Abstract
The recognition of microbial components by Toll-like receptors (TLRs) is an event central to the activation of innate and adaptive immune systems. TLR activation triggers the induction of downstream target genes, wherein the TLR-interacting adaptor molecule MyD88 recruits various signaling molecules and transcription factors. Two members of the IFN regulatory factor (IRF) family of transcription factors, IRF-5 and IRF-7, interact with MyD88 and induce proinflammatory cytokines and type I IFNs, respectively. Here, we show that IRF-4 also interacts with MyD88 and acts as a negative regulator of TLR signaling. IRF-4 mRNA is induced by TLR activation, and IRF-4 competes with IRF-5, but not with IRF-7, for MyD88 interaction. The TLR-dependent induction of proinflammatory cytokines is markedly enhanced in peritoneal macrophages from mice deficient in the Irf4 gene, whereas the induction is inhibited by the ectopic expression of IRF-4 in a macrophage cell line. The critical function of IRF-4 in TLR signaling in vivo is underscored by the observation that Irf4-deficient mice show hypersensitivity to DNA-induced shock, with elevated serum proinflammatory cytokine levels. This study may provide an insight into the complex regulatory mechanisms of MyD88 signaling by IRFs.
Keywords: IFN regulatory factor, macrophage, MyD88
Conserved microbial structures, termed pathogen-associated molecular patterns, are recognized by innate immunity receptors, such as Toll-like receptors (TLRs), the activation of which is central to the regulation and orchestration of innate and adaptive immune systems against pathogens (1, 2). The hallmark of TLR signaling is the induction of various cytokines and chemokines. In most cases, the induction of cytokines and chemokines depends on a common molecular pathway that is anchored by an adaptor protein termed MyD88 (1–3). MyD88 contains a structural region called the death domain, which allows it to associate with molecules that further transmit signals (3, 4). These MyD88-interacting molecules include the IL-1-receptor-associated kinase (IRAK)1 and IRAK4, both of which also contain a death domain and the adaptor molecule TNF-receptor-associated factor (TRAF) 6, and these signaling molecules are critical for the activation of the NF-κB and AP-1 transcription factors (1, 2, 5, 6).
In addition, recent studies have revealed the role of IFN regulatory factors (IRFs) in MyD88-dependent TLR signaling. Among nine members of the IRF family (7), IRF-5 and IRF-7 have been shown to interact with MyD88 to regulate the TLR-dependent induction of proinflammatory cytokines and type I IFNs (IFN-α/β), respectively (8–11). These reports, therefore, suggested the complexity of the MyD88 signaling pathways, and we proposed that the MyD88 complex serves as the cytoplasmic transductional–transcriptional processor, functioning as a cytoplasmic point of an organized array of signaling molecules and transcription factors that dynamically determines the specificity, strength, and longevity of the input signal to the output of transcriptional events (8).
In this study, we analyzed the possible involvement of other IRF family members and found that IRF-4 also forms a complex with MyD88. We show that IRF-4 binds to the same region of MyD88 as IRF-5, which is distinct from that for IRF-7 binding. We also provide evidence that IRF-4 competes with IRF-5, but not with IRF-7, for MyD88 interaction. The induction of proinflammatory cytokines by TLRs is markedly enhanced in peritoneal macrophages derived from Irf4-deficient (Irf4-/-) mice, and, conversely, the induction is inhibited by the ectopic expression of IRF-4 in a macrophage cell line RAW264.7. The DNA-binding domain of IRF-4 is not involved in this inhibition. On the other hand, the IRF-7-dependent induction of IFN-α in plasmacytoid dendritic cells (pDCs) from Irf4-/- mice remained the same as that in wild-type pDCs. Finally, Irf4-/- mice are highly sensitive to unmethylated DNA-induced shock, which is mediated by MyD88-dependent signaling. Considering that IRF-4 mRNA is induced upon TLR activation, IRF-4 may participate in the negative-feedback regulation of TLR signaling. This study adduces evidence for a unique facet of the role of IRF-4: It functions not only as a transcription factor in lymphocyte differentiation but also as a negative regulator of TLR signaling.
Materials and Methods
Mice. C57BL/6 mice were purchased from CLEA Japan, Osaka. The generation of Irf5-/- mice and Irf4-/- mice is described in refs. 10 and 12.
Plasmid Construction. The expression vectors for hemagglutinin (HA)-tagged mouse IRF-3, IRF-5, and IRF-7, FLAG-tagged full-length mouse MyD88, and the series of deletion mutants of MyD88, cyan fluorescent protein (CFP)-tagged MyD88 and yellow fluorescent protein (YFP)-tagged IRF-5 were described in refs. 8 and 10. The fragments of the full-length mouse IRF-4, IRF-8, and IRF-9 and the DNA-binding domain-deleted mutant IRF-4 (IRF-4ΔDBD) and the regulatory-domain-deleted mutant IRF-4 (IRF-4ΔRD) were obtained by RT-PCR and independently cloned into the pCAGGS-HA, pCAGGS-YFP, or pCAGGS-red fluorescent protein vector (8, 10). The expression vector for FLAG-tagged TRAF6 was kindly provided by J. Inoue (University of Tokyo).
Reagents. Unmethylated CpG-oligodeoxynucleotides (CpG-ODNs), ODN1668 (13) and ODN-D19 (14), were synthesized as described in ref. 11. Lipopolysaccharide (LPS), polyuridylic acid [Poly(U)], Leptomycin B, and d-galactosamine were purchased from Sigma. Poly(U) was complexed with DOTAP (Roche Diagnostics) as described in ref. 11. Peptideglycan was purchased from Fluka.
