Abstract
BRIT1 [BRCT-repeat inhibitor of hTERT expression], a repressor of human telomerase function, is implicated in cellular immortalization. Here, we find that BRIT1 acts as a regulator of both the intra-S and G2/M checkpoints. When BRIT1 expression is depleted, cells lose the ionizing radiation (IR)-induced cell cycle arrest and become IR sensitive. BRIT1 is a chromatin-associated protein that forms irradiation-induced nuclear foci that colocalize with γ-H2AX foci. BRIT1 is also required for the expression of both BRCA1 and the checkpoint kinase Chk1 and phosphorylation of Nbs1. Thus, the checkpoint defects in the absence of BRIT1 are likely to result from its regulation of Nbs1, BRCA1, and Chk1. BRIT1 is identical to the recently discovered MCPH1 gene, found mutant in patients with primary microcephaly. The ataxia telangiectasia mutated-Rad3 related (ATR)–Chk1 pathway is defective in Seckel syndrome, another microcephaly disorder. We propose that the microcephaly observed in patients with MCPH1 deficiencies is due to disruption of the ATR–BRCA1–Chk1 signaling pathway that is also disrupted in Seckel syndrome patients.
Keywords: BRCA1, BRCT, Chk1
BRIT1 (BRCT-repeat inhibitor of hTERT expression) is a gene we previously identified in a genetic screen for transcriptional repressors of hTERT, the catalytic subunit of human telomerase (1). The sequence of BRIT1 was derived from a hypothetical protein that later matched to a putative disease gene called microcephalin (MCPH1), one of at least six loci implicated in the autosomal recessive disease primary microcephaly (2).
When the protein structure of BRIT1 was analyzed by the simple modular architecture research tool (smart) program, it revealed that BRIT1 contained three BRCT domains: one in its N terminus and two in its C terminus. BRCT domains have been found predominantly in proteins involved in cell cycle checkpoint functions responsive to DNA damage. This finding suggested that, in addition to its role in hTERT repression, BRIT1 may play a role in DNA damage responses.
The DNA damage response involves the sensing of DNA damage followed by transduction of the damage signal to a network of cellular pathways, including cell cycle checkpoints, DNA repair, and the apoptotic pathway. In this network, two phosphatidylinositol-3-related kinases, ATM (ataxia telangiectasia mutated) and ATR (ATM-Rad3 related), are located at the top of checkpoint signal cascades, which phosphorylate and activate a variety of molecules to execute the DNA damage response (3–5). ATM is activated primarily by double-strand breaks induced by ionizing radiation (IR), whereas ATR also responds to UV or stalled replication forks (5). When phosphorylated by ATM or ATR, the p53 protein is activated and stabilized, resulting in cell cycle arrest in G1 (3, 5, 6). The BRCA1 tumor suppressor plays a role in homologous recombination and may function in DNA repair by serving as a scaffold for ATM and ATR, thereby facilitating phosphorylation of downstream targets (5, 7, 8). BRCA1 is also involved in the intra-S and G2/M checkpoints (9). The two effector kinases Chk1 and Chk2 are phosphorylated and activated by ATM and ATR and phosphorylate and negatively regulate the Cdc25 family of phosphatases that promote cell cycle transitions. Inhibition and destruction of these proteins leads to cell cycle arrest and execution of the G1/S, intra-S, and G2/M checkpoints (5, 10, 11).
Because BRIT1 protein contains three BRCT domains, we suspected that BRIT1 might also play important roles in the DNA damage response. In this study, we show that BRIT1 is required for intact intra-S and G2/M checkpoints after IR and that these activities may result from its regulation of the expression or activation of at least three other checkpoint regulators, Chk1, BRCA1, and NBS1.
Methods
Cells. U2OS cells were purchased from the American Type Culture Collection and maintained in McCoy's 5A medium supplemented with 10% FBS, glutamine, and penicillin and streptomycin. All other cell lines were maintained in DMEM with 10% FBS.
