Abstract
The 26S proteasome plays a major role in eukaryotic protein breakdown, especially for ubiquitin-tagged proteins. Substrate specificity is conferred by the regulatory particle (RP), which can dissociate into stable lid and base subcomplexes. To help define the molecular organization of the RP, we tested all possible paired interactions among subunits from Saccharomyces cerevisiae by yeast two-hybrid analysis. Within the base, a Rpt4/5/3/6 interaction cluster was evident. Within the lid, a structural cluster formed around Rpn5/11/9/8. Interactions were detected among synonymous subunits (Csn4/5/7/6) from the evolutionarily related COP9 signalosome (CSN) from Arabidopsis, implying a similar quaternary arrangement. No paired interactions were detected between lid, base or core particle subcomplexes, suggesting that stable contacts between them require prior assembly. Mutational analysis defined the ATPase, coiled-coil, PCI and MPN domains as important for RP assembly. A single residue in the vWA domain of Rpn10 is essential for amino acid analog resistance, for degrading a ubiquitin fusion degradation substrate and for stabilizing lid–base association. Comprehensive subunit interaction maps for the 26S proteasome and CSN support the ancestral relationship of these two complexes.
Keywords: COP9 signalosome/26S proteasome/proteolysis/ubiquitin/yeast two-hybrid
Introduction
The 26S proteasome is a 2 MDa ATP-dependent protease responsible for the bulk of non-lysosomal proteolysis in eukaryotes, often using the covalent modification of proteins by ubiquitylation to assist in target recognition (Voges et al., 1999). It consists of a 20S proteolytic core particle (CP) and a 19S regulatory particle (RP). The CP is an ATP-independent peptidase, containing post-acidic, post-basic and post-hydrophobic hydrolyzing activities. Its cylindrical shape is created by the assembly of four stacked heptameric rings: the internal rings are each composed of seven related β-subunits and the outer rings are each composed of seven related α-subunits arranged in a α1–7/β1–7/β1–7/α1–7 configuration (Voges et al., 1999). The proteolytic active sites involving the β1, β2 and β5 subunits reside within the central chamber. A small channel formed by the α-subunit rings restricts access to this chamber such that only unfolded proteins may enter. In yeast and probably other eukaryotes, it appears that the channel is gated by flexible N-terminal extensions in the α-subunits to control substrate entry and product exit (Glickman, 2000).
Appended to either or both ends of the CP is the RP that confers both ubiquitin and ATP dependence to the 26S proteasome (Voges et al., 1999). Its proposed functions are to recognize and help unfold substrates, open the channel and translocate the substrates into the CP for proteolysis. The RP can be dissociated further into two subcomplexes, the base that directly associates with the CP and a peripheral lid. The base is composed of three non-ATPase subunits (Rpn1, 2 and 10) and six ATPase subunits (Rpt1–6) that are members of the AAA-ATPase family. It is presumed that the Rpt subunits assemble into a six-membered ring similar to the ATP-dependent ClpAP and HslVU proteases, and likewise use ATP hydrolysis to facilitate channel opening and target unfolding (Strickland et al., 2000). The lid is a 400 kDa complex, assembled from at least eight additional Rpn subunits (Rpn3, 5–9, 11 and 12) (Glickman, 2000). Its role in 26S proteasome function remains unclear. At least in vitro, the lid is necessary for proper degradation of polyubiquitylated proteins. Presumably, the lid helps recognize appropriate targets and remove the covalently bound ubiquitins before transport of the target into the CP by the base-related activities. These functions are supported by genetic analyses showing that some lid subunits have substrate-specific roles (e.g. van Nocker et al., 1996; Bailly and Reed, 1999).
One RP base subunit, Rpn10, appears to have multiple functions. It was first identified by its ability to bind polyubiquitin chains, thus implicating it as a polyubiquitin-protein receptor (see van Nocker et al., 1996). However, only short-lived proteins degraded by the ubiquitin fusion degradation (UFD) pathway [e.g. ubiquitin-proline β-galactosidase (Ub-Pro-βgal)] are stabilized in yeast rpn10Δ strains, suggesting that Rpn10 has a substrate-specific function (van Nocker et al., 1996). rpn10Δ strains from various species are hypersensitive to amino acid analogs, implicating Rpn10 in abnormal protein removal (van Nocker et al., 1996; Girod et al., 1999). Rpn10 also has a role in RP stability. Loss of Rpn10 weakens association of the lid and base, suggesting that it is necessary for proper contacts between the two subcomplexes (Glickman et al., 1998). Whereas binding of polyubiquitin chains requires a hydrophobic patch in the C-terminal half of Rpn10, the N-terminal portion of the polypeptide appears crucial for in vivo functions that confer amino acid analog resistance and Ub-Pro-βgal degradation (Fu et al., 1998).
The RP lid is structurally similar to the newly discovered regulatory complex, the COP9 signalosome (CSN) that is present in most eukaryotes except Saccharomyces cerevisiae (Deng et al., 2000). The CSN contains eight core subunits that assemble into a 450 kDa particle. The core CSN subunits show a remarkable one-to-one sequence correspondence with those of the RP lid, suggesting a common ancestry and architecture (Glickman et al., 1998; Deng et al., 2000). The CSN plays an essential role in a number of developmental processes, including Arabidopsis photomorphogenesis and Drosophila embryogenesis by affecting the turnover of numerous 26S proteasome substrates (Deng et al., 2000). The biochemical function(s) of the CSN is not yet clear. Several activities have been detected, including a protein kinase activity (Bech-Otschir et al., 2001) and a hydrolase activity that removes Nedd8/Rub1, a ubiquitin-related modifier that becomes attached to the SCF E3 ubiquitin ligase complex (Lyapina et al., 2001). Interactions between subunits of the RP and CSN have been reported (see Kim et al., 2001) and mutations in the CSN were found to affect assembly of the RP (Peng et al., 2001), suggesting that the CSN and RP overlap functionally as well as structurally.
To help understand how the RP and CSN function, structural resolution of these complexes would be instrumental. At present, such analyses have been hampered by low purification yields, low stability, potential subunit heterogeneity and flexible shape. Electron microscopy has provided crude three-dimensional pictures of the 26S proteasome (Walz et al., 1998), and yielded the first structural comparison of the lid and CSN (Kapelari et al., 2000). Cross-linking and far-western blotting have provided limited insights into the RP base. They identified several interacting pairs including those involving Rpt subunits (Richmond et al., 1997; Gorbea et al., 2000; Hartmann-Petersen et al., 2001).
As an alternative strategy to map RP and CSN topology, we exploited the yeast two-hybrid (Y2H) method to help identify interacting subunits. Previous Y2H studies using a limited subset of RP subunits identified a few interacting partners (see Ferrell et al., 2000; Uetz et al., 2000; Hartmann-Petersen et al., 2001; Ito et al., 2001). Here, we completed a comprehensive paired interaction analysis among all 17 principal subunits of the yeast RP and seven CP α-subunits, and among all eight subunits of the CSN. Remarkably similar interactions among synonymous subunits of the CSN and the RP support a related structural organization for these particles. Mutational analysis located structural motifs/residues that are required for contacts between individual RP subunits, and between the lid and base subcomplexes. For Rpn10 in particular, Asp11 appears critical for in vivo protein turnover by the RP and for binding of the lid to the base, probably by participating in a salt bridge that stabilizes a vWA-like protein contact fold.
Results
Y2H system and tested combinations
To help define interactions among the RP subunits, we tested all possible combinations of the 17 yeast RP subunits (Rpt1–6, and Rpn1–3 and 5–12) by a GALl4-based Y2H method (Phizicky and Fields, 1995). Rpn4 is not considered to be part of the RP complex and, indeed, we did not find that Rpn4 interacts with any other RP subunits (data not shown). We assembled a library of Y2H constructions, which included 17 that express RP subunits as C-terminal fusions to the GAL4 binding domain (BD; designated with a BD prefix) and 34 that express RP subunits as C- or N-terminal fusions to the GAL4 activation domain (AD; designated with an AD prefix or suffix, respectively). By using both AD orientations, we hoped to eliminate potential interference by the AD domain. We also attempted to detect interactions using RP subunits fused to the GAL4 BD N-terminus. However, none of these BD fusions succeeded (data not shown), possibly because the N-terminal appendages were detrimental to BD activity.
