Abstract
In cells with reduced mitochondrial function, RTG1, 2 and 3 are required for expression of genes involved in glutamate synthesis. Glutamate negatively regulates RTG-dependent gene expression upstream of Rtg2p, which, in turn, acts upstream of the bHLH/Zip transcription factors, Rtg1p and Rtg3p. Here we report that some mutations [lst8-(2–5)] in LST8, an essential gene encoding a seven WD40-repeat protein required for targeting of amino acid permeases (AAPs) to the plasma membrane, bypass the requirement for Rtg2p and abolish glutamate repression of RTG-dependent gene expression. The lst8-1 mutation, however, which reduces plasma membrane expression of AAP, cannot bypass the Rtg2p requirement, but still suppresses glutamate repression of RTG target gene expression. We show that Lst8p negatively regulates RTG gene function, acting at two sites, one upstream of Rtg2p, affecting glutamate repression of RTG-dependent gene expression through Ssy1p, an AAP-like sensor of external amino acids, and the other between Rtg2p and Rtg1p–Rtg3p. These data, together with genome-wide transcription profiling, reveal pathways regulated by glutamate, and provide insight into the regulation of cellular responses to mitochondrial dysfunction.
Keywords: LST8/mitochondria/RTG genes/WD40 repeat/yeast
Introduction
In the budding yeast, Saccharomyces cerevisiae, mitochondrial dysfunction, such as respiratory deficiency or blocks in the tricarboxylic acid (TCA) cycle, results in increased expression of a subset of nuclear genes involved in metabolism, small molecule transport pathways, peroxisomal biogenesis and stress responses (Liao et al., 1991; Chelstowska and Butow, 1995; Traven et al., 2001; Epstein et al., 2001b). This pathway of inter-organelle communication, called retrograde regulation, accommodates cells to their mitochondrial defect. For example, many of the reactions catalyzed by target genes in the retrograde pathway serve, directly or indirectly, to maintain the supply of key TCA cycle intermediates, e.g. oxaloacetate, that would otherwise become severely limiting in cells with mitochondrial deficiencies. The expression of some of the genes in the retrograde pathway, such as CIT2 (encoding a glyoxylate cycle isoform of citrate synthase), DLD3 (encoding a d-lactate dehydrogenase) and PDH1 (a gene involved in propionate metabolism) is directly dependent on three regulatory genes, RTG1, RTG2 and RTG3 (Liao and Butow, 1993; Jia et al., 1997; Chelstowska et al., 1999).
In cells that are respiratory deficient or have reduced respiratory activity, the RTG genes also regulate the expression of CIT1, ACO1, IDH1 and IDH2 (Liu and Butow, 1999), which encode enzymes catalyzing the first three steps of the TCA cycle that lead to the synthesis of α-ketoglutarate, the precursor of glutamate. In cells with robust mitochondrial respiratory activity, expression of those TCA cycle genes is under the control of the HAP transcription complex (Forsburg and Guarente, 1989; Gangloff et al., 1990; Rosenkrantz et al., 1994). Glutamate homeostasis, one of the central functions of RTG-dependent gene expression, exemplifies how the retrograde pathway serves to adapt cells to mitochondrial dysfunction. The levels of glutamate regulate the retrograde response via a negative feedback loop, whereby low glutamate levels activate and high glutamate levels repress RTG-dependent gene expression (Liu and Butow, 1999). Indeed, in cells with compromised mitochondrial function, mutations in any one of the RTG genes result in glutamate auxotrophy (Liao and Butow, 1993; Jia et al., 1997).
Although the exact mechanism of glutamate signaling is unclear, in respiratory-competent cells or in cells grown in the presence of high levels of glutamate in the medium, the basic helix–loop–helix/leucine zipper (bHLH/Zip) transcription factors, Rtg1p and Rtg3p, are present as an inactive complex in the cytoplasm (Sekito et al., 2000). In respiratory-deficient cells or in cells grown in medium lacking glutamate, these factors accumulate in the nucleus by processes requiring Rtg2p, a novel cytoplasmic protein with an N-terminal ATP-binding domain similar to the hsp70/actin/sugar kinase superfamily of ATP-binding proteins (Bork et al., 1992). Rtg2p thus acts as a proximal sensor of mitochondrial dysfunction, perhaps via glutamate levels, to regulate the subcellular localization of Rtg1p and Rtg3p (Sekito et al., 2000).
Recent studies have shown that the RTG pathway also responds to the quality of the nitrogen source in the medium and to the target of rapamycin (TOR) kinase pathway (Komeili et al., 2000; Shamji et al., 2000). In particular, when cells are grown in a poor nitrogen source, or when the TOR kinase pathway is inhibited by rapamycin, Rtg1p and Rtg3p translocate from the cytoplasm to the nucleus in an Rtg2p-dependent manner to activate target gene expression, similarly to the Rtg2p-dependent nuclear accumulation of Rtg1p and Rtg3p in cells with dysfunctional mitochondria (Sekito et al., 2000). Together, these studies show that pathways of carbohydrate and nitrogen metabolism are linked together via RTG-dependent gene expression.
To identify additional regulatory genes in the RTG pathway, we searched for mutations that could bypass the requirement of RTG2 for CIT2 expression. This search led to the identification of LST8, an essential gene encoding a seven WD40-repeat protein that has been implicated in amino acid permease transport from the Golgi to the cell surface (Roberg et al., 1997a). We show that some, but not all, mutations in LST8 can bypass the requirement of Rtg2p for Rtg1p/Rtg3p-dependent gene expression. We conclude that Lst8p functions as a negative regulator of RTG-dependent gene expression, acting at least at two sites, one upstream of Rtg2p at the level of external glutamate sensing and the other between Rtg2p and Rtg1p–Rtg3p.
Results
Isolation of RTG2 bypass mutants
To identify new genes in the retrograde pathway, we carried out a genetic screen to identify mutants that could bypass the requirement of Rtg2p for CIT2 expression. Using an rtg2Δ strain with a CIT2-lacZ reporter gene integrated at the URA3 locus, cells were mutagenized and screened for colonies that appeared blue on X-gal plates. This screen yielded 14 candidates that were selected as potential rtg2Δ bypass mutants, designated rtb (Rtg two bypass). Northern blot analysis revealed that all of the rtb isolates also had high levels of expression of the endogenous CIT2 mRNA (data not shown), indicating that the rtg2Δ bypass phenotype was not peculiar to the reporter gene. Standard genetic tests showed that the rtg2Δ bypass phenotype of each isolate was recessive and due to a single gene mutation. Further, the isolates could be placed into two complementation groups, rtb1 and rtb2. Five independent rtb2 mutants were obtained, and the properties of some of them are described here; details of the rtb1 complementation group will be described elsewhere.
