Abstract
Some species of ciliates undergo massive DNA elimination and genome rearrangement to construct gene-sized “chromosomes” in their somatic nucleus. An example is the extensively scrambled DNA polymerase α gene that is broken into 48 pieces and distributed over two unlinked loci in Stylonychia. To understand the emergence of this complex phenomenon during evolution, we examined DNA polymerase α genes in several earlier diverging species, representing evolutionary intermediates. Mapping these data onto an evolutionary tree suggests that this gene became extensively fragmented and scrambled over evolutionary time through a series of steps, each leading to greater complexity. Our results also suggest a possible mechanism for intron loss by deletion of intron sequences as DNA during development of the somatic nucleus.
Keywords: ciliate, DNA polymerase, hypotrich, intron loss, spirotrich
Ciliates comprise a diverse group of microbial eukaryotes, or protists. These organisms contain two types of nuclei: a somatic macronucleus that is active during vegetative growth and a germ-line micronucleus that is quiescent except during sexual recombination (1). During sexual conjugation, the two mating cells exchange haploid micronuclei, after which the old macronucleus degrades, and the new diploid micronucleus develops into the new macronucleus. Extensive loss of DNA from the long micronuclear chromosomes, coupled to DNA rearrangement, removes transposons and most intergenic spacer DNA between genes, as well as intragenic spacer DNA, known as internal eliminated segments (IESs). In spirotrichous (formerly hypotrichous) ciliates, the remaining DNA sequences, known as macronuclear destined segments (MDSs), can occupy as little as 2-5% of the germ-line genome. These mostly coding regions assemble together to form ≈24,000 types of DNA molecules in the somatic nucleus. Because their average size is just 2.2 kbp and they usually contain only one gene, they are sometimes called “nano-chromosomes.” Short telomeres are added to both ends before these molecules are amplified several-thousand-fold (1-3).
Furthermore, the germ-line order of the coding regions in 20-30% of the genes in stichotrichous ciliates (a subset of spirotrichous ciliates) are permuted, or scrambled, relative to their linear order in the macronucleus. Although the mechanism of reordering these segments is not well understood, short direct repeats, called pointers, may guide the unscrambling process (1, 4). Invariably, one copy of a repeated word pair is present at the 3′ end of germ-line segment n and at the 5′ end of segment n + 1, providing the information to link them together, with only one copy of the repeated word retained in the final DNA molecule (1). Although fragmented genes do occur in other organisms [examples include V(D)J recombination in the vertebrate immune system and split ribosomal RNA genes in the mitochondria of fungi (5) and protists (6, 7)], rarely are they sewn back together at the DNA level.
Three different scrambled genes, actin I (8-10), α telomerebinding protein (α-TBP) (11, 12), and DNA polymerase α (pol α) (4, 13), have been studied in ciliates. The DNA pol α gene is particularly beguiling for two reasons: it has the highest density of scrambled segments (by a factor of 3), and both PCR linkage analysis and population genetic evidence suggest that these segments are distributed over two unlinked loci in Stylonychia lemnae, representing the related ciliates known as oxytrichids (4, 14).
One hypothesis repeated in the literature has been that scrambled genes arise in two main events, first by fragmentation of a gene into many pieces, followed by permuting the order of these segments, possibly by recombination between the noncoding DNA sequences (IESs) that separate them (15, 16). To test this hypothesis, we set out to describe and compare the germ-line and somatic architectures of the DNA pol α gene from several ciliates that diverged before the oxytrichids, by using this complex case as a model system to probe the evolutionary origin of a scrambled gene. We found that the evolutionary transition from nonscrambled to scrambled forms displays striking increases in complexity. Our results also suggest that extensive fragmentation of a gene is not a prerequisite for scrambling or permuting the order of its germ-line segments over evolutionary time. Another observation from our study is that introns may be lost by eliminating them as DNA instead of RNA, along with the deletion of other noncoding DNA segments that interrupt genes (IESs). This observation could help explain the reported relative paucity of introns in ciliates (1).
Materials and Methods
Ciliate Culture and DNA Extraction. Ciliates isolated from lakes and soils in the Princeton, NJ, area were characterized by morphology to the genus level. Holosticha sp. is morphologically similar to Holosticha kessleri (17) and Uroleptus sp. is similar to Uroleptus gallina. Species of interest were isolated and grown as described (17). Macronuclei and micronuclei were separated before DNA extraction as described (17). U. grandis DNA was a generous gift from David Prescott (University of Colorado, Boulder). This species is difficult to find, and RNA was unavailable.
rDNA Amplification and Phylogenetic Construction. Small subunit ribosomal DNA sequences were PCR amplified with universal primers (18) and deposited into the GenBank database (Holosticha sp., AY294647; Uroleptus sp., AY294646; Paraurostyla weissei, AY294648). The rDNA sequence from locally isolated P. weissei was 100% identical to a sequence in the database from the same species (AF164127), confirming its identification, whereas that of the other two species did not match any previously reported sequence.
