Abstract
The Ser/Arg-rich (SR) proteins constitute a family of highly conserved nuclear phosphoproteins that are involved in many steps of mRNA metabolism. Previously, we demonstrated that shuttling SR proteins can associate with translating ribosomes and enhance translation of reporter mRNAs both in vivo and in vitro. Here, we show that endogenous, cytoplasmic splicing factor 2/alternative splicing factor (SF2/ASF) associated with the translation machinery is hypophosphorylated, suggesting that the phosphorylation state of the Arg-Ser-rich (RS) domain may influence the role of SF2/ASF in cytoplasmic RNA processing. In agreement, we show that mutations mimicking a hypophosphorylated RS domain strongly increased SF2/ASF binding to cytoplasmic mRNA and its activity in translation. We also demonstrate that, whereas the RS domain is not required for the function of SF2/ASF in mRNA translation in vivo or in vitro, its second RNA recognition motif (RRM)2 plays a critical role in this process. Taken together, these data suggest that RS-domain phosphorylation may influence the association of SF2/ASF with mRNA, whereas RRM2 may play an important role in mediating protein–protein interactions during translation. These data are consistent with a model whereby reversible protein phosphorylation differentially regulates the subcellular localization and activity of shuttling SR proteins.
Keywords: splicing factors, translation, subcellular localization, RNA binding
The Ser/Arg-rich (SR) proteins are a family of phylogenetically conserved, structurally related, splicing factors that have dual roles in pre-mRNA splicing affecting both constitutive and alternative splicing (1). SR family proteins have a modular structure consisting of one or two copies of an N-terminal RNA-recognition motif (RRM) followed by a C-terminal domain rich in alternating Ser and Arg residues, known as the Arg-Ser-rich (RS) domain. The RRMs determine RNA binding specificity, whereas the RS domain functions as a protein–protein interaction module by recruiting components of the core splicing apparatus to promote splice site pairing (reviewed in ref. 2).
SR proteins are primarily localized to the nuclear speckles (reviewed in ref. 3), and a subset of SR proteins shuttle continuously between the nucleus and the cytoplasm (4). Within the cell, the RS domain acts as a nuclear localization signal by mediating the interaction with the SR protein nuclear import receptor transportin-SR (5–7) and also influences the nucleocytoplasmic shuttling of individual SR proteins (4). The shuttling ability of a subset of SR proteins suggested additional roles in mRNA transport, and/or in cytoplasmic events, such as mRNA localization, stability, or regulation of translation. In agreement with this hypothesis, two shuttling SR proteins, SRp20 and 9G8, have been shown to promote mRNA export of intronless RNAs (8) and also act as adapter proteins for TAP-dependent mRNA export (9). SR proteins have also been implicated in RNA stability and quality control. For instance, splicing factor 2/alternative splicing factor (SF2/ASF) promotes the degradation of the PKCI-1 mRNA in chicken fibroblasts (10). In addition, overexpression of various SR proteins strongly enhanced nonsense-mediated decay in an RS-domain-dependent manner (11). Finally, the shuttling protein SF2/ASF is associated with polyribosomes in cytoplasmic extracts and enhances translation of a luciferase reporter in an enhancer-dependent manner, both in vivo and in vitro (12).
The RS domain of SR proteins is extensively phosphorylated on Ser residues, playing an important role in regulating their nuclear activities. Phosphorylation of the RS domain of SF2/ASF has been shown to enhance protein–protein interactions with other RS-domain-containing splicing factors, such as the U1 small nuclear ribonucleoprotein (snRNP)-specific protein U1–70K, playing an important role in driving spliceosome assembly (13). However, dephosphorylation of SR proteins and RS-domain-containing proteins is necessary for splicing catalysis to proceed (14, 15). Thus, a dynamic cycle of phosphorylation and dephosphorylation is required for constitutive splicing.
The Saccharomyces cerevisiae RNA-binding protein Npl3p, the closest orthologue to mammalian SR proteins, shuttles between the nucleus and the cytoplasm (16). Its subcellular distribution is also regulated by a cycle of phosphorylation and dephosphorylation. In the nucleus, Npl3p is phosphorylated on C-terminal Ser residues and interacts with both the pre-mRNA and components of the nuclear RNA processing machinery. The activity of a protein phosphatase Glc7p that dephosphorylates Npl3p in the nucleus is required for recruiting the mRNA export receptor Mex67p to mRNA, allowing mRNA export to proceed (17). In the cytoplasm, rephosphorylation of Npl3p by the protein kinase Sky1p may act to promote both the release of Npl3p from mRNA and interactions with the cognate nuclear import receptor Mtr10p (17–19). In mammalian cells, splicing-dependent dephosphorylation of the shuttling SR protein 9G8 functions as a molecular signal to recruit the mRNA export factor TAP (20). Thus, cycles of nuclear dephosphorylation and cytoplasmic rephosphorylation may control the flow of shuttling RNA-binding proteins in both yeast and man.
Here, we have investigated how the interaction between SF2/ASF and cytoplasmic mRNA is regulated. Our findings suggest that the phosphorylation of the RS domain functions to regulate the association of SF2/ASF with mRNA. Moreover, we present evidence that the domains responsible for the nuclear activities of SF2/ASF function differently when participating in cytoplasmic mRNA metabolism. Altogether, our data suggest that reversible protein phosphorylation differentially affects the nuclear and cytoplasmic functions of SF2/ASF.