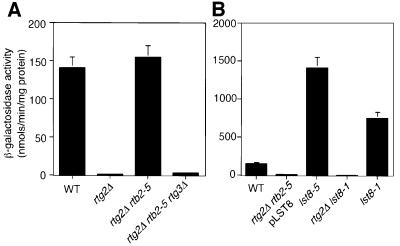
Preliminary characterization of one of the rtb2 isolates (rtb2-5) showed that the block in CIT2-lacZ expression due to the rtg2Δ mutation was completely reversed by the rtb2-5 mutation (Figure 1A). Similar results were obtained for the other rtb2 mutants. To exclude the possibility that the rtb2 mutations activated an alternate pathway for CIT2 expression, we introduced an rtg3 null mutation into rtg2Δ rtb2-5 mutant cells. CIT2-lacZ expression was abolished in the triple mutant (Figure 1A), suggesting that the rtg2Δ bypass does not involve the recruitment of alternative transcriptional activators.
Fig. 1. RTB2/LST8 is a negative regulator of CIT2 expression. (A) RTB2 regulates CIT2 expression upstream of RTG3, and downstream of RTG2. Wild-type (PSY142), rtg2Δ (PSY142-rtg2), rtg2Δ lst8-5 (RBY426) and rtg2Δ rtb2-5 rtg3Δ (RBY427) cells were grown in YPD medium to mid-log phase and collected for β-galactosidase activity analysis to determine transcriptional activation of an integrated CIT2-lacZ reporter gene. β-galactosidase assays on whole-cell extracts were carried out in triplicate as described in Materials and methods. (B) RTB2 is LST8. Wild-type PSY142 cells and rtg2Δ lst8-5 transformed with pRS416-LST8, lst8-5, rtg2Δ lst8-1 and lst8-1 derivatives were grown in YNBcasD medium plus uracil (omitted in cells transformed with pRS416-LST8) to mid-log phase, and CIT2-lacZ reporter gene expression was determined by β-galactosidase activities.
RTB2 is LST8
To identify the RTB2 gene, rtg2Δ rtb2-5 mutant cells were transformed with a centromere-based wild-type yeast genomic library and screened for transformants that restored CIT2-lacZ expression to the low level observed in otherwise wild-type rtg2Δ cells. Complementing plasmids were obtained, and subsequent subcloning yielded a single gene, LST8, which was necessary and sufficient to abolish reporter gene expression in each of the rtg2Δ rtb2 isolates, as is shown for rtg2Δ rtb2-5 cells (Figure 1B). To verify that RTB2 and LST8 are the same, we carried out complementation tests by examining CIT2-lacZ expression in diploids obtained from a cross between the rtg2Δ rtb2-5 strain and an rtg2Δ lst8Δ::LEU2 strain carrying a wild-type copy of LST8 on a centromeric plasmid. The resulting diploids were cured of the plasmid and tested for CIT2-lacZ expression. We found that CIT2-lacZ expression was high in such diploids (data not shown), indicating that RTB2 is LST8. Finally, as described in the next section, we identified point mutations in the LST8 coding region for each of the rtb2 mutant alleles identified in our screen. Hereafter, we will refer to RTB2 as LST8. When the lst8 mutant alleles were examined singly, as is shown for lst8-5 (Figure 1B), CIT2-lacZ expression was nearly 10-fold higher than its expression in wild-type cells. Altogether, these data show that LST8 is a negative regulator of RTG gene function.
The lst8-1 mutant allele does not bypass the rtg2Δ mutation
LST8 was shown previously to encode an essential seven WD-repeat protein that functions in the delivery of Gap1p, and possibly other amino acid permeases, to the cell surface (Roberg et al., 1997a). In that study, a mutant allele of LST8 (lst8-1) was identified as a synthetic lethal with the sec13-1 mutation. To determine whether the lst8-1 mutation had similar effects on RTG-dependent gene activity, the lst8-1 allele was transplaced into the LST8 locus of wild-type and rtg2Δ cells and the resultant transformants assayed for CIT2-lacZ expression (Figure 1B). Surprisingly, unlike the lst8 mutants we obtained, the lst8-1 allele was unable to bypass the rtg2Δ mutation, although by itself it resulted in an ∼4-fold activation of CIT2-lacZ expression. These findings raise the possibility that Lst8p has separable functions, a notion that will be developed later in this report.
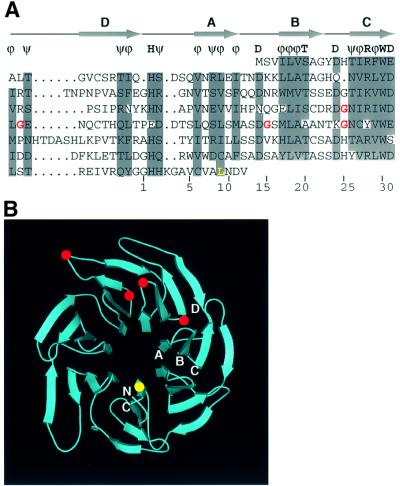
Sequencing of the five lst8 mutants we isolated showed that each contained a single missense mutation of a glycine residue, one in WD repeat 4 and the remainder in WD repeat 5 (Figure 2A). Two of the mutants contained the identical mutation at position 181 in WD repeat 5. Based on the design of our rtg2Δ bypass screen (see Materials and methods), those mutants were likely to have arisen independently. All of the mutated glycine residues were localized to tight turns in the protein. WD repeat 5 is the most divergent among the seven, lacking the conserved His2, Ser/Thr20 and Asp24, and containing a four-residue insertion in the linker region leading to WD repeat 6. We also determined the mutation in the lst8-1 allele identified by Roberg et al. (1997a), and found that there was a leucine to serine change at position 300, a residue conserved among all seven WD repeats of Lst8p, as well as in the WD repeats of the G protein β-subunit (Sondek et al., 1996). We have designated the different lst8 mutant alleles obtained in this study lst8-2 through lst8-5, and collectively as lst8-(2–5). Structural modeling of Lst8p suggests that the lst8-(2–5) mutations are clustered and localized on the surface of the protein, whereas lst8-1 would be at an internal position on the opposite side of the protein (Figure 2B).
Fig. 2. Analysis of lst8 mutations. (A) Alignment of the seven WD repeats in Lst8p. Five or more conserved residues among seven WD repeats are highlighted in gray, and the consensus sequence formed is shown above the alignment; ϕ designates any hydrophobic residue and ψ designates any hydrophilic residue; T designates serine or threonine. Based on the alignment between Lst8p and the G protein β1 subunit (ter Haar et al., 1998), each WD repeat is likely to consist of four β-strands, indicated by arrows A, B, C and D. The last row gives a canonical numbering system for residues of a WD repeat. The red type indicates mutations identified in our screen: lst8-2, Gly146 to glutamate; lst8-3, Gly138 to aspartate; lst8-4 and lst8-6, Gly181 to aspartate; lst8-5, Gly171 to aspartate. The yellow type indicates the lst8-1 mutation identified as a synthetic lethal with sec13 (Roberg et al., 1997a). (B) Three-dimensional model of Lst8p. The model was created by Swiss-Model (Peitsch, 1996), an automated comparative protein modeling server, based on the homology between Lst8p and G protein β1 subunit. The image was prepared using programs Gl_render (http://www.hhmi.swmed.edu/external/Doc/G1_render.html), BOBSCRIPT (Esnouf, 1997), MOLSCRIPT (Kraulis, 1991) and Raster3D (Meritt, 1997). The positions of four β-strands A–D from WD repeat 6 are indicated. N and C designate the N- and C-terminal ends of Lst8p. Locations of the lst8-(2–5) mutations are indicated by the red dots, and the lst8-1 mutation by the yellow dot.