Small Interfering RNA (siRNA). The siRNA duplexes were 19 base pairs with a two-base deoxythymidine overhang (Dharmacon Research, Lafayette, CO). The sequences of BRIT1 siRNA1 and siRNA2 oligonucleotides are AGGAAGUUGGAAGGAUCCAdTdT and CUCUCUGUGUGAAGCACCUdTdT, respectively. The control luciferase siRNA has the sequence UAAGGCUAUGAAGAGAUACdTdT. Cells were transfected with siRNA duplexes by using Oligofectamine (Invitrogen), following the manufacturer's instructions.
Antibodies. The BRIT1 antibody was directed against a GST-BRIT1 fusion protein generated by Proteintech (Chicago). Anti-Chk1 and anti-Orc2 (C-18) were purchased from Santa Cruz Biotechnology. Anti-phospho-Chk1 and anti-phospho-NBS1 were purchased from Cell Signaling Technology (Beverly, MA). The BRCA1 antibody was purchased from Oncogene Science, and anti-γH2AX was a monoclonal antibody purchased from Upstate Biotechnology (Lake Placid, NY).
Cell Survival Assays. U2OS cells were transfected with siRNAs two times with a 24-h interval and, 48 h after the second transfection, were plated at low density and irradiated with various doses of IR. Cells were incubated for 2–3 weeks to allow colonies to form. Colonies were detected by staining with 2% methylene blue/50% ethanol.
Radioresistant DNA Synthesis (RDS) Assay. The RDS assay was performed as described in ref. 12. Briefly, U2OS cells were transfected with siRNAs twice. After the second transfection, cells were incubated in McCoy's 5A medium containing 10 nCi/ml (1 Ci = 37 GBq) [14C]thymidine (NEN) overnight. The medium was then replaced with normal McCoy's 5A medium and incubated for another 24 h. Cells were irradiated, incubated for 30 min at 37°C, and then pulse-labeled with 2.5 μCi/ml [3H]thymidine (NEN) for 15 min. Cells were harvested, washed twice with PBS, and fixed in 70% methanol for 30 min. After cells were transferred to Whatman filters and fixed sequentially with 70% and 95% methanol, the filters were air-dried, and the radioactivity was assayed in a liquid scintillation counter. The resulting ratio of 3H cpm to 14C cpm, corrected for cpm that resulted from channel crossover, was a measure of DNA synthesis.
G2/M Checkpoint Assay. U2OS cells were transfected with siRNAs two times with 24-h intervals and, 48 h after the second transfection, were exposed to 3 Gy of IR. One hour later, cells were fixed and stained with propidium iodide and antibody against phospho-histone H3 (Cell Signaling Technology), followed by FITC-conjugated secondary antibody (Jackson ImmunoResearch). The percentage of M phase cells was determined by flow cytometry.
Generation of BRIT1 Mutants. Wild-type BRIT1 cDNA cloned in pCR-Blunt II-Topo (Invitrogen) was used to generate BRIT1 mutant by changing three nucleotides (TTG to CTC and GAA to GAG) in the BRIT1 siRNA1 sequence by using the Gene Tailor Site Directed Mutagenesis System from Invitrogen. Mutant clones were sequenced to confirm the changed nucleotides in BRIT1 cDNA, and the right clone was then subcloned into pMSCV-puro retroviral vector (Clontech).
Generation of Mutant BRIT1 and Control pMSCV Stable Pools. 293 T cells (1 × 109) plated the day before were cotransfected with 5 μg each of mutated BRIT1 cDNA or empty vector pMSCV along with PCG-gag pol and vesicular stomatitis virus glycoprotein envelop expression constructs by FuGENE 6 (Roche Diagnostics). Viral supernatant was collected 48–72 h posttransfection, filtered through a 0.45-μm membrane (Millipore), and directly used to infect U2OS cells with a 1.5 multiplicity of infection of virus without drug selection.