Materials and Methods
cDNA Constructs. The mammalian expression vectors pCG T7-SF2/ASF and the SF2/ASF variants deleting the RS domain (ΔRS) or including phosphomimetic mutations in the RS domain have been described in ref. 21. The pLCS-EDA and EDAmt reporter plasmids have also been described in ref. 12. The SF2/ASF-mutant protein harboring a deletion of the RS domain and a triple point mutation in a conserved motif within RRM2 (SF2/ASF ΔRS WDK→AAA) was cloned by PCR amplification using the full-length SF2/ASF AAA cDNA as a template (a gift of G. Biamonti, Istituto di Genetica Molecolare, Pavia, Italy). Primer sequences are available upon request. The plasmids expressing SR protein kinase (SRPK)2 and the kinase-inactive mutant (22) were a gift of M. Hagiwara (Medical Research Institute, Tokyo Medical and Dental University, Tokyo).
Cell Culture and Transfection. HeLa and 293T HEK cells were grown in Dulbecco's MEM (Life Technologies), supplemented with antibiotics and 10% FBS (HyClone). Lipofectamine 2000 (Invitrogen) was used to transiently transfect cultured cells according to the manufacturer's instructions.
Antibodies and Western Blotting. The following antibodies and dilutions were used: anti-SC35 (mAb 1:500, Abcam), anti-SF2/ASF (mAb 96, 1:500) (23), antiphosphorylated SR proteins (mAb 104 undiluted cell-culture supernatant, American Type Culture Collection). T7 epitope-tagged proteins were detected with a monoclonal anti-T7 antibody (1:10,000, Novagen). Protein extracts were resolved by 12% SDS/PAGE and transferred to either hybond P (Amersham Pharmacia) or protran BA85 nitrocellulose (Schleicher and Schuell) membranes by using the Genie Blotter system, following the manufacturer's instructions (Idea Scientific, Corvallis, OR). Secondary antibodies conjugated to horseradish peroxidase and West Pico super signal detection reagent were obtained from Perbio (Cramlington, Northumberland, U.K.).
Sucrose-Gradient Fractionation. Ten percent to 50% sucrose gradients were used to resolve cytoplasmic RNP complexes as described in ref. 12.
In Situ UV Crosslinking mRNP-Capture Assay. In situ UV crosslinking mRNP capture was performed as described in ref. 24. Briefly, cells were scraped from 60-mm plates in 400 μl of RSB 100 [10 mM Tris·HCl, pH 7.5/100 mM NaCl/2.5 mM MgCl2/digitonin at a final concentration of 20 μg/ml and minicomplete EDTA-free protease inhibitor (Roche Diagnostics)]. The cells were fractionated, and extracts were denatured by the addition of an equal volume of 2× binding buffer (20 mM Tris·HCl, pH 7.5/1.0 M NaCl/1% SDS/0.2 mM EDTA), and nuclear extract was sonicated briefly. Oligo(dT) cellulose (≈25-μl packed-bed volume) equilibrated in 1× binding buffer was added to each fraction. The extracts were mixed with oligo(dT) cellulose for 1 h at room temperature on a rotating wheel and washed three times with 1.0 ml of 1× binding buffer. Captured mRNPs were eluted from the resin with 400 μl of elution buffer (10 mM Tris·HCl, pH 7.5/1 mM EDTA, minicomplete EDTA-free protease inhibitor) and 4 μl of RNase A/T1 mixture (Ambion, Huntingdon, U.K.) for 30 min at 37°C. Liberated mRNA-binding proteins were precipitated (by adding an equal volume of 20% TCA and incubating on ice for 20 min) and pelleted in a refrigerated microcentrifuge for 20 min at 12,000 × g. The precipitated proteins were washed in ice-cold acetone and resuspended in 40 μl of SDS/PAGE sample buffer. Captured mRNA-binding proteins were resolved by 12% SDS/PAGE and analyzed by Western blotting.
In Vitro and in Vivo Translation Assays. In vivo translation assays were as described in ref. 12. In vitro translation assays using HeLa-cell-based extracts were performed by using in vitro-transcribed pLCS-3x EDA reporter mRNA as described in ref. 12. Recombinant SF2/ASF wild-type and ΔRS proteins were purified from transiently transfected 293T cells as described in ref. 25. Recombinant hnRNP A1 was a gift of A. Krainer (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY).
Results
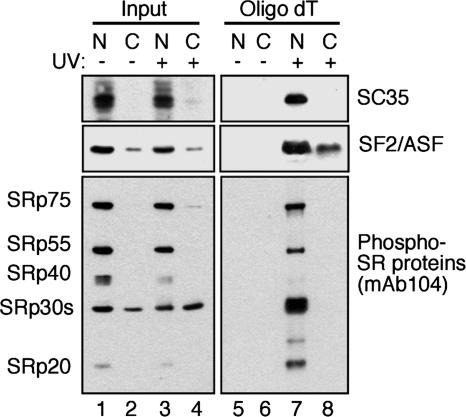
SF2/ASF Is Directly Bound to Nuclear and Cytoplasmic mRNA. Despite the well characterized nucleocytoplasmic shuttling activity of SF2/ASF and its proposed role in postsplicing activities, such as mRNA export, stability, and translation, it has not yet been directly shown whether SF2/ASF is bound to mRNA in the cytoplasm. To address this issue, we used in situ UV crosslinking and denaturing oligo(dT) selection to purify messenger RNP (mRNP) particles from either nuclear or cytosolic fractions (24). To control for the integrity of the nuclear and cytosolic fractions, we blotted equivalent amounts of extracts before oligo(dT) selection (Fig. 1, Input) with antibodies against the nonshuttling SR protein SC35 or the shuttling SR protein SF2/ASF. As expected, SC35 is highly enriched in the nuclear fraction, whereas little or no signal is detectable in the cytoplasmic fraction (Fig. 1 Top Left). By contrast, SF2/ASF can be readily detected in both fractions (Fig. 1 Middle Left). These data indicate that nuclear leakage of SR proteins is not a significant source of contamination within the cytoplasmic fraction.