lst8-5, but not lst8-1, rescues the glutamate auxotrophy of an rtg2Δ mutant
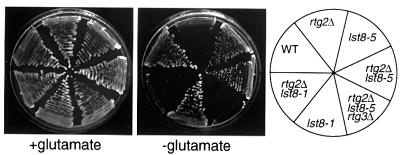
A hallmark of rtg mutant cells is that they are glutamate auxotrophs when their mitochondrial function is reduced or compromised (Liao and Butow, 1993; Liu and Butow, 1999). Given the striking difference in the ability of the lst8-(2–5) mutant alleles versus that of the lst8-1 allele to suppress the block in CIT2 expression in rtg2Δ cells, we asked whether there was also a difference among these lst8 mutants in their ability to reverse the glutamate auxotrophy of an rtg2Δ strain grown in glucose medium. As shown for lst8-5 (Figure 3), all of the lst8-(2–5) mutant alleles restored glutamate prototrophy to rtg2Δ cells, which also depended on the presence of RTG3. In contrast, the lst8-1 mutation was unable to restore glutamate prototrophy to rtg2Δ cells, consistent with the inability of that lst8 allele to bypass the loss of CIT2 expression in rtg2Δ cells. In all cases when CIT2 reporter gene expression was high, Rtg3p was localized in the nucleus (data not shown). These findings not only underscore the difference between the lst8-(2–5) and lst8-1 mutants, but suggest that a site of negative regulation of RTG-dependent gene expression by Lst8p is between Rtg2p and the Rtg1p–Rtg3p transcription complex.
Fig. 3. The glutamate auxotrophy of an rtg2Δ mutation is rescued by the lst8-5 but not the lst8-1 mutation. Wild-type PSY142 cells, and rtg2Δ, lst8-5, rtg2Δ lst8-5, rtg2Δ rtb2-5 rtg3Δ, lst8-1 and rtg2Δ lst8-1 derivatives were streaked on YNBD medium supplemented or not with 0.02% glutamate as indicated, and with uracil, leucine and lysine, and incubated at 30°C for 2–3 days.
Both lst8-5 and lst8-1 mutant cells are largely insensitive to glutamate repression
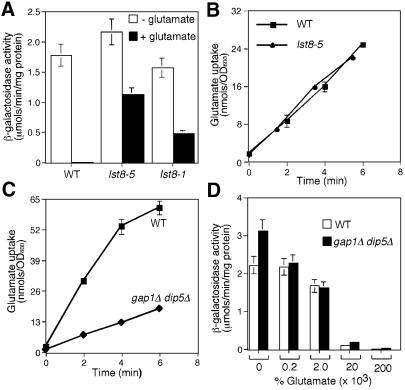
To investigate further the regulation of RTG-dependent gene expression by Lst8p, we examined the effects of the addition of glutamate to the growth medium on CIT2-lacZ expression in LST8, lst8-5 and lst8-1 cells. In LST8 cells grown in minimal medium, CIT2-lacZ expression was repressed >100-fold by the addition of 0.2% glutamate to the medium (Figure 4A). In lst8-5 mutant cells, however, CIT2-lacZ expression was largely insensitive to glutamate repression, decreasing by only ∼2-fold by the addition of 0.2% glutamate. This loss of sensitivity to glutamate repression was not the result of any bulk impairment of glutamate uptake, because lst8-5 mutant cells showed the same kinetics of glutamate uptake as wild-type cells (Figure 4B). Interestingly, although the lst8-1 mutation did not bypass the requirement for Rtg2p, it greatly reduced the glutamate repression of CIT2-lacZ expression.
Fig. 4. The insensitivity of glutamate repression of CIT2-lacZ reporter gene expression in lst8-5 mutant cells is not the result of a defect in glutamate uptake. (A) Lst8-5 and lst8-1 mutants are largely insensitive to glutamate repression of CIT2-lacZ reporter gene expression. Wild-type (PSY142), lst8-1 and lst8-5 mutant strains with an integrated copy of CIT2-lacZ reporter gene were grown in YNB5%D medium with or without 0.2% glutamate. Whole-cell extracts were prepared and β-galactosidase activities were determined as described in Materials and methods. (B) Glutamate uptake in wild-type and lst8-5 mutant cells. Wild-type (PSY142) and lst8-5 mutant strains were grown in YNBD medium and glutamate uptake was assayed as described in Materials and methods. Glutamate was present at a final concentration of 0.01%. (C) Glutamate uptake into wild-type and gap1Δ dip5Δ mutant strains. Wild-type (S288C) and a gap1Δ dip5Δ derivative strain were grown in YNBD medium, and the glutamate uptake assay was carried out as described in (B). (D) CIT2-lacZ reporter gene expression is still repressed by glutamate in gap1Δ dip5Δ mutant cells. Wild-type (S288C) and a gap1Δ dip5Δ mutant derivative strain were grown in YNB5%D medium with or without glutamate as indicated. Whole-cell extracts were prepared and β-galactosidase activities were determined as described in Materials and methods.
We next asked whether glutamate would still repress CIT2-lacZ reporter gene expression in mutant cells that are defective in glutamate transport. In yeast, there are two major pathways for glutamate uptake, one involving the general amino acid permease encoded by GAP1 and the other a specific glutamate/aspartate permease encoded by the DIP5 gene (Regenberg et al., 1998). Accordingly, we examined the effects of inactivation of these genes on glutamate repression of CIT2-lacZ expression. In a gap1Δ dip5Δ double mutant, the rate of glutamate uptake was inhibited by ∼85% (Figure 4C). Nevertheless, CIT2-lacZ expression in those double mutant cells was as sensitive to glutamate repression as in wild-type cells (Figure 4D). These data suggest that glutamate repression of RTG-dependent gene expression is not strictly dependent on glutamate uptake, and raise the possibility that glutamate repression may involve external glutamate sensing.
An Ssy1p–Ptr3p signal transduction cascade mediates glutamate repression of CIT2 expression
One candidate for an external glutamate sensor is Ssy1p, a plasma membrane protein with a predicted structure similar to that of amino acid permeases (Didion et al., 1998; Iraqui et al., 1999; Klasson et al., 1999). Previous studies have shown that amino acid induction of AGP1, which encodes a broad-specificity amino acid permease, as well as several other genes encoding amino acid permeases, was abolished in ssy1 mutant cells (Didion et al., 1998; Iraqui et al., 1999). These amino acid inductive effects have been shown also to require Ptr3p, a peripheral plasma membrane protein, and it was proposed that Ptr3p interacts with Ssy1p as part of a signal transduction cascade for external amino acid sensing. In addition, induction probably requires Shr3p, a protein whose function is to facilitate the exit of Ssy1p from the endoplasmic reticulum (Klasson et al., 1999) and, hence, its targeting to the plasma membrane.