Imaging. Cells were cultured on glass-bottom 35-mm tissue-culture dishes (Matsunami Glass, Osaka) and were transfected with expression vectors for fluorescent-protein-tagged fusion proteins by using FuGENE 6 reagent (Roche Diagnostics) or SuperFect transfection reagent (Qiagen, Valencia, CA). Confocal microscopic analysis was performed by using an Olympus FV-1000 confocal microscope. Dual-color images were acquired in the sequential acquisition mode to avoid cross-excitation. Time-lapse imaging, fluorescence resonance energy transfer (FRET) analysis and the calculation of corrected FRET (FRETc) are described in detail in ref. 8.
Immunoprecipitation and Immunoblotting. Immunoprecipitation and immunoblotting were carried out as described in ref. 8. Anti-HA and anti-FLAG monoclonal antibodies were purchased from Sigma.
Reporter Assay. Human embryonic kidney (HEK) 293T cells seeded on 24-well plates were transiently cotransfected with 100 ng of a reporter plasmid [p55C1B-Luc (15) or p125-Luc (16)], together with expression plasmids for IRFs, MyD88, and TRAF6 by using FuGENE 6 reagent (Roche Diagnostics). The total amounts of DNA were kept constant by supplementation with an empty vector [pcDNA3.1 (Invitrogen)]. Twenty-four hours after transfection, the cells were harvested, and luciferase activity was measured as described in ref. 8.
Measurement of Cytokine Production. Resident peritoneal macrophages were obtained by peritoneal lavage. Splenic pDCs (B220+CD11cintermediate cells) were prepared as described in ref. 11. To obtain bone-marrow-derived macrophages (BMMs), bone marrow cells were cultured for 6 days with 20 ng/ml M-CSF (Genzyme). The full-length or mutant IRF-4-transfected RAW 264.7 cells (5 × 107 cells per ml) were prepared by electroporation of expression vectors (3 μg) by using a Nucleofector device (Amaxa, Gaithersburg, MD). Cells were seeded into 96-well plates at 2 × 105 cells per ml and stimulated with 0.3 μM ODN1668, 3 μM ODN-D19, 1 μg/ml LPS, and 5 μg/ml poly(U) complexed with DOTAP or 10 μg/ml peptideglycan for the indicated periods. Where indicated, 30 units/ml IFN-γ (Genzyme) was added to the culture medium. The concentrations of IL-12 p40, IL-6, and TNF-α in culture supernatants were determined by using ELISA kits (R & D Systems). The ELISA kit for mouse IFN-α was purchased from PBL Biomedical Laboratories (Piscataway, NJ). For RNA analysis, total RNA was extracted by using Sepasol-RNA I Super (Nacalai Tesque, Kyoto), and quantitative real-time RT-PCR analysis was performed by using a LightCycler and SYBR Green system (Roche Diagnostics). Data were normalized by the level of β-actin expression in each sample. The primers for IL-12p40, IL-6, TNF-α, and β-actin were described in ref. 10. The following primers for IRF-4, IκBα, FLICE-inhibitory protein (FLIP), vascular cell adhesion molecule-1 (VCAM-1), and macrophage inflammatory protein (MIP)2α were used: IRF-4, 5′-GCCCAACAAGCTAGAAAG-3′ and 5′-TCTCTGAGGGTCTGGAAACT-3′; IκBα, 5′-TTGGTGACT T TGGGTGCT-3′ and 5′-TGACATCAGCCCCACATTT-3′; FLIP, 5′-ACAGAGTGAGGCGGTTTGAC-3′ and 5′-GCTCTCACCAATCTCCATCAG-3′; VCAM-1, 5′-CCGTCATTGAGGATATTGG-3′ and 5′-TCATTGTCACAGCACCAC-3′; MIP2α, 5′-GTGCCTCGCTGTCTGAGAGTT-3′ and 5′-AGTTCCCAACTCACCCTCTCC-3′.
Results
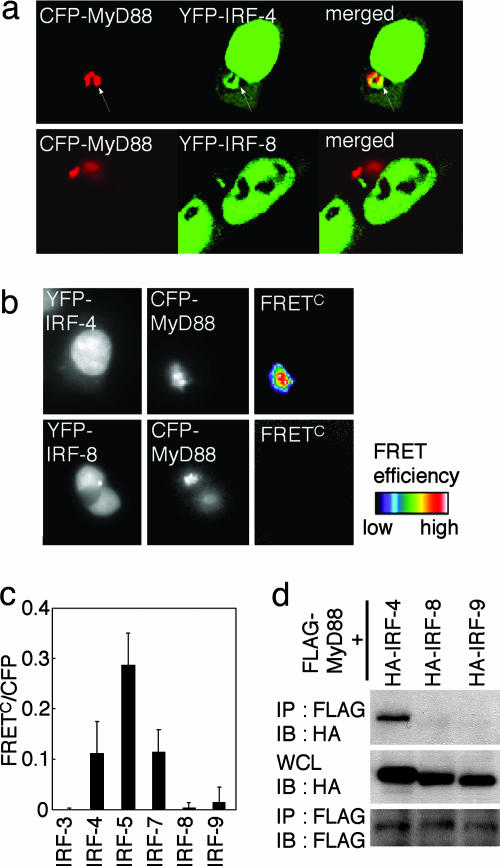
Interaction of IRF-4 with MyD88. We previously reported that two IRF members, IRF-5 and IRF-7, interact with MyD88 and that these IRFs are critical in the TLR-dependent induction of proinflammatory cytokines and type I IFNs, respectively (8–11). These findings raised an issue of whether other IRF members also participate in TLR–MyD88 signaling. We examined the interaction of IRF-4 and IRF-8, both of which are predominantly expressed in cells of hematopoietic origin (7, 17–19), with MyD88. We first independently expressed IRF-4 and IRF-8, each tagged with YFP (referred to as YFP-IRF-4 and YFP-IRF-8), together with MyD88 tagged with CFP (CFP-MyD88) in HEK293T cells and subjected the cells to fluorescence microscopic analysis. As shown in Fig. 1a, YFP-IRF-4 was predominantly expressed in the nucleus, but a significant fraction was also expressed in the cytoplasm and colocalized with CFP-MyD88. In contrast, YFP-IRF-8 failed to colocalize with CFP-MyD88. Furthermore, FRETc images revealed a strong energy transfer from CFP-MyD88 to YFP-IRF-4 but not to YFP-IRF-8, indicating that MyD88 and IRF-4 are in direct contact with each other (Fig. 1 b and c). Consistently, HA-tagged IRF-4 (HA-IRF-4), but not HA-IRF-8, coimmunoprecipitated with FLAG-tagged MyD88 (FLAG-MyD88), when coexpressed in HEK293T cells (Fig. 1d). Another IRF member, IRF-9, which participates in IFN signaling (7), showed no sign of interactions in these assays (Fig. 1 c and d).