To determine the phylogenetic relationships among these new isolates and other ciliates, we manually aligned these rDNA sequences with those from seven other spirotrichous ciliates and one heterotrichous ciliate, Nyctotherus ovalis, taking into account the secondary structure of the RNA in the rRNA database (19). We used N. ovalis as an outgroup, because this anaerobic ciliate is paraphyletic to spirotrichs (20). Phylogenetic analyses were conducted by using paup 4.0 (21) for optimality criteria maximum parsimony (MP) and minimum evolution (ME) and treepuzzle 5.0 (22) for quartet-puzzling (QP) tree reconstruction. Of 1,607 sites compared, 120 were parsimony-informative. For QP tree reconstruction, we used modeltest 3.06 (23) in conjunction with paup and found TrN+I+Gamma the best model to perform this test. The consensus MP tree is shown in Fig. 1 Left, and branch supports derived from the MP (10,000 bootstrap replicates), ME (10,000 bootstrap replicates), and QP (10,000 replicates) analyses are reported under branching nodes. rDNA sequences from the database are as follows: N. ovalis AJ222678, Euplotes aediculatus X03949, U. grandis AF164129, Uroleptus pisces AF164131, U. gallina AF164130, Sterkiella histriomuscorum (Oxytricha trifallax) AF164121, S. lemnae AF164124, Sterkiella nova (Oxytricha nova) X03948, and Stylonychia pustulata X03947.
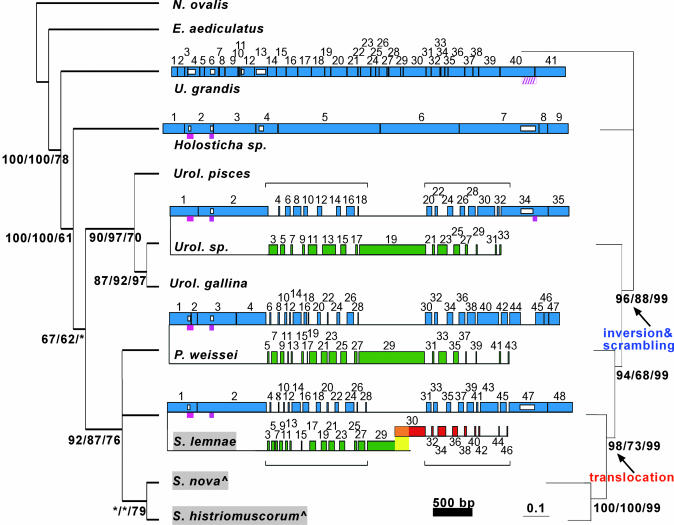
Fig. 1.
Phylogenetic analysis of the scrambled DNA pol α gene. (Left) Consensus cladogram of 19,501 most-parsimonious trees constructed with small subunit rDNA sequences for 10 spirotrichs and one heterotrich, N. ovalis. Numbers under internal branches give branch supports for maximum parsimony/minimum evolution/quartet-puzzling; branches <60% collapsed or labeled *.^, S. nova = O. nova; S. histriomuscorum = O. trifallax.(Right) Unrooted phylogram inferred from concatenated protein sequences of actin I, α-TBP, DNA pol α, and eukaryotic release factor I (eRFI). Branch supports shown for maximum parsimony/quartet-puzzling/Bayesian analyses. In the center, comparison of germ-line DNA pol α orthologs in U. grandis, Holosticha, Uroleptus, and P. weissei with S. lemnae, representing the Oxytrichidae (highlighted gray) as its sequence is the most complete; the germ-line maps for these three orthologs are very similar (4, 15, 28). Large boxes are MDSs (to scale) numbered based on somatic order; horizontal lines are IESs (not to scale); and black vertical lines indicate pointers. Confirmed introns are magenta boxes; and the putative U. grandis intron is hatched. Small white boxes represent alignment gaps ≥40 bp, for example, the absence of, or length variation in, an intron or MDS (Fig. 4). Green MDSs are upstream/inverted relative to blue MDSs; red MDSs on an unlinked locus. Horizontal brackets highlight two regions of extensive scrambling. The yellow region in S. lemnae is a 199-bp overlap between major/minor loci (4). Tables 1-4 provide MDS/IES lengths and pointer sequences.
Concatenated Protein Sequences and Phylogenetic Analyses. Alignments of eukaryotic release factor I, DNA pol α, α-TBP, and actin-I protein sequences were carried out by using clustalx, Ver. 1.81 (24), with default parameters, and individually refined by eye. The four protein sequences from seven species were concatenated for different phylogenetic analyses (Holosticha sp., U. grandis, Uroleptus sp., P. weissei, S. lemnae, S. nova, and S. histriomuscorum; 2,543-aa positions). Maximum parsimony analyses were carried out with the heuristic search method, using random stepwise additions of the sequences and 10,000 bootstrap replicates. Quartet-puzzling trees were calculated by using the blosum 62-aa substitution model (25) and Gamma-distributed rate variation model. The program mrbayes 3.0 (26) was used to calculate the Bayesian inference of phylogeny. The JTT+Gamma model was used with 10,000 generations of Markov chain Monte Carlo. GenBank accession nos. for eukaryotic release factor I sequences (27) are Holosticha sp. AY517523, U. grandis. AY517522, Uroleptus sp. AY517521, P. weissei AY517520, S. lemnae AF317834, S. nova AF188150, and S. histriomuscorum AF317832; GenBank accession nos. for α-TBP (L. C. Wong and L.F.L., unpublished results) are Holosticha sp. AY493350, Uroleptus sp. AY493354, P. weissei AY493352, S. lemnae AF190703, S. histriomuscorum AF067831, and S. nova M31310; actin I are U. grandis AF508054, Holosticha sp. AY644730, Uroleptus sp. AY644729, P. weissei AY644728, S. lemnae AY046534, S. nova AF134156, and S. histriomuscorum U18940; DNA pol α accession numbers are below.