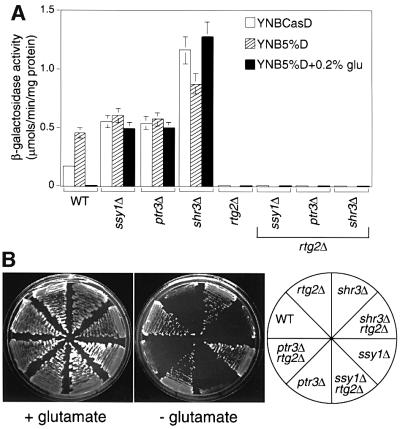
To investigate whether Ssy1p might mediate glutamate repression of RTG target gene expression, we first determined whether glutamate could still repress the activity of a CIT2-lacZ reporter gene in ssy1Δ, ptr3Δ and shr3Δ mutant cells. Figure 5A shows that the strong repression of CIT2-lacZ activity by glutamate in wild-type cells was essentially blocked in each of the mutant strains. The lack of significant glutamate repression of CIT2-lacZ expression in ssy1Δ, ptr3Δ and shr3Δ mutant cells is similar to that of the lst8 mutants. However, unlike the lst8-(2–5) mutations, the loss of CIT2 reporter gene expression in rtg2Δ cells was not reversed by the ssy1Δ, ptr3Δ or shr3Δ mutations (Figure 5A), and the double mutants rtg2Δ ssy1Δ, rtg2Δ ptr3Δ and rtg2Δ shr3Δ remained glutamate auxotrophs (Figure 5B). These data suggest that, like the lst8-1 mutation, the ssy1Δ, ptr3Δ and shr3Δ mutations affect RTG gene function upstream of RTG2.
Fig. 5. Effects of shr3Δ, ssy1Δ and ptr3Δ mutations on glutamate repression, rtg2Δ suppression and glutamate auxotrophy. (A) CIT2-lacZ expression. Transformants of wild-type (PLY126), shr3Δ (PLY151-ura3), ssy1Δ (HKY20), ptr3Δ (HKY31), rtg2Δ (PLY126-rtg2), shr3Δ rtg2Δ (PLY151-rtg2), ssy1Δ rtg2Δ (HKY20-rtg2) and ptr3Δ rtg2Δ (HKY31-rtg2) strains containing a CIT2-lacZ reporter gene on the centromere-based plasmid pCIT2-lacZ were grown to mid-log phase in the media indicated in the figure, and β-galactosidase activities were determined in extracts as described in Materials and methods. Strains rtg2Δ (PLY126-rtg2), shr3Δ rtg2Δ (PLY151-rtg2), ssy1Δ rtg2Δ (HKY20-rtg2) and ptr3Δ rtg2Δ (HKY31-rtg2) were not tested for CIT2-lacZ expression when grown in YNB5%D medium because they are glutamate auxotrophs. (B) The shr3Δ, ssy1Δ and ptr3Δ mutations cannot rescue glutamate auxotrophy of rtg2Δ cells. Cells were streaked on YNBD medium with or without 0.02% glutamate and incubated at 30°C for 2–3 days.
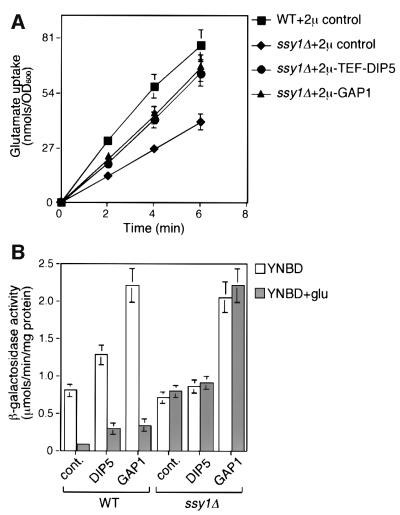
We next measured the rate of glutamate uptake in ssy1Δ cells, and found that glutamate uptake was inhibited by only ∼50% (Figure 6A). These data are in agreement with a previous report showing a modest decrease in glutamate uptake in ssy1 mutant cells (Klasson et al., 1999). To provide additional evidence that glutamate repression of CIT2-lacZ expression is not a function of glutamate uptake, we overexpressed Dip5p or Gap1p in ssy1Δ cells and determined the rate of glutamate uptake in those cells. Overexpression of these amino acid permeases in ssy1Δ cells restored rates of glutamate uptake to ∼80% of the wild-type rate (Figure 6A), but CIT2-lacZ expression remained insensitive to glutamate repression (Figure 6B). Although the level of glutamate repression of CIT2-lacZ expression in wild-type cells overexpressing Dip5p and Gap1p was somewhat less than we observed in wild-type control cells, possibly because the high level of expression of these plasma membrane proteins competes for Ssy1p targeting, it is clear that in ssy1Δ cells overexpressing these amino acid permeases, glutamate completely fails to repress CIT2-lacZ reporter gene expression. Altogether, these data support the view that sensing via the Ssy1p–Ptr3p signal transduction pathway, rather than glutamate uptake, is the major factor responsible for glutamate repression of RTG-dependent gene expression.
Fig. 6. The loss of glutamate repression of CIT2-lacZ reporter gene expression in ssy1Δ cells is uncoupled from glutamate uptake. (A) Glutamate uptake in ssy1Δ cells overexpressing Dip5 and Gap1. ssy1Δ (HKY20) cells were transformed with the 2µ control plasmid (empty vector), 2µ-TEF-DIP5 or 2µ-GAP1 plasmids, and the resultant transformants, including wild-type cells transformed with the 2µ control plasmid, were analyzed for glutamate uptake as described in Figure 4B. (B) Glutamate repression of CIT2-lacZ reporter gene expression in ssy1Δ cells overexpressing Dip5p and Gap1p. The various transformants described in (A) above were transformed with the pCIT2-lacZ reporter gene construct and the transformants were grown in YNBD medium with or without 0.2% glutamate. Whole-cell extracts were prepared and β-galactosidase activities were determined as described in Materials and methods.
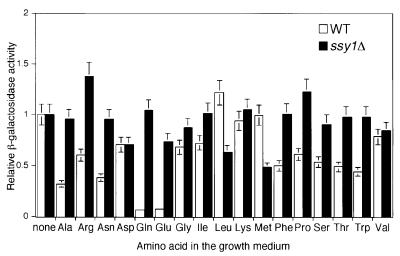
We next determined the amino acid specificity for repression of CIT2 reporter gene expression in wild-type cells. CIT2-lacZ expression was repressed most strongly in cells grown in the presence of glutamate or glutamine, whereas its expression was reduced <3-fold in the presence of other amino acids (Figure 7). In an ssy1Δ mutant, repression of CIT2-lacZ expression by glutamate and glutamine was eliminated and, in most instances, the modest repression by the other amino acids was also eliminated. These data suggest that specific and non-specific repression of RTG target gene expression is effected through Ssy1p.