Immunofluorescence Staining. Cells cultured on coverslips were washed twice in PBS, incubated in cytoskeleton buffer {Pipes [piperazine-N,N′-bis(2-ethanesulfonic acid)], pH 6.8/100 mM NaCl/300 mM sucrose/3 mM MgCl2/1 mM EGTA/0.5% Triton X-100} for 3 min on ice. The cells were washed with ice-cold PBS three times and then incubated in stripping buffer (10 mM Tris·HCl, pH 7.4/10 mM NaCl/3 mM MgCl2/1% Tween 20/0.25% sodium deoxycholate) for 3 min on ice. After washing with ice-cold PBS three times, cells were fixed with 4% paraformaldehyde at 4°C for 30 min. After fixation, the cells were permeabilized in 1% Triton X-100 and 0.5% Nonidet P-40 for 30 min. Samples were blocked with 1% BSA and then incubated with primary antibody for 2 h and secondary (fluorescein isothiocyanate or rhodamine) antibody for 1 h. Cells were then stained with DAPI to visualize nuclear DNA. The coverslips were mounted onto glass slides with VectaShield antifade (Vector Laboratories) and visualized by using a Leica DM LB fluorescence microscope. Images were captured with a Kodak digital imaging system.
Chromatin Isolation. A total of ≈4 × 106 cells was washed with PBS and resuspended in 200 μl of solution A (10 mM Hepes, pH 7.9/10 mM KCl/1.5 mM MgCl2/0.34 M sucrose/10% glycerol/1 mM DTT/10 mM NaF/1 mM Na2Vo3/protease inhibitors). Triton X-100 was added to a final concentration of 0.1%, and the cells were incubated for 5 min on ice. Cytoplasmic proteins were separated from nuclei by low-speed centrifugation (4 min at 1,300 × g at 4°C). Isolated nuclei were washed once with solution A and lysed in 200 μl of solution B (3 mM EDTA/0.2 mM EGTA/1 mM DTT/protease inhibitor). Insoluble chromatin was collected by centrifugation (4 min at 1,700 × g at 4°C), washed once in buffer B, and centrifuged again at high speed (10,000 × g for 1 min). The final chromatin pellet was resuspended in 200 ml of Laemmli buffer and sonicated for 15 s. To digest chromatin with micrococcal nuclease, nuclei were resuspended in solution A containing 1 mM CaCl2 and 50 units of micrococcal nuclease (Sigma). After incubation at 37°C for 1 min, the nuclease reaction was stopped by the addition of 1 mM EGTA. Nuclei were collected by low-speed centrifugation and lysed according to the chromatin isolation protocol described above.
Results
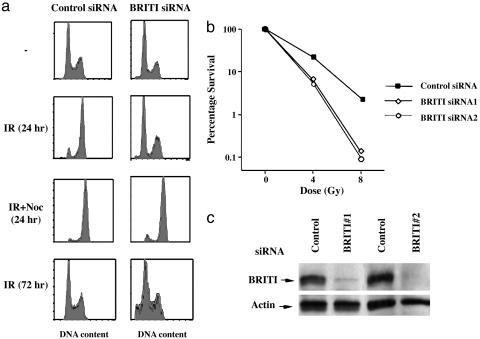
BRIT1 Knock-Down Leads to Escape from Cell Cycle Arrest and Increases Sensitivity to DNA Damage. The presence of BRCT repeats in BRIT1 led us to the hypothesis that, in addition to repressing hTERT expression, BRIT1 may play an additional role in the DNA damage response. To test this hypothesis, we transfected U2OS cells twice with two independent siRNAs against two nonoverlapping sequences in BRIT1. Forty-eight hours after the second transfection, cells were challenged with IR (10 Gy). As shown in Fig. 1a, 24 h after exposure to 10 Gy, cells treated with control siRNA (siRNA against luciferase) were arrested with a G2 DNA content, indicating the presence of an intact DNA damage checkpoint in this cell line. In contrast, BRIT1 knockdown cells showed no significant alteration in cell cycle distribution after exposure to IR. Because BRIT1 siRNA-treated cells could be arrested with a G2 DNA content by nocodazole (Fig. 1a), an agent that disrupts spindle assembly, the failure of BRIT1 knock-down cells to arrest with a G2 DNA content after IR most likely resulted from a defect in the DNA damage checkpoint rather than a pause in cell cycle progression, a point we address below. The BRIT1 knock-down cells also demonstrated an increase in the sub-G1 (apoptotic) population 72 h after irradiation (Fig. 1a Bottom), a finding that is compatible with the lack of an intact DNA damage response. In colony-forming assays, the BRIT1 siRNA-treated cells were significantly more sensitive to IR than were control siRNA cells (Fig. 1b). In all of the BRIT1 knock-down experiments, we confirmed BRIT1 depletion by Western blot analysis (Fig. 1c). We conclude that BRIT1 knockdown compromises the ability of cells to respond to DNA damage and increases cell sensitivity to IR.