Fig. 1.
Endogenous SF2/ASF binds directly to cytoplasmic mRNA and is partially dephosphorylated. Shown is in situ UV crosslinking of 293T HEK cells. Following cell fractionation, mRNP complexes were purified by oligo(dT) chromatography under denaturing conditions. Lanes 1–4 (Input) contain nuclear (lanes 1 and 3) and cytosolic (lanes 2 and 4) extracts before oligo(dT) selection of mRNPs, whereas lanes 5–8 (oligo dT) contain purified nuclear (lanes 5 and 7) and cytosolic (lanes 6 and 8) mRNPs eluted from oligo(dT) cellulose. mAbs against SC35 and SF2/ASF were used to visualize a nonshuttling and a shuttling SR protein, respectively, by Western blots. The same samples were also blotted with mAb 104, which recognizes a conserved phosphoepitope within the RS domain of SR proteins.
To investigate the phosphorylation status of cytoplasmic SR proteins, we used a monoclonal antibody directed against the phosphorylated RS domain of SR proteins (mAb 104) (26). Blotting the same fractions with this antibody revealed that the majority of phosphorylated SR proteins are enriched in the nucleus (Fig. 1 Bottom Left). Interestingly, mAb 104 also detects phosphorylated 30-kDa SR proteins (presumably SF2/ASF and/or 9G8) in the cytoplasmic inputs (Fig. 1 Bottom Left). Affinity selection of mRNPs by oligo(dT) cellulose clearly demonstrates that both the shuttling SR protein SF2/ASF and the nonshuttling SR protein SC35 are bound to nuclear mRNA (Fig. 1, lane 7). Interestingly, a subset of SF2/ASF, but not SC35, is directly bound to mRNA in the cytoplasm as well. These data provide direct evidence that SF2/ASF remains associated with mRNA after nuclear pre-mRNA processing and mRNA export. Fig. 1 also clearly shows that mAb 104 detects SR proteins bound to mRNA from the nuclear fraction; however, little, if any, reactivity is detected in captured cytoplasmic mRNPs, despite the presence of phosphorylated 30-kDa SR proteins (SF2/ASF and/or 9G8) in the cytoplasmic inputs (Fig. 1 Bottom Left). Thus, despite the presence of phosphorylated SR proteins in the cytoplasm, few, if any, are detected in a complex with mRNA. Taken together, these data indicate that the SF2/ASF protein that is bound to cytoplasmic mRNA is at least partially dephosphorylated.
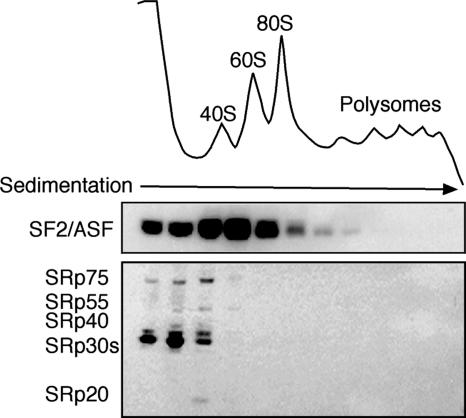
Dephosphorylated SF2/ASF Is Associated with the Translation Machinery. We next analyzed the phosphorylation state of SF2/ASF associated with the translation machinery. Cytoplasmic extracts prepared from HeLa cells transfected with epitope-tagged SF2/ASF were fractionated across 10–50% sucrose gradients, and the distribution of SF2/ASF was analyzed by Western blotting. As previously reported, we observed that a subset of SF2/ASF cosediments with the 80S ribosome and light polyribosomes (Fig. 2 Upper). Use of mAb 104 revealed that phosphorylated SR proteins do not cosediment with ribosomes and are restricted to the top of the gradient (Fig. 2 Lower). Similar results were obtained when the endogenous SF2/ASF protein from nontransfected cells was analyzed, as shown in ref. 12 (data not shown).
Fig. 2.
Partially dephosphorylated SF2/ASF cosediments with the translation machinery. Cytoplasmic extracts prepared from HeLa cells transfected with epitope-tagged SF2/ASF were fractionated across 10–50% sucrose gradients. Fractions were analyzed by Western blotting against the T7 epitope tag (Upper) and mAb 104 (Lower).