Fig. 7. Effects of different amino acids on CIT2-lacZ reporter gene expression in wild-type and ssy1Δ cells. Wild-type (PLY126) and ssy1Δ (HKY20) strains transformed with a centromere-based plasmid pCIT2-lacZ were grown to mid-log phase in YNB5%D medium supplemented with 0.1% of the indicated amino acids. Whole-cell extracts were prepared and β-galactosidase activities were determined as described in Materials and methods.
Genome-wide transcription analysis of genes regulated by glutamate
Because glutamate can act as both a repressor and an activator of gene expression (Iraqui et al., 1999), with both effects operating through the Ssy1p–Ptr3p signal transduction pathway, we wished to obtain a more global view of glutamate-regulated genes. For this purpose, we carried out microarray analysis of ∼6200 yeast genes to determine how their relative mRNA abundance is affected by the addition of glutamate to the medium. A number of studies have shown that different quality nitrogen sources give rise to different patterns of gene expression (Cardenas et al., 1999; Hardwick et al., 1999; Komeili et al., 2000). Assessment of the quality of different nitrogen sources has been complicated by the fact that there is variation among strains in their response to the utilization of different nitrogen sources, including ammonia and glutamate. Accordingly, as we have done for all of the experiments described here on the effects of glutamate on RTG-dependent gene expression, glutamate was added to medium already containing ammonia, so as to avoid comparing different, sole nitrogen sources.
Strain S288C was grown to mid-log phase in YNB5%D medium with or without supplementation with 0.2% glutamate. mRNA was isolated, and labeled cDNAs were prepared by reverse transcription in the presence of Cy3- or Cy5-labeled nucleotides. These labeled cDNAs were then mixed and hybridized in duplicate with both dye configurations to yeast whole-genome arrays. Applying a stringent cut-off of an average of a 2.5-fold difference for individual genes, we found 45 genes whose expression was affected by glutamate (Table I); of these, 31 genes were repressed and 14 were induced. Among the repressed genes, 12 are known to be subject to nitrogen catabolite repression (NCR), including MEP2, encoding an ammonia permease, and GAP1, encoding a general amino acid permease. The repression of these NCR genes by glutamate suggests that ammonia is not a preferred nitrogen source for strain S288C, and that the repression of the NCR genes by glutamate is likely to be due to the combined effect of glutamate and ammonia to generate glutamine, which is a preferred nitrogen source. As expected, three RTG target genes, ACO1, CIT2 and IDH2, were among the glutamate-repressible genes.
Table I. Genome-wide effects of glutamate on gene expression.
| Gene expression | ORF | Gene | Gene product/function | Fold change |
|---|---|---|---|---|
| Repressed NCR target genes | ||||
| YNL142W | MEP2 | ammonia permease of low capacity and high affinity | 14.3 | |
| YKR039W | GAP1 | general amino acid permease | 14.0 | |
| YKR034W | DAL80 | negative regulator of multiple nitrogen catabolic genes | 12.4 | |
| YLR158C | ASP3C | l-asparaginase II, extracellular | 9.0 | |
| YIR032C | DAL3 | ureidoglycolate hydrolase | 7.7 | |
| YLR155C | ASP3A | l-asparaginase II, extracellular | 7.5 | |
| YLR157C | ASP3B | l-asparaginase II, extracellular | 7.4 | |
| YLR160C | ASP3D | l-asparaginase II, extracellular | 5.8 | |
| YJR152W | DAL5 | allantoate permease | 5.1 | |
| YIR031C | DAL7 | malate synthase 2 | 4.5 | |
| YLR142W | PUT1 | proline oxidase | 3.3 | |
| YOR348C | PUT4 | putative proline-specific permease | 2.7 | |
| RTG target genes | ||||
| YLR304C | ACO1 | aconitase, mitochondrial | 6.5 | |
| YCR005C | CIT2 | non-mitochondrial citrate synthase | 3.3 | |
| YOR136W | IDH2 | NAD+-dependent isocitrate dehydrogenase | 3.1 | |
| Others | ||||
| YKR033C | protein of unknown function | 11.4 | ||
| YLR348C | DIC1 | mitochondrial dicarboxylate transport protein | 5.4 | |
| YLR349W | protein of unknown function | 4.4 | ||
| YBR182C | SMP1 | probable transcription factor | 4.3 | |
| YBR294W | SUL1 | probable sulfate transport protein | 4.2 | |
| YPL058C | PDR12 | multidrug resistance transporter | 3.6 | |
| YNL208W | protein of unknown function | 3.2 | ||
| YOR135C | protein of unknown function | 2.9 | ||
| YJL172W | CPS1 | carboxypeptidase yscS | 2.9 | |
| YGL045W | protein of unknown function | 2.7 | ||
| YML131W | putative NAD-dependent oxidoreductase | 2.7 | ||
| YLR161W | protein of unknown function | 2.7 | ||
| YER124C | protein of unknown function | 2.6 | ||
| YLL053C | aquaporin water channel protein | 2.6 | ||
| YIL121W | member of MFS-MDR family of multidrug permeases | 2.5 | ||
| YGL028C | SCW11 | soluble cell wall protein | 2.5 | |
| Induced | ||||
| Amino acid permeases | ||||
| YDR046C | BAP3 | valine transporter | 22.9 | |
| YDR068 | BAP2 | probable amino acid permease for leucine, isoleucine and valine | ||
| YDR | 20.0 | |||
| YCL025C | AGP1 | amino acid permease | 12.1 | |
| YBR069C | VAP1 | probable amino acid transport protein | 12.0 | |
| YDR508C | GNP1 | high-affinity glutamine permease | 6.1 | |
| YGR055W | MUP1 | high-affinity methionine permease | 4.3 | |
| Proteases | ||||
| YIR039C | YPS6 | GPI-anchored aspartic protease | 6.6 | |
| YOL154W | protein with similarity to zinc metalloproteinases | 2.8 | ||
| Other | ||||
| YDR509W | protein of unknown function | 4.8 | ||
| YNL160W | YGP1 | glycoprotein synthesized in response to nutrient limitation | 3.8 | |
| YMR011W | HXT2 | high-affinity hexose transporter-2 | 3.6 | |
| YBR092C | PHO3 | acid phosphatase, constitutive | 3.2 | |
| YOL158C | ENB1 | enterobactin transporter | 3.0 | |
| YNL065W | member of MFS-MDR family of multidrug permeases | 2.7 |
Four independent cultures of strain S288C were grown to mid-log phase in YNB5%D with or without 0.2% glutamate. After isolation of poly(A)+ RNA from each, labeled cDNAs were made with either Cy3 or Cy5 and mixed with the oppositely labeled cDNA from treated or untreated samples. The indicated fold changes in gene expression were determined as described in Epstein et al. (2001a) using a cut-off of 2.5-fold change in the average of the expression ratios from two independent RNA preparations, each repeated with reversal of Cy3 and Cy5 labeling.
