Abstract
The AML1–CBFβ transcription factor complex is the most frequent target of specific chromosome translocations in human leukemia. The MOZ gene, which encodes a histone acetyltransferase (HAT), is also involved in some leukemia-associated translocations. We report here that MOZ is part of the AML1 complex and strongly stimulates AML1-mediated transcription. The stimulation of AML1-mediated transcription is independent of the inherent HAT activity of MOZ. Rather, a potent transactivation domain within MOZ appears to be essential for stimulation of AML1-mediated transcription. MOZ, as well as CBP and MOZ–CBP, can acetylate AML1 in vitro. The amount of AML1–MOZ complex increases during the differentiation of M1 myeloid cells into monocytes/macrophages, suggesting that the AML1–MOZ complex might play a role in cell differentiation. On the other hand, the MOZ–CBP fusion protein, which is created by the t(8;16) translocation associated with acute monocytic leukemia, inhibits AML1-mediated transcription and differentiation of M1 cells. These results suggest that MOZ–CBP might induce leukemia by antagonizing the function of the AML1 complex.
Keywords: AML1/histone acetylation/leukemia/MOZ
Introduction
Specific chromosome translocations are found frequently in human leukemia and often result in the expression of fusion gene products (Rabbitts, 1994). The genes encoding the AML1–CBFβ transcription factor complex are the most frequent targets of these translocations (Look, 1997). AML1 (Runx1) forms a complex with CBFβ (PEBP2β) to regulate transcription of certain hematopoietic cell-specific genes by binding to the specific DNA sequence TGT/cGGT (Ogawa et al., 1993; Wang et al., 1993). Both AML1 and CBFβ are essential for all lineages of definitive hematopoiesis in mouse fetal liver (Okuda et al., 1996; Sasaki et al., 1996; Wang et al., 1996a,b; Niki et al., 1997). AML1 functions cooperatively with certain co-activators, including p300/CBP histone acetyltransferases (HATs), to stimulate gene transcription and cell differentiation (Kitabayashi et al., 1998b). AML1 also represses transcription through interaction with the Groucho and mSin3A co-repressors (Imai et al., 1998; Levanon et al., 1998; Lutterbach et al., 2000). The leukemia-associated fusion AML1–MTG8(ETO), a product of t(8;21), forms complexes with histone deacetylases (HDACs) through an interaction with N-CoR/SMRT and/or MTGR1/MTG16 (Gelmetti et al., 1988; Kitabayashi et al. 1998a; Lutterbach et al., 1998; Wang et al., 1998; our unpublished data). Leukemia-associated translocations also involve genes that encode HAT proteins such as CBP, p300, TIF2 and monocytic leukemia zinc finger protein (MOZ) (Borrow et al., 1996; Ida et al., 1997; Satake et al., 1997; Sobulo et al., 1997; Taki et al., 1997; Carapeti et al., 1998; Liang et al., 1998; Chaffanet et al., 2000; Kitabayashi et al., 2001; Panagopoulos et al., 2001). The t(8;16) translocation results in the expression of the MOZ–CBP fusion protein (Borrow et al., 1996).
Histone and non-histone proteins associate with DNA to form nucleosomes and higher order structures in eukaryotes. The N-termini of histones can be modified by acetylation. Acetylation of specific lysine residues of histones is associated with gene activity and may play a fundamental role in transcriptional regulation. The level and state of histone acetylation are regulated by HATs and HDACs. Some HATs and HDACs interact with specific DNA-binding transcription activators and repressors, which strongly suggests that they modulate the transcriptional activity of specific promoters by regulating local histone acetylation (Grunstrein, 1997; Pazin and Kadonaga, 1997; Wade et al., 1997; Struhl, 1998). It was reported that bromodomains, which are found in several eukaryotic transcription factors, specifically interact with acetyl-lysines in histones H3 and H4 (Dhalluin et al., 1999). This finding suggests that bromodomain-containing proteins may be involved in the regulatory effects of histone acetylation through their binding to acetyl-lysines.
MOZ shows HAT activity (Champagne et al., 2001; see Figure 5), which suggests that it might be involved in the regulation of transcription and chromatin function. However, how MOZ acts remains unclear. Here we present evidence to show that MOZ interacts with the AML1 transcription factor and functions as a potent co-activator of AML1. In contrast, the leukemia-associated MOZ–CBP fusion inhibits AML1-mediated transcription. Our results suggest that MOZ–CBP targets the AML1 complex, which contains both MOZ and p300/CBP, to induce leukemia.
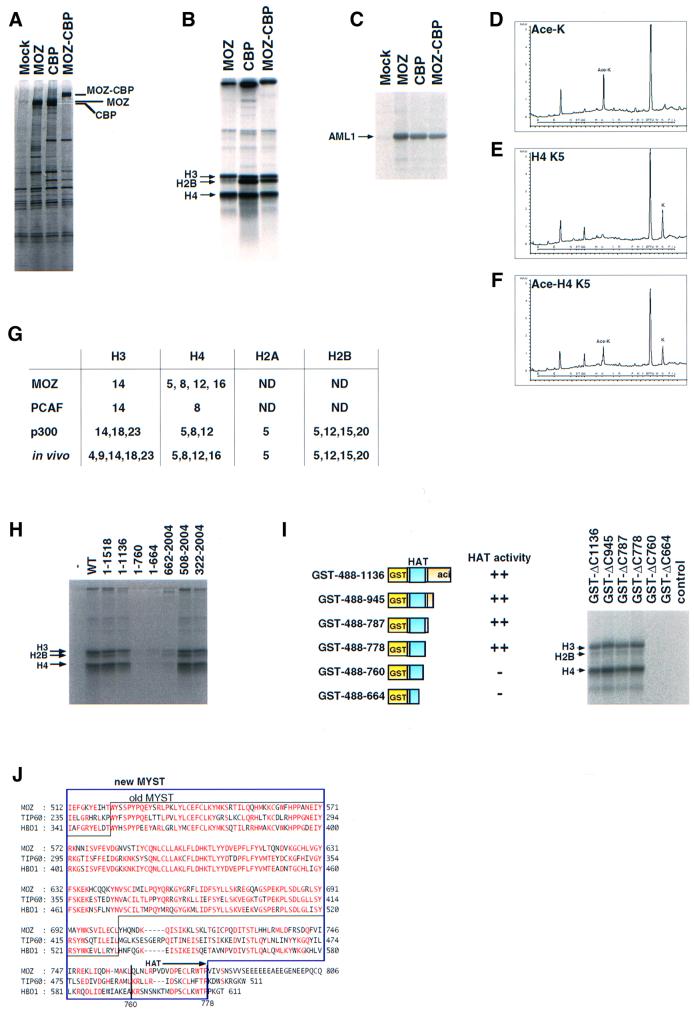
Fig. 5. MOZ acetylates histones H3 and H4. (A) Purification of MOZ, CBP and MOZ–CBP. Bosc23 cells were transfected with 10 µg of either the pLNCX vector (–), pLNCX-FLAG-MOZ (MOZ), pLNCX-FLAG-CBP (CBP) or pLNCX-FLAG-MOZ–CBP (MOZ–CBP) in a 10 cm dish. Proteins were immunoprecipitated using anti-FLAG antibody (M2)-conjugated beads and eluted by incubation with 0.2 mg/ml FLAG peptide. The protein constituents of the eluates were separated by 5–20% SDS–PAGE and stained with silver. (B and C) Acetylation of histones and AML1 by MOZ, CBP and MOZ–CBP. The affinity-purified MOZ, CBP and MOZ–CBP proteins were assayed for acetylation activity with histones H2B, H3 and H4 (B) or FLAG-AML1 (C). (D) HPLC charts of standard acetylated lysine. (E and F) Untreated and MOZ-acetylated histones H3 and H4 were subjected to N-terminal microsequencing. HPLC charts of fifth cycles for untreated (D) and MOZ-acetylated (E) histones H4 are shown. (G) Comparison of lysine residues acetylated in vivo, or in vitro by MOZ, PCAF or p300. The results of PCAF- and p300-acetylating residues were adopted from Schiltz et al. (1999). (H) HAT activity of MOZ mutants. Bosc23 cells were transfected with deletion mutants of MOZ as described above. The cell lysates were immunoprecipitated with anti-FLAG antibody and eluted by incubation with 0.2 mg/ml FLAG peptide. The eluates were analyzed for HAT activity. (I) HAT activity of GST–MOZ. GST–MOZ fusion proteins were produced in Escherichia coli, purified by using glutathione–Sepharose beads and analyzed for HAT activity. (J) Comparison of amino acid sequences for the HAT domains of MOZ, TIP60 and HBO1. Amino acids that are identical in more than two proteins are indicated in red.