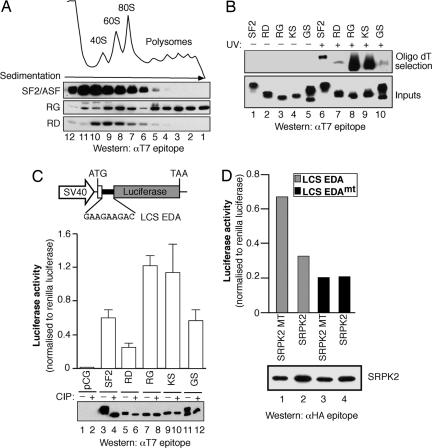
Dephosphorylation of the RS Domain Enhances mRNA-Binding and Translational Activity. We then asked whether phosphorylation of the RS domain influenced the cytoplasmic functions of SF2/ASF. Previously, we constructed variants of human SF2/ASF with artificial domains replacing the natural C-terminal RS domain. In these variant proteins, Ser residues within RS or SR dipeptide repeats of the RS domain of SF2/ASF (residues 198–248) were substituted to generate a RG or RD domain to mimic a hypo- or hyperphosphorylated RS domain, respectively (21). Sucrose-gradient fractionation of cytoplasmic extracts prepared from transfected HeLa cells revealed that, whereas wild-type SF2/ASF or the RD-mutant version had similar distributions within the gradient, the SF2/ASF-mutant protein harboring a RG domain was shifted to the much heavier polysome-containing fractions (Fig. 3A). Importantly, the sedimentation of the RG mutant depended on the integrity of ribosomes, because treatment of the extracts with EDTA shifted the distribution of RG away from the polyribosomes (data not shown).
Fig. 3.
The phosphorylation state of the RS domain influences the cytoplasmic activities of SF2/ASF. (A) Sucrose-gradient analyses of cytoplasmic extracts were analyzed by Western blotting with an mAb directed against the epitope tag. (B) In situ UV crosslinking mRNP capture assay from cells transfected with epitope-tagged wild-type SF2/ASF or variants of SF2/ASF, with artificial domains replacing the natural C-terminal RS domain (RD, RG, KS, and GS mutants). Captured mRNA-binding proteins were visualized by Western blotting, with antibodies directed against the epitope tag. Expression of the transiently expressed constructs was confirmed by Western blotting against the T7 epitope tag (Bottom). (C) Schematic diagram of the pLCS reporter system. HeLa cells were cotransfected with pLCS-EDA and empty vector pCGT7-SF2/ASF or the RD-, RG-, KS-, or GS-domain-mutant cDNAs. Expression of the transiently expressed constructs was confirmed by Western blotting against the T7 epitope tag. Phosphorylation of the RS-domain mutants was confirmed by treating the lysates with calf intestinal phosphatase (Bottom, lanes 2, 4, 6, 8, 10, and 12). (D) HeLa cells were transiently transfected with pLCS-EDA or pLCS-EDAmt and wild-type SRPK2 or kinase-inactive mutant (SRPK2 MT). Expression of SRPK2 was confirmed by Western blotting against the HA epitope tag.
One possible explanation of these results is that phosphorylation of the RS domain may play a role in regulating the extent of mRNA binding. To test this hypothesis, crosslinked mRNPs were purified under denaturing conditions by oligo(dT) chromatography from cytoplasmic extracts prepared from 293T cells transfected with epitope-tagged SF2/ASF containing either a wild-type or the RD, RG, KS, and GS mutations in their RS domains (Fig. 3B). Western blotting for the T7 epitope tag revealed that similar levels of overexpressed proteins were present in the input lysates before affinity selection (Fig. 3B Lower). We found that the RG-mutant protein bound mRNA to a far greater extent then either wild-type SF2/ASF or the RD-mutant protein (Fig. 3B Upper, lanes 6–8). To determine whether the positive charge of Arg residues within the RS domain is required to enhance mRNA binding, we also analyzed SF2/ASF containing a KS domain (Arg→Lys), which conserves the positive charge throughout the RS domain but is not phosphorylated in vivo, or a GS domain (Arg→Gly), which lacks the positively charged side chain but is phosphorylated in vivo (Fig. 3C Lower, lanes 9–12). Interestingly, the KS-mutant protein displays enhanced cytoplasmic mRNA binding, whereas the binding activity of the GS-mutant protein is dramatically reduced (Fig. 3B Upper, lanes 9–10). These data indicate that the positive charge of the Arg residues throughout the RS domain play an import role in cytoplasmic mRNA binding.