Fig. 1.
BRIT1 deficiency leads to checkpoint defects and increases cellular sensitivity to IR. (a) U2OS cells were transfected twice with control or BRIT1 siRNA1s, and, 48 h after the second transfection, cells were treated with IR (10 Gy) with or without 1 mg/ml nocodazole. Then, 24 or 72 h after the irradiation, the DNA content of cells was analyzed by FACS. (b) U2OS cells were transfected with siRNAs as described above, plated at low density, and irradiated, and the colonies were counted 2 weeks later. (c) The cell lysates prepared from the siRNA-transfected cells were subjected to Western blot analysis and probed with antibodies against either BRIT1 or actin.
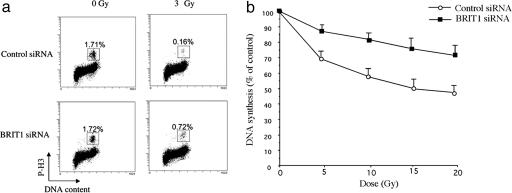
BRIT1 Knock-Down Results in Defects at Intra-S and G2/M DNA Damage Checkpoints. To assay directly for checkpoint defects, we treated cells with BRIT1-specific siRNA, irradiated them, and labeled them with H3 (anti-phospho-histone) antibody, a marker for cells in M phase (9). In contrast to control cells, which arrested in G2, a significantly higher proportion of BRIT1-depleted cells entered mitosis, indicating that BRIT1 is required for G2 cell cycle arrest in response to damage (Fig. 2a).
Fig. 2.
BRIT1 is required for the IR-induced G2/M checkpoint and the intra-S checkpoint. (a) IR-induced G2/M checkpoint analysis. U2OS cells were first transfected with control or BRIT1 siRNA1 twice. Forty-eight hours after the second transfection, cells were either left untreated or irradiated with 3 Gy and then incubated for 1 h before fixation. Cells in mitosis were determined by staining with propidium iodide and phospho-histone H3 antibody followed by FITC-conjugated secondary antibody. The percentage of M phase cells was determined by FACS for phospho-histone H3. (b) IR-induced intra-S phase checkpoint. DNA synthesis was assessed 30 min after various doses of IR in U2OS cells twice-transfected with BRIT1 siRNA1 or the control siRNA.
We also examined whether BRIT1 played a role in the intra-S phase checkpoint. IR is well known to trigger a reduction in DNA synthesis through activation of this checkpoint (13). Thus, we examined DNA synthesis in response to IR in BRIT1-deficient cells. BRIT1-deficient cells showed a significant radioresistant DNA synthesis phenotype (Fig. 2b), indicating that BRIT1 also participates in the intra-S phase checkpoint.