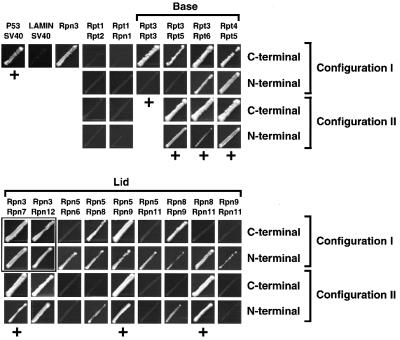
The RP constructions were expressed in all possible paired combinations of GAL4 BD–AD fusions and assessed for subunit interaction by the HIS3 and LacZ reporters using the yeast YRG2 strain. The LacZ reporter appears to be a more stringent detector, since all of the LacZ-positive combinations displayed histidine auxotrophic growth, but not vice versa. In each case, binding activity was compared with a known interacting pair, p53 and SV40 T-antigen, and a non-interacting pair, lamin C and SV40 T-antigen. When each AD and BD fusion was expressed individually, only BD:Rpn3 activated the HIS3 reporter (Figure 1 and data not shown), but it did not elicit significant LacZ activity, suggesting a weak transactivation activity. Paired assortment of the RP subunits gave rise to 136 hetero-interacting combinations and 17 self-interacting combinations. Each of the hetero-interacting combinations was tested in two configurations. Configur ation I refers to BD–AD fusions based on an ascending order of Rpt1–6 to Rpn1–12 subunits (e.g. BD:Rpt1– AD:Rpt2) whereas configuration II refers to BD–AD fusions based on a descending order of subunits (e.g. BD:Rpt2–AD:Rpt1). In total, 578 combinations ([(136 × 2) + 17] × 2) were examined.
Fig. 1. RP subunit interactions of the yeast 26S proteasome as detected by Y2H. The 17 yeast RP subunits were tested for potential interaction in all possible paired BD–AD configurations (I and II, see text) by histidine auxotrophic growth. A subset of non-interacting pairs and all interacting pairs are shown with all tested configurations. Brackets: group interacting pairs within the base and lid. p53–SV40 (SV40 T-antigen) and LAMIN (lamin C)–SV40 represent known interacting and non-interacting protein partners. As shown, BD:Rpn3 can self-activate the HIS3 reporter. Interaction pairs with positive LacZ activity are indicated by a ‘+’ (see Table I). Boxes show interacting pairs involving BD:Rpn3, which were confirmed using different testing configurations and/or by LacZ assay.
Rpt subunit interactions suggest a minimal base cluster involving Rpt4/5/3/6
RP subunit pairs that showed a positive reaction when assayed for histidine auxotrophic growth are shown in Figure 1. Surprisingly, none of the lid subunits showed detectable affinity for any of the base subunits, and vice versa. Seventeen positive pairs involved the BD fusion of Rpn3 (Rpn3 × Rpt1–6, Rpn1–3, 5–12), which could not be concluded as interacting due to self-activation by BD:Rpn3 (Figure 1 and data not shown). The remaining interacting pairs involved four subunits within the base and eight subunits within the lid.
Within the base complex, the Rpt3/3, Rpt3/5, Rpt3/6 and Rpt4/5 pairs appeared to interact. In most cases, binding was evident using either BD–AD configuration (I or II) and either N- or C-terminal AD fusion (Figure 1). Self-interactions were observed only with Rpt3 in the BD:Rpt3–AD:Rpt3 orientation. Since we presume that the six Rpt subunits assemble as a heteromeric ring (Glickman, 2000), the in vivo significance of this self-interaction is unclear. With the exception of BD:Rpt4– AD:Rpt5, these base interactions were confirmed by assaying LacZ activities that were 2- to 20-fold higher than the negative control (Table I, lamin C–SV40 T-antigen). Collectively, the interactions suggest a minimal base cluster involving Rpt4/5/3/6.
Table I. Yeast RP subunit interaction detected by the two-hybrid method using the LacZ reportera.
| Interacting pair | BD/AD configuration Ib |
BD/AD configuration IIb |
||
|---|---|---|---|---|
| C-terminalc | N-terminalc | C-terminalc | N-terminalc | |
| p53–SV40 | 308.3 ± 30.4 | ndd | nd | nd |
| Lamin C–SV40 | 30.1 ± 1.2 | nd | nd | nd |
| Rpn3e | 20.7 ± 1.9 | |||
| Rpt1–Rpt2f | 16.3 ± 0.1 | nd | nd | nd |
| Rpt1–Rpn1f | 17.0 ± 1.5 | nd | nd | nd |
| Rpt3–Rpt3 | 194.7 ± 16.6 | nd | nd | nd |
| Rpt3–Rpt5 | 130.2 ± 7.3 | nd | 443.0 ± 21.1 | 104.3 ± 17.8 |
| Rpt3–Rpt6 | 613.2 ± 10.9 | 242.0 ± 32.7 | 381.2 ± 40.4 | 61.4 ± 10.4 |
| Rpt4–Rpt5 | 17.7 ± 1.9 | 66.4 ± 7.6 | 557.3 ± 25.3 | 186.3 ± 45.6 |
| Rpn3–Rpn7 | 1165.9 ± 106.2 | 645.9 ± 114.4 | 362.9 ± 30.6 | 653.7 ± 135.9 |
| Rpn5–Rpn9 | 144.2 ± 15.2 | 302.6 ± 30.6 | 148.1 ± 12.7 | 76.3 ± 9.9 |
| Rpn8-Rpn11 | 11.9 ± 0.8 | 278.4 ± 57.3 | 89.0 ± 19.3 | 223.3 ± 45.8 |
aLacZ activity is expressed in nmol o-nitrophenol/h/mg protein; ± indicates standard error.
bIn the order of Rpt1–6–Rpn1–12, each pair of 17 × 17 RP subunits (except 17 self-interacting pairs) were constructed separately with a BD or AD domain, respectively, in either an ascending (configuration I) or descending (configuration II) manner (see text).
cRP subunits are fused to either the C- or N-terminal end of the GAL4 AD domain.
dnd, not determined.
eHIS-positive YRG2 cells expressing the BD fusion of Rpn3 alone were assayed for LacZ activity.
fRpt1/2 and Rpt1/Rpn1 are representatives having background LacZ activities.
A cluster of lid subunit interactions involves Rpn5, 8, 9 and 11
Within the lid, we identified nine interacting pairs: Rpn3/7, Rpn3/12, Rpn5/6, Rpn5/8, Rpn5/9, Rpn5/11, Rpn8/9, Rpn8/11 and Rpn9/11 (Figure 1). Except for one configuration (BD:Rpn8–AD:Rpn11), three of these partners, Rpn3/7, Rpn5/9 and Rpn8/11, were positive by both the HIS3 and LacZ reporters in all four configurations, suggesting strong affinity (Figure 1 and Table I). Since BD:Rpn3 by itself grew on histidine-minus medium, the Rpn3/7 interaction could not be demonstrated by the HIS3 reporter alone. However, a strong interaction was supported by significantly higher LacZ activities (31- and 56-fold, respectively) for the BD:Rpn3–AD:Rpn7 and BD:Rpn3–Rpn7:AD combinations as compared with BD:Rpn3 alone (Table I).
Six pairs, Rpn3/12, Rpn5/6, Rpn5/8, Rpn5/11, Rpn8/9 and Rpn9/11, were detected by HIS3 (Figure 1) but not by the LacZ reporter. Moreover, only some of the four configurations were HIS3 positive, suggesting that the interactions within these combinations were weak or substantially affected by the AD–BD appendages. The exception was the Rpn3/12 pair that showed robust growth in all four combinations, including those containing Rpn3 fusions to AD, which alone did not elicit growth. Most of the lid interaction pairs involved Rpn5, 8, 9 and 11 (seven out of nine), suggesting a localized structural cluster containing these four subunits.
Individual interactions between RP and CP subunits were not detected
In an attempt to identify which α-subunits interact with which RP subunits, we subjected all seven of the yeast α-subunits to Y2H analysis in combination with the 17 RP subunits. The α-subunits were tested either as C-terminal fusions to BD or N-terminal fusions to AD, whereas the RP subunits were tested as both N- and C-terminal AD fusions and C-terminal BD fusions, resulting in 238 BD:α1–7 × 34 AD–RP fusions and 119 α1–7:AD × 17 BD–RP fusions. When the α-fusions were expressed individually, only BD:α5 activated the HIS3 reporter (data not shown). Except for the 34 pairs involving the BD:α5 construction, none of the combinations appeared to interact.