Fig. 1.
Interaction of IRF-4 with MyD88. (a) Confocal images of HEK293T cells transiently expressing YFP-IRF-4 or YFP-IRF-8 with CFP-MyD88. Arrows indicate the colocalization of IRF-4 with MyD88. (b and c) The analysis of inter-molecular FRET between CFP-MyD88 and YFP-IRFs was performed by using HEK293T cells. FRETc was calculated and demonstrated by using pseudocolor images (b) or FRETc/CFP values (c). (d) Cell lysates prepared from HEK293T cells transiently transfected with a combination of FLAG-MyD88 and HA-IRFs were immunoprecipitated (IP) with the anti-FLAG antibody and subjected to immunoblot (IB) analysis by using the anti-HA or anti-FLAG antibody, as indicated. WCL, whole-cell extracts.
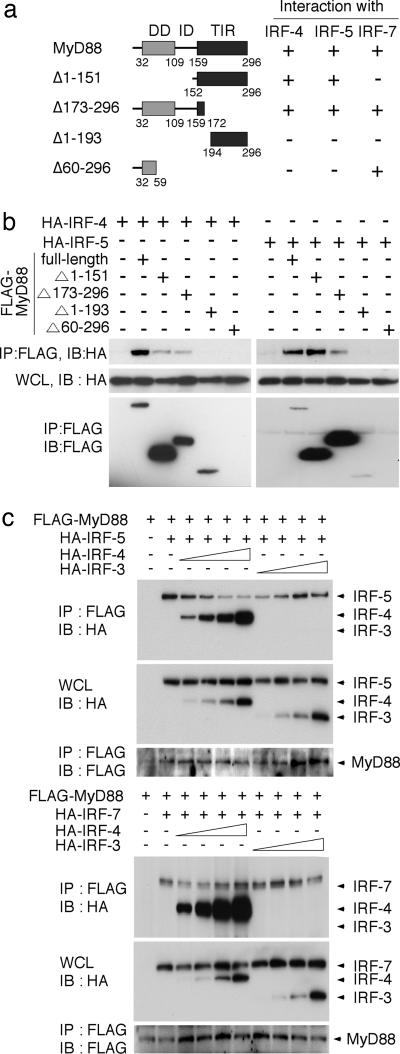
Region of MyD88 Interacting with IRF-4. We next examined the region of MyD88 that is responsible for its interaction with IRF-4 by generating deletion mutants of MyD88, each tagged with the FLAG epitope (Fig. 2a) (8). Each FLAG-MyD88 mutant was coexpressed with HA-IRF-4 in HEK293T cells and subjected to coimmunoprecipitation analysis. As shown in Fig. 2b, FLAG-MyD88(Δ1–151) and FLAG-MyD88(Δ173–296) interacted with H A-IRF-4, but FLAG-MyD88(Δ1–193) and FLAG-MyD88(Δ60–296) failed to interact. Interestingly, a similar result was obtained when we analyzed the region of FLAG-MyD88 interacting with HA-IRF-5 (Fig. 2b), suggesting that IRF-4 and IRF-5 compete with each other for interaction with the middle region of MyD88 (the intermediary domain and part of the Toll/IL-1 receptor domain).
Fig. 2.
Competition of IRF-4 with IRF-5 for MyD88 interaction. (a) Schematic diagram of MyD88 truncated mutants. +, positive interaction with IRFs; -,no interaction, assessed by immunoprecipitation assay. (b) Each FLAG-MyD88 mutant was coexpressed with HA-IRF-4 (Left) or HA-IRF-5 (Right) in HEK293T cells and subjected to coimmunoprecipitation analysis. (c) HEK293T cells were transfected with fixed amounts of the HA-IRF-5 (1.0 μg) or HA-IRF-7 (2.0 μg) expression vector and FLAG-MyD88 expression vector (1 μg) and increasing amounts of the HA-IRF-4 or HA-IRF-3 expression vector (0, 0.1, 0.2, 0.5, or 1.0 μg). Cell lysates were immunoprecipitated (IP) with the anti-FLAG antibody and subjected to immunoblot (IB) analysis by using the anti-HA or anti-FLAG antibody, as indicated.