DNA pol α Genes. Partial macronuclear DNA pol α genes were amplified with degenerate PCR primers (4, 28). The 5′ and 3′ ends of the chromosomes were amplified by using telomere suppression PCR (17, 29). At least three clones were sequenced to minimize PCR errors. More were sequenced to recover two alleles. See Fig. 4, which is published as supporting information on the PNAS web site, for an annotated alignment based on predicted protein sequences and Fig. 5, which is published as supporting information on the PNAS web site, for detail in one region.
Partial germ-line DNA pol α sequences were initially determined by PCR from gel-purified micronuclear DNA using primers derived from the macronuclear sequence. After obtaining the first IES sequences, we designed and used IES-specific primers in conjunction with macronuclear-based primers to recover complete micronuclear DNA pol α genes for each species (see Figs. 6-9, which are published as supporting information on the PNAS web site). Several long-range PCRs confirmed that MDSs and IESs were located on the same locus. Primer sequences are in Figs. 6-9.
Typical micronuclear PCRs were 25 μl (0.2 μM each primer; 0.2 mM dNTPs; 1 unit Roche Taq polymerase, Indianapolis) 40 cycles (94°C, 30 s; 50°C, 1 m; 72°C, 2-3 m). Expand Long Template PCR (Roche): 50 μl (0.3 μM each primer; 0.35 mM dNTPs/Roche Buffer 1/3.75 units polymerase mix) 10 cycles (94°C, 10 s; 50°C, 30 s; 68°C, 6 m) then 30 cycles (94°C, 15 s; 50°C, 30 s; 68°C, 6 m and an additional 20 s each cycle).
Macro- and micronuclear DNA pol α sequences were compared to determine boundaries and features of MDSs, IESs, and pointers (see Tables 1-4, which are published as supporting information on the PNAS web site) with the aid of the wisconsin package Ver. 10.1 (GCG) and gene unscrambler (30). GenBank accession nos.: U. grandis macronuclear type I, AY008387; type II, AY008386; micronuclear type I, AY008389; type II, AY008388; Holosticha sp. macronuclear, AY293851; micronuclear, AY293853; Uroleptus sp. macronuclear, AY293852; micronuclear, AY293850; P. weissei macronuclear, AY293806; micronuclear, AY293805.
RNA Isolation and RACE. Ciliates were fed green algae 1 day before harvesting RNA. Cells were filtered through a 10-μm sieve (Sefar American, Depew, NY), and RNA was extracted by using TRIzol LS (Invitrogen). 5′- and 3′-RACE used the FirstChoice RLM-RACE kit (Ambion, Austin, TX).
Other Computational Analyses. To test the significance of overlapping pointer repeat locations in scrambled and nonscrambled DNA pol α orthologs, we performed a computational simulation of pointer distribution, using conserved regions of the alignment. While maintaining pointer locations in U. grandis or Holosticha as the reference sequence, we randomly assigned the locations of pointer repeats in Uroleptus (34 pointers) or P. weissei (44 pointers) and then scored the numbers of pointer nucleotides that overlapped between these species. This process was repeated 1,000 times to generate random background distribution. Based on this distribution, we determined the statistical significance of the number of overlapping pointer nucleotides in the actual data set relative to the background distribution. We also ran the same test on U. grandis (39 pointers sampled) against Holosticha as the reference.
We determined the expected occurrence (upper limit) of a particular repeated word in an IES in P. weissei, given its presence in the counterpart MDS flanked by the same pair of pointers. Let lengths of the IES and repeated word be N and M, respectively. Expected occurrence = base frequencies of the word × (N - M + 1). We used the base composition of the micronuclear DNA pol α gene of P. weissei to represent frequencies of each of the four nucleotides (A = 0.372, T = 0.358, G = 0.138, and C = 0.132).