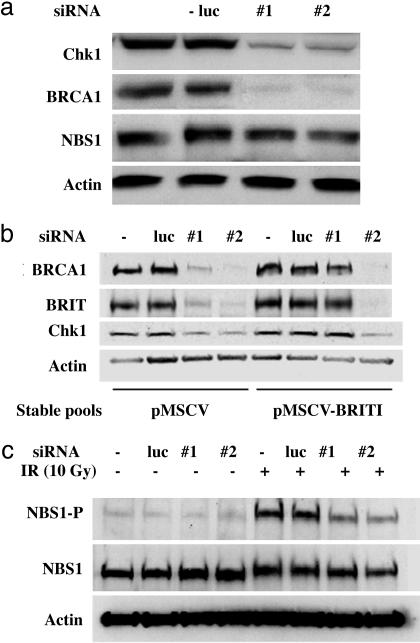
BRIT1 Knock-Down Leads to Reduced Expression of BRCA1 and Chk1. To explore the position within the damage signaling pathway in which BRIT1 might act, we examined the phosphorylation status of key checkpoint-regulated molecules required for cell cycle arrest in response to DNA damage. BRCA1 and Chk1 are both key regulators in the control of intra-S and G2/M checkpoints. While investigating the phosphorylation status of these proteins, Western blot analysis revealed that the expression levels of both BRCA1 and Chk1 were significantly reduced in BRIT1 knockdown cells (Fig. 3a). These effects were specific, because the same effects had been induced by two pairs of siRNA against two different BRIT1 sequences but not by control siRNAs. As further evidence of specificity, BRIT1 siRNA did not alter the expression of NBS1, another DNA damage checkpoint protein, or the expression of actin. Because BRIT1 knock-down alone did not affect cell cycle distribution (see Fig. 1a), the reduction in BRCA1 and Chk1 levels was unlikely to have been due to an effect on cell cycle progression. We also demonstrated that BRCA1 and Chk1 expression could be restored when siRNA-resistant BRIT1 was ectopically expressed (Fig. 3b). For the rescue experiment, a BRIT1 mutant (pMSCV-mBRIT1) that is resistant to siRNA1 but not siRNA2 was created by replacing the siRNA1 target in the pMSCV expression vector with mutations on three nucleotides in which third-codon wobble was used to introduce silent sequence changes. We found that expression defects of both BRCA1 and Chk1 in BRIT1 knock-down cells were rescued in a stable cell pool that expressed mutant BRIT1 (Fig. 3b). Thus, BRIT1 is required for the expression of both BRCA1 and Chk1, and the abrogation of the DNA damage checkpoints may be, at least in part, a consequence of regulation of the expression of these two checkpoint regulators.
Fig. 3.
BRIT1 regulates the expression of Chk1, BRCA1, and IR-dependent NBS1 phosphorylation. (a) U2OS cells were mock-transfected or transfected with luciferase or two different BRIT1 siRNAs twice. Forty-eight hours after the second transfection, cells were harvested for Western blotting and probed with antibodies against the indicated proteins. (b) Cells infected with a virus expressing siRNA1-resistant BRIT1 (pMSCV-BRIT1) or the vector control cells (pMSCV) were mock-transfected or transfected with luciferase or two different BRIT1 siRNAs once. Forty-eight hours after the transfection, cells were harvested for Western blot analysis and probed with the indicated antibodies. (c) U2OS cells were mock-transfected or transfected with luciferase or two different BRIT1 siRNAs twice. Forty-eight hours after the second transfection, cells were either left unirradiated or irradiated with IR (10 Gy). Two hours after irradiation, cells were harvested for Western blotting and probed with pS343-NBS1, NBS1, or actin antibodies.
BRIT1 Is Required for NBS1 Phosphorylation upon IR. In addition to BRCA1 and Chk1, NBS1 is another checkpoint regulator required for intact intra-S phase checkpoint. It has been shown that NBS1 is phosphorylated by ATM at Ser-343 after IR, and this phosphorylation is required for S phase checkpoint. Thus, we sought to determine whether BRIT1 depletion also affected IR-dependent NBS1 phosphorylation. U2OS cells were transfected with control or BRIT1 siRNAs and subsequently treated with 10 Gy of IR 48 h after the second transfection. As shown in Fig. 3c, BRIT1 depletion significantly inhibited IR-induced NBS1 phosphorylation without affecting its protein level, indicating a defect in the ATM signaling pathway.