Competition of IRF-4 with IRF-5 for MyD88 Interaction. Because a distinct region, that is, the N-terminal region, of MyD88 is responsible for its interaction with IRF-7 (8), IRF-4 may compete with IRF-5, but not with IRF-7, for MyD88 interaction. To test this possibility, we designed a competition assay in which the binding of IRF-5 or IRF-7 to MyD88 was assessed by increasing the expression level of IRF-4: HEK293T cells were transfected with fixed amounts of the HA-IRF-5 or HA-IRF-7 expression vectors, the FLAG-MyD88 expression vector, and increasing amounts of the HA-IRF-4 expression vector, and the IRF–MyD88 interaction was monitored by coimmunoprecipitation analysis. In parallel, HA-IRF-3, which cannot interact with MyD88 (8) was also included as a control. As shown in Fig. 2c, IRF-4 inhibited the interaction of IRF-5 with MyD88 in a dose-dependent manner (Upper), whereas IRF-4 expression had no effect on the IRF-7 and MyD88 interaction (Lower).
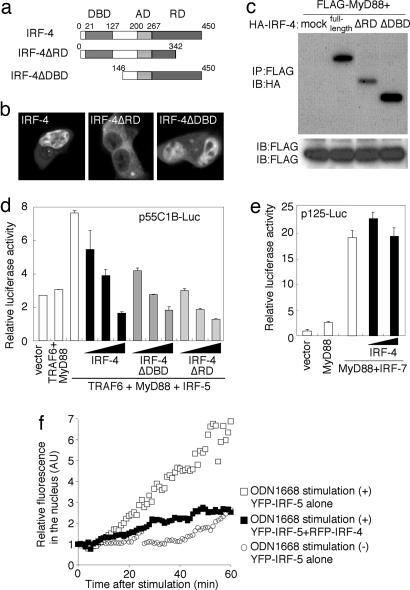
Inhibition of IRF-5-Dependent Gene Induction by IRF-4. As an approach to further examine the significance of IRF-4, which selectively competes with IRF-5 for MyD88 interaction, we next generated two mutants, one lacking the regulatory domain (IRF-4ΔRD) (17, 20) and the other lacking the DNA-binding domain (IRF-4ΔDBD), tagged with YFP (YFP-IRF-4ΔRD and YFP-IRF-4ΔDBD, respectively; Fig. 3a). In contrast to YFP-IRF-4 and YFP-IRF-4ΔDBD, both of which were predominantly expressed in the nucleus, YFP-IRF-4ΔRD was predominantly expressed in the cytoplasm (Fig. 3b). As shown in Fig. 3c, both IRF-4 mutants still interacted with MyD88.
Fig. 3.
Negative regulation of the MyD88-dependent IRF-5 activation by IRF-4. (a) Schematic diagram of IRF-4-truncated mutants. DBD, DNA-binding domain; AD, activation domain; RD, regulatory domain (17, 20). (b) Confocal images of HEK293T cells transiently expressing YFP-IRF-4, YFP-IRF-4ΔDBD, or YFP-IRF-4ΔRD. (c) HEK293T cells were transfected transiently with the indicated combination of FLAG-tagged MyD88 and HA-tagged full-length IRF-4 or deletion mutants of IRF-4 and subjected to an immunoprecipitation assay. (d) The effect of IRF-4 expression on MyD88-TRAF6-dependent IRF-5 activation. HEK293T cells were transiently cotransfected with p55C1B-Luc and the expression vectors for the indicated combinations of MyD88 (25 ng), TRAF6 (25 ng), IRF-5 (25 ng), and full-length IRF-4 or mutants of IRF-4 (0, 1, 5, or 15 ng). Luciferase activity was measured 24 h after transfection. (e) The effect of IRF-4 expression on MyD88-dependent IRF-7 activation. HEK293T cells were cotransfected transiently with p125-Luc and the expression vectors for the indicated combinations of MyD88 (25 ng), IRF-7 (25 ng), and full-length IRF-4 (0, 5, or 15 ng). Luciferase activity was measured 24 h after transfection. (f) The effect of IRF-4 on the nuclear translocation of IRF-5 induced by ODN1668 stimulation. RAW264.7 cells expressing YFP-IRF-5 alone or both YFP-IRF-5 and RFP-IRF-4 were placed on a time-lapse microscope, and images were obtained at 1-min intervals. Cells were stimulated or left unstimulated with 1 μM ODN1668 and incubated for up to 60 min in the presence of 10 ng/ml leptomycin B. The normalized YFP intensities of nuclear regions were plotted with time.
We had shown that IRF-5 is activated by MyD88 and TRAF6 (10): The coexpression of IRF-5 with MyD88 and TRAF6 results in the activation of the p55C1B-Luc reporter gene that contains multiple IFN-stimulated response elements (ISRE) (15). The above results prompted us to examine whether IRF-4 and its mutants interfere with IRF-5-mediated ISRE activation. As shown in Fig. 3d, the activation of the reporter gene by the coexpression of IRF-5, MyD88, and TRAF6 was suppressed by the expression of full-length IRF-4 in a dose-dependent manner. Furthermore, a similar suppressive effect was still observed with the HA-IRF-4ΔRD and HA-IRF-4ΔDBD mutants (Fig. 3d), supporting the notion that IRF-4 exerts its suppressive effect on IRF-5 in the cytoplasm. As one may expect from the observation that IRF-4 fails to compete with IRF-7 for MyD88 (Fig. 2c), IRF-4 expression had no effect on the MyD88-IRF-7-dependent activation of the IFN-β promoter (p125-Luc) (Fig. 3e).