N-terminal coiled-coil domains are involved in Rpt subunit interactions
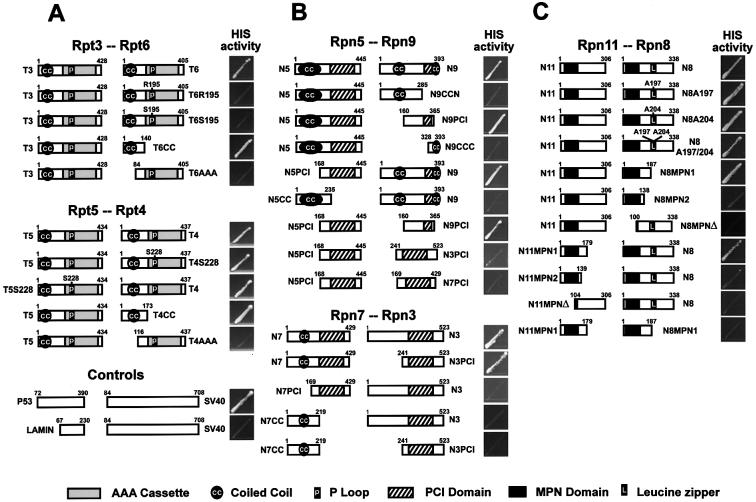
Several motifs have been identified within the RP subunits that may be important for activity, structure and/or protein–protein interactions. To examine their roles in the subunit interactions described above, we tested various site-directed and deletion mutants. All six Rpt subunits contain an N-terminal coiled-coil and an ATPase cassette that defines them as members of the AAA-ATPases superfamily (Glickman et al., 1998). The P-loop present within the ATPase cassette contains an invariant lysine necessary for ATP hydrolysis (Rubin et al., 1998). As shown in Figure 2A, the Rpt3/6 and Rpt4/5 interactions were maintained with the C-terminal deletion mutants of Rpt6 (T6CC) and Rpt4 (T4CC) missing the AAA-ATPase cassette, but abrogated with the N-terminal deletion mutants (T6AAA and T4AAA) missing the coiled-coil, indicating that the coiled-coil is important. For the Rpt4/5 pair, the invariant Lys228 in the P-loop was not required for the interaction. However, substitution of the invariant Lys195 in the P-loop of Rpt6 with either arginine (T6R195) or serine (T6S195) abolished the Rpt3/6interaction, indicating that the P-loop of Rpt6 has a role in the context of the full-length protein (Figure 2A).
Fig. 2. Detection of structural domains involved in various RP subunit interactions by Y2H using histidine auxotrophic growth. The organization of the various deletions and amino acid substitutions is indicated next to each protein. (A) Coiled-coil domains of Rpt4 (T4) and Rpt6 (T6) and the invariant K195 of Rpt6 are critical for Rpt3/6 or Rpt4/5 interaction. (B) PCI domains are essential for Rpn5/9 and Rpn3/7 interaction. (C) The sequences encompassing the MPN domains are involved in Rpn8/11 interaction.
PCI and MPN domains are required for specific lid subunit interactions
Defined structural domains in the lid subunits include the PCI domain (proteasome, COP9, eIF3) found in six subunits (Rpn3, 5–7, 9 and 12), the MPN domain (Mpr1, Pad1 N-terminal) found in Rpn8 and 11, putative coiled-coils in Rpn5, 7 and 9, and a potential leucine zipper in Rpn8 (Glickman et al., 1998). Analysis of deletion mutants indicated that the PCI domains and not the coiled-coils are essential for interaction of the Rpn5/9 pair (Figure 2B). Rpn5 and 9 mutants (N5PCI and N9PCI) containing the PCI domain but not the coiled-coil maintained their interaction, whereas Rpn5 and 9 mutants missing the PCI domain but containing the coiled-coil (N9CCN, N9CCC and N5CC) did not (Figure 2B). In fact, BD–AD fusions containing just the PCI domains (N5PCI/N9PCI) interacted, indicating that this domain alone is sufficient for the Rpn5/9 binding. Interaction of PCI domains between Rpn5 and 9 appeared to be subunit specific since the PCI domain from neither Rpn3 (N3PCI) nor Rpn7 (N7PCI) could substitute for that of Rpn9 in its interaction with Rpn5 (Figure 2B).
For Rpn3/7 interaction, the PCI domain of Rpn3 was found to be critical, but the motif required in Rpn7 was not obvious. The Rpn3 deletion mutant (N3PCI) containing the PCI maintained its interaction with Rpn7, but a similar deletion in Rpn7 (N7PCI) failed to interact with Rpn3 (Figure 2B). The coiled-coil domain of Rpn7 alone was also insufficient, as the N-terminal half of Rpn7 (N7CC) encompassing this domain did not bind to either full-length Rpn3 or N3PCI.
The roles of the MPN and leucine zipper domains were tested in the Rpn8/11 interactions. Alanine substitution mutants of Rpn8, changing key leucines at positions 197 and 204 within the leucine zipper, either singly or in tandem (N8A197, N8A204 and N8A197/204), failed to disrupt its association with Rpn11, indicating that the leucine zipper is not essential (Figure 2C). In contrast, mutants (N8MPN1 and N11MPN1) containing the MPN domain interacted with their wild-type partners whereas the complementary mutants missing their MPN domains (N8MPNΔ and N11MPNΔ) did not, indicating that the MPN domain is involved (Figure 2C). Deletions of Rpn8 (N8MPN2) and Rpn11 (N11MPN2) removing additional sequences near the MPN domains failed to associate with their wild-type partners, as did segments containing just the regions surrounding the MPN domains (N11MPN1 and N8MPN1). Thus, it is possible that the amino acids flanking the C-terminus of the MPN domains contain essential contacts or are needed for proper folding of the MPN domains.
Conserved interactions among the synonymous subunits from the Arabidopsis CSN complex
Amino acid sequence alignments between subunits of the RP lid and the CSN suggest that these two complexes arose from a common progenitor (Glickman et al., 1998; Deng et al., 2000). In particular, Rpn3, 5, 6, 7, 8, 9, 11 and 12 appear to be synonymous with Csn3, 4, 2, 1, 6, 7, 5 and 8, respectively (Table II). To test whether the synonymous CSN subunits also share protein–protein interactions similar to those of the lid, we subjected all eight proteins from the Arabidopsis CSN to Y2H analysis. Previous studies (see Deng et al., 2000) and our database searches showed that most CSN subunits are encoded by single genes in Arabidopsis. The exceptions are Csn5 and Csn6, which are both encoded by two genes (designated A and B) that share ∼86–88% amino acid sequence identities (Table II). The presence of cDNAs for all the Csn genes in various Arabidopsis expressed sequence tag (EST) collections indicates that all are actively expressed (Table II). Csn7 is unique because it encodes two protein isoforms, Csn7i and Csn7ii. They are derived by alternative splicing of the seventh intron such that the Csn7i protein contains two additional amino acids at its C-terminus and the Csn7ii protein has a 37 amino acid C-terminal extension (data not shown). Two Csn7 proteins differing by several kilodaltons were detected in Arabidopsis, which may reflect these two isoforms (Karniol et al., 1999). The amino acid sequence similarities between orthologous CSN subunits from Arabidopsis and human are ∼45–75% as compared with ∼32–54% similarities between synonymous subunits of the lid and CSN from Arabidopsis (Table II).
Table II. Characterization of Arabidopsis genes encoding the subunits of the COP9 signalosome (CSN) complex.
| Genea | Chromosome locationb | Peptide length (aa)/mol. wt (kDa) | No of EST hits | Similarity of AtCSN homologsc |
Identity/similarity to human CSN and Arabidopsis lid subunitsc |
||
|---|---|---|---|---|---|---|---|
| Nucleotide | Peptide | Human CSN | Arabidopsis lid | ||||
| AtCSN1 | III, cM 96–98 | 441/50.6 | 8 | 49/61 (433) | Rpn7 22/34 (312) | ||
| AtCSN2 | II, cM 56–57 | 439/51.2 | 4 | 61/73 (433) | Rpn6 24/39 (374) | ||
| AtCSN3 | V, cM 24–26 | 429/47.7 | 7 | 40/48 (407) | Rpn3 28/38 (206) | ||
| AtCSN4 | V, cM 90–93 | 397/45.0 | 3 | 52/63 (389) | Rpn5 27/40 (328) | ||
| AtCSN5a | I, cM 120–122 | 358/40.3 | 1 | 84 | 86/90 (357) | 68/73 (341) | Rpn11 43/53 (206) |
| AtCSN5b | I, cM 37–39 | 357/39.7 | 11 | 68/75 (336) | Rpn11 42/54 (206) | ||
| AtCSN6a | V, cM 120–121 | 317/35.7 | 14 | 86 | 88/91 (317) | 42/54 (308) | Rpn8 28/41 (295) |
| AtCSN6b | IV, cM 86–87 | 317/35.4 | 4 | 41/53 (308) | Rpn8 28/42 (305) | ||
| AtCSN7i | I, cM 0–3 | 225/25.5 | 11 | 35/45 (211) | Rpn9 27/40 (173) | ||
| AtCSN7ii | 260/29.5 | 11 | 35/45 (211) | Rpn9 27/40 (173) | |||
| AtCSN8 | IV, cM 29 | 197/22.6 | 1 | 34/46 (163) | Rpn12 21/32 (71) | ||
aNomenclature is adapted from Deng et al. (2000).
bMap locations determined by AtDB’s Seq Map (http://www.arabidopsis.org/cgi-bin/maps/Schrom).
cPercentage peptide sequence identity/similarity was determined by UW-GCG program BESTFIT; numbers in parentheses are lengths of the best matched polypeptides for each comparison pair; similarities between homologs are compared with both nucleotide (identity) and peptide sequences.
