusing certain proteins to ODC may supplant the need for AZ. In this regard, the AZ protein is known to induce a conformational change in ODC that exposes a proteasome-binding domain in its C terminus (33), raising the possibility that some fusion partners mimic this effect of AZ.
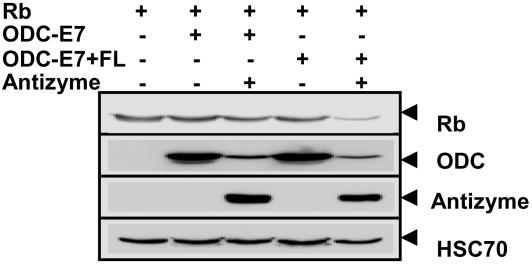
We empirically determined that inclusion of a FL between ODC and the protein-interaction domain can be critical for successful degradation of cellular substrates, with the [Gly-Gly-Gly-Gly-Ser]3 linker yielding best results. Fig. 2 provides an example, comparing the efficiency of a ODC fusion containing a Rb-binding fragment (amino acids 2–34) expressed with or without a FL, with respect to degradation of cellular target Rb.
Fig. 2.
Requirement of FL for targeted degradation of Rb by ODC-F7 peptide. HEK293T cells were transiently transfected with 0.2 μg of plasmid encoding HA-Rb (0.5 μg), ODC (0.5 μg), ODC-E7 peptide(0.5 μg), ODC-E7 peptide plus FL (GGGGS) (0.5 μg), or myc-AZ (0.5 μg), in various combinations as indicated (total DNA amount normalized). After 24 h, cell lysates were prepared from duplicate dishes of transfectants, normalized for total protein content (20 μg per lane), and analyzed by SDS/PAGE/immunoblotting using antibodies specific for HA (Rb), Myc (ODC or AZ), or HSC70 (as a control) with enhanced-chemiluminescence-based detection.
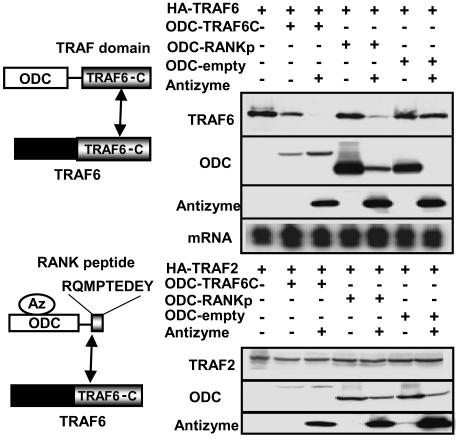
Analysis of Specificity of AZ-Based Protein Target Degradation. We performed a variety of experiments to explore the specificity of the ODC/AZ system for inducing target-protein reductions. For example, the target protein TRAF6 is a member of a family of six mammalian adapter proteins with differential specificity for peptidyl motifs located in the cytosolic domains of TNF-family receptors (18). A C-terminal region in these proteins (C-TRAF domain) is responsible for TNFR binding. These adapter proteins also form homotrimers through the proximal portion of their TRAF domains (34). We therefore contrasted the levels of TRAF6 and TRAF2 proteins in cells expressing ODC chimeric fusion proteins containing either the TRAF domain of TRAF6 or a TRAF6-binding peptidyl motif from the cytosolic domain of RANK (“RANKp”) (Fig. 3). ODC nonfusion protein served as a negative control. As shown in Fig. 3, coexpressing ODC-C-TRAF6 caused reductions in the levels HA-TRAF6 but not HA-TRAF2 protein. Cotransfection of AZ further decreased levels of HA-TRAF6 but not HA-TRAF2. Levels of HA-TRAF6 were reduced in cells expressing ODC-RANKp only when AZ was coexpressed. ODC-RANKp did not affect levels of HA-TRAF2, confirming the specificity of these results. ODC control protein also did not alter levels of HA-TRAF6 or HA-TRAF2.
Fig. 3.