We also examined the effect of IRF-4 expression on the nuclear translocation of YFP-tagged IRF-5 in RAW264.7 cells in response to TLR9 activation by the CpG-ODN termed ODN1668 (13, 21). As shown in Fig. 3f, a significant fraction of YFP-IRF-5 became detectable in the nucleus upon ODN1668 stimulation, and this nuclear translocation was suppressed when IRF-4 was coexpressed.
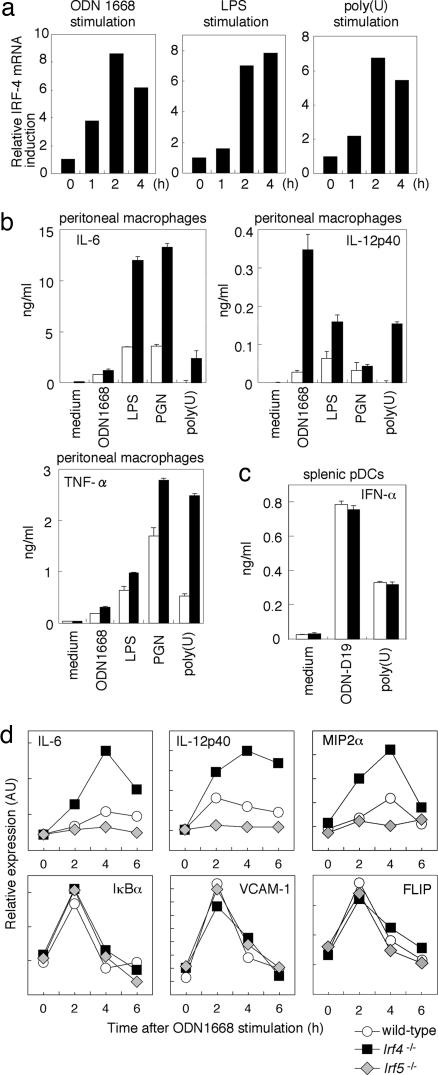
Hyperresponsiveness to TLR Stimuli in IRF4-Deficient Macrophages. IRF-4 was originally identified as a T cell- and B cell-specific member of the IRF family that functions in the regulation of lymphocyte activation and differentiation (12, 17, 18), but the above results, in toto, indicate a heretofore unknown function of IRF-4 in TLR signaling. We examined the induction of IRF-4 mRNA in peritoneal macrophages from wild-type mice stimulated with various TLR ligands, namely, ODN1668 (for TLR9), LPS (for TLR4), and a synthetic single-stranded RNA [Poly(U)] (for TLR7), which all activate the IRF-5 pathway for proinflammatory cytokine production (10). As shown in Fig. 4a, IRF-4 mRNA was induced by these stimuli, suggesting that IRF-4-expression levels are increased after TLR stimulation, wherein IRF-4 may act as a negative-feedback regulator. Interestingly, Irf4-/- peritoneal macrophages showed 2- to 3-fold-higher levels of induction of IL-6, IL-12 p40, and TNF-α when stimulated with various TLR stimuli than did the wild-type macrophages (Fig. 4b). On the other hand, IFN-α induction by TLR9 activation by IFN-inducing CpG-ODN, ODN-D19 (14) in pDCs from Irf4-/- mice was normal (Fig. 4c). This result is consistent with the previous and above results indicating that the function of IRF-7, which is essential for IFN-α induction (11), is not affected by IRF-4.
Fig. 4.
Hyperresponsivenass to TLR stimuli in Irf-4-/- peritoneal macrophages. (a) Resident peritoneal macrophages from wild-type mice were stimulated with ODN1668, LPS, or poly(U) for indicated periods. Total RNA was prepared and analyzed for IRF-4 mRNA expression by quantitative real-time RT-PCR. (b) Resident peritoneal macrophages derived from wild-type or Irf4-/- mice were stimulated with the indicated TLR ligands in the presence of IFN-γ for 24 h. The concentrations of IL-12p40, IL-6, and TNF-α in culture supernatants were measured by ELISA. Results shown are the means (±SD) of triplicate determinations. (c) Splenic pDCs prepared from wild-type and Irf4-/- mice were stimulated with ODN-D19 or poly(U) for 24 h. IFN-α concentration was measured by ELISA. (d) Induction of mRNAs of proinflammatory cytokines, chemokines, and NF-κB-inducible genes. Resident peritoneal macrophages from wild-type, Irf4-/-, or Irf5-/- mice were stimulated with ODN1668 for indicated periods. Total RNA was prepared and subjected to quantitative real-time RT-PCR analysis of indicated genes.
It is noteworthy that Irf4-/- macrophages showed an mRNA induction profile that was a mirror image of Irf5-/- macrophages in response to ODN1668 stimulation, as determined by quantitative RT-PCR analysis: Regarding those genes for which mRNA induction by TLR9 depended on IRF-5 (IL-12 p40, IL-6, and MIP2α), induction levels were elevated in the absence of IRF-4, whereas, for other genes for which IRF-5 was not required for mRNA induction, the mRNA induction levels (IκBα, FLIP, and VCAM-1) in the Irf4-/- macrophages remained the same as those in the wild-type macrophages (Fig. 4d). Because the latter genes depend on the NF-κB transcription factor (22), our results imply that the TLR-NF-κB pathway is not affected by IRF-4, at least under our experimental conditions.