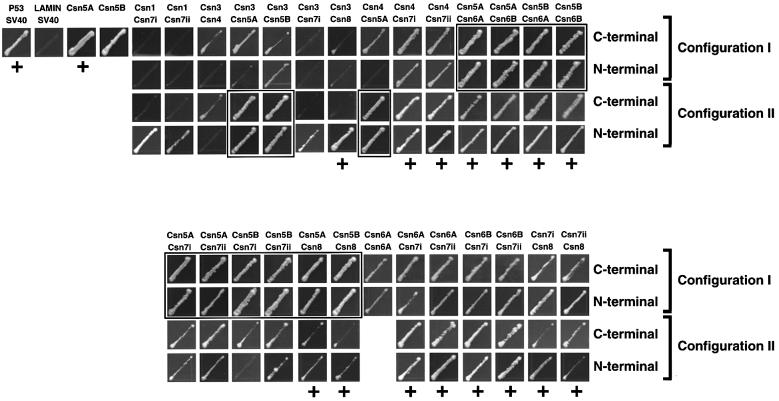
The Arabidopsis CSN was tested by Y2H either as C- and N-terminal AD fusions or as C-terminal BD fusions. When each of the AD and BD fusions was expressed alone, only BD:Csn5A and BD:Csn5B self-activated the HIS3 reporter (Figure 3) and only BD:Csn5A slightly activated the LacZ reporter (Table III). The 11 CSN subunits (including isoforms of Csn5, 6 and 7) gave rise to 55 hetero-interacting combinations and 11 self-interacting combinations. When assembled in both ascending (configuration I) and descending (configuration II) configurations, a total of 242 combinations ([(55 × 2) + 11] × 2) were examined.
Fig. 3. Interactions of Arabidopsis CSN subunits as detected by Y2H using histidine auxotrophic growth. Interacting pairs are shown with all tested configurations. Interactions with positive LacZ activity are indicated by a ‘+’ (see Table III). Boxes show interactions involving self-activated BD:Csn5A or BD:Csn5B, which were confirmed using different testing configurations and/or by LacZ assay.
Table III. Arabidopsis CSN subunit interaction detected by the two-hybrid method using the LacZ reportera.
| Interacting pair | BD/AD configuration-Ib |
BD/AD configuration-IIb |
||
|---|---|---|---|---|
| C-terminalc | N-terminalc | C-terminalc | N-terminalc | |
| p53–SV40 | 547.1 ± 17.0 | ndd | nd | nd |
| Lamin C–SV40 | 50.1 ± 2.3 | nd | nd | nd |
| BD:Csn5Ae | 109.2 ± 15.3 | |||
| BD:Csn5Be | 53.4 ± 2.2 | |||
| Csn5B–Csn7if | 53.7 ± 11.3 | 61.4 ± 6.5 | 16.7 ± 0.4 | 13.7 ± 0.9 |
| Csn5B–Csn7iif | 48.5 ± 4.5 | 48.4 ± 6.0 | 16.5 ± 1.6 | 12.0 ± 0.7 |
| Csn3–Csn8 | nd | nd | nd | 1460.6 ± 156.8 |
| Csn4–Csn7i | 240.8 ± 19.4 | 396.6 ± 19.4 | 369.2 ± 19.5 | 243.0 ± 100.2 |
| Csn4–Csn7ii | 132.3 ± 42.1 | 296.4 ± 78.5 | 435.7 ± 32.3 | 368.7 ± 166.6 |
| Csn5A–Csn6A | 711.4 ± 44.1 | 732.0 ± 8.6 | 234.4 ± 33.8 | 341.4 ± 42.2 |
| Csn5A–Csn6B | 1089.0 ± 30.9 | 873.9 ± 21.5 | 628.6 ± 102.4 | 844.9 ± 108.7 |
| Csn5B–Csn6A | 1019.6 ± 40.7 | 1605.7 ± 58.1 | 506.8 ± 35.8 | 844.6 ± 72.0 |
| Csn5B–Csn6B | 524.6 ± 5.9 | 1692.3 ± 32.0 | 845.3 ± 67.4 | 939.3 ± 53.9 |
| Csn5A–Csn8 | 227.0 ± 28.4 | 104.0 ± 8.3 | 95.7 ± 8.6 | 165.0 ± 49.5 |
| Csn5B–Csn8 | 47.2 ± 2.9 | 47.6 ± 3.4 | 77.1 ± 2.2 | 130.0 ± 42.9 |
| Csn6A–Csn7i | 86.5 ± 8.2 | 237.42 ± 22.1 | 435.1 ± 45.0 | 420.0 ± 68.8 |
| Csn6A–Csn7ii | 80.1 ± 5.9 | 434.2 ± 54.9 | 476.9 ± 39.2 | 1116.2 ± 152.5 |
| Csn6B–Csn7i | 119.0 ± 12.2 | 115.6 ± 7.5 | 769.3 ± 64.4 | 762.8 ± 89.1 |
| Csn6B–Csn7ii | 45.4 ± 4.7 | 202.2 ± 21.3 | 899.2 ± 84.1 | 886.5 ± 309.4 |
| Csn7i–Csn8 | 50.6 ± 3.4 | 57.8 ± 13.4 | 99.6 ± 3.9 | 370.4 ± 242.9 |
| Csn7ii–Csn8 | 58.6 ± 5.9 | 69.7 ± 22.3 | 225.6 ± 12.2 | 161.0 ± 60.5 |
aLacZ activity is expressed in nmol o-nitrophenol/h/mg protein; ± indicates standard error.
bIn the order of Csn1–8, each pair of 11 × 11 Csn subunits (except 11 self-interacting pairs) were constructed separately with the BD or AD domain, respectively, in either an ascending (configuration I) or descending (configuration II) manner (see text).
cCsn subunits are fused to either the C- or N-terminal end of the GAL4 AD domain.
dnd, not determined.
eHIS-positive YRG2 cells expressing a BD fusion of Csn5A or Csn5B were assayed for LacZ activity.
fCsn5B/Csn7i and Csn5B/Csn7ii are representatives having background LacZ activities.
When assayed by the HIS3 reporter, 27 CSN pairs showed an interaction with at least one configuration (Figure 3). In general, the two homologs of Csn5 and Csn6 and the two isoforms of Csn7 gave similar results. Compilation of the data identified 13 interacting pairs: Csn1/7, Csn3/4, Csn3/5, Csn3/7, Csn3/8, Csn4/5, Csn4/7, Csn5/6, Csn5/7, Csn5/8, Csn6/6, Csn6/7 and Csn7/8. Interactions involving Csn5/6, Csn5/7 and Csn5/8 could only be confirmed in configuration II due to self-activation of the BD:Csn5A and BD:Csn5B fusions (Figure 3). The subunit pairs, Csn1/7, Csn3/4, Csn3/5, Csn3/7, Csn4/5, Csn5/7 and Csn6/6 showed a modest interaction by the HIS3 reporter but failed by the LacZ reporter, suggesting weak associations. In contrast, the collective data for the Csn3/8, Csn4/7, Csn5/6, Csn5/8, Csn6/7 and Csn7/8 pairs implied strong binding. Interactions were identified in all or most of the four potential configurations with the HIS3 reporter and elicited significant LacZ activities with at least one configuration and more often with 2–4 configurations (Table III). Remarkably, most of these Csn partners (Csn3/8, Csn4/5, Csn4/7, Csn5/6, Csn5/7 and Csn6/7) were synonymous to interacting pairs identified within the RP lid (Rpn3/12, Rpn5/11, Rpn5/9, Rpn11/8, Rpn11/9 and Rpn8/9, respectively).
Asp11 of Rpn10 is critical for amino acid analog resistance
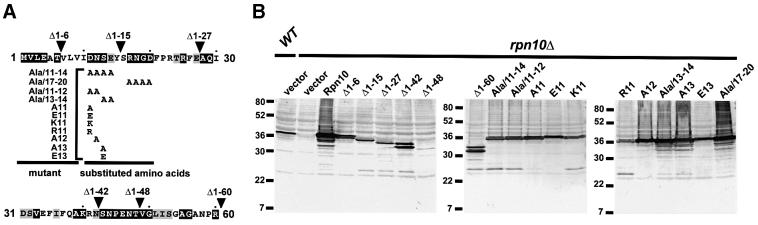
Mutagenic studies indicated that the N-terminal 60 residues of Rpn10 contain a domain important for conferring amino acid analog resistance, Ub-Pro-βgal degradation and stable association of the RP lid with the base (Fu et al., 1998; Glickman et al., 1998). To define further the essential N-terminal residues, a series of deletion and substitution mutants was examined (Figure 4A). Using growth sensitivity to the arginine and phenylalanine analogs, canavanine (CAN) and p-fluorophenylalanine (PFP), respectively, deletion analysis identified residues 7–15 as being important for analog resistance (Figure 5). Similarly to previous studies (Fu et al., 1998), the growth of the yeast rpn10Δ strain is highly sensitive to the addition of CAN/PFP to the medium, which can be rescued by reintroducing wild-type Rpn10 (Figure 5). Whereas, Rpn10 deletion Δ1–6 also restored growth on CAN/PFP medium, larger N-terminal deletions (Δ1–15, Δ1–27, Δ1–42, Δ1–48 and Δ1–60) failed even though high levels of the truncated proteins were expressed (Figures 4B and 5).