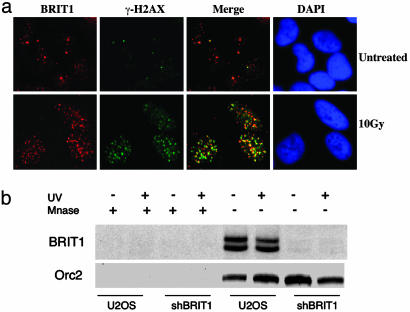
BRIT1 Is a Chromatin-Binding Protein That Forms Irradiation-Induced Nuclear Foci That Colocalize with γ-H2AX. Several BRCT domain-containing proteins such as 53BP1, MDC1, and BRCA1 are targets for ATM- or ATR-mediated phosphorylation and participate in transmitting the DNA damage signal to downstream targets such as Chk1 and Chk2 (14, 15). After DNA damage, those proteins are recruited to the sites of DNA damage and phosphorylated by ATM and/or ATR and form discrete irradiation-induced nuclear foci (14, 16). Because BRIT1 is a BRCT-containing molecule, we were interested in knowing whether BRIT1 also formed irradiation-induced foci. Immunofluorescence staining indicated that BRIT1 showed discrete nuclear foci even without irradiation (Fig. 4a). However, the number of BRIT1 foci significantly increased 2 h after exposure to 10 Gy of IR. Also, BRIT1 foci colocalized with γ-H2AX foci, a hallmark of the damaged DNA loci in cells (16, 17). We also found that BRIT1 associated with chromatin both before and after UV treatment, and the amounts of BRIT1 on chromatin did not change in damaged cells (Fig. 4b), a very similar pattern as previously shown for ATR (18). The chromatin association of BRIT1 was verified by micrococcal nuclease digestion and short hairpin RNA-specific knock-down (Fig. 4b). These results suggest that, in addition to regulating the expression of BRCA1 and Chk1, BRIT1 may have a direct role in transmitting DNA damage signals.
Fig. 4.
BRIT1 is a chromatin-associated protein that forms irradiation-induced nuclear foci that colocalize with γ-H2AX foci. (a) U2OS cells were untreated or treated with 10 Gy of IR. Two hours after irradiation, cells were fixed and stained with the polyclonal BRIT1 antibody and the monoclonal γ-H2AX, washed, and subsequently stained with rhodamine- or FITC-conjugated antibodies. DAPI staining indicated the location of the nuclei. (b) U2OS cells with or without BRIT1 knock-down by short hairpin RNA were left untreated or treated with 50 J/m2. Two hours after irradiation, chromatin-enriched sediment and the sediment from micrococcal nuclease-treated nuclei (Mnase) were subjected to Western blot analysis and probed with antibodies against BRIT1 or Orc2 as a loading control.
Discussion
In this report, we investigate the role of BRIT1 in DNA damage checkpoints. BRIT1 was originally identified as a hTERT repressor from genetic screens. Based on the domain analysis on this protein, we suspected that BRIT1 might exert additional functions in DNA damage checkpoints or repair. As shown in our studies, BRIT1 controls multiple checkpoint regulators and is required for both intra-S and G2/M checkpoints. In addition to forming irradiation-induced nuclear foci, the effects of BRIT1 on Chk1 and BRCA1 expression are particularly noteworthy. Most participants in the DNA damage response regulate the response by affecting the phosphorylation cascades within the signaling network to control the activity of the downstream effectors. BRIT1 may affect DNA damage checkpoints by both directly transmitting DNA damage signals and controlling the expression levels of other checkpoint regulators. The mechanism of this regulation remains to be determined. We should also note that during the course of our studies, a study on BRIT1 was published that confirms some of the findings presented here (19).
BRIT1 may play two distinct roles in preventing cell transformation: suppressing hTERT activation and maintaining intact checkpoints. Because BRIT1 affects the expression of its downstream targets, it is possible that BRIT1 may function as a transcriptional regulator that positively regulates the expression of BRCA1 and Chk1 but negatively regulates hTERT expression. Alternatively, the inhibitory effect of BRIT1 on hTERT expression may be an indirect event as a consequence of its regulation of BRCA1. BRCA1 has recently been identified as a negative regulator of hTERT expression (20). BRIT1 may therefore inhibit hTERT expression through its positive regulation of BRCA1 expression.