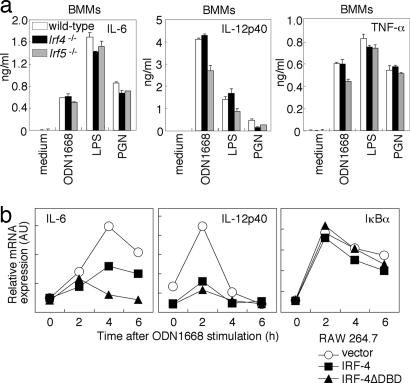
Cell-Type-Specific Action of IRF-5 and IRF-4. We also examined proinflammatory cytokine induction in M-CSF-cultured BMMs. Curiously, we observed normal proinflammatory cytokine-induction levels in BMMs lacking either IRF-5 or IRF-4 (Fig. 5a). The reason for this differential requirement of IRF-5 in TLR signaling is currently unknown, but we also obtained similar results with dendritic cells cultured in vitro by GM-CSF or Flt3 ligand (data not shown). Therefore, we postulate that the requirement of IRF-5 in TLR signaling differs, depending on the type and/or differentiation status of cells; it is possible that these cells may have undergone in vitro differentiation that may not be the same as those differentiated in vivo. Whatever the mechanism, our results suggest that IRF-4 exerts its inhibitory effect in cells in which the function of the MyD88-IRF-5 pathway is critical.
Fig. 5.
Cell-type-specific contribution of IRF-4 and IRF-5 system. (a) BMMs from wild-type, Irf5-/-,or Irf4-/- mice were stimulated with indicated stimuli in the presence of IFN-γ. The concentrations of IL-12p40, IL-6, and TNF-α in culture supernatants were measured by ELISA. (b) RAW264.7 cells were transiently transfected with the control, full-length IRF-4, or IRF-4ΔDBD expression vector by electroporation. After 12 h, cells were stimulated with ODN1668 for indicated periods. Total RNA was prepared and subjected to quantitative real-time RT-PCR analysis.
As an approach to further assessing the negative regulation of TLR signaling by IRF-4, we transiently transfected an IRF-4 expression vector into RAW264.7 cells, in which, otherwise, IRF-4 is expressed at a low level (data not shown) and analyzed cytokine mRNA induction upon ODN1668 stimulation. As shown in Fig. 5b, the induction of IL-6 and IL-12 p40 mRNAs was markedly suppressed in IRF-4-expressing cells. It is noteworthy that the induction of these cytokine genes in RAW264.7 cells was also suppressed by the expression of IRF-4ΔDBD, an observation consistent with the above reporter assay (Fig. 3d). On the other hand, the induction of IκBα mRNA, which is independent of IRF-5 but depends on NF-κB (Fig. 4d), occurred normally in both IRF-4- and IRF-4ΔDBD-expressing cells (Fig. 5b). These results, in toto, support the view that IRF-4 functions as a negative regulator by selectively competing with IRF-5.
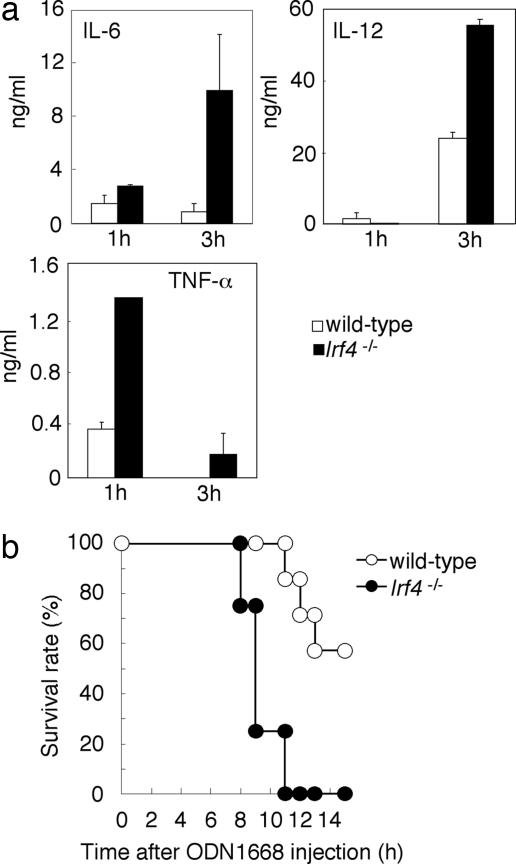
In Vivo Role of IRF-4 in TLR Signaling. To assess the function of IRF-4 in vivo, we intravenously injected wild-type and Irf4-/- mice with the ODN1668. As shown in Fig. 6a, Irf4-/- mice showed a more potent inflammatory response than did the wild-type mice, as evidenced by a considerably increased serum cytokine level after a lethal ODN1668 challenge (21). Indeed, Irf4-/- mice succumbed more rapidly (between 8 and 10 h after injection) than did the wild-type control mice (Fig. 5b). The induction levels of IL-12 p40 and IL-6 mRNAs are consistently elevated in both spleen and liver cells from Irf4-/- mice (data not shown). These results underscore the role of IRF-4 in vivo as a critical, negative regulator of TLR signaling.
Fig. 6.
Role of IRF-4 in vivo. Age-matched wild-type (n = 7) and Irf4-/- (n = 4) mice were i.p. injected with ODN1668 (10 nmol) and d-GalN (20 mg). (a) The concentrations of serum IL-6, IL-12p40, and TNF-α were measured by ELISA. Results shown are the means (±SD) of serum samples. (b) The survival of these mice was monitored for 15 h.