Results
AML1 forms complexes with MOZ
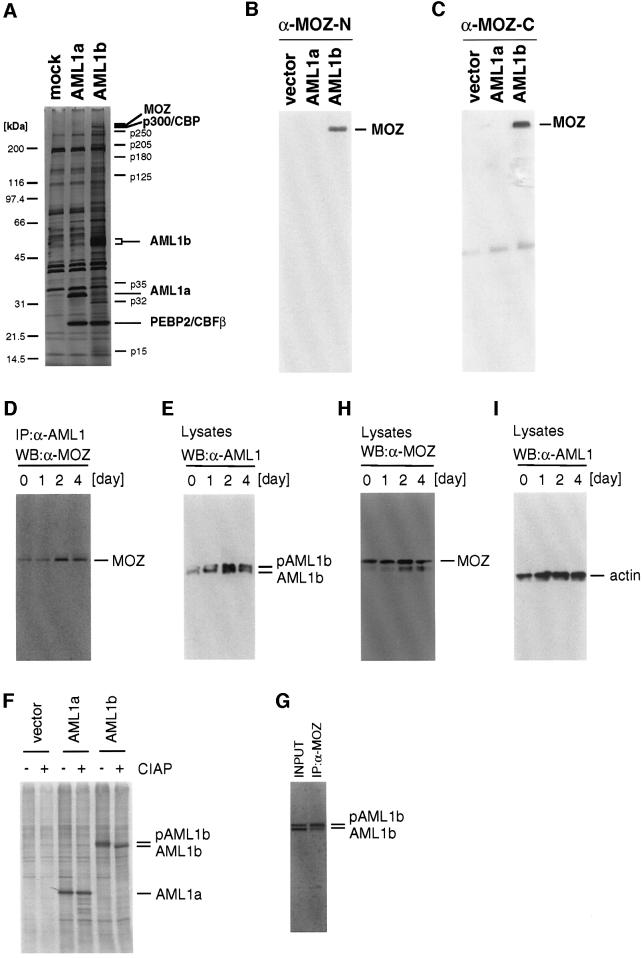
To identify AML1-interacting proteins, we purified AML1 complexes. Flag-tagged full-length AML1 (AML1b) and a splicing variant lacking a C-terminal transcriptional activation domain (AML1a) were stably expressed in mouse myeloid L-G cells using a retrovirus vector as described previously (Kitabayashi et al., 1998b). The complexes of AML1a and AML1b were partially purified using affinity beads conjugated with an anti-Flag antibody M2 (Figure 1A). The heterodimeric partner CBFβ protein, which binds to the N-terminal runt domain of AML1, was co-purified abundantly with both AML1a and AML1b. The AML1b fraction contained multiple proteins that were absent from both the AML1a fraction and the mock-purified fraction, suggesting that they interact specifically with AML1b. To identify these proteins in the AML1b fraction, specific proteins were analyzed by mass spectrometry. The spectrum obtained with peptides from the ∼270 kDa band identified MOZ. Immunoblot analysis using anti-MOZ antibodies showed that MOZ was co-purified readily with AML1b (Figure 1B and C). MOZ was present neither in the AML1a fraction nor in the mock-purified fraction, which suggests that MOZ specifically interacts with the C-terminal region of AML1b.
Fig. 1. MOZ forms a complex with AML1b. (A) FLAG-tagged AML1a and AML1b proteins were partially purified from lysates of infectants and vector control cells (mock) using anti-FLAG antibody (M2). (B and C) The fractions in (A) were analyzed by immunoblotting using polyclonal antibodies against the N- (B) and C-terminal region of MOZ (C). (D, E, H and I) AML1–MOZ complex during the differentiation of M1 cells. M1 cells were exposed to IL-6 for the indicated number of days. The cell lysates were prepared and immunoprecipitated using anti-AML1 antibody. The immunoprecipitates (D) or the cell lysates (E, H and I) were analyzed by immunoblotting with anti-MOZ-N (D and E), anti-AML1 antibodies (F) and anti-actin antibodies (I). (F) The M1 infectants that expressed Flag-tagged AML1a or AML1b were exposed to IL-6 for 4 days and labeled with [35S]methionine. The cell lysates were immunoprecipitated with anti-FLAG antibody. The immunoprecipitates were then incubated with (+) or without (–) calf intestine alkaline phosphatase (CIAP) and analyzed by 10% SDS–PAGE followed by autoradiography. (G) The M1 infectants that expressed Flag-tagged AML1b were exposed to IL-6 for 2 days. The cell lysates were immunoprecipitated with anti-MOZ antibody. The cell lysates (INPUT) and the immunoprecipitates (IP:α-MOZ) were analyzed by immunoblotting using horseradish peroxidase-conjugated anti-FLAG antibody.
Increase in the level of the AML1–MOZ complex during M1 cell differentiation
To monitor the level of the intrinsic complex of AML1 and MOZ, AML1 proteins were immunoprecipitated before and after differentiation of M1 cells, which can be induced to differentiate into monocytes/macrophages in response to interleukin-6 (IL-6). Western blot analysis of the immunoprecipitates indicated that the amounts of MOZ proteins in the immunoprecipitates increased during the differentiation of M1 cells in response to IL-6 (Figure 1D). Western blot analysis using anti-AML1 antibody indicated that IL-6 treatment resulted in a decrease in the electrophoretic mobility of AML1 (Figure 1E). In undifferentiated M1 cells, a faster migrating species predominated, but this species was replaced by a more slowly migrating species after 1–2 days exposure to IL-6. Phosphatase treatment of the upper, slowly migrating AML1b protein resulted in its migration to the lower position (Figure 1F). No changes in mobility of AML1a proteins were observed after treatment with phosphatase. These results suggested that the difference in mobility was due mainly to differences in the phosphorylation state of the C-terminal region. Western blot analysis of the MOZ immunocomplex indicated that, although MOZ interacted with both highly phosphorylated and underphosphorylated AML1, phosphorylated AML1 was the preferred interaction partner (Figure 1G). Total amounts of AML1 proteins also increased 1–2 days after exposure to IL-6. The levels of MOZ and control actin in cell lysates did not change strongly (Figure 1H and I). Thus, increased levels of the AML1–MOZ complex are associated with AML1 phosphorylation and/or increased amounts of AML1 proteins.
Two AML1-interacting domains on MOZ
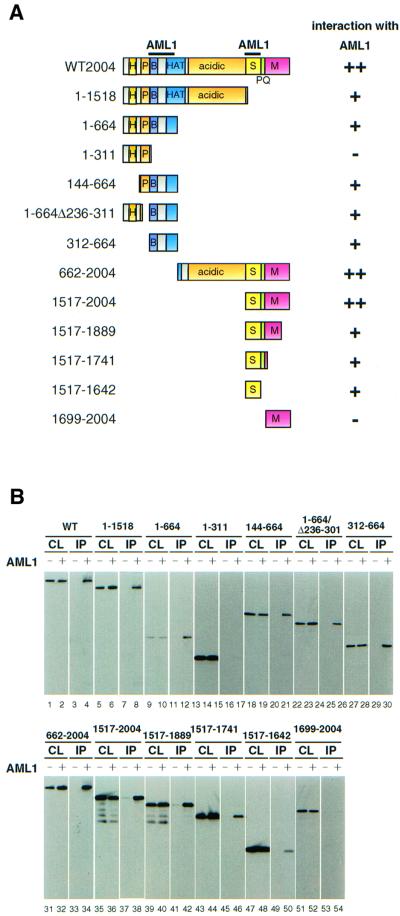
To determine the AML1-interacting domains on MOZ, the interaction between MOZ and AML1 was examined by immunoprecipitation (IP)–western analysis using deletion mutants of MOZ. Bosc23 cells, whose transfection efficiency is very high, were transfected with FLAG-tagged AML1b and hemagglutinin (HA)-tagged wild-type MOZ or HA-tagged MOZ mutants. The expression of MOZ mutants and the results of the IP–western analysis are shown in Figure 2B. The results are summarized in Figure 2A. Deletion mutants lacking the C-terminal region up to amino acids 1518 (1–1518) and 664 (1–664) as well as wild-type MOZ were co-precipitated with AML1b. However, further deletion to amino acid 311 strongly reduced AML1-binding activity, suggesting that the region between 312 and 664 is important for MOZ interaction with AML1. Indeed, a mutant containing only the region between 312 and 664 could still bind to AML1. Sur prisingly, however, the truncated derivatives 662–2004 and 1517–2004 were also able to bind to AML1, suggesting the presence of another AML1-interacting domain in the C-terminal region of MOZ. Further deletion analysis indicated that the region between 1517 and 1642 also had AML1-binding activity. These results suggest that there are at least two AML1-interacting domains in MOZ, one of which lies in the basic domain between 312 and 664 and the other of which lies in the serine-rich region between 1517 and 1642.