Selective degradation of TRAF6 but not TRAF2 by ODC chimeric fusion proteins. Duplicate cultures of HEK293T cells were transiently transfected with 0.2 μg of plasmid encoding HA-TRAF6 (0.5 μg) (Upper), HA-TRAF2 (0.5 μg) (Lower), ODC (0.5 μg), ODC-C-TRAF6 (0.5 μg), ODC-RANKp (0.5 μg), or myc-AZ (0.5 μg), in various combinations as indicated (total DNA amount normalized). After 24 h, cell lysates were prepared for analysis of either protein or mRNA. Immunoblot analysis was performed by using detergent lysates normalized for total protein content (20 μg per lane) by SDS/PAGE/immunoblotting using antibodies specific for HA (TRAF6; TRAF2), Myc (ODC or AZ) with enhanced-chemiluminescence-based detection. The relative levels of TRAF6 mRNA were analyzed by Northern blotting using sample normalized for total RNA content (10 μg per lane). Ethidium bromide staining verified loading of equivalent amounts of RNA for each sample (data not shown).
Immunoblot analysis confirmed production of the ODC-chimeric fusion proteins and AZ in the transfected cells. Note that accumulation of ODC-C-TRAF6 was markedly reduced, compared with ODC-RANKp, suggesting that fusing C-TRAF6 to ODC promotes its proteasome-dependent degradation independent of AZ. As expected, reductions in ODC-RANKp were induced by coexpressing AZ, consistent with AZ-dependent degradation of this ODC chimeric fusion protein. Thus, we surmise that some ODC chimeric fusion proteins spontaneously associate with and are efficiently degraded by the 26S proteasome (e.g., ODC-C-TRAF6), whereas others (e.g., ODC-RANKp) require AZ as a cofactor for their degradation, like wild-type ODC.
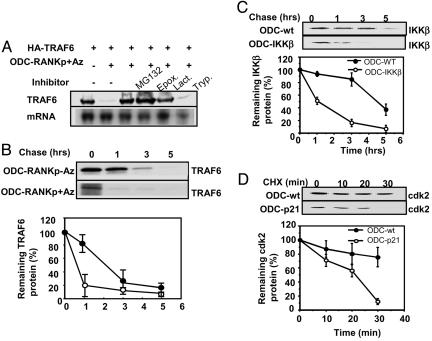
AZ/ODC System Increases Rate of Target Protein Degradation. Next, we undertook experiments to determine the mechanism by which target protein reductions were achieved when using the ODC/AZ system. First, we determined the effect of ODC chimeric fusion proteins on the level of mRNA encoding target proteins, anticipating that mRNA levels should be unchanged. Analysis of TRAF6 mRNA levels in cells transfected with plasmids encoding AZ and either ODC-C-TRAF6 or ODC-RANKp fusion proteins confirmed no effect on expression at the mRNA level (Fig. 3). Second, we explored the effects of pharmacological inhibitors of the 26S proteasome. Fig. 4A shows an example where HEK293T cells were cotransfected with a plasmid encoding HA-TRAF6 alone or in combination with plasmids encoding AZ and an ODC chimeric fusion protein containing a TRAF6-binding peptide from the cytsolic domain of RANK (RANKp). Coexpression of ODC-RANKp and AZ with HA-TRAF6 resulted in profound reductions in the steady-state levels of HA-TRAF6 protein, as determined by immunoblotting. Culturing these transfected cells with proteasome inhibitors MG132, epoximycin, or lactacystin restored HA-TRAF6 levels. In contrast, a trypsin inhibitor, used here as a control, was ineffective (Fig. 4A). These data demonstrate that ODC/AZ-induced degradation of target proteins is proteasome-dependent. Third, we determined the effects of the ODC/AZ system on protein half-life using 35S-l-methionine pulse–chase methods. Fig. 4B shows results comparing the half-life of HA-TRAF6 in cells cotransfected with ODC-RANKp, with or without AZ. In cells expressing AZ, the half-life of HA-TRAF6 was reduced from ≈2 hto <1 h, consistent with target protein degradation occurring via an AZ-dependent mechanism. We conclude, therefore, that the ODC/AZ system induced proteasome-dependent degradation of target proteins without affecting mRNA expression. Pulse–chase experiments were also performed for IKKβ, comparing cells transfected with plasmids encoding ODC versus ODC-IKKβ. The starting levels of IKKβ were lower in cells expressing ODC-IKKβ before initiating the chase, suggesting ongoing degradation. Cold l-methionine chase revealed that, indeed, the rate of degradation of IKKβ was faster in cells expressing ODC-IKKβ, compared with ODC control (Fig. 4C). Fourth, we also used another approach to gauge the rates of target-protein degradation where cells were transfected with ODC-expressing plasmids and then protein synthesis was shut off a day later by adding cycloheximide to cultures. Using ODC-p21 as an example, we compared the rate of degradation of the p21 target Flag-tagged Cdk2 in HEK293T cells transfected with ODC-control or ODC-p21 plasmids. Before cycloheximide treatment, steady-state levels of Flag-tagged Cdk2 were lower in the ODC-p21-expressing cells compared with ODC-control cells (Fig. 4D), suggesting ongoing degradation. After addition of cycloheximide, the rate of decline in Cdk2 protein levels was faster in ODC-p21-expressing cells, as determined by densitometric quantification of immunoblot data developed by using an anti-Flag antibody with chemiluminescent detection.