Results and Discussion
Phylogenetic Relationships of Spirotrichs. To study the natural history of an extremely scrambled gene and to understand the emergence of scrambling as a complex feature in genomes, we sequenced the germ-line, somatic, and RNA versions of the DNA pol α genes from three early diverging ciliates, Holosticha sp., Uroleptus sp., and P. weissei. We also sequenced germ-line and somatic versions of this gene from Urostyla grandis, the earliest known ciliate among the stichotrichs (28). To allow mapping of germ-line events onto an evolutionary timescale, we inferred the relationships among these four species and other ciliates from their small subunit ribosomal RNA sequences (Fig. 1, left) (31) and also from four concatenated protein sequences: actin I, α-TBP, DNA pol α, and eukaryotic release factor I (eRFI) (Fig. 1, right). Both trees share similar topology; moreover, unresolved groups in the rDNA tree were fully resolved in the concatenated protein tree with strong statistical support. Based on these two trees, particularly the concatenated protein tree (Fig. 1), E. aediculatus represents the earliest diverging spirotrichous ciliate, followed by U. grandis, Holosticha sp., Uroleptus sp., P. weissei, and finally a major group including the oxytrichids with extensively scrambled genes (4, 9-11, 13, 32) (Fig. 1).
Character Mapping of Germ-Line DNA Rearrangements. A comparison of the germ-line and somatic DNA pol α sequences revealed that the germ-line architectures in the earliest diverging species, U. grandis and Holosticha sp., are not scrambled but vary greatly in level of fragmentation. The U. grandis gene contains 40 or 41 segments in each of two alleles (Figs. 1 and 4). However, the orthologous gene in Holosticha is fragmented into only nine segments (Fig. 1), suggesting either massive accumulation of noncoding DNA sequences (IESs) that interrupt the gene in U. grandis and/or massive loss of such noncoding segments from Holosticha after these two species diverged. The shorter root-to-tip length of the Holosticha lineage in the tree based on concatenated proteins (Fig. 1, right) or just DNA pol α (not shown) may favor accumulation of noncoding DNA along the Urostyla lineage.
The germ-line DNA pol α gene is extensively scrambled in Uroleptus and P. weissei. Thirty-one of 35 segments are scrambled in Uroleptus, making it the least-scrambled intermediate species, and 39 of 47 MDSs are scrambled in P. weissei. Whereas the germ-line segments in the oxytrichid species S. lemnae, S. nova (formerly Oxytricha nova), and S. histriomuscorum (formerly O. trifallax) appear distributed on two separate loci (4, 13-15), with one additional segment missing in both S. lemnae and S. histriomuscorum (4, 15), the four new species in this study contain all their coding segments on one locus with no missing sequence, consistent with their evolutionary positions as intermediates.
Furthermore, the breakpoint distance, a measure of the number of steps required to rearrange scrambled segments, originally used to infer evolutionary history from genome rearrangement events (33, 34), corresponds precisely here to the minimum number of pointer repeats (see below) between scrambled segments in a micronuclear/macronuclear gene pair. If we use breakpoint distance to rank the complexity of DNA pol α germ-line architectures, the nonscrambled Holosticha and Urostyla orthologs have a score of zero, because they require no permutation. Uroleptus ' score is 32, P. weissei is 40, and S. lemnae and the other oxytrichids are 45 and higher. These numbers highlight the expansion of complexity during the evolution of scrambled genes in Fig. 1.
Evolution of Pointer Locations. The germ-line architecture of the DNA pol α genes in Uroleptus sp., P. weissei, S. lemnae, S. nova, and S. histriomuscorum is largely conserved (Figs. 1 and 4), suggesting that this nonrandomly scrambled order with an inversion arose in a common ancestor. Despite apparent conservation (defined as overlap by at least one nucleotide) of a small number of pointer repeats and despite similar levels of fragmentation in some species pairs, we found no significant overlap of pointers between U. grandis and Holosticha (P = 1) or between either of these genes and their scrambled orthologs (see Materials and Methods). This suggests either that the pointer distributions evolved independently in these lineages, or that they rapidly shifted after divergence from a common ancestor. For example, the overlap in pointer distributions between either U. grandis and Uroleptus or Holosticha and P. weissei was not significantly different from a simulated dataset of randomly reassigned pointer locations (P = 0.58 and 0.12, respectively), whereas the same test performed between two scrambled orthologs, P. weissei and Uroleptus, was highly significant (P < 0.001). Furthermore, two regions containing large nonscrambled sections in the most scrambled orthologs, S. lemnae (segments 1, 2, and 29, 30) (4), S. nova, and S. histriomuscorum (13, 15), are heavily fragmented into 13 and 10 segments in U. grandis. There are a few notable exceptions. For example, both of segment 21's pointers in U. grandis occur at similar locations among the five scrambled genes (positions 2401 and 2560 in Fig. 4), suggesting that these boundaries could be ancient. This region may have been delineated in a nonscrambled ancestor and then translocated upstream of the original 5′ end of the gene; however, the absence of these boundaries from Holosticha suggests that the matches between U. grandis and its scrambled relatives could be due to chance, unless both IESs flanking segment 21 in U. grandis were lost from the Holosticha lineage. In addition, the left boundaries of both extensively scrambled regions in all scrambled genes (horizontal brackets in Fig. 1) overlap the locations of pointer repeats between segments 13-14 and 30-31 in U. grandis (positions 1380 and 3454 in Fig. 4). This suggests that these boundaries may be ancient, which could point to some of the early germ-line recombination events that scrambled this gene. Given the large number of pointer repeats, however, a few at coincident locations between nonscrambled and scrambled genes may be due to chance, consistent with their absence from Holosticha. Holosticha shares just one conserved boundary (between segments 4 and 5; position 1501 in Fig. 3) with a scrambled pointer in four orthologs.