Fig. 2. AML1 interacts with two regions of MOZ. (A) Schematic representation of the structure of MOZ deletion mutants used for determining the AML1-interacting domains. The H15 domain (H), the PHD-type zinc finger domain (P), the basic domain (B), the histone acetyltransferase (HAT) domain, the serine-rich region (S), the proline- and glutamine-rich region (PQ), the methionine-rich region (M) and the regions that bind to AML1 (bars) are indicated. (B) Regions of MOZ required for interaction with AML1. Bosc23 cells were transfected with deletion mutants of MOZ together with (+) or without (–) pLNCX-FLAG-AML1b. The cell lysates were immunoprecipitated with anti-FLAG antibody. The immunoprecipitates (IP) and the cell lysates (CL) were separated by SDS–PAGE and subjected to immunoblot analysis using the anti-HA antibody.
The activation domain of AML1 is required for its interaction with MOZ
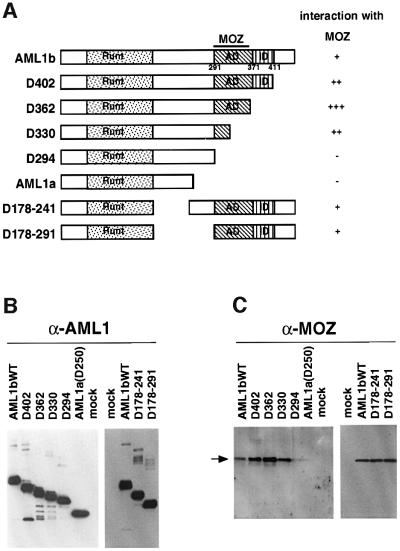
To determine the region of AML1 required for binding to MOZ, IP–western analysis was done using a series of deletion mutants of AML1 (Figure 3A). An equivalent amount of each AML1 mutant was immunoprecipitated by the anti-FLAG antibody M2 (Figure 3B). The level of MOZ increased in the immunoprecipitates when AML1 was deleted of its C-terminal region up to residue 362 (Figure 3C). However, further deletion to residue 294 resulted in a loss of MOZ from the immunoprecipitates. An internal deletion of the region between amino acids 178 and 241 (D178–241) or of the region between 178 and 291 (D178–291) did not affect the interaction. Thus the region of AML1 between positions 362 and 295 is responsible for the interaction with MOZ. This region corresponds to the domain required for transcription activation (AD) (T.Kanno et al., 1998). This suggests that MOZ may play an important role in AML1-mediated transcriptional activation.
Fig. 3. MOZ interacts with the transactivation domain of AML1. (A) Schematic representation of the structure of AML1 deletion mutants. The runt homology domain (Runt), transcription activation domain (AD) and transcription inhibition domain (ID) are indicated. (B and C) Regions of AML1 required for interaction with MOZ. Bosc23 cells were transfected with FLAG-tagged AML1 mutants. The lysates of the transfectants were immunoprecipitated with the anti-FLAG antibody M2. The immunoprecipitates were analyzed by immunoblotting using anti-AML1 (B) and anti-MOZ (C) antibodies.
MOZ activates AML1-mediated transcription
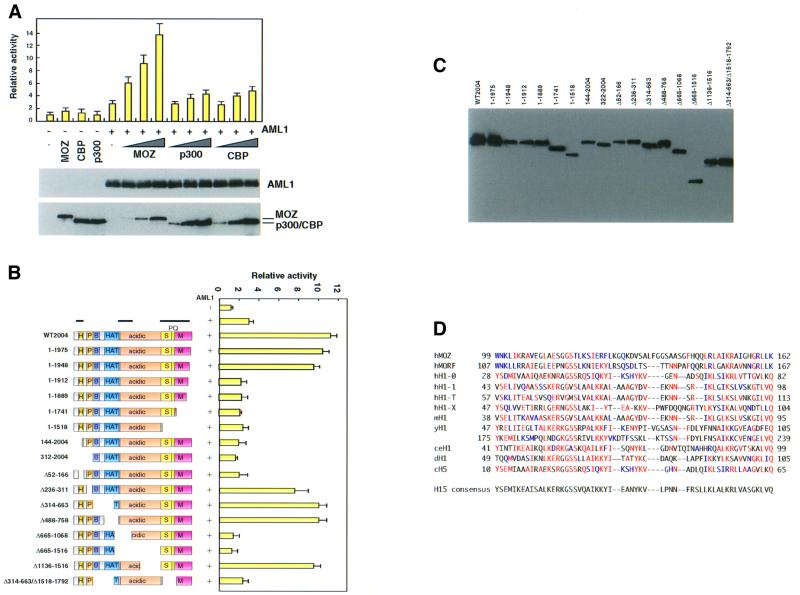
To monitor AML1-mediated transcription, we used reporter plasmids under the control of the MPO or CCP1 promoter that contained AML1-binding sequences. MOZ strongly stimulated AML1-mediated activation of both reporters in a dose-dependent manner (Figure 4A and data not shown). p300/CBP also significantly stimulated AML1-mediated transcription activation. These results suggest that MOZ functions as a strong transcription co-activator of AML1.
Fig. 4. MOZ strongly activates AML1-mediated transcription. (A) Activation of AML1 by MOZ. P19 cells were transfected with 50 ng of MPO-luc, 200 ng of pLNCX-AML1b and either 30 (lanes 6, 9 and 12), 100 (lanes 7, 10 and 13) or 350 ng (lanes 2–4, 8, 11 and 14) of pLNCX-HA-MOZ (lanes 2 and 6–8), pLNCX-HA-p300 (lanes 3 and 9–11) or pLNCX-HA-CBP (lanes 4 and 12–14) and 25 ng of pRL-tk in 24-well plates. Luciferase activity was assayed as described in Materials and methods. The lysates of the cells transfected as described above were also analyzed by immunoblotting with anti-AML1 and anti-HA antibodies (for MOZ, p300 and CBP). Essentially similar results were obtained using the CCP1-luc reporter instead of MPO-luc. Note that the expression of intrinsic AML1 could not be detected in P19. (B) Regions of MOZ required for activation of AML1. Saos-2 cells were transfected with 50 ng of MPO-luc, 200 ng of pLNCX-AML1b, 350 ng of pLNCX-HA-MOZ mutants and 25 ng of pRL-tk. The left panel shows a schematic representation of the structure of the deletion mutants. Bars above the panel indicate the region essential for stimulation of AML1-mediated transactivation. Essentially similar results were obtained using 3T3 cells. H, H15; P, PHD; B, basic; HAT, histone acetyltransferase; S, serine-rich; PQ, proline and glutamine; M, methionine-rich. (C) Expression of MOZ mutants. The Saos2 cells were transfected with HA-tagged MOZ mutants and the cell lysates were analyzed by immunoblotting with anti-HA antibody. (D) Comparison of amino acid sequences for the H15 domain of human MOZ (hMOZ), human MORF (hMORF) and linker histone H1 and H5 species. Amino acids that are identical and homologous to the H15 consensus are indicated in red and blue, respectively.
To determine the region of MOZ required for co-activation, AML1-dependent transcription was tested using deletion mutants of MOZ (Figure 4B). Progressive N- and C-terminal deletion analyses revealed that the first 144 amino acids and the 36 amino acids (1913–1948) close to the C-terminus were required for the activation of AML1-mediated transcription. Internal deletion of the region between amino acids 52 and 166 completely abolished activity. BLAST homology searches revealed that this region is homologous to a domain (H15) found in linker histones, such as histones H1 and H5 (Figure 4D). Deletion of the PHD finger domain had only a slight effect on activity. A mutant lacking the N-terminal AML1-interacting domain still activated transcription. However, deletion of both the N- and C-terminal AML1-interacting domains completely abolished activity. Interestingly and surprisingly, the co-activation function of MOZ was not affected by deletion of the evolutionarily conserved MYST sequence, a putative HAT domain, in this system. Deletion of the region containing the N-terminal part, but not the C-terminal half, of the acidic domain completely blocked activity. These results suggest that the co-activation function of MOZ requires various regions such as the H15 domain, a part of the acidic region, the AML1-interacting domains and the C-terminal region.