Fig. 4.
AZ-dependent, proteasome-dependent degradation of target protein induced by ODC chimeric fusion protein. (A) Proteasome-dependent degradation of TRAF6 by ODC-RANKp peptide. HEK293T cells were transiently transfected with plasmids encoding HA-TRAF6 (0.5 μg), ODC-RANKp (0.5 μg), or myc-AZ (0.5 μg) in various combinations, as indicated (total DNA amount normalized). After 24 h, cells were cultured with or without 1 μM MG132, 1 nM epoximycin (Epox), 10 μM lactacystin (Lact), or 1 μM Trypsin inhibitor (Tryp) for 6 h. Cell lysates were prepared from duplicated dishes of transfectants, normalized for total protein content (20 μg per lane), and analyzed by SDS/PAGE/immunoblotting using antibodies specific for HA. (B) Pulse–chase analysis of TRAF6 protein-degradation rate in ODC-RANKp transfected cells. HEK239T cells were transiently cotransfected with plasmids encoding HA-TRAF6 and ODC-RNAKp, with or without myc-AZ. After 24 h, cells were pulse-labeled with [35S]methionine and [35S]cysteine in methionine/cysteine-free medium, and chased with media lacking the labeled amino acids. Cells were lysed at the indicated times, and HA-TRAF6 was recovered by immunoprecipitation by using HA antibody. Immunecomplexes were subjected to SDS/PAGE, and dried gels were analyzed by PhosphorImager (Upper). Data from pulse–chase analysis are presented as the average (±SE) from duplicate experiments (Lower). The blots shown are representative of duplicate experiments. (C) Pulse–chase analysis of IKKβ protein-degradation rate. HEK293T cells were transiently cotransfected with plasmids encoding HA-IKKβ, with ODC or ODC-IKKβ. After 24 h, cells were pulse-labeled with [35S]methionine and [35S]cysteine in methionine/cystein-free medium and chased with media lacking the labeled amino acids. Cells were lysed at the indicated times, and HA-IKKβ was recovered by immunoprecipitation by using HA antibody. Immunecomplexes were subjected to SDS/PAGE, and dried gels were analyzed by PhosphorImager (Upper). Data from pulse–chase analysis are presented as the average (±SE) from duplicate experiments (Lower). The blots shown are representative of duplicate experiments. (D) The analysis of Flag-Cdk2 protein-degradation rate after cycloheximide treatment. HEK293T cells (six wells) were transiently cotransfected with plasmids encoding Flag-Cdk2 (0.5 μg), with ODC (2 μg) or ODC-p21 (2 μg). After 24 h, cells were treated with cycloheximide (50 μg/ml), lysed at the indicated times and analyzed by SDS/PAGE/immunoblotting using antibodies specific for Flag (Upper). Data are presented as the average (±SE) from duplicate experiments (Lower). The blots shown are representative of duplicate experiments.