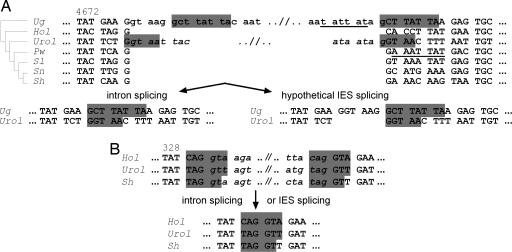
Fig. 3.
Examples of introns flanked by direct repeats. (A) Partial sequence alignment of DNA pol α genes from seven spirotrichs (U. grandis, Holosticha sp., Uroleptus sp., P. weissei, S. lemnae, S. nova, and S. histriomuscorum) surrounding the 3′ intron of this gene in Uroleptus. Confirmed intron sequences are shown in lowercase and italics. The 168-/198-bp putative 3′ intron in U. grandis (see text) is shown in lowercase. Direct repeats flanking introns are highlighted, and pointer sequences are underlined. The product after hypothetical IES splicing in U. grandis is six nucleotides longer than the product after putative intron removal; thus, the reading frame would be preserved. The phylogeny on the left is from Fig. 1. (B) Sequence surrounding the first 5′ intron in three species. Products after intron removal in these three cases would be identical to products after hypothetical IES splicing.
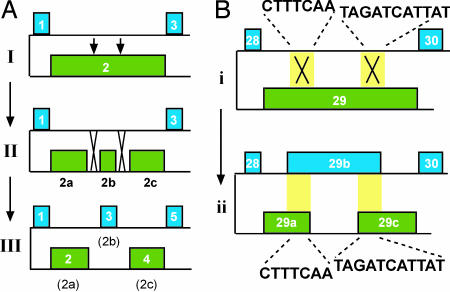
Scrambled Gene Origins. Several models have been put forth to explain the origins of scrambled genes, particularly the emergence of the strikingly nonrandom segregation of odd- and even-numbered segments (4, 12, 15). Prescott et al. (12) suggest that extensive recombination between AT-rich DNA and an ancestral α-TBP gene led to the nonrandom distribution of segments in this gene. Hoffman and Prescott (15) proposed a general model for the evolution of scrambling, in which two noncoding segments (IESs) invade a coding region (MDS) and then subsequently recombine with another IES to permute the order of the new segments, creating three scrambled MDSs (Fig. 2A). However, this model predicts an evolutionary intermediate containing a nonscrambled segment in a scrambled region (MDS 2b in Fig. 2 A), which we did not observe. Rather, whole regions of the gene are either scrambled or nonscrambled (Fig. 1). We therefore consider an alternative model in which homologous or nonhomologous germ-line recombination occurs between coding and noncoding regions, MDSs and IESs, homologous at chance matches between juxtaposed sequences flanked by the same pair of pointers (Fig. 2B) to generate odd/even split architectures. For example, P. weissei has an 11-bp sequence (TAGATCATTAT) present in both segment 29 and the IES between segments 28 and 30, and a 13-bp duplicate sequence (AAACAGCATGTAT) in both segment 30 and the counterpart IES between segments 29-31, in addition to numerous small repeats. These short repeats might be an early stage in the formation of a new coding segment via the process we propose in Fig. 2B, or they could just be chance occurrences of a repeated word (expected occurrence <10-4 and <10-6, respectively, for the 11- and 13-bp words; see Materials and Methods), but they are a representative length for scrambled pointers. Such a mechanism for acquiring newly scrambled regions differs from the acquisition of more broken but nonscrambled regions, which presumably involves invasion of noncoding DNA in the germ-line (35).
Fig. 2.
Two models for the steps involved in gene scrambling. (A) IES invasion model (15). Two IESs invade an MDS, as indicated by arrows (I). Then, these IESs recombine with another IES nearby, taking advantage of their similarity in AT-rich noncoding sequence (II). The result is the fragmentation of one MDS, number 2, into three new scrambled MDSs, 2-4 (III); however, no examples of step II have been observed. (B) IES-MDS recombination model. We propose that IESs recombine directly with MDSs at occasional repeated or similar word sequences, as shown in i. The duplicate sequences are actual sequences representing the germ-line DNA pol α gene of P. weissei (see text). These repeats, highlighted in yellow, then become the pointers defining three new MDSs (ii). MDS numbers in A are hypothetical but in B represent P. weissei to show the positions of example repeats; green MDSs are inverted relative to blue MDSs.
Because the germ-line DNA pol α gene in U. grandis shares a similar degree of fragmentation with its scrambled homologs in other ciliates, it is also possible that intense fragmentation of the gene preceded scrambling. However, an unlikely series of events would have had to cluster most odd- and even-numbered segments on either side of an inversion, and/or some selective pressure must have driven the introduction of such a peculiar pattern, as observed in modern scrambled DNA pol α genes.