Fig. 4. Yeast rpn10Δ expressing various N-terminal mutants of Rpn10. (A) A schematic of the N-terminal 60 amino acids of Rpn10 showing the various N-terminal deletion and substitution mutants. Black and gray boxes denote residues that are identical among Rpn10 proteins from yeast, human, Drosophila, Arabidopsis and Physcomitrella patens or identical in two or more species, respectively. Arrowheads indicate the beginning of each deletion mutant. The substitution mutants are shown below with the substitutions indicated under the corresponding wild-type residues. (B) Expression of various N-terminal mutations of Rpn10 in yeast. Crude extracts (15 µg) from wild-type (WT), rpn10Δ (vector) and rpn10Δ strains expressing various Rpn10 mutations were subjected to SDS–PAGE and immunoblotted with Arabidopsis Rpn10 antibodies.

Fig. 5. Growth sensitivity to amino acid analogs of yeast rpn10Δ strains expressing various Rpn10 N-terminal mutants. The Rpn10 mutants are shown in Figure 4. Wild-type (WT, vector), rpn10Δ (rpn10Δ, vector) and rpn10Δ strains expressing various N-terminal mutants were grown to the same cell density (A600 = 1.0), and spotted as 10-fold serial dilutions (left to right) onto SC media containing the amino acid analogs. Colony growth was observed after incubation at 30°C for 6 days.
To locate the critical amino acids, a series of substitution mutants was then tested with an emphasis on the Asp-Asn-Ser-Glu sequence at positions 11–14 that is highly conserved among Rpn10 orthologs (Figure 4A). Replacing these four residues with alanines (Ala/11–14) abrogated analog resistance conferred by Rpn10, indicating that one or more of these four were important (Figure 5). In contrast, a similar alanine scan of conserved residues at positions 17–20 generated an Rpn10 protein with near wild-type activity. Additional substitution mutants pinpointed Asp11 as the critical residue. Any individual alanine mutant that altered this residue failed to rescue analog resistance even though high levels of the mutant Rpn10 proteins were present (Figures 4B and 5). Its acidic nature appeared important as the conservative substitution of Asp11 for glutamate generated an active protein whereas substitutions of Asp11 with lysine or arginine did not (Figures 4B and 5).
Asp11 of Rpn10 is essential for Ub-Pro-βgal degradation and RP stability
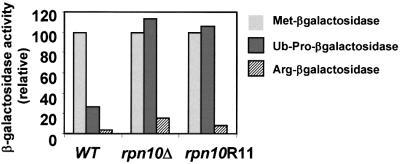
Whereas the half-life of many short-lived proteins is unaffected by Rpn10 N-terminal deletions, the UFD pathway substrate, Ub-Pro-βgal, is stabilized remarkably (van Nocker et al., 1996; Fu et al., 1998). To test whether Asp11 is critical for selective breakdown of Ub-Pro-βgal, we determined the steady-state levels of βgal in the rpn10Δ strains as an indirect measure of half-life (Figure 6). Whereas, Met-βgal is a stable protein, the N-end rule substrate Arg-β-gal and the UFD substrate Ub-Pro-βgal are short lived in wild-type yeast, with half-lives of ∼2 and ∼6 min, respectively (Johnson et al., 1992). As a consequence, rpn10Δ expressing wild-type Rpn10 accumulated high levels of Met-βgal but only low levels of Arg-βgal and Ub-Pro-βgal (Figure 6). In rpn10Δ strains, the steady-state level of Ub-Pro-βgal but not Arg-βgal increased to that matching Met-βgal, indicating that the Ub-Pro-βgal protein was stabilized selectively. A similar selective increase was observed for the rpn10Δ strain expressing the rpn10R11 protein. Thus, Asp11 in Rpn10 is essential for the degradation of UFD pathway substrates, but not N-end rule substrates.
Fig. 6. Steady-state levels of β-galactosidase derivatives in yeast rpn10Δ strains expressing wild-type Rpn10 (WT) or the Asp11 to arginine mutant rpn10R11. Steady-state levels of Met-βgal (light gray), Ub-Pro-βgal (dark gray) or Arg-βgal (striped) were quantified enzymatically. As shown here, elevated levels of β-galactosidase activity indicate that Ub-Pro-βgal is stabilized specifically in rpn10Δ and rpn10R11 mutants.
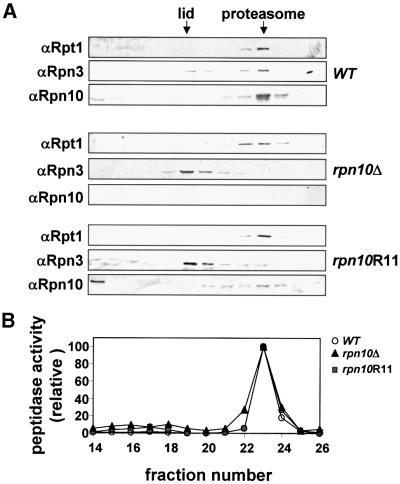
Next, we analyzed the stability of the 26S proteasome isolated from rpn10Δ or rpn10Δ expressing wild-type Rpn10 or rpn10R11. As can be seen in Figure 7, peptidase activity, Rpt1 and Rpn3 (indicators of CP, base and lid subcomplexes, respectively) co-eluted when 26S proteasome preparations from wild-type yeast were analyzed by anion exchange chromatography. Only a small amount of lid (as detected by the presence of Rpn3) eluted earlier in the NaCl gradient, indicating that most of the 26S proteasomes remained intact (Figure 7A, top panel). A substantial percentage of the lid dissociated from the 26S complex isolated from the rpn10Δ strain. This was observed by the majority of Rpn3 eluting much earlier than peptidase activity and Rpt1 (Figure 7A, middle panel). A similar effect was observed for preparations obtained from the rpn10Δ strain expressing rpn10R11, i.e. a substantial percentage of Rpn3 eluted earlier than the rest of the 26S proteasome (Figure 7A, lower panel). Thus, it appears that substitution of this single aspartate in Rpn10 was sufficient to weaken binding of the RP lid to the base. As discussed below, this negative charge appears to help stabilize an internal fold in Rpn10 as opposed to directly participating in lid–base contacts. Following separation of the lid and base, rpn10R11 appeared to remain associated with the base since the protein co-fractionated with the peptidase activity and Rpt1 (Figure 7A, lower panel).
Fig. 7. Integrity of 26S proteasomes from rpn10Δ strains expressing Rpn10 variants. Partially purified 26S proteasomes were subjected to Mono Q FPLC using an NaCl gradient. (A) Fractions analyzed by SDS–PAGE and immunoblot analysis with antibodies against Rpn10 and Rpt1 from the RP base and Rpn3 from the RP lid. (B) Fractions tested for the CP by a peptidase assay using the fluorogenic peptide suc-LLVY-AMC (circles, WT; triangle, rpn10Δ, squares, rpn10R11). Whereas the lid, base and CP co-eluted when extracted from WT (A; top panel), the lid eluted at lower salt concentrations than the proteasome complex when extracted from a rpn10Δ strain or a rpn10R11 mutant (A; middle and lower panels).
Discussion
By the extensive use of Y2H, we developed interaction maps for both the 19S RP of the yeast proteasome and the Arabidopsis CSN, revealing a remarkably similar core structure. Collectively, our results confirmed a number of interactions identified by other methods (Table IV) and detected several new interactions to yield a more complete topology for the 26S proteasome and CSN (Figure 8). However, in some cases, we failed to detect interactions described previously by other methods, suggesting that only through a combination of approaches can the complete organization of the RP and CSN be elucidated.