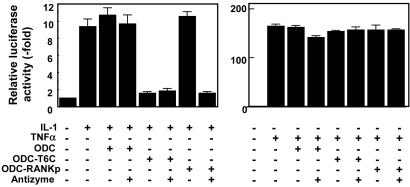
Modulating Cellular Pathways by Using AZ-Based Targeted Protein Degradation. Finally, we explored whether the ODC/AZ system could be used to successfully ablate the function of endogenous proteins. First, we examined the effects of ODC-C-TRAF6 and ODC-RANKp on induction of NFκB activity in HEK293T cells exposed to either TRAF6-dependent (e.g., IL-1) or independent (e.g., TNFα) cytokines (Fig. 5). IL-1 induced marked increases in NFκB activity in HEK293T cells, as determined by reporter gene assays, which were reduced to near baseline levels by expression of ODC-C-TRAF6 or by coexpression of ODC-RANKp with AZ. As expected, ODC control protein had no effect on NFκB induction by IL-1, confirming the specificity of these results. In contrast to the effects of ODC-C-TRAF6 and ODC-RANKp chimeric fusion proteins on IL-1 signaling, induction of NFκB activity by TNFα was unimpaired, consistent with the differential use of TRAF-family adapter proteins by IL-1 (e.g., TRAF6) and TNFα (e.g., TRAF2). These data thus parallel the differential effects of these ODC chimeric fusion proteins on TRAF6 and TRAF2 protein levels (Fig. 3).
Fig. 5.
Functional ablation of endogenous TRAF6 by ODC/AZ system. HEK293T cells were transiently transfected with a reporter gene plasmid (0.1 μg) that contains a NFκB-responsive element cloned upstream of a luciferase reporter gene, with 0.01 μg of pCMVβ-gal as a transfection-efficiency control and 0.1 μg of the indicated plasmids encoding ODC, ODC-TRAF6C, ODC-RANKp, or AZ, in various combinations as indicated (total DNA amount normalized). After 24 h, cells were treated with 50 ng/ml IL-1 (Left) or 10 ng/ml TNFα (Right) for an additional 24 h, and luciferase activity was measured in cell lysates and normalized relative to β-galactosidase (mean ± SD; n = 3).
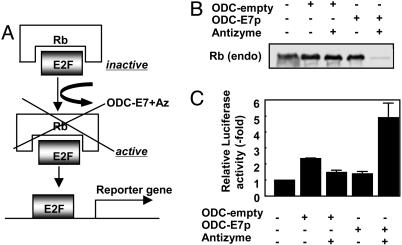
We extended these studies of effects of the ODC/AZ system on endogenous proteins to the tumor suppressor Rb. The Rb protein binds and suppresses E2F-family transcription factors, thus preventing them from activating target genes (35). We therefore expressed in cells an ODC chimeric fusion protein containing a Rb-binding peptide from the E7 protein, alone or in combination with AZ (Fig. 6A). Analysis of the levels of endogenous Rb protein by immunoblotting showed that the combination of ODC-E7p and AZ induced nearly complete disappearance of the Rb protein. In contrast, Rb protein levels were unaffected either by expressing ODC-E7p without AZ, or by expressing ODC control protein with or without AZ (Fig. 6B). Measurements of E2F activity by using reporter gene assays demonstrated a marked increase in cells coexpressing ODC-E7p and AZ-1 (Fig. 6C), consistent with the observed loss of Rb protein. We conclude, therefore, that the ODC/AZ system is capable of ablating the function of endogenous target proteins in cells.
Fig. 6.
Ablation of endogenous Rb expression by using ODC/AZ system. (A) Scheme for E2F activation by AZ-assisted degradation of Rb proteins by ODC-E7p. (B) Degradation of endogenous Rb protein by ODC-E7p. HEK293T cells (100-mm dish) were transiently transfected with 2 μg of plasmid encoding ODC (2 μg), ODC-E7p (2 μg), or myc-AZ (2 μg) in various combinations, as indicated (total DNA amount normalized). After 48 h, lysates were normalized for total protein content and subjected to immunoprecipitation by using 1 μg of anti-Rb monoclonal antibody. The resulting immunecomplexes were analyzed by SDS/PAGE/immunoblotting using an anti-Rb monoclonal antibody with enhanced-chemiluminescence-based detection. (C) Effect of ODC-E7p on E2F transcriptional activity. HEK293T cells were transiently transfected with a reporter gene plasmid (0.1 μg) that contains an E2F responsive element cloned upstream of a luciferase reporter gene, 0.01 μg of pCMVβ-galactosidase as an transfection-efficiency control, and 0.1 μg of the indicated plasmids encoding ODC, ODC-E7p, or AZ, in various combinations as indicated (total DNA amount normalized). Luciferase activity was measured in cell lysates 24 h later and normalized relative to β-galactosidase (mean ± SD; n = 3).