Among 14 genes induced by glutamate, six encode amino acid permeases, five of which, BAP2, BAP3, AGP1, GNP1 and VAP1/TAT1, belong to the Cluster I amino acid permease family that shows a broad substrate specificity (Regenberg et al., 1999). Induction of these genes by external amino acids is Ssy1p dependent (Iraqui et al., 1999), underscoring the conclusion that glutamate can effect both induction and repression of gene expression.
Discussion
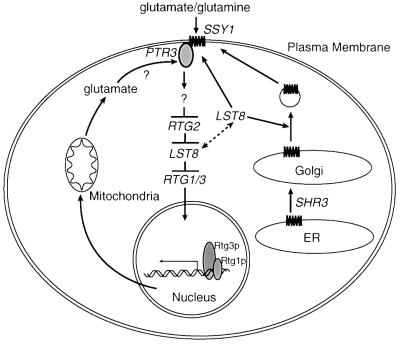
To identify additional components of the RTG pathway, we sought mutants that could bypass the dependence on Rtg2p, but still require the Rtg1p–Rtg3p transcriptional activators for target gene expression. The current work shows that certain mutations in the seven WD40-repeat protein, Lst8p, can bypass the requirement for Rtg2p, and establishes that in wild-type cells, Lst8p is a negative regulator of RTG-dependent gene expression. As diagrammed in the model of Figure 8, we suggest that Lst8p can regulate RTG-dependent gene expression at two sites, one downstream of Rtg2p and the other upstream at the level of external glutamate repression of the RTG pathway.
Fig. 8. Model for dual regulation of RTG gene functions by Lst8p. Once on the cell surface, Ssy1p binds to glutamate or glutamine and sends an inhibitory signal to Rtg2p through Ptr3p. As revealed by the lst8-1 mutation, Lst8p might affect targeting or assembly of the Ssy1p–Ptr3p system, or its signal transduction function as suggested by Roberg et al. (1997a), and thus act upstream of Rtg2p. Lst8p is also proposed to function downstream of Rtg2p, as revealed by the lst8-(2–5) mutations. Intracellular glutamate is hypothesized to be sensed by Ptr3p (Klasson et al., 1999), or some additional component of the Ssy1p signaling pathway. Repressive signals generated by glutamate would lock Lst8p in its ‘RTG inhibition’ state so that less Lst8p would be available for its function in Ssy1p–Ptr3 signaling; this would have the effect of attenuating glutamate repression of RTG gene functions.
Because in both lst8-1 and lst8-(2–5) mutant cells, expression of the CIT2-lacZ reporter gene was constitutive and insensitive to repression by glutamate, one simple way that Lst8p could regulate the RTG pathway is by controlling the cells’ ability to take up glutamate. Indeed, a previous study demonstrated that the general amino acid permease, Gap1p, failed to be targeted to the plasma membrane in lst8-1 mutant cells (Roberg et al., 1997a). Our results suggest, however, that glutamate repression of RTG-dependent gene expression is more likely to occur through glutamate sensing involving the Ssy1p–Ptr3p signal transduction pathway rather than by processes dependent on glutamate uptake. First, inactivation of the Ssy1p–Ptr3p pathway eliminated glutamate repression of RTG-dependent gene expression, but glutamate uptake in ssy1Δ cells was inhibited only by ∼50%; the latter result is in agreement with a previous study (Klasson et al., 1999). Secondly, glutamate repression of the RTG pathway was largely unaffected by inactivation of both the general (Gap1) and glutamate/aspartate-specific (Dip5p) amino acid permeases. Finally, overexpression of Gap1p and Dip5p in ssy1Δ cells restored the rate of glutamate uptake to ∼80% of that of wild-type cells, yet CIT2 expression, as in ssy1Δ cells alone, was insensitive to glutamate repression. These data suggest that, through Ptr3p-coupled glutamate signaling, the binding of glutamate to Ssy1p would send a signal (inhibitory) to the RTG pathway, as has been proposed for the effects of other amino acids in the Ssy1p/Ptr3p-dependent activation of gene expression in the amino acid inductive pathway (Didion et al., 1998; Klasson et al., 1999; Iraqui et al., 1999). Despite the loss of glutamate repression of the RTG pathway in ssy1Δ, ptr3Δ, shr3Δ as well as in lst8-1 cells, none of these mutations can bypass the requirement of Rtg2p for CIT2 expression as can the lst8(2–5) mutations.
In considering the effect of glutamate on RTG-dependent gene expression, it is important to distinguish between the sensing of extra- and intracellular glutamate. In cells with compromised mitochondrial function, intracellular glutamate supplies are maintained by RTG regulation of expression of the four TCA cycle genes responsible for the synthesis of α-ketoglutarate (Liu and Butow, 1999), by increased expression of genes encoding enzymes that function in anaplerotic pathways, small molecule transport and peroxisomal activities that converge to supply acetyl-CoA, oxaloacetate and citrate to the TCA cycle, and by increased expression of genes that enable cells to utilize poor nitrogen sources (Epstein et al., 2001b). In contrast, when glutamate is available in the medium, the RTG-dependent gene expression is down-regulated by the SSY1–PTR3 signal transduction pathway.
Although the RTG pathway is also insensitive to glutamate repression in lst8-(2–5) mutant cells, it is quite possible that the Lst8p regulatory site(s) downstream of Rtg2p may have no direct connection to glutamate regulation of the pathway. The WD40-repeat motif was identified originally in the β-subunit of heterotrimeric G proteins (Fong et al., 1986) and subsequently has been found in a wide spectrum of regulatory proteins, where it functions in mediating protein–protein interactions. Thus Lst8p could be a component of a complex that tethers Rtg1p–Rtg3p in the cytoplasm. In this model, the loss of glutamate repression of RTG-dependent gene expression in lst8-(2–5) mutant cells would be a consequence of the constitutive localization of the Rtg1p–Rtg3p complex in the nucleus. An alternative model is that Lst8p could bind to and regulate the activity of a kinase or phosphatase, thus effecting a change in the phosphorylation state of Rtg3p; this, in turn, would dictate Rtg3p’s nuclear entry or exit. It remains to be determined how internal glutamate is sensed by the RTG pathway and how Rtg2p functions to transduce glutamate signals to the Rtg1p–Rtg3p transcription complex.