Table IV. Subunit interactions within the 26S proteasome and COP9 signalosome.
| Paira | Speciesb | Domainc | Methodsd | Referencese |
|---|---|---|---|---|
| Within RP base | ||||
| t1–t2 | Hs, Sc | t2: cc | P: fb, ss, cc, 2h | 1, 2, 3 |
| t1–t3 | Hs | P: cc, 2h | 1, 4 | |
| t2–t6 | Hs | P: cc | 1 | |
| t3–t4 | Sc | P: 2h | 3 | |
| t3–t5 | Hs, Sc | P: cc, 2h | 1, 3, 4, 5, this work | |
| t3–t6 | Hs, Sc | t6: cc*, K195* | P: fb, ss, 2h | 2, 3, 4, this work |
| t4–t5 | Hs, Sc, Ce | t4: cc* | P: fb, ss, cc, 2h | 1, 2, 5, 6, this work |
| t4–t6 | Hs, Sc | P/G: cc, sl, 2h | 1, 4 | |
| t1–n1 | Hs | t1: Ct | P: fb | 7 |
| t2–n1 | Hs, Sp, Ce | t2: Nt | P/G: fb, sl, pd, 2h | 4, 6, 7 |
| t4–n2 | Hs | P: fb, ss | 2 | |
| t6–n2 | Hs | P: cc | 1 | |
| n1–n2 | Hs | P: fb | 7 | |
| n1–n10 | Sp | G: sl | 8 | |
| Within RP lid | ||||
| n3–n7 | Sc, Ce | n3: PCI* | P: 2h | 5, 6, this work |
| n3–n12 | Sc | P/G: su, 2h | 4, this work | |
| n5–n6 | Sc | P: 2h | this work | |
| n5–n8 | Sc | P: 2h | this work | |
| n5–n9 | Sc | n5/n9: PCI* | P: 2h | this work |
| n5–n11 | Sc | P: 2h | this work | |
| n8–n9 | Sc, Ce | P: 2h | 6, this work | |
| n8–n11 | Sc, Ce | n8/n11: MPN* | P: 2h | 3, 6, this work |
| n9–n11 | Sc, Ce | P: 2h | 6, this work | |
| n11–n12 | Sp | G: su, sl | 4 | |
| Between base and lid | ||||
| t1–n12 | Sc | G: sl | 4 | |
| t3–n10 | Ce | P: 2h | 6 | |
| n1–n11 | Sp | G: su | 4 | |
| n2–n12 | Sc | G: sl, su | 4 | |
| n9–n10 | Sc | P: 2h | 4 | |
| n10–n11 | Sp | G: sl | 8 | |
| n10–n12 | Sp, Sc | P/G: sl, su, pd | 4, 8 | |
| Between CP and RP | ||||
| α1–t6 | Sc | G: su | 1 | |
| α2–t6 | Ss | P: cc | 1 | |
| α4–t2 | Hs | P: 2h, pd | 1 | |
| α6–t4 | Hs | P: cc | 1 | |
| CSN | ||||
| N1–N2 | Sp, Hs | N1: PCI | P: 2h | 1, 9 |
| N1–N3 | Hs | N1/N3: PCI | P: fb, 2h | 1, 9 |
| N1–N4 | At, Hs | N1: PCI | P: 2h | 1 |
| N1–N5 | At, Hs | P: 2h, fb | 1, 9 | |
| N1–N7 | At | P: 2h | 1, this work | |
| N2–N3 | Hs | N3: PCI | P: fb | 9 |
| N2–N5 | Dm, Hs | P: 2h, fb | see 1, 9 | |
| N2–N6 | Hs | P: fb | 9 | |
| N2–N7 | Dm, Hs | N7: PCI | P: 2h, fb | see 1, 9 |
| N3–N4 | At | P: 2h | this work | |
| N3–N5 | At | P: 2h | this work | |
| N3–N7 | At | P: 2h | this work | |
| N3–N8 | At | P: 2h | this work | |
| N4–N5 | At | P: 2h | 1, this work | |
| N4–N7 | At, Dm | P: 2h | 1, this work | |
| N4–N8 | At | P: 2h | 1 | |
| N5–N6 | At | P: 2h | this work | |
| N5–N7 | Hs | P: fb | 9, this work | |
| N5–N8 | At | P: 2h | this work | |
| N6–N7 | Hs | P: fb | 9, this work | |
| N7–N8 | At, Hs | P: 2h, fb | 1, 9, this work |
aSingle lower case letters n and t are employed to indicate Rpn and Rpt subunits for the 26S proteasome, and upper case letter N to indicate CSN subunits. See also Figure 8.
bSpecies abbreviation: At, Arabidopsis thaliana; Ce, Caenorhabditis elegans, Dm, Drosophila melanogaster; Hs, Homo sapiens; Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Ss, Sus scrofa.
cDomains reported to be involved in interactions: cc, coiled-coil; Ct, C-terminal; Nt, N-terminal; PCI, proteasome, COP9 and eIF3; MPN, Mpr1 and Pad1 N-terminal; K195, invariant lysine of AAA-ATPase P-loop. Asterisks indicate domains for interactions that were identified from this study.
dSpecific methods used: P, protein–protein interaction; G, genetic evidence; 2h, yeast two-hybrid; cc, chemical cross-linking; fb, filter binding; pd, GST pull-down; sl, synthetic lethal; ss, co-sedimentation on sucrose gradient; su, suppressor.
eReferences: 1, Hartmann-Petersen et al. (2001); 2, Richmond et al. (1997); 3, Uetz et al. (2000); 4, Ferrell et al. (2000); 5, Ito et al. (2001); 6, Davy et al. (2001); 7, Gorbea et al. (2000); 8, Wilkinson et al. (2000); 9, Kapelari et al. (2000).
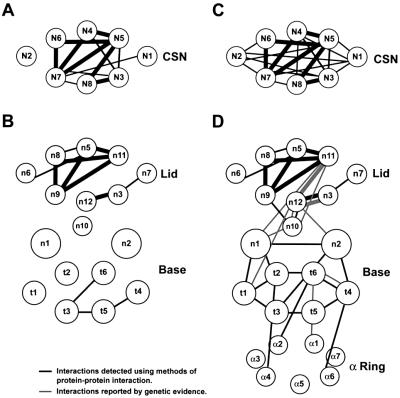
Fig. 8. Subunit interaction maps of the CSN complex (A and C) and the 26S proteasome (B and D). Synonymous lid and CSN subunits (see Table II) are plotted at identical positions. (A and B) Subunit interactions detected here by Y2H. (C and D) Subunits interactions identified here in combination with those determined previously (Table IV). Black lines denote interactions detected directly by protein–protein interacting assays. Gray lines indicate interactions implied from various genetic studies (Table IV). Bold lines indicate conserved interactions between synonymous pairs in the RP lid and CSN.
For the RP, we identified two interaction clusters. One involves four base subunits Rpt3, 4, 5 and 6 arranged in a Rpt4/5/3/6 configuration and the other involves the lid subunits Rpn5, 8, 9 and 11, which is potentially arranged in a ring held together by a lattice of contacts (Figure 8B). Surprisingly, Y2H analysis failed to detect binding between any of the lid and base subunits and between any of the base subunits and the α-subunits from the CP. While it remains possible that the AD–BD appendages block these interactions, a more likely scenario, based on the facile dissociation of the RP from the CP under various in vitro conditions, is that stable interactions between these subcomplexes require prior assembly.
The interactions observed here for the base Rpt subunits suggest a minimal tetrameric structure with a Rpt4/5/3/6 organization. Taken together with previously reported binding partners among Rpt subunits (Table IV), we propose that the Rpt subunits are organized in a hexameric Rpt1/2/6/4/5/3 ring (Figure 8D). Although we did not determine binding partners of Rpn1 and 2 by Y2H, far-western blotting and cross-linking suggest that Rpn1 binds to both Rpt1 and 2, and Rpn2 binds to Rpt4 and Rpt6 to form the RP base (Table IV; Figure 8D). Based on previous studies, the Rpt ring then interacts with the CP using contacts that include α1–Rpt6, α2–Rpt6, α4–Rpt2 and α6–Rpt4 pairs (see Ferrell et al., 2000; Hartmann-Petersen et al., 2001). Due to the symmetry mismatch (seven α-subunits versus six Rpt subunits), a one-to-one correspondence of Rpt to CP α-subunits is not possible and thus a loose interface between pre-assembled CP and RP may exist.
It has been suggested that the coiled-coils of the Rpt subunits interact either with proteolytic substrates or with each other to define their place within the RP (Glickman, 2000). Here, we found that these domains are essential for the interactions between the Rpt3 and 6 and between Rpt4 and 5. Both this study showing that the P-loop in Rpt6 is important for its interactions with Rpt3 and a previous one showing that the P-loop in Rpt2 is essential for overall proteasome stability (Rubin et al., 1998) suggest that the ATPase cassettes also confers critical structural information. Given that attachment of the RP to the CP is ATP-dependent (Voges et al., 1999), one possible role is to couple ATP hydrolysis to conformational changes in the Rpt subunits that favor RP–CP association or interactions among the Rpt subunits. The Rpt subunits presumably help open the α-subunit channel and translocate substrates into the CP. In a proposed hexameric ring, the Rpt3/6 interaction observed here and elsewhere (Table IV) would seem to span the pore and block this function. However, our observation that the ATPase domain of Rpt6 is involved in the Rpt3/6 interaction raises an intriguing possibility that gating is controlled by multiple Rpt subunits besides the regulation by Rpt2–α3 proposed previously (Glickman, 2000).