With respect to its role in the DNA damage response, BRIT1 appears to function both in the ATR branch of the pathway through Chk1 and BRCA1 regulation and in the ATM branch by means of regulation of Nbs1 phosphorylation. Because Nbs1 phosphorylation is an early step in the ATM pathway, it is likely that BRIT1 functions very early in this pathway at a step yet to be determined.
BRIT1 has been found to be the MCPH1 gene, one of the genes responsible for primary microcephaly. Microcephaly is a genetic disorder in which affected individuals have a head circumference <3 standard deviations below the age- and sex-related mean (2). Microcephaly has been observed in other genetic diseases such as Seckel syndrome (21) and Nijmegen breakage syndrome (22). Like BRIT1/MCPH1-deficient cells, both Seckel and Nijmegen breakage syndrome cells are defective in checkpoint signaling. All three of these microcephaly disorders have defects in Chk1 regulation. ATR mutations, as well as the defects in its downstream signaling pathway, have recently been reported in cells derived from patients with Seckel syndrome (21, 23), and Chk1 is the key downstream target of ATR (24). The ATM pathway also regulates Chk1, and NBS1, the gene defective in Nijmegen breakage syndrome, is required for an activating phosphorylation of Chk1 in response to IR (25). Other recent studies implicate NBS1 in control of the ATR pathway in response to UV (26). Our studies of Chk1 (24, 27) and studies of ATR (28) have determined that the ATR–Chk1 pathway is essential for cellular viability. Even loss of one copy of Chk1 has severe consequences for cells (27). Given this fact, we propose that all three of these microcephaly disorders arise through defective regulation of Chk1. This defect in Chk1 function leads to increased cellular lethality in neural lineages and severely decreased brain and head size. In addition, these results suggest that other genes responsible for microcephaly might also be deficient in Chk1 regulation. Why these lineages are so sensitive to Chk1 function is not clear and remains an important area for investigation.
Acknowledgments
This work was supported by National Cancer Institute Grant R01 CA112291-01A1, a University of Texas M. D. Anderson Cancer Center institutional research grant (to S.-Y.L.), and a National Institutes of Health grant (to S.J.E). S.J.E. is a Howard Hughes Medical Institute Investigator.
Author contributions: S.-Y.L. and S.J.E. designed research; S.-Y.L., R.R., K.L., and Z.-X.X. performed research; S.-Y.L. and S.J.E. analyzed data; and S.-Y.L. and S.J.E. wrote the paper.
Abbreviations: IR, ionizing radiation; ATM, ataxia telangiectasia mutated; ATR, ATM-Rad3 related; siRNA, small interfering RNA.
References
- 1.Lin, S. & Elledge, S. (2003) Cell 113, 881–889. [DOI] [PubMed] [Google Scholar]
- 2.Jackson, A., Eastwood, H., Bell, S., Adu, J., Toomes, C., Carr, I., Roberts, E., Hampshire, D., Crow, Y., Mighell, A., et al. (2002) Am. J. Hum. Genet. 71, 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou, B. B. & Elledge, S. J. (2000) Nature 408, 433–439. [DOI] [PubMed] [Google Scholar]
- 4.Shiloh, Y. (2001) Curr. Opin. Genet. Dev. 11, 71–77. [DOI] [PubMed] [Google Scholar]