What could be the molecular basis for the proposed dual sites of action of Lst8p as diagrammed in the model of Figure 8? We suggest that there are at least two domains of Lst8p, one functioning in the assembly or activity of the Ssy1p–Ptr3p signal transduction complex and the other in the negative regulation of RTG-dependent gene expression downstream of Rtg2p. In addition to the genetic evidence already summarized, clearly defining sites of Lst8p action both upstream and downstream of Rtg2p, it is striking that the lst8-(2–5) mutations are clustered and localize to a region of Lst8p that is predicted to be on a side of the protein opposite to that of the lst8-1 mutation (Figure 2B). WD40-repeat proteins usually adopt a β-propeller structure, which can use one or two blades to interact with other proteins without affecting the other blades (Goodman et al., 1997; ter Haar et al., 1998). We propose that the distinctive phenotypes of two types of lst8 mutations are due to defects in the interaction of the protein with different factors. The dual functionality of Lst8p may facilitate glutamate homeostasis by participating in a negative feedback loop for regulating glutamate levels. In the presence of high concentrations of external glutamate, signals generated by Ssy1p could ‘lock’ Lst8p in a form that functions to sequester the Rtg1p–Rtg3p complex in the cytoplasm. Thus, less Lst8p would be available for the Ssy1p signal transduction system, where it plausibly might function in targeting or post-translational modification of one or more components of that system. This model could explain the observation that glutamate blocks the targeting of Gap1p to the plasma membrane (Roberg et al., 1997a,b), and further suggests a means for attenuating glutamate repression as the concentration of glutamate in the medium decreases. Consistent with this ‘switching’ model, we have observed that in rtg mutant cells grown in rich medium, there is an ∼10-fold induction of AGP1 expression (data not shown). This finding could indicate that Lst8p has been released from its site(s) downstream of Rtg2p so that more of the protein is available for the SSY1–PTR3 pathway. Altogether, these findings imply that Lst8p might be localized to more than one cellular compartment. Although the specific intracellular location of Lst8p has not been established, preliminary cell fraction studies suggest that Lst8p may indeed be localized to different cellular compartments (unpublished observations). Further studies will be required to clarify these issues.
Ssy1p has been shown to be involved in the amino acid inductive expression of some amino acid permeases (Didion et al., 1998; Iraqui et al., 1999), a peptide transporter, Ptr2p (Didion et al., 1998), and, as shown here, in glutamate repression. These findings prompted us to carry out a genome-wide transcriptional analysis on the effects of glutamate on gene expression. Microarray experiments revealed three major pathways that were affected by glutamate: NCR; the RTG pathway; and the amino acid inductive pathway. Interestingly, Ssy1p appears to be involved in all three of these pathways (Didion et al., 1998; Iraqui et al., 1999; Klasson et al., 1999). One set of genes whose expression was affected by glutamate in our microarray analysis was amino acid-inducible genes, BAP2, BAP3, GNP1, VAP1/TAT1 and AGP1. Their transcription has been shown to be dependent on Ssy1p (Iraqui et al., 1999). Although, compared with hydrophobic amino acids, glutamate and glutamine are strong repressors of RTG gene function, they are weak inducers for AGP1 expression. We consistently observed that the addition of 0.01% glutamate to the growth medium resulted in a 4- to 5-fold greater induction of AGP1 compared with addition of 0.2% glutamate (data not shown), further supporting the Lst8p switching model discussed above. The amino acid induction of PTR2 transcription involves a transcriptional repressor Cup9p and a N-end rule pathway component, Ubr1p (Goodman et al., 1997; Byrd et al., 1998). Is there any cross-regulation among these pathways? We found in preliminary experiments (data not shown) that ubr1, ure2 or gln3 deletion mutations have no significant effect on the expression of the RTG target gene, CIT2, and, conversely, that rtg mutations do not affect expression of the NCR target gene, GLN1. These findings suggest that there is little or no direct cross-regulation of these pathways downstream of Ssy1p–Ptr3p.
Recent work has shown that the TOR (target of rapamycin) kinase signaling pathway, which is involved in nutrient sensing, also affects RTG-dependent gene expression in the same manner as the retrograde pathway, namely by affecting the subcellular localization of Rtg1p and Rtg3p in an Rtg2p-dependent manner (Komeili et al., 2000). Inhibition of the TOR kinase pathway by treating cells with rapamycin or by growing them in a poor nitrogen source promotes the dissociation of Ure2p from a cytoplasmic, phosphorylated form of a positive regulator of the NCR pathway, Gln3p, resulting in Gln3’s dephosphorylation and nuclear accumulation (Beck and Hall, 1999; Cardenas et al., 1999; Hardwick et al., 1999). Dephosphorylation of Gln3p is believed to be controlled by the type 2A-related phosphatase, Sit4p, and its inhibitor, Tap42p, an effector protein of the TOR pathway (Beck and Hall, 1999). An sit4 deletion mutation, however, does not affect CIT2 expression (unpublished data), suggesting that different TOR effectors may serve to regulate the NCR and RTG pathways.
Materials and methods
Strains
Strains used in this study are listed in Table II.
Table II. Saccharomyces cerevisiae strains used.
| Strain | Genotype | Source |
|---|---|---|
| PSY142 | MATα ura3-52 leu2 lys2 ura3::CIT2-lacZ | R.Butow |
| PSY142-rtg2 | MATα ura3-52 leu2 lys2 ura3::CIT2-lacZ rtg2Δ::ura3 | R.Butow |
| RBY346 | MATα ura3-52 leu2::LEU2-RTG2 lys2 ura3::CIT2-lacZ rtgΔ::ura3 lst8-5 | this study |
| RBY426 | MATα ura3-52 leu2 lys2 ura3::CIT2-lacZ rtg2Δ::ura3 lst8-5 | this study |
| RBY427 | MATα ura3-52 leu2 lys2 ura3::CIT2-lacZ rtg2Δ::ura3 lst8-5 rtg3Δ::LEU2 | this study |
| RBY254 | MATα ura3-52 leu2 lys2 ura3::CIT2-lacZ lst8-1 | this study |
| RBY264 | MATα ura3-52 leu2 lys2 ura3::CIT2-lacZ lst8-1 rtg2Δ::kanMX4 | this study |
| RBY418 | MATa ura3 leu2 lys2 | this study |
| RBY428 | MATa/α ura3/ura3 leu2/leu2 lys2/lys2 LST8/lst8Δ::LEU2 [pLST8] | this study |
| RBY419 | MATa ura3 leu2 lys2 rtg2Δ::kanMX4 | this study |
| RBY420 | MATa ura3 leu2 lys2 lst8Δ::LEU2 pLST8 | this study |
| RBY421 | MATa ura3 leu2 lys2 lst8Δ::LEU2 rtg2Δ::kanMX4 pLST8 | this study |
| RBY422 | MATa/α ura3/ura3::CIT2-lacZ leu2/leu2 lys2/lys2 RTG2/rtg2Δ::ura3 LST8/lst8-5 | this study |
| RBY423 | MATa/α ura3/ura3::CIT2-lacZ leu2/leu2 lys2/lys2 rtg2Δ::ura3/rtg2Δ::kanMX4 LST8/lst8-5 | this study |
| RBY424 | MATa/α ura3/ura3::CIT2-lacZ leu2/leu2 lys2/lys2 rtg2Δ::ura3/rtg2Δ::kanMX4 lst8Δ::LEU2/lst8-5 | this study |
| ECY408 | MATa ura3 lst8-1 | C.Kaiser |
| S288C | MATa ura3-52 | |
| M4276 | MATa ura3 gap1Δ dip5Δ::kanMX | M.Kielland-Brandt |
| PLY126 | MATa ura3-52 lys2Δ201 | P.Ljungdahl |
| PLY151-ura3 | MATa ura3-52 lys2Δ201 ade2 shr3Δ1::ura3 | P.Ljungdahl |
| HKY20 | MATa ura3-52 lys2Δ201ssy1Δ13 | P.Ljungdahl |
| HKY31 | MATa ura3-52 lys2Δ201 ptr3Δ15 | P.Ljungdahl |
| PLY126-rtg2 | MATa ura3-52 lys2Δ201 rtg2::kanMX4 | this study |
| PLY151-rtg2 | MATa ura3-52 lys2Δ201 alde2 shr3Δ1::ura3 rtg2::kanMX4 | this study |
| HKY20-rtg2 | MATa ura3-52 lys2Δ201 ssy1Δ13 rtg2::kanMX4 | this study |
| HKY31-rtg2 | MATa ura3-52 lys2Δ201 ptr3Δ15 rtg2::kanMX4 | this study |
Growth media, growth conditions
Yeast strains were grown at 30°C in YPD (1% yeast extract, 2% bactopeptone and 2% dextrose), YNBcasD (0.67% yeast nitrogen base, 1% casamino acids and 2% dextrose) or minimal YNB medium (0.67% yeast nitrogen base) supplemented with 2 or 5% glucose (YNBD or YNB5%D, respectively), with or without Na glutamate (as indicated in the text and figures), and supplemented with the required nutrients.