Among lid subunits, numerous interactions were evident by Y2H, especially involving the Rpn5/8/9/11 cluster. When combined with genetic evidence (Table IV), the lid appears to be held together by a complex lattice of interactions involving all Rpn subunits (Figure 8D). Our mutational analyses support the proposal that the PCI and MPN domains in the lid Rpn subunits are protein contact sites (Aravind and Ponting, 1998; Hofmann and Bucher, 1998) by showing that the Rpn5/9 and Rpn3/7 pairs require the PCI domains and the Rpn8/11 pair needs sequences surrounding the MPNs for binding. How the lid associates with the base remains unclear. There is ample evidence for genetic interactions between lid and base subunits, including Rpt1/Rpn12, Rpn1/11, Rpn2/12, Rpn10/11 and Rpn10/12 (Table IV). However, reports on direct protein–protein interaction are limited to Rpn10/12 and Rpn9/10 (Table IV). Certainly, Rpn10 plays a role, as the lid is released more easily from purified 26S proteasome when this subunit is absent (Glickman et al., 1998; this study). That Rpn10 helps tether the lid to the base is also supported by the observations that in some instances, Rpn10 co-fractionated with the base (Glickman et al., 1998; this study), and in other cases it co-fractionated with the lid (Saeki et al., 2000). Thus, in addition to its potential role as a ubiquitin receptor, Rpn10 has another role in maintaining a stable lid–base contact. However, its role is not essential as rpn10 knockout mutants in S.cerevisiae, Schizosaccharomyces pombe, Physcometrilla patens and Arabidopsis are not only viable but also capable of assembling functional 26S proteasomes (van Nocker et al., 1996; Girod et al., 1999; Wilkinson et al., 2000; J.Smalle and R.D.Vierstra, unpublished).
For the CSN, an interaction cluster similar to that in the RP lid was detected. Strong interactions were identified involving Csn3, 8, 4, 5, 6 and 7, with the latter four potentially organized in a very similar fashion to their synonymous lid Rpn subunits (Figure 8A and B). Like the lid, the PCI and MPN domains in the CSN probably promote complex assembly. In fact, recent work of Tsuge et al. (2001) showed that the PCI domain of Csn1 is required for its interactions with Csn2, 3 and 4. This conservation further supports the sequence comparisons showing that the RP lid and the CSN share a common evolutionary ancestry, a similar structural scaffold and possibly overlapping functions. This conserved internal structure is striking when compared with electron microscopic analysis showing that the surface topologies of the RP and CSN complexes are substantially different (Kapelari et al., 2000). This divergence may reflect the distinct biological functions of the respective particles: the RP involved in recruiting proteins for degradation by the CP (Voges et al., 1999) and the CSN involved in protein phosphorylation and protein modification by Nedd8/Rub1 (Bech-Otschir et al., 2001; Lyapina et al., 2001).
Whereas the polyubiquitin receptor function of Rpn10 has been localized to a ubiquitin interacting motif (UIM) at its C-terminus (Fu et al., 1998; Hofmann and Falquet, 2001), stable lid–base contacts require the N-terminal portion of Rpn10. Structural predictions suggest that the N-terminal 150 residues of Rpn10 contain a motif related to the von Willebrand factor A domain (vWA), a protein contact motif found multiple times in the von Willebrand factor (vWF) (Hofmann and Falquet, 2001). In vWF, all vWA domains contain a conserved aspartate in a DXS motif that appears by X-ray structural analysis to be buried within the protein and required to maintain the vWA fold by electrostatic interactions (Matsushita and Sadler, 1995; Emsley et al., 1998). Likewise, the Asp11 critical for Rpn10 function is also in a DXS motif similarly positioned in a vWA-like domain (Figure 4A). That glutamate but not alanine, lysine or arginine can substitute for Asp11 supports its role in forming a salt bridge. Taken together, a likely possibility is that the salt bridge formed by Asp11 helps to maintain the structural integrity of the vWA in Rpn10; the vWA in turn helps promote association of the lid with the base. In a remarkable resemblance to loosening of lid–base association upon substitution of Asp11 in Rpn10, a single alanine substitution of Asp520 in the vWF-A1 DXS motif attenuates binding of vWF to its partner glycoprotein Ib, probably by relaxation of the vWA fold (Matsushita and Sadler, 1995; Emsley et al., 1998). Given that rpn10R11 protein appears to maintain its association with the base upon separation of the base from the lid, it is possible that a properly folded vWA is responsible for direct interactions of Rpn10 with the lid.
Asp11 mutations of Rpn10 also enhance the sensitivity of yeast to amino acid analogs and selectively stabilize the UFD substrate Ub-Pro-βgal, suggesting that these other two rpn10 phenotypes are caused by loss of appropriate lid–base contacts. This destabilization could attenuate the capacity of the 26S proteasome to degrade certain types of substrates and the turnover of abnormal polypeptides caused by analog incorporation. Furthermore, since Rpn10 is the only yeast proteasome subunit found in significant amounts as a free form not associated with the proteasome (van Nocker et al., 1996), one could easily imagine Rpn10 binding reversibly to the 26S proteasome in a way that increases the efficiency of the protease by stabilizing the lid–base contacts. In support of this, one intriguing property of Rpn10 is that rpn10Δ yeast strains become more sensitive to amino acid analogs when complemented with Rpn10 mutant proteins affected at Asp11 (Figure 5; Fu et al., 1998). This effect suggests that the Asp11 mutant proteins are not neutral but in fact can be detrimental to lid–base contacts within the RP, and thus capable of generating an even more inefficient protease.
Materials and methods
Yeast strains, media and Y2H techniques
The haploid yeast rpn10Δ strain (MATa) derived from DF5 was described previously (van Nocker et al., 1996). Yeast transformation and yeast rich (YPD) and synthetic complete (SC) media were as described (Fu et al., 1998). For the amino acid analog sensitivity, arginine and phenylalanine were omitted from SC medium and their corresponding analogs, CAN and PFP, were added to 1.5 and 25 µg/ml, respectively. The Y2H analysis was performed according to the manufacturer’s recommendations (Stratagene).
Isolation of genes encoding 26S proteasome and CSN subunits
Full-length coding sequences for the RP and the CP α-subunits were amplified by PCR using PfuTurbo™ (Stratagene) from S.cerevisiae DF5 (MATa) genomic DNA. By using human CSN subunits as queries, Arabidopsis CSN genes previously unreported were identified from the Arabidopsis Genome Initiative database (http://www.arabidopsis.org/home.html; see Table II). The intron/exon organization was annotated by comparison with corresponding full-length CSN cDNAs. The full-length cDNA for CSN3 was an EST (161P1) from the Arabidopsis Biological Resource Center (ABRC, Ohio State University). The remaining full-length CSN cDNAs were isolated by PCR from the cDNA libraries CD4-13 and CD4-14 from ABRC. All Arabidopsis CSN cDNAs have been deposited in the DDBJ/EMBL/GenBank database under accession Nos AF395057–AF395067.
Construction of GAL4 BD and GAL4 AD fusions
For each of the 26S proteasome and CSN subunits, the 5′ and 3′ amplification primers were designed to add appropriate restriction sites to the ends to facilitate subsequent cloning in the Y2H vectors. For insertion as N-terminal fusions to GAL4 AD, the 3′ primers were also designed to substitute restriction sites for the stop codon to allow the synthesis of the protein as an in-frame fusion. For C-terminal GAL4 AD and BD fusion constructions, the Y2H vectors, pAD-GAL4-2.1 and pBD-GAL4 Cam (Stratagene), were used, respectively. For N-terminal GAL4 AD fusion constructs, the Y2H vector pADCT was constructed from pAD-GAL4-2.1 (Stratagene) to allow expression of proteins as N-terminal fusions with the GAL4 AD. In this vector, a GAL4 AD cassette (ADCT) was inserted between the KpnI and PstI sites of pAD-GAL4-2.1. It contained (from 5′ to 3′), a 398 bp fragment of the alcohol dehydrogenase promoter, a region containing the unique restriction sites NdeI, EcoRI and SalI (NES), and the GAL4 AD coding sequence (AD), encompassing codons 768–881 followed by an engineered stop codon. Structural domains were identified using analysis programs from ExPASy Molecular Biology Server (Swiss Institute of Bioinformatics, http://www.expasy.ch/). Deletion and substitution mutants were generated using PCR strategies. The sequences of all constructions were verified as correct by DNA sequence analysis. Sequences of the PCR primers and organization of the final GAL4 BD–AD fusion vectors will be made available upon request (H.Fu, unpublished).
Mutational analysis of Rpn10
Deletion mutants of Rpn10 were constructed by PCR amplification from the wild-type gene with appropriate primers designed to add an NdeI site at the deletion point and an EcoRI site at the 3′ end. The constructions were moved into a 2µ plasmid, pRS424-RPN10ΔP, which was modified from pRS424 to include a 552 bp RPN10 promoter fragment immediately followed by NdeI and EcoRI sites (Fu et al., 1998).
Preparation of yeast crude extracts, SDS–PAGE and immunoblot analyses were described previously (Fu et al., 1998). Steady-state levels of the Met-, Arg- and Ub-Pro-βgal fusion proteins were measured by testing LacZ activity according to Johnson et al. (1995). 26S proteasomes were purified by the method of Glickman et al. (1998). Fractions were assayed for peptidase activity and for the Rpt1, Rpn10 and Rpn3 subunits by immunoblot analysis. Peptidase activity of the CP was determined according to Glickman et al. (1998) using Suc-LLVY-AMC as a substrate.