Discussion
The appropriate regulation of activating signaling cascades constitutes a crucial aspect in the immune system, wherein the selective use of negative regulators of TLR signaling might contribute to determining the specificity, magnitude, and longevity of inflammatory responses (23). There are several negative regulators of TLR signaling, most of which seem to target the MyD88-dependent signaling cascade, including MyD88s (the short form of MyD88), IRAK-M, SOCS1, and Toll-interacting protein (Tollip), which might participate in the complex, depending on the cell type and the nature of the stimuli (23). In our present study, we adduced evidence that IRF-4 is a negative regulator of MyD88 signaling. Indeed, MyD88 has been shown to form a multimolecular complex consisting of IRAK1, IRAK4, TRAF6, IRF-5, and IRF-7 for the induction of a variety of genes, and we previously referred to this complex as the cytoplasmic transductional–transcriptional processor (CTTP); in an analogy to computing terminology, in which the processor is central to converting the input to generate the output, CTTP may dynamically determine the output of transcriptional events (8, 10). IRF-4 may be another member of CTTP, selectively controlling the gene subsets among MyD88-dependent genes in a cell-type-specific manner.
Because IRF-4 mRNA is induced by various TLR ligands, we infer the following scenario. IRF-4 expression is induced upon TLR signaling, and the induced IRF-4 binds to MyD88 in a region overlapping with that of IRF-5 and inhibits the further binding of IRF-5 to MyD88, thereby attenuating the MyD88-dependent activation of IRF-5. Such attenuation will not occur for IRF-7, because IRF-4 does not compete with IRF-7, leaving IFN-α induction in pDCs unaffected. It has been reported that IRF-4 could function as a repressor by binding to an IFN-stimulated response element (ISRE) of some genes, such as those encoding MHC class I, IFN-stimulated gene 15 (ISG15) and TRAIL (17, 24, 25). Although further work will be required to rigorously assess the detailed mechanism underlying IRF-4-mediated gene suppression, the inhibitory effect of IRF-4 on MyD88 signaling was also observed with an IRF-4 mutant lacking the DNA-binding domain or a mutant whose subcellular localization is shifted to the cytoplasm. Thus, we postulate a unique facet of IRF-4 function, in that IRF-4 exerts its inhibitory effect on TLR signaling, not by binding to the ISRE in the nucleus but by binding to MyD88 in the cytoplasm.
The gene induction profile of Irf4-/- peritoneal macrophages showed a mirror image of Irf5-/- cells in response to CpG-ODN stimulation, that is, aberrantly high induction levels of mRNAs for IL-12, IL-6, and MIP2α and normal induction levels of mRNAs for IκBα, FLIP, and VCAM-1, which all depend on NF-κB (22), strongly suggesting that IRF-4 selectively regulates the IRF-5-dependent genes. Notably, in contrast to the case of peritoneal macrophages, both IRF-4 and IRF-5 contribute little in M-CSF-cultured BMMs. Although the reason for this cell-type-specific contribution of IRF-4 and IRF-5 is currently unclear, it has been known that many genes are differentially controlled by transcription factors, depending on the type and/or differentiation status of cells (26). In this regard, it is interesting that IRF-4 may affect the NF-κB pathway during TLR4 signaling, as evidenced by an increased DNA-binding activity of NF-κB upon LPS stimulation in peritoneal macrophages elicited by 2,6,10,14-tetramethyl pentadecane (pristane) (27). It is possible that the difference between our results and these results may come from the usage of different macrophage populations, and/or IRF-4 may exert its suppressive effect on TLR signaling by dual mechanisms.
The negative regulators of TLR-dependent signaling can be categorized into distinct subgroups according to the level of control, inhibiting either the entire TLR signaling pathway or a selective pathway (23). Because IRF-4 selectively inhibits IRF-5-dependent proinflammatory genes activated by TLRs, this study may contribute to the development of a proper therapeutic strategy for inflammatory diseases.
Acknowledgments
We thank T. Koga and S. Ueno for valuable discussion and advice and A. Miyawaki (Brain Science Institute, RIKEN, Saitama, Japan) for Venus (referred to as YFP). This work was supported by a special grant for Advanced Research on Cancer, a Grant-In-Aid for Scientific Research on a Priority Area from the Ministry of Education, Culture, Sports, Science and Technology of Japan, and the Uehara Memorial Foundation.
Author contributions: H.N., Y.O., T.T., and K. Honda designed research; H.N., Y.O., H.Y., A.T., and K. Honda performed research; K. Honma, K.Y., and T.M. contributed new reagents/analytic tools; H.N., Y.O., and K. Honda analyzed data; and H.N., T.T., and K. Honda wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: BMMs, bone-marrow-derived macrophages; CFP, cyan fluorescent protein; CpG-ODN, CpG-oligodeoxynucleotide; FRET, fluorescence resonance energy transfer; FRETc, corrected FRET; HA, hemagglutinin; HEK, human embryonic kidney; IRAK, IL-1-receptor-associated kinase; IRF, IFN regulatory factor; pDCs, plasmacytoid dendritic cells; poly(U), polyuridylic acid; TLR, Toll-like receptor; TRAF, TNF-receptor-associated factor; YFP, yellow fluorescent protein.