HAT domain of MOZ
Since the MYST domain is not required for co-activation by MOZ, we wondered if the MYST domain might be responsible for HAT activity. To test this, we examined the HAT activity of MOZ and identified the region required for HAT activity. We initially examined HAT activity using partially purified Flag-tagged derivatives of MOZ, CBP and MOZ–CBP which were obtained from the extracts of Bosc23 transfectants by pull-down with anti-Flag antibody-conjugated beads (Figure 5A). As shown in Figure 5B, MOZ shows acetyltransferase activity for histones H3 and H4. MOZ–CBP and CBP had acetylation activity for histones H2B, H3 and H4. In addition, MOZ, as well as CBP and MOZ–CBP, acetylated themselves efficiently. Interestingly, MOZ, CBP and MOZ–CBP could acetylate AML1 (Figure 5C), suggesting that these HATs may regulate AML1 function through acetylation. Analyses of the N-terminal amino acids of unmodified and MOZ-acetylated histones H3 and H4 suggested that Lys14 of histone H3 and Lys5, Lys8, Lys12 and Lys16 of histone H4 could be acetylated by MOZ in vitro (Figure 5G). Deletion analysis indicated that deletion of the C-terminus to residue 1137 did not affect HAT activity; however, a further deletion to residue 761 completely abolished HAT activity (Figure 5H). Deletion from the N-terminus to residue 507 did not reduce HAT activity. A further deletion to residue 661, which removes a large part of the MYST domain, strongly reduced HAT activity, which suggests that the MYST domain is required for internal HAT activity. However, since this mutant still showed a weak HAT activity, MOZ may interact with other HATs through its C-terminal region.
GST–MOZ fusion proteins also acetylated histones H3 and H4 (Figure 5I). Deletion analysis indicated that the region between 488 and 778, which contains the previously defined MYST domain (Borrow et al., 1996) and its C-terminally flanking region, is necessary and sufficient for internal HAT activity. The flanking region also shows significant homology with other mammalian MYST domain proteins, such as TIP60 and HBO1 (Figure 5J). MOZ recently has been shown to have HAT activity (Champagne et al., 2001).
Transactivation domain
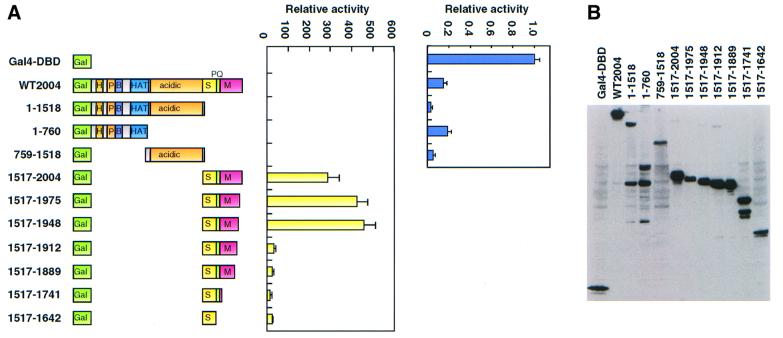
The lack of any requirement for the HAT domain of MOZ for transcriptional co-activation suggests that, besides internal HAT activity, MOZ may have another domain for transcriptional activation. To test this possibility, various regions in MOZ were fused to the Gal4 DNA-binding domain and the fusion constructs were co-transfected with a reporter plasmid carrying Gal4-binding sites. As shown in Figure 6A, the C-terminal region of MOZ showed strong transactivation activity. Deletion analysis indicated that the region between positions 1517 and 1948 was sufficient for strong transactivation. Importantly, the C-terminal boundary (1948) of this transactivation domain corresponds to the boundary of the regions required for co-activation of AML1-dependent transcription (see Figure 4B). MOZ has been reported recently to have transactivation activity (Champagne et al., 2001).
Fig. 6. Transactivation domain of MOZ. (A) Transactivation by GAL4-MOZ. Saos-2 cells were transfected with 250 ng of Gal4-luc (pFR-luc), 50 ng of GAL4-MOZ mutants and 25 ng of pRL-tk. The luciferase assay was performed as described in Materials and methods. Results are given as relative activity, based on the control activity for GAL4-DBD (arbitrarily set at 1). (B) Expression of GAL4-MOZ mutants. The Saos2 cells were transfected with GAL4-MOZ mutants and the cell lysates were analyzed by immunoblotting with anti-GAL4 antibody.
Surprisingly, the full-length MOZ fusion inhibited transactivation activity in this Gal4-fusion system (Figure 6A, right panel). This suggested that MOZ might contain domain(s) that repress transcription. Deletion of the C-terminal transactivation domain enhanced transcription repression, whereas deletion of the acidic domain alleviated transcription repression. The acidic domain by itself showed strong inhibitory activity.
The H15 domain is implicated in nuclear localization
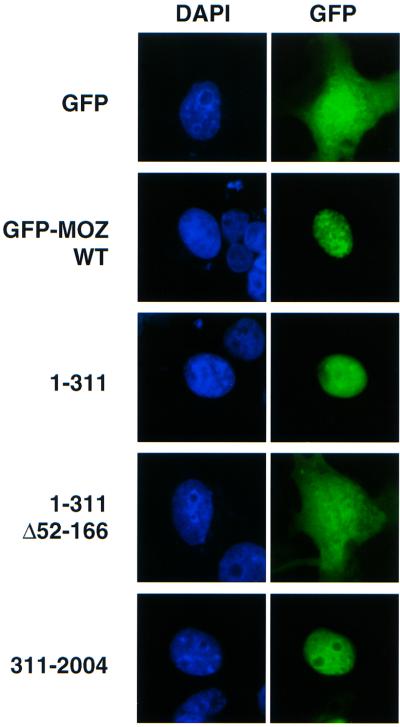
Analysis of the subcellular location of a MOZ–green fluorescent protein (GFP) fusion protein showed that MOZ is a nuclear protein that co-localizes to specific regions within the nucleus containing a lot of dot-like structure (Figure 7). Deletion of the C-terminal region of MOZ to position 311 still resulted in nuclear localization, but the staining pattern became diffuse. An additional deletion of the region containing the H15 domain resulted in the loss of nuclear localization. These results suggest that the H15 domain is important for nuclear localization. However, it is likely that MOZ has other nuclear targeting signal(s), because a mutant lacking 310 amino acids from the N-terminus was still localized to the nucleus.
Fig. 7. The H15 domain is important for nuclear localization of MOZ. COS7 cells were transfected with the wild-type and deletion mutants of GFP–MOZ fusion constructs. Left and right panels show DAPI staining and GFP fluorescence, respectively.
MOZ–CBP blocks AML1-dependent transcription
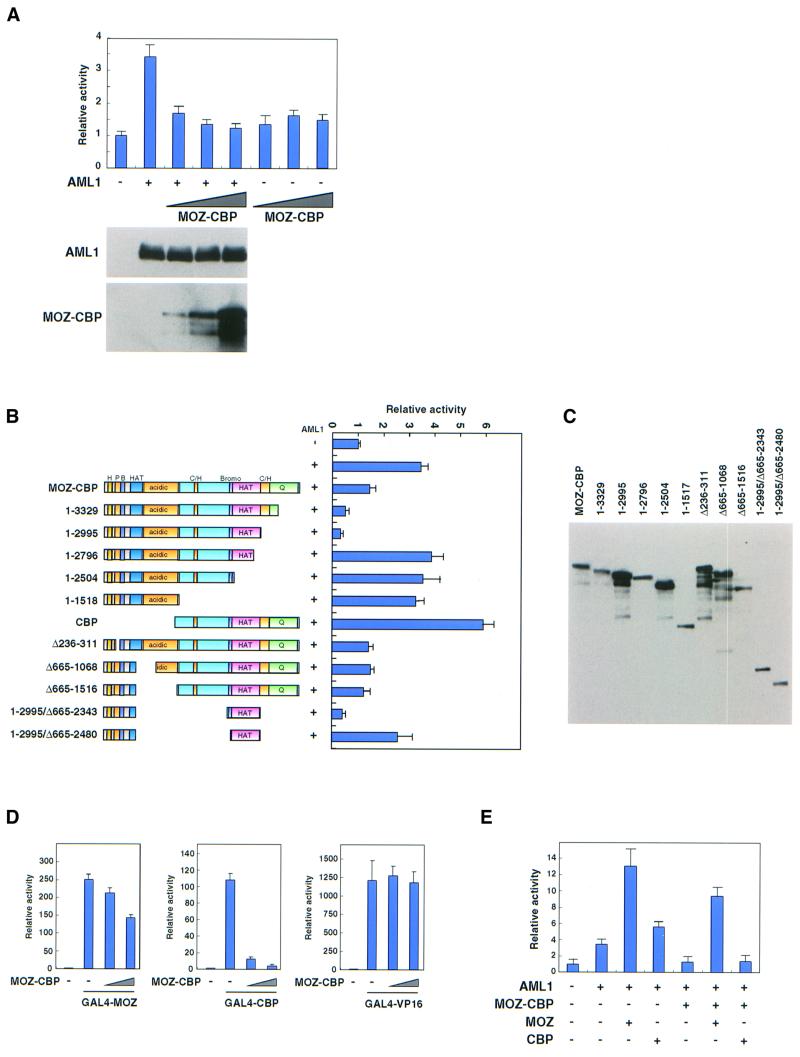
Since MOZ and CBP function as co-activators of AML1, we tested the effects of MOZ–CBP on AML1-dependent transcription. As shown in Figure 8A, MOZ–CBP strongly inhibited AML1-dependent but not AML1-independent transcription in a dose-dependent manner. Deletion from the C-terminus to amino acid 2995 enhanced inhibition (Figure 8B). However, further deletion of the HAT domain of the CBP portion abolished repression. Internal deletion analysis indicated that mutants of MOZ–CBP that lacked the PHD finger domain, the HAT domain of MOZ or the acidic domain were still inhibitory. However, a mutant lacking the bromodomain in the CBP portion showed only weak repression. These results suggest that bromo- and HAT domains in the CBP portion are important for the inhibition of AML1-mediated transcription by MOZ–CBP.