Discussion
Previous studies have demonstrated that ODC is degraded by the 26S proteasome through a ubiquitin-independent mechanism, whereby AZ binding induces exposure of the C terminus of ODC and accelerates its degradation by 50- to 100-fold. Normally, this pathway is induced in response to polyamines (spermine, spermidine, and putresine), which triggers AZ production, thus providing a negative feedback loop for maintaining appropriate intracellular levels of these molecules (reviewed in ref. 14). We exploited the ODC/AZ system for targeting degradation of selected proteins in cells. The ODC/AZ system affords the advantage over most ubiquitin-pathway-based strategies that posttranslational modification of the target protein (by ubiquitination) is not required, thus providing a more direct means of delivering ligand/target complexes to the proteasome for degradation. Indeed, compared with a variety of ubiquitin-pathway-based approaches examined, we found the ODC/AZ system to be more effective at achieving degradation of a test set of 12 target proteins for which interacting proteins are known. However, successful degradation was achieved in only 5 of 12 test cases, suggesting that some proteins are recalcitrant to this targeting approach. Multiple explanations could account for the intractability of certain protein targets, including (i) insufficient affinity interactions of the protein ligands with their cellular target proteins; (ii) dissociation of ligand and target during digestion of the ODC-ligand fusion by the proteasome, thus stripping the target protein off; and (iii) impeded entry of the target protein into the pore of the proteasome because of rigid protein structure, necessitating protein unfolding. Thus, the tractability of specific protein targets to degradation by the ODC/AZ system must be empirically determined.
A potential concern with the ODC/AZ system is that expression of ODC-fusion proteins or AZ in cells may alter polyamine levels, leading to artifactual changes in cell growth, chromatin structure, or other cellular events. Measurement of cell-division rates for HeLa and HEK293T cells used for our experiments revealed no apparent effect of ODC-fusion proteins or AZ (data not shown), suggesting that at least some types of cells are not particularly sensitive to these manipulations. However, more subtle changes in cells overexpressing ODC fusion proteins and AZ conceivably may occur and therefore should be considered in interpreting data derived from use of the ODC/AZ-based approach to targeted protein degradation. The suitability of this approach may also be dependent on the endogenous levels of AZ and AZ inhibitors in particular types of primary cells or cell lines.
In this regard, although the findings reported here provide proof-of-concept evidence that the ODC/AZ system can be exploited for targeted protein degradation, a variety of future improvements on the basic system can be envisioned. For instance, mutant versions of ODC that lack enzymatic activity but which preserve proteasome-dependent degradation could obviate untoward effects on polyamine synthesis, particularly nondimerizing mutants which cannot bind endogenous ODC. Similarly, production of complementary pairs of ODC and AZ mutants that bind each other but fail to interact with their endogenous (wild-type) counterparts would also provide a means to avoid effects on polyamine synthesis. It should be noted, however, that AZ may have additional cellular targets besides ODC, a possibility that must be considered, including cyclin D1 and Smad1, at least in certain type of cells (36, 37). One possible advantage of the ability of polyamines to induce AZ expression is that it might be possible to forego transfection of AZ-encoding plasmid by adding polyamines or polyamine analogues to the cell cultures.
Results obtained with the ODC/AZ system and other previously reported approaches for inducing proteasome-dependent degradation of specific proteins (2, 3, 5–8) should be interpreted with understanding that the particular protein ligand chosen may have multiple cellular targets, including unknown or unanticipated protein targets in addition to known interacting proteins intended for targeted degradation. Thus, phenotypes created by these targeted protein-degradation methods could potentially reflect the loss of expression of several interacting proteins. When searching for functions of gene products where more specific methods such as antisense or small interfering RNA have failed to yield phenotypes, ablating the expression of the interrogated gene product's interacting partners may provide clues for eventually understanding its function. Moreover, targeted protein-degradation methods that attack interacting proteins afford an approach for dealing with multigene families, where a particular protein ligand may interact with multiple members of a family of homologous gene products, thereby ablating expression simultaneously of several redundant members and revealing phenotypes that would be undetected by nucleic-acid-based methods for silencing gene expression at the mRNA.