- 5.Osborn, A., Elledge, S. & Zou, L. (2002) Trends Cell Biol. 12, 509–516. [DOI] [PubMed] [Google Scholar]
- 6.Delia, D., Fontanella, E., Ferrario, C., Chessa, L. & Mizutani, S. (2003) Oncogene 22, 7866–7869. [DOI] [PubMed] [Google Scholar]
- 7.Wang, Q., Zhang, H., Fishel, R. & Greene, M. (2000) Oncogene 19, 6152–6158. [DOI] [PubMed] [Google Scholar]
- 8.De la Torre, C., Pincheira, J. & Lopez-Saez, J. (2003) Histol. Histopathol. 18, 225–243. [DOI] [PubMed] [Google Scholar]
- 9.Xu, B., Kim, S. & Kastan, M. (2001) Mol. Cell. Biol. 21, 3445–3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez, Y., Wong, C., Thoma, R. S., Richman, R., Wu, Z., Piwnica-Worms, H. & Elledge, S. J. (1997) Science 277, 1497–1501. [DOI] [PubMed] [Google Scholar]
- 11.Matsuoka, S., Huang, M. & Elledge, S. J. (1998) Science 282, 1893–1897. [DOI] [PubMed] [Google Scholar]
- 12.Wang, B., Matsuoka, S., Carpenter, P. & Elledge, S. (2002) Science 298, 1435–1438. [DOI] [PubMed] [Google Scholar]
- 13.Painter, R. & Young, B. (1980) Proc. Natl. Acad. Sci. USA 77, 7315–7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sancar, A., Lindsey-Boltz, L. A., Unsal-Kaccmaz, K. & Linn, S. (2004) Annu. Rev. Biochem. 73, 39–85. [DOI] [PubMed] [Google Scholar]
- 15.Motoyama, N. & Naka, K. (2004) Curr. Opin. Genet. Dev. 14, 11–16. [DOI] [PubMed] [Google Scholar]
- 16.Lisby, M. & Rothstein, R. (2004) Curr. Opin. Cell. Biol. 16, 328–334. [DOI] [PubMed] [Google Scholar]
- 17.Rogakou, E. P., Boon, C., Redon, C. & Bonner, W. M. (1999) J. Cell Biol. 146, 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou, L., Cortez, D. & Elledge, S. J. (2002) Genes Dev. 16, 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu, X., Lee, J. & Stern, D. F. (2004) J. Biol. Chem. 279, 34091–34094. [DOI] [PubMed] [Google Scholar]
- 20.Xiong, J., Fan, S., Meng, Q., Schramm, L., Wang, C., Bouzahza, B., Zhou, J., Zafonte, B., Goldberg, I. D., Haddad, B. R., et al. (2003) Mol. Cell. Biol. 23, 8668–8690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Driscoll, M., Ruiz-Perez, V., Woods, C., Jeggo, P. & Goodship, J. (2003) Nat. Genet. 33, 497–501. [DOI] [PubMed] [Google Scholar]
- 22.Varon, R., Vissinga, C., Platzer, M., Cerosaletti, K., Chrzanowska, K., Saar, K., Beckmann, G., Seemanova, E., Cooper, P., Nowak, N., et al. (1998) Cell 93, 467–476. [DOI] [PubMed] [Google Scholar]
- 23.Alderton, G., Joenje, H., Varon, R., Borglum, A., Jeggo, P. & O'Driscoll, M. (2004) Hum. Mol. Genet. 13, 3127–3138. [DOI] [PubMed] [Google Scholar]
- 24.Liu, Q., Guntuku, S., Cui, X. S., Matsuoka, S., Cortez, D., Tamai, K., Luo, G., Carattini-Rivera, S., DeMayo, F., Bradley, A., et al. (2000) Genes Dev. 14, 1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 25.Gatei, M., Sloper, K., Sorensen, C., Syljuasen, R., Falck, J., Hobson, K., Savage, K., Lukas, J., Zhou, B. B., Bartek, J. & Khanna, K. K. (2003) J. Biol. Chem. 278, 14806–14811. [DOI] [PubMed] [Google Scholar]
- 26.Stiff, T., Reis, C., Alderton, G. K., Woodbine, L., O'Driscoll, M. & Jeggo, P. A. (2005) EMBO J. 24, 199–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam, M. H., Liu, Q., Elledge, S. J. & Rosen, J. M. (2004) Cancer Cell 6, 45–59. [DOI] [PubMed] [Google Scholar]
- 28.Brown, E. J. & Baltimore, D. (2003) Genes Dev. 17, 615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]