Fig. 8. MOZ–CBP inhibits AML1-mediated transcription. (A) Effects of MOZ–CBP on AML1 activity. Saos-2 cells were transfected with 50 ng of MPO-luc, 200 ng of pLNCX-AML1b (lanes 2–5), and either 30 (lanes 3 and 6), 100 (lanes 4 and 7) or 350 ng (lanes 5 and 8) of pLNCX-HA-MOZ–CBP and 25 ng of pRL. Luciferase assays were performed as described in Materials and methods. The lysates of the cells transfected as above were also analyzed by immunoblotting with anti-AML1 and anti-HA antibodies (for MOZ–CBP). (B) Effects of deletion mutants of MOZ–CBP on AML1 activity. Saos-2 cells were transfected with 50 ng of MPO-luc, 200 ng of pLNCX-AML1b, 350 ng of pLNCX-HA-MOZ–CBP mutants and 25 ng of pRL-tk. Luciferase assays were performed. (C) Expression of deletion mutants of MOZ–CBP. The Saos-2 cells were transfected with HA-tagged MOZ–CBP mutants and the cell lysates were analyzed by immunoblotting with anti-HA antibody. (D) Effects of MOZ–CBP on transactivation by using GAL4 fusions. Saos-2 cells were transfected with 250 ng of Gal4-luc (pFR-luc), 50 ng of either GAL4-MOZ (lanes 2–4), GAL4-CBP (lanes 6–8) or GAL4-VP16 (lanes 10–12), 50 (lanes 3, 7 and 11) or 200 ng (lanes 4, 8 and 12) of MOZ–CBP and 25 ng of pRL. Luciferase assays were performed. (E) Effects of MOZ–CBP on co-activation by MOZ and CBP. Saos-2 cells were transfected with 50 ng of MPO-luc, 200 ng of pLNCX-AML1b (lanes 2–7), 100 ng of pLNCX-HA-MOZ–CBP (lanes 5–7), 250 ng of either pLNCX-HA-MOZ (lanes 3 and 6) or pLNCX-HA-CBP (lanes 4 and 7) and 25 ng of pRL. A luciferase assay was performed.
To determine whether MOZ–CBP inhibits either MOZ-dependent or CBP-dependent transactivation, we tested the effect of MOZ–CBP on transactivation by Gal4-MOZ(1517–2004) and Gal4-CBP(267–2442) fusions, which strongly activated the reporter-containing GAL4-binding sites. As shown in Figure 8D, MOZ–CBP clearly inhibited transactivation by GAL4-CBP. Transactivation by GAL4-MOZ was only weakly affected and that by GAL4-VP16 was not affected. Thus, MOZ–CBP mainly inhibits CBP-mediated transactivation. Con sistent with this, MOZ but not CBP could activate AML1-dependent transcription even in the presence of MOZ–CBP (Figure 8E).
MOZ–CBP inhibits differentiation of M1 cells to macrophages
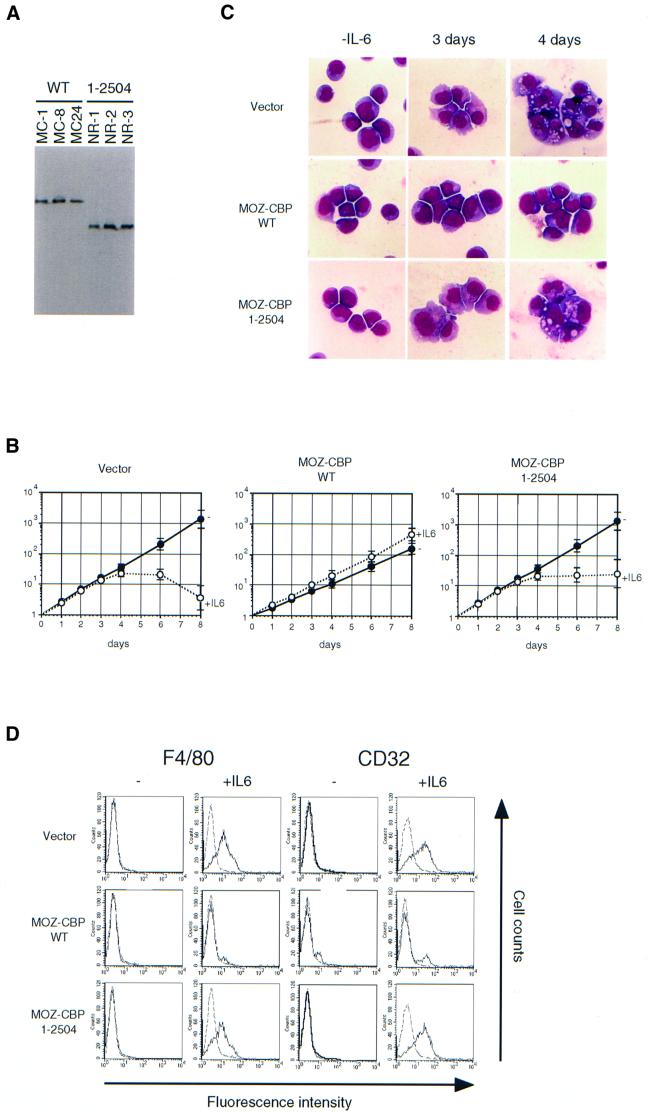
The t(8;16) translocation, which results in the expression of the MOZ–CBP fusion, is associated with acute monocytic leukemia, suggesting that MOZ–CBP may affect the growth and/or differentiation of monocytic lineages. Therefore, we examined the effects of forced expression of MOZ–CBP on murine myeloid M1 cells, which can be induced to differentiate into monocytes/macrophages in response to IL-6. M1 cells were transfected with wild-type or mutant MOZ–CBP (1–2504), or an empty vector, and the transfected cells were selected with G418. Stable clones expressing high levels of MOZ–CBP and mutant MOZ–CBP (Figure 9A) were cultured in the presence or absence of IL-6. The vector-transfected control cells stopped growing 3–4 days after exposure to IL-6 (Figure 9B) and underwent differentiation into macrophages, as shown by characteristic changes in morphology (Figure 9C). Fluorescence- activated cell sorting (FACS) analysis indicated that the cells expressed macrophage-specific markers such as F4/80 and CD32 after exposure to IL-6 (Figure 9D). The cells underwent apoptosis 6–8 days after exposure to IL-6. Compared with the control cells, wild-type MOZ–CBP-expressing cells showed growth retardation in the absence of IL-6. Even in the presence of IL-6, the MOZ–CBP-expressing cells did not stop growing and proliferated exponentially with a slightly faster growth rate without differentiating into macrophages, as shown by the absence of both morphological changes and expression of F4/80 and CD32. However, the cells that expressed the mutant MOZ–CBP protein, which lacks the HAT domain in the CBP portion, stopped growing and underwent differentiation into macrophages. Thus, expression of MOZ–CBP inhibited differentiation of M1 cells into macrophages, and the region containing the HAT domain of the CBP portion is required for this inhibition.
Fig. 9. MOZ–CBP inhibits the differentiation of M1 cells. (A) Expression of wild-type and a deletion mutant (1–2504) of MOZ–CBP. M1 cells were transfected with linearized LNCX, LNCX-HA-MOZ–CBP or LNCX-HA-MOZ–CBP(1–2504) and selected with 0.8 mg/ml G418. Selected colonies were cloned. Cell lysates of M1 clones were separated by 5% SDS–PAGE and analyzed by western blotting using anti-HA antibody (3F10). (B) Growth curve of the M1 infectants in response to IL-6. Each of the three M1 clones in (A) was cultured in the presence or absence of 5 ng/ml of IL-6. Results represent the average values of the relative numbers of viable cells for these clones. (C) Morphology of the cells. The M1 clones, which expressed wild-type MOZ–CBP (MC-1) or deletion mutant (NR-1), were exposed to IL-6 for 3 or 4 days and stained with May–Gruenwald and Giemsa solutions. Similar results were obtained with other clones. (D) The expression of surface antigens. The M1 infectants were exposed to IL-6 for 3 days and the expression of F4/80 and CD32 was analyzed by the indirect immunofluorescent method. Representative results for MC-1 (wild-type) and NR-1 (1–2504) are shown.