We previously reported that SF2/ASF activates translation of a luciferase reporter harboring an exonic splicing enhancer (ESE) derived from the EDA alternative exon of the fibronectin gene, which is known to recruit SF2/ASF and 9G8 (12). This ESE sequence was inserted in either a wild-type or a mutant version that lacked binding sites for SF2/ASF (pLCS-EDA or pLCS-EDAmt, respectively). Here, we show that overexpression of wild-type SF2/ASF strongly activated translation of the pLCS-EDA reporter mRNA, as reported in ref. 12 (Fig. 3C). Interestingly, overexpression of SF2/ASF containing an RD domain was less active than wild-type SF2/ASF. The RG- and KS-mutant proteins were more robust activators of the pLCS-EDA reporter, whereas the GS-mutant protein behaved like wild-type SF2/ASF (Fig. 3C Middle). Importantly, direct comparison of the RG and RD mutants demonstrates that the RD mutant was ≈4.5-fold less active than the RG mutant in the in vivo translation assay, even though the proteins are expressed to similar levels (Fig. 3C Bottom). Importantly, Western blot analysis with the T7 epitope demonstrated that similar levels of proteins were expressed in each experiment (Fig. 3C Bottom). Moreover, treatment of the cell lysates with calf intestinal phosphatase demonstrated that, like SF2/ASF, the GS mutant is phosphorylated in vivo, whereas the RD, RG, and KS mutants are not (Fig. 3C Bottom). Previous work showed that phosphomimetic mutations in the RS domain, such as RG, RE, KS, and GS and, to a lesser extent, RD domains, exhibited aberrant cytoplasmic localization (21). However, despite the increased cytoplasmic accumulation of RS-domain-mutant proteins, only the RG- and KS-mutant proteins are highly active in translation. By contrast, the GS-mutant protein binds mRNA very weakly and is less active in the translation assay. Moreover, the RD-mutant protein, which mimics a hyperphosphorylated RS domain, is less active than wild-type SF2/ASF in translation, despite an increased cytoplasmic localization. These data, combined with the absence of endogenous SR proteins in a phosphorylated state from monosomes and polysomes (Fig. 2), strongly suggest that the phosphorylation state of the RS domain, more than the relative abundance of cytoplasmic SF2/ASF, is what determines the activity of SF2/ASF in translation. Thus, SR-protein phosphorylation does not necessarily reduce SF2/ASF translational activity by reducing its cytoplasmic levels; rather, phosphorylation results in decreased binding to cytoplasmic mRNA, and this finding correlates well with a reduction in translational activity.
To determine whether SRPKs can modulate the activity of SF2/ASF in mRNA translation, we cotransfected the translation reporters with wild-type or kinase-inactive SRPK2 or SRPK2 MT, respectively (27). Interestingly, expression of SRPK2, but not of the mutant protein, decreased expression of pLCS-EDA, whereas expression of the pLCS EDAmt remained unchanged (Fig. 3D Upper). Western blot analysis of the transfected kinases demonstrated that the WT and MT isoforms are expressed to similar levels (Fig. 3D Lower). Interestingly, expression of a primarily nuclear SRPK Clk/Sty had a very similar effect to that of SRPK2 (data not shown). These data suggest that hyperphosphorylation of SF2/ASF SRPK2 negatively regulates the cytoplasmic functions of SF2/ASF.
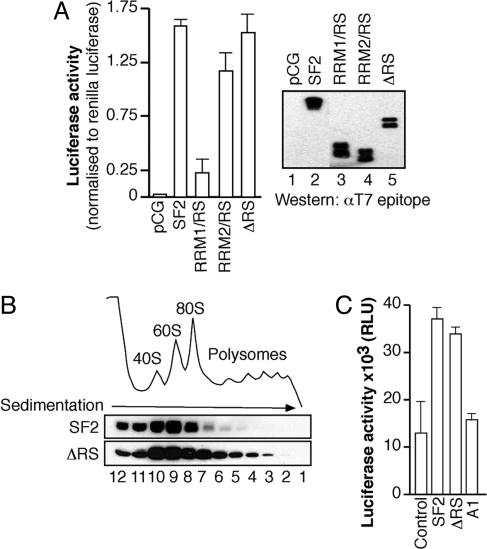
The RS Domain Is Not Required for the Role of SF2/ASF in Translation. To determine whether the RS domain is directly involved in stimulating translation of the reporter mRNA, we coexpressed the pLCS-EDA reporter along with wild-type SF2/ASF or a mutant protein lacking the RS domain (ΔRS, Fig. 4A). Surprisingly, overexpression of the ΔRS-mutant protein robustly activated translation of the reporter mRNA, demonstrating that the RS domain is not required to stimulate translation in vivo. Interestingly, deletion of the second RRM of SF2/ASF (RRM1/RS) significantly decreased the expression of the pLCS EDA reporter, whereas deletion of RRM1 (RRM2/RS) had only a modest effect (Fig. 4A). Importantly, Western blot analysis of the transfected proteins with an antibody against the T7 epitope tag revealed that similar levels of each construct were expressed (Fig. 4A Right). Sucrose-gradient analysis of cytoplasmic extracts prepared from cells transiently transfected with wild-type SF2/ASF or the ΔRS variant revealed that the latter was more strongly associated with polyribosomes than wild-type SF2/ASF (Fig. 4B). Moreover, the ΔRS protein was also more strongly associated with cytoplasmic mRNA than wild type SF2/ASF (data not shown). Taken together, these data indicate that the RS domain is not required for SF2/ASF-mediated stimulation of mRNA translation in vivo and that RRM2 of SF2/ASF has a role in this function (see below).
Fig. 4.
The RS domain is not required to stimulate mRNA translation either in vivo or in vitro. (A) HeLa cells were transiently transfected with the pLCS-EDA reporter and either empty vector (control), wild-type SF2/ASF, or SF2/ASF cDNAs lacking RRM2 (RRM1/RS), RRM1 (RRM2/RS), or the RS domain (ΔRS), respectively (Left). Expression of the transiently expressed constructs was confirmed by Western blotting against the T7 epitope tag (Right). (B) Cytoplasmic extracts prepared from HeLa cells transfected with epitopetagged wild-type SF2/ASF or the ΔRS variant were fractionated across 10–50% sucrose gradients. Fractions were analyzed by Western blotting against the T7 epitope tag. (C) HeLa-cell-free in vitro translation assay comparing the ability of 5 pmol of exogenous SF2/ASF, ΔRS, or hnRNP A1 recombinant proteins to stimulate translation of in vitro-transcribed pLCS 3x EDA reporter mRNA.