In summary, we prefer the model (4) that step-wise recombination between AT-rich sequences and the ancestral DNA pol α gene created the complex odd/even segregated pattern, because of the model's simplicity and parsimony. In this model, the predicted lightly fragmented ancestral state of the gene is similar to the pattern observed in Holosticha. Combined with the lack of a significant correlation between the pointer distributions in U. grandis and its scrambled relatives, we favor the view that extensive fragmentation arose independently in U. grandis and was probably not a first step toward scrambling, which differs from other published views (16).
Fission and Fusion of Coding Segments. Although the positions of most coding segments are conserved in the five scrambled genes, some fission and fusion of coding segments have occurred; thus, tracing the changes in scrambled regions then becomes an additional tool to infer evolutionary events in this lineage. Linkages of coding segments, similar to the use of fused genes (36) or gene order (33, 34), may serve as an independent marker to trace the evolutionary history of species harboring complex scrambled genes. A parsimony tree based on changes in the scrambling pattern (not shown) generally agrees with the topology of the concatenated protein tree; however, it includes fewer taxa.
New appearances of noncoding DNA segments (IESs) were common in this study, but we also infer the possible loss of an IES and fusion of three coding segments (Figs. 1, 4, and 5), an unprecedented phenomenon: the region homologous to segment 10 in S. nova is split into three scrambled segments in P. weissei (segments 11-12-13), S. lemnae (segments 11-12-13), and S. histriomuscorum (Figs. 1, 4, and 5) (15). This region could simply be incongruous on the tree, perhaps derived from paralogous coding regions, or segment 10 in S. nova may be a fusion of three segments produced by germ-line recombination. Alternatively, this split region could have arisen independently in P. weissei and in S. histriomuscorum and S. lemnae. The latter two species share closely overlapping pointers in this region (Fig. 5), indicating a recent common ancestor, but differences from P. weissei could be due to pointer sliding along the DNA sequence (35).
The dynamics of germ-line recombination (during meiosis or mitosis of the micronucleus) that create new scrambled segments (fission) or join segments (fusion) are unknown. Although homologous recombination at pointers can both eliminate IESs and erase segment boundaries, we expect that such precise fusions in the germ line are rare, in part because the scanRNA mechanism (see below), thought to provide some of the specificity for IES elimination, should target the developing macronucleus but not the germ line. Furthermore, that most scrambled segments are conserved (Fig. 1) indicates that fusion, which would erase conserved boundaries, might occur less often between scrambled segments than fission. IES locations in the nonscrambled genes in this study are not conserved, but genealogies of these IESs are obscured by rapid sequence divergence. Therefore, gain or loss of IESs, i.e., fission or fusion of MDSs in nonscrambled regions, remains open.
A Streamlined Scrambled Gene. Also surprising, P. weissei contains a modestly more streamlined germ-line gene, with several tiny IESs between scrambled segments, including two “0-bp IESs” flanked by conventional 8- to 9-bp pointer repeats (Table 4). Only one copy of these repeats and no other DNA sequence is eliminated.
Direct Repeats Flanking Introns. We detected a 192-/162-bp insert containing in-frame stop codons at the 3′ end of the DNA pol α gene in both alleles of U. grandis. This region is flanked by the ciliate intron consensus GTaag... tAG (37), and it has a 1:3 ratio of silent/replacement nucleotide substitutions between the two alleles, compared with a ratio of 17:1 in the rest of the gene. Together, these data suggest that this region is an intron (Figs. 3A and 4). Curiously, however, this region is also flanked by a pair of octamer direct repeats, GCTTATTA, suggesting the possibility of either a mechanism for removal similar to IES elimination, the ability to remove this sequence as an IES, which was undetected, an evolutionary stage toward becoming an IES, or, alternatively, a recent history of this region as an IES. Like intron splicing, IES excision at the octamer repeat would restore the reading frame (Figs. 3A and 4); thus, we propose there may be selection to turn wayward IESs into introns if they fail to be eliminated as DNA sequences during macronuclear development. This provides a second opportunity to remove the intervening sequence when there is inefficient IES removal. By therefore relieving the constraint on efficient DNA excision, a sequence may eventually become fixed as an intron.
Intron-IES Conversion Model. Like IESs, introns are noncoding AT-rich sequences interrupting both protein-coding and noncoding DNA. Whereas IESs are removed during macronuclear development, introns are maintained in the DNA and spliced as RNA. In 50% (6 of 12) of the confirmed introns we characterized in DNA pol α orthologs, direct repeats surround the introns (Figs. 3B and 4), resembling pointers flanking IESs. Surprisingly, in all cases but one, removal of the region as an IES instead of as an intron, with one copy of the repeat retained, would still preserve the reading frame (Figs. 3B and 4). Furthermore, the position of the IES between segments 44 and 45 in P. weissei overlaps the 3′ intron in Uroleptus (Figs. 1, 3A, and 4). This intron is also present in the human homolog (GenBank accession no. NC_000023) and most likely in U. grandis (Figs. 1, 3A, and 4). These observations suggest that this intron is ancient. That P. weissei lacks this intron but has an IES at the same location leads us to conjecture it may have become an IES in this species instead. In support of our hypothesis, the recent discovery of scanRNAs in Tetrahymena (38) and Paramecium (39) suggests a mechanism for evolutionary loss of introns. In the scanRNA model, double-stranded RNA corresponding to micronuclear-limited sequence is processed by dicer-like proteins into small RNAs, which in their single-stranded form resemble processed intron fragments. The small RNAs most likely bind to homologous sequence and, through a series of steps (39-41), lead to elimination of those DNA sequences during development. Once a DNA sequence for an intron is deleted, its absence from the macronucleus predicts that it will also be deleted as an IES at the next round of conjugation, leading to stable conversion to IES. One possible evolutionary advantage of converting an intron into an IES is that the eliminated sequence no longer needs to be copied in the multiploid macronuclear genome. The extreme fragmentation that typifies the species in this study may reflect strong selection to eliminate most noncoding DNA from the somatic nucleus. Our new hypothesis that introns may be converted to IESs during evolution by this model of epigenetic inheritance could help explain the reported relative paucity of introns in ciliate genes (1).