Acknowledgments
Acknowledgements
We thank Dr Kay Hofmann for helpful analysis, Dr Charles Papa for reading the manuscript, Hung-Cheng Hsieh, Clarissa Hew and Tzuning Ho for technical assistance, and Drs Carl Mann and Aki Toh-e for supplying the Rpt1 and Rpn3 antibodies. This work was supported by grants from the NSC (892311B001125 and 902311B001067), Academia Sinica, Taiwan and the Li Foundation (USA) to H.F.; the Israel Ministry of Science, the Israel Science Foundation, the ICRF, the Foundation for Promotion of Research at the Technion and the Vice President of the Technion Research Fund to M.G.; and the USDA-NRICGP (00-3501-9040) to R.D.V.
References
- Aravind L. and Ponting,C.P. (1998) Homologues of 26S proteasome subunits are regulators of transcription and translation. Protein Sci., 7, 1250–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailly E. and Reed,S.I. (1999) Functional characterization of rpn3 uncovers a distinct 19S proteasomal subunit requirement for ubiquitin-dependent proteolysis of cell cycle regulatory proteins in budding yeast. Mol. Cell. Biol., 19, 6872–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech-Otschir D., Kraft,R., Huang,X., Henklein,P., Kapelari,B., Pollmann,C. and Dubiel,W. (2001) COP9 signalosome-specific phosphorylation targets p53 to degradation by the ubiquitin system. EMBO J., 20, 1630–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davy A. et al. (2001) A protein–protein interaction map of the Caenorhabditis elegans 26S proteasome. EMBO Rep., 2, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X.W. et al. (2000) Unified nomenclature for the COP9 signalosome and its subunits: an essential regulator of development. Trends Genet., 16, 202–203. [DOI] [PubMed] [Google Scholar]
- Emsley J., Cruz,M., Handin,R. and Liddington,R. (1998) Crystal structure of the von Willebrand factor A1 domain and implications for the binding of platelet glycoprotein Ib. J. Biol. Chem., 273, 10396–10401. [DOI] [PubMed] [Google Scholar]
- Ferrell K., Wilkinson,C.R.M., Dubiel,W. and Gordon,C. (2000) Regulatory subunit interactions of the 26S proteasome, a complex problem. Trends Biochem. Sci., 25, 83–88. [DOI] [PubMed] [Google Scholar]
- Fu H., Sadis,S., Rubin,D.M., Glickman,M., van Nocker,S., Finley,D. and Vierstra,R.D. (1998) Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26S proteasome subunit Mcb1. J. Biol. Chem., 273, 1970–1989. [DOI] [PubMed] [Google Scholar]
- Girod P.A., Fu,H., Zryd,J.P. and Vierstra,R.D. (1999) Multiubiquitin chain binding subunit MCB1 (RPN10) of the 26S proteasome is essential for developmental progression in Physcomitrella patens. Plant Cell, 11, 1457–1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M.H. (2000) Getting in and out of the proteasome. Semin. Cell Dev. Biol., 11, 149–158. [DOI] [PubMed] [Google Scholar]
- Glickman M.H., Rubin,D.M., Coux,O., Wefes,I., Pfeifer,G., Cjeka,Z., Baumeister,W., Fried,V.A. and Finley,D. (1998) A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9/signalosome and eIF3. Cell, 94, 615–623. [DOI] [PubMed] [Google Scholar]
- Gorbea C., Taillandier,D. and Rechsteiner,M. (2000) Mapping subunit contacts in the regulatory complex of the 26S proteasome. S2 and S5b form a tetramer with ATPase subunits S4 and S7. J. Biol. Chem., 275, 875–882. [DOI] [PubMed] [Google Scholar]
- Hartmann-Petersen R., Tanaka,K. and Hendil,K.B. (2001) Quaternary structure of the ATPase complex of human 26S proteasomes determined by chemical cross-linking. Arch. Biochem. Biophys., 386, 89–94. [DOI] [PubMed] [Google Scholar]
- Hofmann K. and Bucher,P. (1998) The PCI domain: a common theme in three multi-protein complexes. Trends Biochem. Sci., 23, 204–205. [DOI] [PubMed] [Google Scholar]
- Hofmann K. and Falquet,L. (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal degradation systems. Trends Biochem. Sci., 26, 347–350. [DOI] [PubMed] [Google Scholar]
- Ito T., Chiba,T., Ozawa,R., Yoshida,M., Hattori,M. and Sakaki,Y. (2001) A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl Acad. Sci. USA, 98, 4569–4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., Bartel,B., Seufert,W. and Varshavsky,A. (1992) Ubiquitin as a degradation signal. EMBO J., 11, 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson E.S., Ma,P.C.M., Ota,I.M. and Varshavsky,A. (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J. Biol. Chem., 270, 17442–17456. [DOI] [PubMed] [Google Scholar]
- Kapelari B., Bech-Otschir,D., Hegerl,R., Schade,R., Dumdey,R. and Dubiel,W. (2000) Electron microscopy and subunit–subunit interaction studies reveal a first architecture of COP9 signalosome. J. Mol. Biol., 300, 1169–1178. [DOI] [PubMed] [Google Scholar]
- Karniol B., Malec,P. and Chamovitz,D.A. (1999) Arabidopsis FUSCA5 encodes a novel phosphoprotein that is a component of the COP9 complex. Plant Cell, 11, 839–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.H., Hofmann,K., von Armin,A.G. and Chamovitz,D.A. (2001) PCI complexes: pretty complex interactions in diverse signaling pathways. Trends Plant Sci., 6, 379–386. [DOI] [PubMed] [Google Scholar]
- Lyapina S. et al. (2001) Promotion of NEDD–CUL1 conjugate cleavage by COP9 signalosome. Science, 292, 1382–1385. [DOI] [PubMed] [Google Scholar]
- Matsushita T. and Sadler,J.E. (1995) Identification of amino acid residues essential for von Willebrand factor binding to platelet glycoprotein Ib. Charged-to-alanine scanning mutagenesis of the A1 domain of human von Willebrand factor. J. Biol. Chem., 270, 13406–13414. [DOI] [PubMed] [Google Scholar]
- Peng Z., Staub,J.M., Serino,G., Kwok,S.F., Kurepa,J., Bruce,B.D., Vierstra,R.D., Wei,N. and Deng,X.W. (2001) The cellular level of PR500, a protein complex related to the 19S regulatory particle of the proteasome, is regulated in response to stresses in plants. Mol. Biol. Cell., 12, 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phizicky E.M. and Fields,S. (1995) Protein–protein interactions: methods for detection and analysis. Microbiol. Rev., 59, 94–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond C., Gorbea,C. and Rechsteiner,M. (1997) Specific interactions between ATPase subunits of the 26S proteasome. J. Biol. Chem., 272, 13403–13411. [DOI] [PubMed] [Google Scholar]
- Rubin D.M., Glickman,M.H., Larsen,C.N., Dhruvakumar,S. and Finley,D. (1998) Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J., 17, 4909–4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki Y., Toh-e,A. and Yokosawa,H. (2000) Rapid isolation and characterization of the yeast proteasome regulatory complex. Biochem. Biophys. Res. Commun., 273, 509–515. [DOI] [PubMed] [Google Scholar]
- Strickland E., Hakala,K., Thomas,P.J. and DeMartino,G.N. (2000) Recognition of misfolding proteins by PA700, the regulatory subcomplex of the 26S proteasome. J. Biol. Chem., 275, 5565–5572. [DOI] [PubMed] [Google Scholar]
- Tsuge T., Matsui,M. and Wei,N. (2001) The subunit of the COP9 signalsome suppresses gene expression through its N-terminal domain and incorporates into the complex through the PCI domain. J. Mol. Biol., 305, 1–9. [DOI] [PubMed] [Google Scholar]
- Uetz P. et al. (2000) A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature, 403, 623–627. [DOI] [PubMed] [Google Scholar]
- van Nocker S., Sadis,S., Rubin,D.M., Glickman,M., Fu,H., Coux,O., Wefes,I., Finley,D. and Vierstra,R.D. (1996) The multiubiquitin chain binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substrate-specific role in protein turnover. Mol. Cell. Biol., 16, 6020–6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voges D., Zwickl,P. and Baumeister,W. (1999) The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem., 68, 1015–1068. [DOI] [PubMed] [Google Scholar]
- Walz J., Erdmann,A., Kania,M., Typke,D., Koster,A.J. and Baumeister,W. (1998) 26S proteasome structure revealed by three-dimensional electron microscopy. J. Struct. Biol., 121, 19–29. [DOI] [PubMed] [Google Scholar]
- Wilkinson C.R.M., Ferrell,K., Penny,M., Wallace,M., Dubiel,W. and Gordon,C. (2000) Analysis of a gene encoding Rpn10 of the fission yeast proteasome reveals that the polyubiquitin-binding site of this subunit is essential when Rpn12/Mts3 activity is compromised. J. Biol. Chem., 275, 15182–15192. [DOI] [PubMed] [Google Scholar]