Next, we assayed the ability of recombinant T7 epitope-tagged SF2/ASF and ΔRS to promote translation of a pLCS reporter mRNA containing three copies of the EDA exonic splicing enhancer (pLCS 3x EDA) in vitro. As previously reported, we found that HeLa cell translation extracts supplemented with 5 pmol of recombinant wild-type SF2/ASF protein showed an increase in translation of the luciferase reporter (12). Interestingly, addition of recombinant SF2/ASF ΔRS-mutant protein was able to stimulate translation of the pLCS 3x EDA reporter to the same extent as SF2/ASF, whereas equivalent amounts of hnRNP A1 had no effect (Fig. 4C). Moreover, Northern blot analysis demonstrated no changes in the stability of the reporter mRNA during the course of this assay (data not shown). Thus, we conclude that the RS domain is not required for the role of SF2/ASF in mRNA translation.
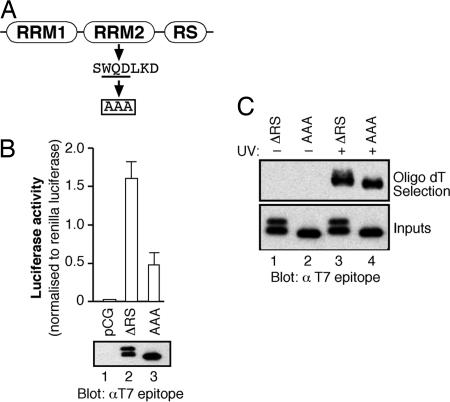
RRM2 Is Required for the Function of SF2/ASF in Translation. The second RRM of SF2/ASF is an atypical RRM that lacks the conserved aromatic residues present in the RNP-1 and RNP-2 motifs that make direct contacts with RNA. RRM2 is characterized by the presence of a phylogenetically conserved heptapeptide, SWQDLKD, which is located in the first α-helix of this domain (28). It has been shown that mutations in this heptapeptide motif affect SF2/ASF functions in pre-mRNA splicing (29) and its recruitment to nuclear stress granules (30). We took advantage of an RRM2 mutant of SF2/ASF in which amino acid substitutions were introduced in this conserved heptapeptide, replacing the WQD residues by three alanines (Fig. 5A, WQD-AAA mutant) (30). We investigated the role of RRM2 in translation by transiently transfecting an SF2/ASF-mutant protein lacking the RS domain but also containing the heptapeptide mutation (WQD-AAA) together with the pLCS-EDA reporter. Fig. 5B shows that, whereas SF2/ASF ΔRS is a potent activator of the pLCS EDA reporter, the AAA mutation strongly decreases the activity of SF2/ASF. Importantly the transiently transfected proteins are expressed at similar levels, as determined by Western blot analysis. Surprisingly, the ΔRS-mutant protein consistently migrated as a doublet when extracts were resolved by SDS/PAGE, whereas the protein harboring the AAA mutation migrates as a single band. We have evidence that SF2/ASF ΔRS is modified by phosphorylation of Ser 129 within RRM2 (data not shown). The conserved heptapeptide motif is unlikely to be directly involved in RNA binding, given its position within the predicted structure of RRM2, but could have a function in protein–protein interactions (29). Fig. 5C clearly shows that both ΔRS and ΔRS AAA proteins can directly bind to cytoplasmic mRNA to the same extent. These data suggest that the heptapeptide motif of RRM2 of SF2/ASF may function to mediate protein–protein interactions, and this activity is important for the role of SF2/ASF in translational regulation.
Fig. 5.
RRM2 is required for SF2/ASF to stimulate mRNA translation in vivo. (A) Schematic of the conserved heptapeptide present in RRM2 of SF2/ASF and point mutations analyzed. (B) In vivo translation assay comparing the activity of ΔRS and ΔRS (WQD-AAA) proteins. Western blot analysis of the transiently transfected constructs with anti-T7 monoclonal antibody (Lower). (C) In situ UV crosslinking mRNP-capture assay comparing cytoplasmic mRNA binding of SF2/ASF ΔRS protein with the RRM2 variant.
Discussion
Here, we have shown that cytoplasmic mRNPs contain partially dephosphorylated SF2/ASF. Moreover, sucrose-gradient analysis of HeLa-cell cytoplasmic extracts suggests that the subset of SF2/ASF cosedimenting with the translation machinery is also partially dephosphorylated. Mutational analysis of the RS domain and RRMs of SF2/ASF revealed a role for the hypophosphorylated RS domain in cytoplasmic mRNA binding and for RRM2 in stimulating translation of a reporter mRNA. These data highlight findings regarding the roles of SF2/ASF in cytoplasmic RNA-processing events.
The cyclic requirement for RS-domain phosphorylation and dephosphorylation during pre-mRNA splicing has been well documented (reviewed in ref. 31). Interestingly, work from several labs, including the data presented here, suggests that splicing-dependent dephosphorylation of shuttling SR proteins differentiates their pre- and postsplicing activities (20, 32). Here, we show that, whereas phosphorylated SR proteins are present in the cytoplasm, they are not bound to mRNA. Perhaps this pool of SF2/ASF has been rephosphorylated by cytoplasmic kinases after their role in translation and is awaiting nuclear import. Our data suggest that dephosphorylation of the RS domain enhances cytoplasmic mRNA binding by SF2/ASF, as shown by mutations mimicking a hypophosphorylated RS domain (RG mutant). It is possible that the dephosphorylated RS domain functions as an ionic anchor, locking the protein to the mRNA.