Conclusion
This study found several different architectures of the DNA pol α gene over a broad range of species, including fragmented ancestral states as well as moderate and more complex evolutionary intermediates, suggesting a path of descent with modification in the history of germ-line rearrangements. We were fortunate to observe these incremental transitions, because the extinction of intermediate forms is a common obstacle to evolutionary studies.
Previous studies of the considerably less-scrambled actin I gene are consistent with an origin of scrambled genes after the divergence of Uroleptus (8) and the oxytrichids from the lineage leading to Urostyla (10). Unlike the DNA pol α gene, none of the early-diverging actin I genes is extensively fragmented. For example, the U. grandis actin I gene possesses only three nonscrambled segments (10), and there are no scrambled intermediate species included in ref. 10. Therefore, we infer that the DNA pol α gene in U. grandis probably accumulated many new noncoding segments after it diverged, and the less-fragmented gene in Holosticha might resemble the nonscrambled ancestral state of this gene.
Although occasional alternative unscrambling (14), like alternative splicing, might confer a selective advantage to organisms that harbor this phenomenon, the acrobatic manipulations of the germ-line genomes in stichotrichous ciliates still leave many questions unanswered, particularly regarding the mechanism for rearrangement, whether it involves genome surveillance as in Tetrahymena (38, 41), the level of accuracy and robustness, and genome stability in the face of massive rearrangements. With the O. trifallax (S. histriomuscorum) genome sequencing project launched (2), we hope that many of the components involved in unscrambling will be unveiled in the next decade.
Supplementary Material
Acknowledgments
We thank David Prescott (University of Colorado, Boulder) for the generous gift of U. grandis DNA and insightful discussion; Li Chin Wong, Ed Curtis, and Victoria Addis for sharing unpublished data; Jingmei Wang, Catherine Lozupone, and Hans Lipps and members of his laboratory for technical assistance and discussion; Andre Calvacanti for analyzing direct repeats flanking introns; and David Ardell for suggesting the presence of introns. This work was supported by National Institute of General Medical Sciences Grant GM59708 and National Science Foundation Grant 0121422.
Author contributions: W.-J.C. and L.F.L. designed research; W.-J.C., P.D.B., and M.K.S. performed research; W.-J.C., P.D.B., H.L., and L.F.L. analyzed data; and W.-J.C., P.D.B., and L.F.L. wrote the paper.
Abbreviations: MDS, macronuclear destined segment; IES, internal eliminated segment; DNA pol α, DNA polymerase α; α-TBP, α telomere-binding protein.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY294646-AY294648, AY493350, AY493352, AY493354, AY644728-AY644730, AY008386-AY008389, AY293850-AY293853, AY293805, and AY293806).