Discussion
MOZ is a potent co-activator for AML1
In this study, we partially purified AML1 complexes from L-G myeloid cells that stably expressed either AML1a or AML1b proteins. As we expected, the dominant component of both AML1a and AML1b complexes was the heterodimeric partner CBFβ. Multiple proteins were also co-purified specifically with AML1b proteins. Since the amount of these co-purified proteins was relatively small compared with that of CBFβ, we propose that AML1b forms ‘regulatory’ complexes with these proteins as well as a ‘core’ complex with CBFβ. Mass analysis and western blot analysis demonstrated that MOZ forms a ‘regulatory’ complex with AML1b. Domain analysis indicated that the transactivation domain of AML1 is required for interaction with MOZ, which suggests that MOZ might play a role in AML1-mediated transcriptional activation. As expected, reporter analysis showed that co-transfection of MOZ with AML1 strongly enhanced the expression of reporters that contain AML1-binding sites. These results suggest that MOZ functions as a strong co-activator of AML1.
MOZ can acetylate histones (Champagne et al., 2001; Figure 5). The catalytic (HAT) domain was mapped to a region between amino acids 488 and 778 of MOZ, which contains a MYST domain. We initially expected that the HAT activity would be very important for the stimulation of certain types of transcription. However, the HAT domain was found not to be essential for AML1-mediated transcription. This suggested that other domain(s) might be involved in transcriptional activation. Domain analysis of MOZ using fusions with the DNA-binding domain of the Gal4 transcription factor indicated that the C-terminal region of MOZ, which contains serine-, proline- and methionine-rich regions, had very strong activity to stimulate transcription of the reporter containing Gal4-binding sites. We also found that the MOZ sequence comprises two AML1-interacting domains. In addition to the AML1-interacting and transactivation domains, the N-terminal region and a part of the acidic region were found to be required for co-activation of AML1-mediated transcription. The functions of the N-terminal region and the acidic region are unclear at this time. Homology searches indicated the presence of a motif (H15) of linker histones, such as histone H1 and H5, in the N-terminal region of MOZ (Figure 4D). Deletion analysis indicated that the H15 domain is essential for co-activation and involved in nuclear localization. Since the H15 domain of histone H5 is implicated in nucleosome binding (Ramakrishnan et al., 1993), the H15 domain of MOZ might also engage in interactions with the nucleosome. Further analysis will be required to clarify the roles of the H15 domain and the acidic region of MOZ.
MOZ–CBP inhibits AML1-mediated transcription
MOZ–CBP lacks the C-terminal transactivation domain of normal MOZ and instead fuses to largely intact CBP. Since MOZ and CBP activate AML1-mediated transcription as shown above and in a previous study (Kitabayashi et al., 1998b), we tested the effects of MOZ–CBP on transcription and found that it inhibited transcription by AML1. The mechanism of transcription inhibition by MOZ–CBP is unclear at present, but there exist several possibilities. MOZ–CBP may prevent MOZ and/or p300/CBP from binding to AML1, since MOZ–CBP as well as MOZ and CBP could interact with AML1. MOZ–CBP lacks the transactivation domain of MOZ and contains the transrepression domain of MOZ. Thus binding of MOZ– CBP may competitively hamper AML1 from recruiting MOZ and/or p300/CBP, resulting in the inhibition of AML1 activity. Alternatively, MOZ–CBP may prevent other cofactors from binding to MOZ and/or p300/CBP. It has been reported that additional proteins such as PCAF, SRC1, ACTR, TIF2 and JMY are important for regulating p300/CBP activity in the presence of certain transcription factors (Yang et al., 1996; Yao et al., 1996; Chen et al., 1997; Torchia et al., 1997; Shikama et al., 1999). Since MOZ–CBP retains a large part of both MOZ and CBP, it would be expected to bind to at least some of these cofactors, which might prevent them from interacting with p300/CBP and/or MOZ. Domain analysis indicated that the bromodomain, which is known to interact with acetyl-lysine, is required for inhibition, suggesting that MOZ– CBP may inhibit transcription by aberrantly binding to acetylated proteins. Finally, MOZ–CBP may regulate the functions of AML1, MOZ, p300/CBP and/or other cofactors by acetylating them. In fact, MOZ–CBP undergoes autoacetylation and can acetylate AML1 as well as histones (Figure 5B and C). Domain analysis indicated that the HAT domain of the CBP portion is also required for inhibition, which suggests that MOZ–CBP may inhibit transcription by aberrantly regulating protein acetylation. It has been reported that p300/CBP acetylates the coactivator ACTR and disrupts complexes of the estrogen receptor and ACTR (Chen et al., 1999). Thus, it is possible that MOZ–CBP acetylates certain cofactor(s), which could disrupt complexes formed by these cofactor(s). Our results indicated that MOZ–CBP strongly and selectively inhibited the transactivation activity of CBP in a reporter expression assay using a fusion to the Gal4 DNA-binding domain. In this respect, the second and third hypotheses described above appear to be more likely. Analysis of MOZ–CBP-interacting proteins and MOZ–CBP-acetylating proteins should lead to a better understanding of the mechanism through which MOZ–CBP induces leukemia.
The AML1 complex: target of chromosome translocation in leukemia
The genes encoding AML1 and its heterodimeric partner, CBFβ, are frequent targets of chromosome translocations in human leukemia (Look, 1997). We and others have found that the genes encoding CBP, p300 and MOZ are disrupted and fused in-frame to other genes by chromosomal translocations in AML and myelodysplastic syndrome (MDS). Thus, AML1 and its associated factors including MOZ, p300/CBP and CBFβ are all targets of chromosomal rearrangements in human leukemia. Since rearrangements of genes for AML1-interacting co-repressors such as mSin3A and TLE/Groucho have not been reported, we propose that active AML1 complexes are the targets of leukemia-associated chromosome aberrations. The involvement of active AML1 complexes suggests that leukemic transformation may be the result of altered expression of genes regulated by AML1. In fact, AML1-mediated transcription is inhibited by leukemia-associated fusion proteins such as AML1-MTG8(ETO), TEL-AML1, AML1-EVI1 and CBFβ-MYH11 (Meyers et al. 1995; Tanaka et al. 1995; Hiebert et al., 1996; Y.Kanno et al., 1998). We show here that MOZ–CBP also represses promoters regulated by AML1. Thus, all these leukemia-associated fusions appear to elicit the leukemic phenotype by antagonizing the activity of AML1 complexes.
Materials and methods
Cells and retroviruses
M1 cells were cultured in minimal essential medium (MEM) supplemented with 10% calf serum and 1% non-essential amino acids. L-G cells were cultured in RPMI1640 medium supplemented with 10% fetal calf serum (FCS), recombinant mouse IL-3 (0.1 ng/ml) (a generous gift from Kirin Brewery) and 50 µM β-mercaptoethanol. Bosc23 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS. For production of retroviruses, Bosc23 cells were transfected with LNSX- and LNCX-derived retrovirus vectors by calcium phosphate precipitation methods, and culture supernatants were recuperated 48 h after transfection. L-G and M1 cells were incubated in the culture supernatant of Bosc23 transfectants for 24 h and then selected with G418 (1 mg/ml for L-G cells and 0.8 mg/ml for M1 cells). For cell growth, viable cells were counted using a Coulter counter (Beckman Coulter Inc.). For cell morphology, cells were stained with May–Gruenwald’s solution and Giemsa’s solution (Merck).
Plasmids
The N-terminal FLAG tag was fused to AML1a and AML1b cDNAs by using the oligonucleotide 5′-agctaggcctaccatggcagactacaaggacgacgatgacaagcgtatccccgtagatgccag-3′ as the upstream primer and 5′-agacagtgatggtcagagtg-3′ as the downstream primer in a PCR. The PCR product was digested with StuI and HindIII, and ligated to the HindIII site near the N-terminus of AML1a and AML1b. Human MOZ and CBP cDNAs were isolated by screening the size-fractionated cDNA library generated from poly(A) RNA of human KG-1 cells. The FLAG-tagged and HA-tagged MOZ and CBP expression vectors were generated by PCR and restriction enzyme digestion as described for FLAG-AML1. MOZ–CBP cDNA was generated by ligation of synthetic DNA coding for the MOZ–CBP fusion region (4942–5155) with MOZ and CBP cDNAs after BamHI and HincII digestion. Deletion mutants of AML1, MOZ and MOZ–CBP were constructed by ligation of the DNA fragments generated by appropriate restriction enzymes and PCRs. The sequences of the above constructs were checked by DNA sequencing.