References
- 1.Janeway, C. A., Jr., & Medzhitov, R. (2002) Annu. Rev. Immunol. 20, 197-216. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S. & Takeda, K. (2004) Nat. Rev. Immunol. 4, 499-511. [DOI] [PubMed] [Google Scholar]
- 3.Medzhitov, R., Preston-Hurlburt, P., Kopp, E., Stadlen, A., Chen, C., Ghosh, S. & Janeway, C. A., Jr. (1998) Mol. Cell 2, 253-258. [DOI] [PubMed] [Google Scholar]
- 4.Wesche, H., Henzel, W. J., Shillinglaw, W., Li, S. & Cao, Z. (1997) Immunity 7, 837-847. [DOI] [PubMed] [Google Scholar]
- 5.Suzuki, N., Suzuki, S. & Yeh, W. C. (2002) Trends Immunol. 23, 503-506. [DOI] [PubMed] [Google Scholar]
- 6.Janssens, S. & Beyaert, R. (2003) Mol. Cell 11, 293-302. [DOI] [PubMed] [Google Scholar]
- 7.Taniguchi, T., Ogasawara, K., Takaoka, A. & Tanaka, N. (2001) Annu. Rev. Immunol. 19, 623-655. [DOI] [PubMed] [Google Scholar]
- 8.Honda, K., Yanai, H., Mizutani, T., Negishi, H., Shimada, N., Suzuki, N., Ohba, Y., Takaoka, A., Yeh, W. C. & Taniguchi, T. (2004) Proc. Natl. Acad. Sci. USA 101, 15416-15421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawai, T., Sato, S., Ishii, K. J., Coban, C., Hemmi, H., Yamamoto, M., Terai, K., Matsuda, M., Inoue, J., Uematsu, S., et al. (2004) Nat. Immunol. 5, 1061-1068. [DOI] [PubMed] [Google Scholar]
- 10.Takaoka, A., Yanai, H., Kondo, S., Duncan, G., Negishi, H., Mizutani, T., Kano, S., Honda, K., Ohba, Y., Mak, T. W. & Taniguchi, T. (2005) Nature 434, 243-249. [DOI] [PubMed] [Google Scholar]
- 11.Honda, K., Yanai, H., Negishi, H., Asagiri, M., Sato, M., Mizutani, T., Shimada, N., Ohba, Y., Takaoka, A., Yoshida, N. & Taniguchi, T. (2005) Nature 434, 772-777. [DOI] [PubMed] [Google Scholar]
- 12.Mittrucker, H. W., Matsuyama, T., Grossman, A., Kundig, T. M., Potter, J., Shahinian, A., Wakeham, A., Patterson, B., Ohashi, P. S. & Mak, T. W. (1997) Science 275, 540-543. [DOI] [PubMed] [Google Scholar]
- 13.Krieg, A. M., Yi, A. K., Matson, S., Waldschmidt, T. J., Bishop, G. A., Teasdale, R., Koretzky, G. A. & Klinman, D. M. (1995) Nature 374, 546-549. [DOI] [PubMed] [Google Scholar]
- 14.Verthelyi, D., Ishii, K. J., Gursel, M., Takeshita, F. & Klinman, D. M. (2001) J. Immunol. 166, 2372-2377. [DOI] [PubMed] [Google Scholar]
- 15.Yoneyama, M., Suhara, W., Fukuhara, Y., Fukuda, M., Nishida, E. & Fujita, T. (1998) EMBO J. 17, 1087-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sato, M., Suemori, H., Hata, N., Asagiri, M., Ogasawara, K., Nakao, K., Nakaya, T., Katsuki, M., Noguchi, S., Tanaka, N. & Taniguchi, T. (2000) Immunity 13, 539-548. [DOI] [PubMed] [Google Scholar]
- 17.Yamagata, T., Nishida, J., Tanaka, S., Sakai, R., Mitani, K., Yoshida, M., Taniguchi, T., Yazaki, Y. & Hirai, H. (1996) Mol. Cell. Biol. 16, 1283-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lohoff, M. & Mak, T. W. (2005) Nat. Rev. Immunol. 5, 125-135. [DOI] [PubMed] [Google Scholar]
- 19.Tamura, T. & Ozato, K. (2002) J. Interferon Cytokine Res. 22, 145-152. [DOI] [PubMed] [Google Scholar]
- 20.Marecki, S. & Fenton, M. J. (2002) J. Interferon Cytokine Res. 22, 121-133. [DOI] [PubMed] [Google Scholar]
- 21.Hemmi, H., Takeuchi, O., Kawai, T., Kaisho, T., Sato, S., Sanjo, H., Matsumoto, M., Hoshino, K., Wagner, H., Takeda, K. & Akira, S. (2000) Nature 408, 740-745. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh, S., May, M. J. & Kopp, E. B. (1998) Annu. Rev. Immunol. 16, 225-260. [DOI] [PubMed] [Google Scholar]
- 23.Liew, F. Y., Xu, D., Brint, E. K. & O'Neill, L. A. (2005) Nat. Rev. Immunol. 5, 446-458. [DOI] [PubMed] [Google Scholar]
- 24.Rosenbauer, F., Waring, J. F., Foerster, J., Wietstruk, M., Philipp, D. & Horak, I. (1999) Blood 94, 4274-4281. [PubMed] [Google Scholar]
- 25.Yoshida, K., Yamamoto, K., Kohno, T., Hironaka, N., Yasui, K., Kojima, C., Mukae, H., Kadota, J. I., Suzuki, S., Honma, K., et al. (2005) Int. Immunol., 10.1093/intimm/dxh324. [DOI] [PubMed]
- 26.Levine, M. & Tjian, R. (2003) Nature 424, 147-151. [DOI] [PubMed] [Google Scholar]
- 27.Honma, K., Udono, H., Kohno, T., Yamamoto, K., Ogawa, A., Takemori, T., Kumatori, A., Suzuki, S., Matsuyama, T. & Yui, K. (2005) Proc. Natl. Acad. Sci. USA 102, 16001-16006. [DOI] [PMC free article] [PubMed] [Google Scholar]