References
- 1.Prescott, D. M. (1994) Microbiol. Rev. 58, 233-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doak, T. G., Cavalcanti, A. R. O., Stover, N. A., Dunn, D. M., Weiss, R., Herrick, G. & Landweber, L. F. (2003) Trends Genet. 19, 603-607. [DOI] [PubMed] [Google Scholar]
- 3.Klobutcher, L. A., Jahn, C. L. & Prescott, D. M. (1984) Cell 36, 1045-1055. [DOI] [PubMed] [Google Scholar]
- 4.Landweber, L. F., Kuo, T. C. & Curtis, E. A. (2000) Proc. Natl. Acad. Sci. USA 97, 3298-3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Forget, L., Ustinova, J., Wang, Z., Huss, V. A. & Lang, B. F. (2002) Mol. Biol. Evol. 19, 310-319. [DOI] [PubMed] [Google Scholar]
- 6.Gillespie, D. E., Salazar, N. A., Rehkopf, D. H. & Feagin, J. E. (1999) Nucleic Acids Res. 27, 2416-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kairo, A., Fairlamb, A. H., Gobright, E. & Nene, V. (1994) EMBO J. 13, 898-905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalby, A. B. & Prescott, D. M. (2004) Chromosoma 112, 247-254. [DOI] [PubMed] [Google Scholar]
- 9.Prescott, D. M. & Greslin, A. F. (1992) Dev. Genet. 13, 66-74. [DOI] [PubMed] [Google Scholar]
- 10.Hogan, D. J., Hewitt, E. A., Orr, K. E., Prescott, D. M. & Muller, K. M. (2001) Proc. Natl. Acad. Sci. USA 98, 15101-15106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitcham, J. L., Lynn, A. J. & Prescott, D. M. (1992) Genes Dev. 6, 788-800. [DOI] [PubMed] [Google Scholar]
- 12.Prescott, J. D., DuBois, M. L. & Prescott, D. M. (1998) Chromosoma 107, 293-303. [DOI] [PubMed] [Google Scholar]
- 13.Hoffman, D. C. & Prescott, D. M. (1996) Nucleic Acids Res. 24, 3337-3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ardell, D. H., Lozupone, C. A. & Landweber, L. F. (2003) Genetics 165, 1761-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffman, D. C. & Prescott, D. M. (1997) Nucleic Acids Res. 25, 1883-1889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jahn, C. L. & Klobutcher, L. A. (2002) Annu. Rev. Microbiol. 56, 489-520. [DOI] [PubMed] [Google Scholar]
- 17.Chang, W.-J., Stover, N. A., Addis, V. M. & Landweber, L. F. (2004) Protist 155, 245-255. [DOI] [PubMed] [Google Scholar]
- 18.Medlin, L., Elwood, H. J., Stickel, S. & Sogin, M. L. (1988) Gene 71, 491-499. [DOI] [PubMed] [Google Scholar]
- 19.Wuyts, J., Van de Peer, Y., Winkelmans, T. & De Wachter, R. (2002) Nucleic Acids Res. 30, 183-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Hoek, A. H., van Alen, T. A., Sprakel, V. S., Hackstein, J. H. & Vogels, G. D. (1998) Mol. Biol. Evol. 15, 1195-1206. [DOI] [PubMed] [Google Scholar]
- 21.Swofford, D. L. (2002) paup (Sinauer, Sunderland, MA), Ver. 4.
- 22.Schmidt, H. A., Strimmer, K., Vingron, M. & von Haeseler, A. (2002) Bioinformatics 18, 502-504. [DOI] [PubMed] [Google Scholar]
- 23.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Henikoff, S. & Henikoff, J. G. (1992) Proc. Natl. Acad. Sci. USA 89, 10915-10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huelsenbeck, J. P., Ronquist, F., Nielsen, R. & Bollback, J. P. (2001) Science 294, 2310-2314. [DOI] [PubMed] [Google Scholar]
- 27.Liang, H., Wong, J. Y., Bao, Q., Cavalcanti, A. R. O. & Landweber, L. F. (2005) J. Mol. Evol. 60, 337-344. [DOI] [PubMed] [Google Scholar]
- 28.Hoffman, D. C. & Prescott, D. M. (1997) J. Mol. Evol. 45, 301-310. [DOI] [PubMed] [Google Scholar]
- 29.Curtis, E. A. & Landweber, L. F. (1999) Ann. N.Y. Acad. Sci. 870, 349-350. [DOI] [PubMed] [Google Scholar]
- 30.Cavalcanti, A. R. O. & Landweber, L. F. (2004) Bioinformatics 20, 800-802. [DOI] [PubMed] [Google Scholar]
- 31.Hewitt, E. A., Muller, K. M., Cannone, J., Hogan, D. J., Gutell, R. & Prescott, D. M. (2003) Mol. Phylogenet. Evol. 29, 258-267. [DOI] [PubMed] [Google Scholar]
- 32.DuBois, M. & Prescott, D. M. (1995) Proc. Natl. Acad. Sci. USA 92, 3888-3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moret, B. M. E., Tang, J., Wang, L.-S. & Warnow, T. (2002) J. Comput. Syst. Sci. 65, 508-525. [Google Scholar]
- 34.Sankoff, D., Leduc, G., Antoine, N., Paquin, B., Lang, B. F. & Cedergren, R. (1992) Proc. Natl. Acad. Sci. USA 89, 6575-6579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prescott, D. M. & DuBois, M. L. (1996) J. Eukaryot. Microbiol. 43, 432-441. [DOI] [PubMed] [Google Scholar]
- 36.Stechmann, A. & Cavalier-Smith, T. (2002) Science 297, 89-91. [DOI] [PubMed] [Google Scholar]
- 37.Prescott, D. M., Prescott, J. D. & Prescott, R. M. (2002) Protist 153, 71-77. [DOI] [PubMed] [Google Scholar]
- 38.Yao, M. C., Fuller, P. & Xi, X. (2003) Science 300, 1581-1584. [DOI] [PubMed] [Google Scholar]
- 39.Garnier, O., Serrano, V., Duharcourt, S. & Meyer, E. (2004) Mol. Cell. Biol. 24, 7370-7379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao, M. C. & Chao, J. L. (2005) Annu. Rev. Genet. 39, 537-559. [DOI] [PubMed] [Google Scholar]
- 41.Mochizuki, K. & Gorovsky, M. A. (2004) Curr. Opin. Genet. Dev. 14, 181-187. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.