Purification of the AML1 complex
L-G cells were infected with LNSX-FLAG-AML1a or LNSX-FLAG-AML1b as described above. The cells (∼1 × 1010) were lysed by incubation at 4°C for 30 min in 500 ml of lysis buffer [20 mM sodium phosphate pH 7.0, 250 mM NaCl, 30 mM sodium pyrophosphate, 0.1% NP-40, 5 mM EDTA, 10 mM NaF, 0.1 mM Na3VO4 and 1 mM phenylmethylsulfonyl fluoride (PMSF)] supplemented with protease inhibitors (Complete, Roche). The lysates were cleared by centrifugation at 30 000 g for 30 min at 4°C and incubated with 2.5 ml of anti-FLAG monoclonal antibody (M2)-conjugated beads with rotation at 4°C for 4 h. The beads with absorbed AML1 immunocomplexes were washed five times with 50 ml of lysis buffer. The AML1 complexes were eluted selectively by incubating twice with 0.2 mg/ml FLAG peptide in 12.5 ml of lysis buffer for 1 h. The eluents were concentrated by using a filtration device (Vivaspin 10K-PES, Sartorius) and separated by 10% SDS–PAGE. Proteins were stained with Coomassie Brilliant Blue, excised, destained with 25 mM ammonium bicarbonate and 50% acetonitrile, dried, digested with sequence grade modified trypsin (Promega) in 50 mM Tris pH 7.6, extracted with 5% trifluoroacetic acid (TFA), 50% acetonitrile, and subjected to LC/MS/MS analysis.
Mass spectorometry
An electrospray mass spectrometer (LCQ-DECA, ThermoQuest Co.) coupled on-line to a capillary HPLC column (Magic 2002, Michrom BioResources) was used to acquire tandem (MS/MS) spectra of peptides. Peptides were subjected to a Magic C18 column (5 µm 200 A, 0.2 × 50 mm) and were eluted with a gradient of 5–65% of buffer B (90% acetonitrile, 0.1% formic acid) and A (2% acetonitrile, 0.1% formic acid) over 20 min at a flow rate of 3 µl/min. The flow was split with a Magic pre-column capillary splitter assembly (Michrom BioResouces) at 50 µl/ml. The mass spectrometer was operated in autodetection mode to acquire MS/MS spectra during HPLC. Proteins were identified by searching sequence databases using TurboSEQUEST software.
Immunoprecipitations, western blotting and antibodies
Immunoprecipitations and western blotting were performed as described previously (Kitabayashi et al., 1998a). The anti-MOZ antibodies were generated by immunizing rabbits with peptides corresponding to residues 441–460 and 1648–1668 of human MOZ, and were affinity purified using antigen-conjugated columns. The anti-AML1 and anti-p300 polyclonal antibodies were described previously (Kitabayashi et al, 1998b). The anti-FLAG monoclonal antibody M2 (Sigma), the anti-HA monoclonal antibody 3F10 (Roche) and the anti-CBP polyclonal antibody (Santa Cruz Biotechnology) were commercially available.
Acetylation of histones and AML1, and identification of acetylation sites
Reactions were made in a 10 µl volume containing 50 mM Tris pH 8.0, 10% glycerol, 1 mM dithiothreitol (DTT), 1 mM PMSF, 10 mM sodium butyrate, 0.5 µl of [14C]acetyl-CoA (50 µCi/ml, Amersham) and 0.5 µg each of histone H2B, H3 and H4 or AML1, and incubated at 37°C for 1 h. Reaction mixtures were analyzed by 18% SDS–PAGE (histones) or 10% SDS–PAGE (AML1), and the gels were subjected to autoradiography. To determine the acetylation sites of histones, acetylated histones were subjected to N-terminal microsequencing using a G1005A protein sequencer (Hewlett Packard). Because acetylated lysine and unmodified lysine are differentially detected in this system (see Figure 5D–F), the relative amounts of acetylated and unmodified lysines before and after acetylation by MOZ were measured for each cycle by HPLC analysis using acetylated lysine (Sigma) as a standard.
Luciferase assay
Saos-2 or P19 cells were transfected by the calcium phosphate precipitation method in 24-well plates, and luciferase activity was assayed 48 h later using a luminometer Lumat LB9507 (Berthold) according to the manufacturer’s protocol (Promega). Results of reporter assays represent the average values for relative luciferase activity generated from three independent experiments that were normalized using the activity of the enzyme from pRL-tk as an internal control.
Flow cytometry
Surface antigens of M1 cells were examined by the indirect immunofluorescent method. M1 cells were incubated with anti-CD32 (PharMingen, San Diego, CA) or with anti-F4/80 at 4°C for 30 min. The cells were rinsed and incubated with fluorescein isothiocyanate (FITC)-conjugated F(ab)′2 goat anti-mouse IgG (Sigma). The cells were then rinsed and analyzed on FACSCalibur (Becton Dickinson). The log fluorescence intensity was determined with reference to the non-specific monoclonal antibody isotype control.
Acknowledgments
Acknowledgements
We thank Dr D.M.Livingston for p300 cDNA and Dr David Baltimore for Bosc23 cells. This work was supported in part by Grant-in-Aids for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology; by a Grant-in-Aid for the Comprehensive 10-year Strategy for Cancer Control and the Grant for Research on Aging and Health from the Ministry of Health, Labor and Welfare; and by the Program for Promotion of Fundamental Studies in Health Sciences of the Organization for Pharmaceutical Safety and Research of Japan.
References
- Borrow J., et al. (1996) The translocation t(8;16)(p11;p13) of acute myeloid leukaemia fuses a putative acetyltransferase to the CREB-binding protein. Nature Genet., 14, 33–41. [DOI] [PubMed] [Google Scholar]
- Carapeti M., Aguiar,R.C.T., Goldman,J.M. and Cross,N.C.P. (1998) A novel fusion between MOZ and the nuclear receptor coactivator TIF2 in acute myeloid leukemia. Blood, 91, 3127–3133. [PubMed] [Google Scholar]
- Chaffanet M., Gressin,L., Preudhomme,C., Soenen-Cornu,V., Birnbaum,D. and Pebusque,M.-J. (2000) MOZ is fused to p300 in an acute monocytic leukemia with t(8;22). Genes Chromosomes Cancer, 28, 138–144. [DOI] [PubMed] [Google Scholar]
- Champagne N., Pelletier,N. and Yang,X.J. (2001) The monocytic leukemia zinc finger protein MOZ is a histone acetyltransferase. Oncogene, 20, 404–409. [DOI] [PubMed] [Google Scholar]
- Chen H., Lin,R.J., Schiltz,R.L., Chakravarti,D., Nash,A., Nagy,L., Privalsky,M.L., Nakatani,Y. and Evans,R.M. (1997) Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell, 90, 569–580. [DOI] [PubMed] [Google Scholar]
- Chen H, Lin,R.J., Xie,W., Wilpitz,D. and Evans,R.M. (1999) Regulation of hormone-induced histone hyperacetylation and gene activation via acetylation of an acetylase. Cell, 98, 675–686. [DOI] [PubMed] [Google Scholar]
- Dhalluin C., Carlson,J.E., Zeng,L., He,C., Aggarwal,A.K. and Zhou,M.M. (1999) Structure and ligand of a histone acetyltransferase bromodomain. Nature, 399, 491–496. [DOI] [PubMed] [Google Scholar]
- Gelmetti V., Zhang,J., Fanelli,M., Minucci,S., Pelicci,P.G. and Lazar,M.A. (1998) Aberrant recruitment of the nuclear receptor corepressor–histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol., 18, 7185–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunstrein M. (1997) Histone acetylation in chromatin structure and transcription. Nature, 389, 349–352. [DOI] [PubMed] [Google Scholar]
- Hiebert S.W. et al. (1996) The t(12;21) translocation converts AML-1B from an activator to a repressor of transcription. Mol. Cell. Biol., 16, 1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ida K., Kitabayashi,I., Taki,T., Taniwaki,M., Noro,K., Yamamoto,M., Ohki,M. and Hayashi,Y. (1997) Adenovirus E1A-associated protein p300 is involved in acute myeloid leukemia with t(11;22)(q23;q13). Blood, 90, 4699–4704. [PubMed] [Google Scholar]
- Imai Y., Kurokawa,M., Tanaka,K., Friedman,A.D., Ogawa,S., Mitani,K., Yazaki,Y. and Hirai,H. (1998) TLE, the human homolog of groucho, interacts with AML1 and acts as a repressor of AML1-induced transactivation. Biochem. Biophys. Res. Commun., 252, 582–589. [DOI] [PubMed] [Google Scholar]