The work presented herein indicates that SF2/ASF participates in nuclear and cytoplasmic steps of mRNA processing by very different mechanisms. During pre-mRNA, splicing phosphorylation of the RS domain stimulates recruitment of SR proteins from interchromatin granule clusters to nascent transcripts (reviewed in ref. 33). Additionally, phosphorylation of the RS domain has been shown to enhance protein–protein interactions between RS-domain-containing splicing factors and to facilitate sequence-specific binding to exonic splicing enhancers by the RRMs, presumably by neutralizing the positively charged Arg residues, thus decreasing nonspecific interactions with pre-mRNA (reviewed in ref. 34). By contrast, in postsplicing mRNPs containing SF2/ASF, the RS domain is dephosphorylated, and this appears to contribute to cytoplasmic mRNA binding. Moreover, we have found that RRM2 of SF2/ASF plays an important but enigmatic role in stimulating translation in the cytoplasm. It is appealing to speculate that RRM2 may also promote unique protein–protein interactions that may be inhibited by the presence of a phosphorylated RS domain. In agreement, recent reports suggest that RRMs are multifunctional domains that can serve as a binding surface for interactions with both RNA and protein partners (35, 36).
Several SRPKs have been implicated in SR-protein localization and spliceosome assembly, including SRPK1, 2 (27, 37), the Clk/Sty family of dual specificity kinases (38, 39), and DNA topoisomerase I (40). Here, we have shown that overexpression of SRPK2 decreases the cytoplasmic activity of SF2/ASF. In addition to the basal phosphorylation cycle described above, SR proteins are also phosphorylated in response to a growing list of cellular and developmental signals (37, 41). Interestingly, regulation of SR protein activities during adenoviral infection occurs through dephosphorylation mediated by the viral E4-ORF4 protein and cellular protein phosphatase 2A (42, 43). Virus-induced dephosphorylation renders SR proteins inactive as both splicing activators and repressors and, thus, alters the alternative splicing of the viral pre-mRNA. In an analogous situation, the herpes simplex virus 1 protein ICP27, modifies SRPK1 activity, resulting in hypophosphorylation of SR proteins, impairing their ability to function in spliceosome assembly (44). Thus, it is tempting to speculate that virus-induced dephosphorylation of shuttling SR proteins during viral infection may play a role in stimulating translation of viral mRNAs.
Thus, the findings presented herein have very interesting implications for the coordination and coupling of different steps of posttranscriptional gene expression and further illustrate the multifunctional nature of shuttling mRNA-binding proteins.
Acknowledgments
We thank G. Biamonti for the gift of the SF2/ASF AAA mutation and M. Hagiwara for SRPK constructs. We are grateful to Sonia Guil (Medical Research Council Human Genetics Unit) for helpful comments on the manuscript. This work was supported by the Medical Research Council (D.C., J.D.E., and J.F.C.) and a postdoctoral fellowship from the Caledonian Research Foundation (to J.R.S.).
Author contributions: J.R.S. and J.F.C. designed research; J.R.S., J.D.E., and D.C. performed research; D.C. contributed new reagents/analytic tools; J.R.S., J.D.E., and J.F.C. analyzed data; and J.R.S. and J.F.C. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: RNP, ribonucleoprotein; mRNP, messenger RNP; RRM, RNA recognition motif; RS, Arg-Ser-rich; SF2/ASF, splicing factor 2/alternative splicing factor; SR, Ser/Argrich; SRPK, SR protein kinase.
References
- 1.Fu, X. D. (1995) RNA 1, 663–680. [PMC free article] [PubMed] [Google Scholar]
- 2.Graveley, B. R. (2000) RNA 6, 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lamond, A. I. & Spector, D. L. (2003) Nat. Rev. Mol. Cell Biol. 4, 605–612. [DOI] [PubMed] [Google Scholar]
- 4.Caceres, J. F., Screaton, G. R. & Krainer, A. R. (1998) Genes Dev. 12, 55–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caceres, J. F., Misteli, T., Screaton, G. R., Spector, D. L. & Krainer, A. R. (1997) J. Cell Biol. 138, 225–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kataoka, N., Bachorik, J. L. & Dreyfuss, G. (1999) J. Cell Biol. 145, 1145–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lai, M. C., Lin, R. I., Huang, S. Y., Tsai, C. W. & Tarn, W. Y. (2000) J. Biol. Chem. 275, 7950–7957. [DOI] [PubMed] [Google Scholar]
- 8.Huang, Y. & Steitz, J. A. (2001) Mol. Cell 7, 899–905. [DOI] [PubMed] [Google Scholar]
- 9.Huang, Y., Gattoni, R., Stevenin, J. & Steitz, J. A. (2003) Mol. Cell 11, 837–843. [DOI] [PubMed] [Google Scholar]