Acknowledgments
We thank Dr. Deveraux (Genomics Institute of the Novartis Research Foundation) for S5a cDNA, R. Newman for helpful discussions, and M. Fariborzi for technical assistance. This work was supported by National Institutes of Health Grant CA69381 and by ISIS Pharmaceuticals (Carlsbad, CA).
Author contributions: S.M. and J.C.R. designed research; M.C., S.M., and T.F. performed research; S.M. and J.C.R. analyzed data; and S.M. and J.C.R. wrote the paper.
Abbreviations: AZ, antizyme; FL, flexible linker; ODC, ornithine decarboxylase.
References
- 1.Varshavsky, A. (1997) Trends Biochem. Sci. 22, 383–387. [DOI] [PubMed] [Google Scholar]
- 2.Stack, J. H., Whitney, M., Rodems, S. M. & Pollok, B. A. (2000) Nat. Biotechnol. 18, 1298–1302. [DOI] [PubMed] [Google Scholar]
- 3.Lindsten, K., Menendez-Benito, V., Masucci, M. G. & Dantuma, N. P. (2003) Nat. Biotechnol. 21, 897–902. [DOI] [PubMed] [Google Scholar]
- 4.Colas, P., Cohen, B., Ko Ferrigno, P., Silver, P. A. & Brent, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13720–13725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou, P., Bogacki, R., McReynolds, L. & Howley, P. M. (2000) Mol. Cell 6, 751–756. [DOI] [PubMed] [Google Scholar]
- 6.Liu, J., Stevens, J., Matsunami, N. & White, R. L. (2004) Biochem. Biophys. Res. Commun. 313, 1023–1029. [DOI] [PubMed] [Google Scholar]
- 7.Sakamoto, K. M., Kim, K. B., Kumagai, A., Mercurio, F., Crews, C. M. & Deshaies, R. J. (2001) Proc. Natl. Acad. Sci. USA 98, 8554–8559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Su, Y., Ishikawa, S., Kojima, M. & Liu, B. (2003) Proc. Natl. Acad. Sci. USA 100, 12729–12734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang, L., Kinnucan, E., Wang, G., Beaudenon, S., Howley, P., Huibregtse, J. & Pavletich, N. (1999) Science 286, 1321–1326. [DOI] [PubMed] [Google Scholar]
- 10.Zheng, N., Wang, P., Jeffrey, P. D. & Pavletich, N. P. (2000) Cell 102, 533–539. [DOI] [PubMed] [Google Scholar]
- 11.Schulman, B. A., Carrano, A. C., Jeffrey, P. D., Bowen, Z., Kinnucan, E. R., Finnin, M. S., Elledge, S. J., Harper, J. W., Pagano, M. & Pavletich, N. P. (2000) Nature 408, 381–386. [DOI] [PubMed] [Google Scholar]
- 12.Zheng, N., Schulman, B. A., Song, L., Miller, J. J., Jeffrey, P. D., Wang, P., Chu, C., Koepp, D. M., Elledge, S. J., Pagano, M., et al. (2002) Nature 416, 703–709. [DOI] [PubMed] [Google Scholar]
- 13.Wu, G., Xu, G., Schulman, B. A., Jeffrey, P. D., Harper, J. W. & Pavletich, N. P. (2003) Mol. Cell 11, 1445–1456. [DOI] [PubMed] [Google Scholar]
- 14.Coffino, P. (2001) Nat. Rev. Mol. Cell Biol. 2, 188–194. [DOI] [PubMed] [Google Scholar]
- 15.Murakami, Y., Matsufuji, S., Kameji, T., Hayashi, S., Igarashi, K., Tamura, T., Tanaka, K. & Ichihara, A. (1992) Nature 360, 597–599. [DOI] [PubMed] [Google Scholar]
- 16.Krek, W., Livingston, D. M. & Shirodkar, S. (1993) Science 262, 1557–1560. [DOI] [PubMed] [Google Scholar]
- 17.Galang, C. K., Der, C. J. & Hauser, C. A. (1994) Oncogene 9, 2913–2921. [PubMed] [Google Scholar]