- Kanno T., Kanno,Y., Chen,L.F., Ogawa,E., Kim,W.Y. and Ito,Y. (1998) Intrinsic transcriptional activation–inhibition domains of the polyomavirus enhancer binding protein 2/core binding factor α subunit revealed in the presence of the β subunit. Mol. Cell. Biol., 18, 2444–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Kanno,T., Sakakura,C., Bae,S.C. and Ito,Y. (1998) Cytoplasmic sequestration of the polyomavirus enhancer binding protein 2 (PEBP2)/core binding factor α (CBFα) subunit by the leukemia-related PEBP2/CBFβ–SMMHC fusion protein inhibits PEBP2/CBF-mediated transactivation. Mol. Cell. Biol., 18, 4252–4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I., Ida,K., Morohoshi,F., Yokoyama,A., Mitsuhashi,N., Shimizu,K., Nomura,N., Hayashi,Y. and Ohki,M. (1998a) The AML1–MTG8 leukemic fusion protein forms a complex with a novel member of the MTG8(ETO/CDR) family, MTGR1. Mol. Cell. Biol., 18, 846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I., Yokoyama,A., Shimizu,K. and Ohki,M. (1998b) Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J., 17, 2994–3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitabayashi I., Aikawa,Y., Yokoyama,A., Hosoda,F., Nagai,M., Kakazu,N., Abe,T. and Ohki,M. (2001) Fusion of MOZ and p300 histone acetyltransferases in acute monocytic leukemia with a t(8;22)(p11;q13) chromosome translocation. Leukemia, 15, 89–94. [DOI] [PubMed] [Google Scholar]
- Levanon D., Goldstein,R.E., Bernstein,Y., Tang,H., Goldenberg,D., Stifani,S., Paroush,Z. and Groner,Y. (1998) Transcriptional repression by AML1 and LEF-1 is mediated by the TLE/Groucho corepressors. Proc. Natl Acad. Sci. USA, 95, 11590–11595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J., Prouty,L., Willians,B.J., Dayton,M.A. and Blanchard,K.L. (1998) Acute mixed lineage leukemia with an inv(8)(p11q13) resulting in fusion of genes for MOZ and TIF2. Blood, 92, 2118–2122. [PubMed] [Google Scholar]
- Look A.T. (1997) Oncogenic transcription factors in human acute leukemia. Science, 278, 1059–1064. [DOI] [PubMed] [Google Scholar]
- Lutterbach B. et al. (1998) ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol., 18, 7176–7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutterbach B., Westendorf,J.J., Linggi,B., Isaac,S., Seto,E. and Hiebert,S.W. (2000) A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J. Biol. Chem., 275, 651–656. [DOI] [PubMed] [Google Scholar]
- Meyers S., Lenny,N. and Hiebert,S.W. (1995) The t(8;21) fusion protein interferes with AML 1B-dependent transcriptional activation. Mol. Cell. Biol., 15, 1974–1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki M. et al. (1997) Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc. Natl Acad. Sci. USA, 94, 5697–5702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa E., Inuzuka,M., Maruyama,M., Satake,M., Naito-Fujimoto,M., Ito,Y. and Shigesada,K. (1993) Molecular cloning and characterization of PEBP2β, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2. Virology, 194, 314–331. [DOI] [PubMed] [Google Scholar]
- Okuda T., van Deursen,J., Hiebert,S.W., Grosveld,G. and Downing,J.R. (1996) AML1, the target of multiple chromosomal translocations in human leukemia, is essential for normal fetal liver hematopoiesis. Cell, 84, 321–330. [DOI] [PubMed] [Google Scholar]
- Panagopoulos I., Fioretos,T., Isaksson,M., Samuelsson,U., Billstrom,R., Strombeck,B., Mitelman,F. and Johansson,B. (2001) Fusion of the MORF and CBP genes in acute myeloid leukemia with the t(10;16)(q22;p13). Hum. Mol. Genet., 10, 395–404. [DOI] [PubMed] [Google Scholar]
- Pazin M.J. and Kadonaga,J.T. (1997) What’s up and down with histone acetylation and transcription? Cell, 89, 325–328. [DOI] [PubMed] [Google Scholar]
- Rabbitts T.H. (1994) Chromosomal translocations in human cancer. Nature, 372, 143–149. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan V., Finch,J.T., Graziano,V., Lee,P.L. and Sweet,R.M. (1993) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature, 362, 219–223. [DOI] [PubMed] [Google Scholar]
- Sasaki K., Yagi,H., Bronson,R.T., Tominaga,K., Matsunashi,T., Deguchi,K., Tani,Y., Kishimoto,T. and Komori,T. (1996) Absence of fetal liver hematopoiesis in mice deficient in transcriptional coactivator core binding factor β. Proc. Natl Acad. Sci. USA, 93, 12359–12363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satake N., Ishida,Y., Otoh,Y., Hinohara,S., Kobayashi,H., Sakashita,A., Maseki,N. and Kaneko,Y. (1997) Novel MLL–CBP fusion transcript in therapy-related chronic myelomonocytic leukemia with a t(11;16)(q23;p13) chromosome translocation. Genes Chromosomes Cancer, 20, 60–63. [PubMed] [Google Scholar]
- Schiltz R.L., Mizzen,C.A., Vassilev,A., Cook,R.G., Allis,C.D. and Nakatani,Y. (1999) Overlapping but distinct patterns of histone acetylation by the human coactivators p300 and PCAF within nucleosomal substrates. J. Biol. Chem., 274, 1189–1192. [DOI] [PubMed] [Google Scholar]
- Shikama N., Lee,C.W., France,S., Delavaine,L., Lyon,J., Krstic-Demonacos,M. and La Thangue,N.B. (1999) A novel cofactor for p300 that regulates the p53 response. Mol. Cell, 4, 365–376. [DOI] [PubMed] [Google Scholar]
- Sobulo O.M. et al. (1997) MLL is fused to CBP, a histone acetyltransferase, in therapy-related acute myeloid leukemia with a t(11;16)(q23;p13.3). Proc. Natl Acad. Sci. USA, 94, 8732–8737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. (1998) Histone acetylation and transcriptional regulatory mechanisms. Genes Dev., 12, 599–606. [DOI] [PubMed] [Google Scholar]
- Taki T., Sako,M., Tsuchida,M. and Hayashi,Y. (1997) The t(11;16)(q23;p13) translocation in myelodysplastic syndrome fuses the MLL gene to the CBP gene. Blood, 89, 3945–3950. [PubMed] [Google Scholar]
- Tanaka T., Mitani,K., Kurokawa,M., Ogawa,S., Tanaka,K., Nishida,J., Yazaki,Y., Shibata,Y. and Hirai,H. (1995) Dual functions of the AML1/Evi-1 chimeric protein in the mechanism of leukemogenesis in t(3;21) leukemias. Mol. Cell. Biol., 15, 2383–2392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torchia J., Rose,D.W., Inostroza.J., Kamei,Y., Westin,S., Glass,C.K. and Rosenfeld,M.G. (1997) The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature, 387, 677–684. [DOI] [PubMed] [Google Scholar]
- Wade P.A., Pruss,D. and Wolffe,A.P. (1997) Histone acetylation: chromatin in action. Trends Biochem. Sci., 22, 128–132. [DOI] [PubMed] [Google Scholar]
- Wang J., Hoshino,T., Redner,R.L., Kajigaya,S. and Liu,J.M. (1998) ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl Acad. Sci. USA, 95, 10860–10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang,Q., Crute,B.E., Melnikova,I.N., Keller,S.R. and Speck,N.A. (1993) Cloning and characterization of subunits of the T-cell receptor and murine leukemia virus enhancer core-binding factor. Mol. Cell. Biol., 13, 3324–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Stacy,T., Binder,M., Marin-Padilla,M., Sharpe,A.H. and Speck,N.A. (1996a) Disruption of the Cbfα2 gene causes necrosis and hemorrhaging in the central nervous system and blocks definitive hematopoiesis. Proc. Natl Acad. Sci. USA, 93, 3444–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q. et al. (1996b) The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell, 87, 697–708. [DOI] [PubMed] [Google Scholar]
- Yang X.-J., Ogryzko,V.V., Nishikawa,J., Howard,B.H. and Nakatani,Y. (1996) A p300/CBP-associated factor that competes with the adenoviral oncoprotein E1A. Nature, 382, 319–324. [DOI] [PubMed] [Google Scholar]
- Yao T.P., Ku,G., Zhou,N., Scully,R. and Livingston,D.M. (1996) The nuclear hormone receptor coactivator SRC-1 is a specific target of p300. Proc. Natl Acad. Sci. USA, 93, 10626–10631. [DOI] [PMC free article] [PubMed] [Google Scholar]