- 10.Lemaire, R., Prasad, J., Kashima, T., Gustafson, J., Manley, J. L. & Lafyatis, R. (2002) Genes Dev. 16, 594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang, Z. & Krainer, A. R. (2004) Mol. Cell 16, 597–607. [DOI] [PubMed] [Google Scholar]
- 12.Sanford, J. R., Gray, N. K., Beckmann, K. & Caceres, J. F. (2004) Genes Dev. 18, 755–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiao, S. H. & Manley, J. L. (1997) Genes Dev. 11, 334–344. [DOI] [PubMed] [Google Scholar]
- 14.Tazi, J., Kornstadt, U., Rossi, F., Jeanteur, P., Cathala, G., Brunel, C. & Luhrmann, R. (1993) Nature 363, 283–286. [DOI] [PubMed] [Google Scholar]
- 15.Cao, W., Jamison, S. F. & Garcia-Blanco, M. A. (1997) RNA 3, 1456–1467. [PMC free article] [PubMed] [Google Scholar]
- 16.Flach, J., Bossie, M., Vogel, J., Corbett, A., Jinks, T., Willins, D. A. & Silver, P. A. (1994) Mol. Cell. Biol. 14, 8399–8407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gilbert, W. & Guthrie, C. (2004) Mol. Cell 13, 201–212. [DOI] [PubMed] [Google Scholar]
- 18.Yun, C. Y. & Fu, X. D. (2000) J. Cell Biol. 150, 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilbert, W., Siebel, C. W. & Guthrie, C. (2001) RNA 7, 302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, Y., Yario, T. A. & Steitz, J. A. (2004) Proc. Natl. Acad. Sci. USA 101, 9666–9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cazalla, D., Zhu, J., Manche, L., Huber, E., Krainer, A. R. & Caceres, J. F. (2002) Mol. Cell. Biol. 22, 6871–6882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koizumi, J., Okamoto, Y., Onogi, H., Mayeda, A., Krainer, A. R. & Hagiwara, M. (1999) J. Biol. Chem. 274, 11125–11131. [DOI] [PubMed] [Google Scholar]
- 23.Hanamura, A., Caceres, J. F., Mayeda, A., Franza, B. R. & Krainer, A. R. (1998) RNA 4, 430–444. [PMC free article] [PubMed] [Google Scholar]
- 24.Pinol-Roma, S. & Dreyfuss, G. (1992) Nature 355, 730–732. [DOI] [PubMed] [Google Scholar]
- 25.Cazalla, D., Sanford, J. R. & Caceres, J. F. (2005) Protein Expression Purif. 42, 54–58. [DOI] [PubMed] [Google Scholar]
- 26.Roth, M. B., Murphy, C. & Gall, J. G. (1990) J. Cell Biol. 111, 2217–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, H. Y., Lin, W., Dyck, J. A., Yeakley, J. M., Songyang, Z., Cantley, L. C. & Fu, X. D. (1998) J. Cell Biol. 140, 737–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Birney, E., Kumar, S. & Krainer, A. R. (1993) Nucleic Acids Res. 21, 5803–5816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dauksaite, V. & Akusjarvi, G. (2002) J. Biol. Chem. 277, 12579–12586. [DOI] [PubMed] [Google Scholar]
- 30.Chiodi, I., Corioni, M., Giordano, M., Valgardsdottir, R., Ghigna, C., Cobianchi, F., Xu, R. M., Riva, S. & Biamonti, G. (2004) Nucleic Acids Res. 32, 4127–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huang, Y. & Steitz, J. A. (2005) Mol. Cell 17, 613–615. [DOI] [PubMed] [Google Scholar]
- 32.Lai, M. C. & Tarn, W. Y. (2004) J. Biol. Chem. 279, 31745–31749. [DOI] [PubMed] [Google Scholar]
- 33.Misteli, T. (2000) J. Cell Sci. 113, 1841–1849. [DOI] [PubMed] [Google Scholar]
- 34.Tacke, R. & Manley, J. L. (1999) Curr. Opin. Cell Biol. 11, 358–362. [DOI] [PubMed] [Google Scholar]
- 35.Shi, H. & Xu, R. M. (2003) Genes Dev. 17, 971–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kielkopf, C. L., Lucke, S. & Green, M. R. (2004) Genes Dev. 18, 1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gui, J. F., Lane, W. S. & Fu, X. D. (1994) Nature 369, 678–682. [DOI] [PubMed] [Google Scholar]
- 38.Colwill, K., Feng, L. L., Yeakley, J. M., Gish, G. D., Caceres, J. F., Pawson, T. & Fu, X. D. (1996) J. Biol. Chem. 271, 24569–24575. [DOI] [PubMed] [Google Scholar]
- 39.Colwill, K., Pawson, T., Andrews, B., Prasad, J., Manley, J. L., Bell, J. C. & Duncan, P. I. (1996) EMBO J. 15, 265–275. [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi, F., Labourier, E., Forne, T., Divita, G., Derancourt, J., Riou, J. F., Antoine, E., Cathala, G., Brunel, C. & Tazi, J. (1996) Nature 381, 80–82. [DOI] [PubMed] [Google Scholar]
- 41.Sanford, J. R. & Bruzik, J. P. (1999) Genes Dev. 13, 1513–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kanopka, A., Muhlemann, O., Petersen-Mahrt, S., Estmer, C., Ohrmalm, C. & Akusjarvi, G. (1998) Nature 393, 185–187. [DOI] [PubMed] [Google Scholar]
- 43.Estmer, N. C., Petersen-Mahrt, S., Durot, C., Shtrichman, R., Krainer, A. R., Kleinberger, T. & Akusjarvi, G. (2001) EMBO J. 20, 864–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sciabica, K. S., Dai, Q. J. & Sandri-Goldin, R. M. (2003) EMBO J. 22, 1608–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]