- 18.Ishida, T., Mizushima, S.-I., Azuma, S., Kobayashi, N., Tojo, T., Suzuki, K., Aizawa, S., Watanabe, T., Mosialos, G., Kieff, E., et al. (1996) J. Biol. Chem. 271, 28745–28748. [DOI] [PubMed] [Google Scholar]
- 19.Ye, H., Arron, J. R., Lamothe, B., Cirilli, M., Kobayashi, T., Shevde, N. K., Segal, D., Dzivenu, O. K., Vologodskaia, M., Yim, M., et al. (2002) Nature 418, 443–447. [DOI] [PubMed] [Google Scholar]
- 20.Rothe, M., Sarma, V., Dixit, V. M. & Goeddel, D. V. (1995) Science 269, 32767–32770. [DOI] [PubMed] [Google Scholar]
- 21.Rothe, M., Xiong, J., Shu, H.-B., Williamson, K., Goddard, A. & Goeddel, D. V. (1996) Proc. Natl. Acad. Sci. USA 93, 8241–8246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Woronicz, J. D., Gao, X., Cao, Z., Rothe, M. & Goeddel, D. V. (1997) Science 278, 866–869. [DOI] [PubMed] [Google Scholar]
- 23.Zhou, P., Chou, J., Olea, R. S., Yuan, J. & Wagner, G. (1999) Proc. Natl. Acad. Sci. USA 96, 11265–11270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takayama, S., Bimston, D. N., Matsuzawa, S., Freeman, B. C., Aime-Sempe, C., Xie, Z., Morimoto, R. J. & Reed, J. C. (1997) EMBO J. 16, 4887–4896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harper, J. W., Adami, G. R., Wei, N., Keyomarsi, K. & Elledge, S. J. (1993) Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- 26.Matsuzawa, S. & Reed, J. C. (2001) Mol. Cell 7, 915–926. [DOI] [PubMed] [Google Scholar]
- 27.Matsuzawa, S., Takayama, S., Froesch, B. A., Zapata, J. M. & Reed, J. C. (1998) EMBO J. 17, 2736–2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boyer, S. N., Wazer, D. E. & Band, V. (1996) Cancer Res. 56, 4620–4624. [PubMed] [Google Scholar]
- 29.Jones, D. L., Thompson, D. A., Suh-Burgmann, E., Grace, M. & Munger, K. (1999) Virology 258, 406–414. [DOI] [PubMed] [Google Scholar]
- 30.Deveraux, Q., Jensen, C. & Rechsteiner, M. (1995) J. Biol. Chem. 270, 23726–23729. [DOI] [PubMed] [Google Scholar]
- 31.Hoedemaeker, F. J., Signorelli, T., Johns, K., Kuntz, D. A. & Rose, D. R. (1997) J. Biol. Chem. 272, 29784–29789. [DOI] [PubMed] [Google Scholar]
- 32.Chang, B. S., Minn, A. J., Muchmore, S. W., Fesik, S. W. & Thompson, C. B. (1997) EMBO J. 16, 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murakami, Y., Matsufuji, S., Hayashi, S.-I., Tanahashi, N. & Tanaka, K. (2000) Biochem. Biophys. Res. Commun. 267, 1–6. [DOI] [PubMed] [Google Scholar]
- 34.Arch, R. H., Gedrich, R. W. & Thompson, C. B. (1998) Genes Dev. 12, 2821–2830. [DOI] [PubMed] [Google Scholar]
- 35.Chellappan, S. P., Hiebert, S., Mudryj, M., Horowitz, J. M. & Nevins, J. R. (1991) Cell 65, 1053–1061. [DOI] [PubMed] [Google Scholar]
- 36.Newman, R. M., Mobascher, A., Mangold, U., Koike, C., Diah, S., Schmidt, M., Finley, D. & Zetter, B. R. (2004) J. Biol. Chem. 279, 41504–41511. [DOI] [PubMed] [Google Scholar]
- 37.Lin, Y., Martin, J., Gruendler, C., Farley, J., Meng, X., Li, B. Y., Lechleider, R., Huff, C., Kim, R. H., Grasser, et al. (2002) BMC Cell Biol. 3, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]