Strain constructions
Standard genetic manipulations were carried out as described in Rose et al. (1990). RBY426 was isolated from the rtg2 bypass screen described below. RBY426 was transformed with a rtg3Δ::LEU2 disruption cassette to form RBY427. RBY254 was an lst8-1 mutant obtained through a two-step gene replacement. RBY418 was derived from PSY142 lacking the integrated CIT2-lacZ reporter gene by mating-type switching of PSY142 cells transformed with the plasmid pGAL-HO (Herskowitz and Jensen, 1991). RBY418 was transformed with an rtg2::kanMX4 disruption cassette to form RBY419. RBY420 is a meiotic segregant obtained following sporulation of the diploid RBY428 transformed with pRS416-LST8. RBY421 was constructed from RBY420 by transforming cells with pUC18-rtg2::kanMX4 digested with PstI. Matings were carried out between RBY426 and RBY418, RBY419 or RBY421 to form RBY422, RBY423 or RBY424, respectively. RTG2 disruptions in the strains PLY126-rtg2, PLY151-rtg2, HKY20-rtg2 and HKY31-rtg2 were carried out by transformation of the rtg2::kanMX4 disruption cassette into their respective parental strains.
Plasmid constructs
Construction of rtg3Δ::LEU2 disruption cassette was described previously (Liu and Butow, 1999). To construct a rtg2Δ::kanMX4 cassette derivative, the primer set 5′-AGGCACACTCTTCTTCAC-3′ and 5′-gactaagcttTGAACAACAAGAAGGTGCCC-3′ was used to amplify RTG2. After digestion with BglII and HindIII, RTG2 DNA was ligated into the BamHI and HindIII sites of pRS416 to form pRS416-RTG2. A 643 bp NdeI fragment of pRS416-RTG2 was cloned into the NdeI site of pUC18 to form pUC18-RTG2. A kanMX4 disruption cassette module was inserted into the SpeI site to form pUC18-rtg2::kanMX4. The primer set 5′-gactactagtACCACCAGGACCAAAGCTTG-3′ and 5′-gactgtcgacCAAAAGCGGGAAGAAGTCA-3′ was used to amplify the 2.1 kbp LST8 sequence. ECY408 genomic DNA was used to amplify the lst8-1 mutant allele. The amplified DNA was digested with SpeI and SalI and ligated into Yip352 or pRs416 to form, respectively, Yip352-lst8-1 and pRS416-lst8-1. Similarly, LST8 was amplified from the PSY142 strain and isogenic rtb2 mutant strains to construct pRS416-LST8, pRS416-lst8-2–6, respectively. pCIT2-lacZ was described previously (Liao et al., 1991; Liu and Butow, 1999). To obtain plasmid 2µ-LYS2, the pRS426 plasmid was digested with EcoRV and StuI to remove part of the URA3 sequence and replaced with a 5.2 kbp LYS2 sequence from SmaI- and PstI-digested YDp-K (Berben et al., 1991). To construct plasmid 2µ-TEF-DIP5, the primer pair 5′-gtcaACTAGTCTCTAAGTAATGAAGATGCC-3′ and 5′-gtcaGTCGACGTGATACCTGTACACTATGG-3′ was used to amplify the DIP5 open reading frame and 413 bp of 3′-untranslated region. The resultant PCR product was digested with SpeI and SalI and ligated into the pRS426-TEF plasmid (Mumberg et al., 1995). The URA3 sequence in the pRS426-TEF-DIP5 was replaced with LYS2, similarly to the construction of 2µ-LYS2. To construct 2µ-GAP1 plasmid, a 3.5 kbp GAP1 sequence was obtained from SpeI- and SalI-digested pPL247 (Ljungdahl et al., 1992) and ligated into pRS426. The resultant pRS426-GAP1 was digested with NcoI and StuI to remove part of the URA3 sequence and replaced with the same LYS2 sequence described above.
Rtg2 bypass mutant screen
Ethyl methanesulfonate mutagenesis was carried out essentially as described previously (Liao and Butow, 1993). Cells were plated on YNBD plus X-gal and amino acid supplements. Fourteen rtb mutants were obtained that could be grouped into two complementation groups, rtb1 and rtb2, as described in the text. The rtb mutants arose at a frequency of ∼1 in 106 cells.
Isolation of the RTB2 (LST8) gene
A URA3 centromeric CIT2-lacZ reporter gene was transformed into a RBY285 (rtg2Δ rtb2-5) derivative lacking the integrated version of the reporter gene. To isolate the RTB2 gene, a wild-type genomic DNA library constructed in the centromeric vector p366 carrying a LEU2 marker was screened for transformants of RBY285 that would abolish CIT2-lacZ expression. Two out of 8000 LEU2+ transformants were obtained that had overlapping restriction maps. Subcloning in the centromeric plasmid pRS415 and retransformation revealed that a fragment containing the LST8 gene was necessary and sufficient to complement the rtb2-5 mutant phenotype.
β-galactosidase assays
β-galactosidase assays were carried out as previously described (Liu and Butow, 1999). For each plasmid–strain combination, assays were conducted in triplicate and independent experiments were carried out two or three times.
Assays for amino acid uptake
Amino acid uptake was assayed as described by Roberg et al. (1997a) except that for glutamate uptake, equal volumes of 0.2 mM 14C-labeled glutamate and 59.1 mM unlabeled glutamate were mixed and diluted 50-fold into the assay mixture. At 2 min intervals, two samples of 100 µl each were diluted into 3 ml of ice-cold 5.9 mM glutamate to quench the reaction, and cells were collected by filtration and washed with 3× 5 ml of ice-cold water. l-[U-14C]glutamic acid (250 mCi/mmol) was obtained from Amersham Pharmacia (Buckinghamshire, UK).
Microarray analysis
A fresh culture of S288C was diluted into 250 ml of YNB5%D with or without 0.2% glutamate. After overnight growth to OD600 0.8, cells were collected and mRNA samples were prepared as described (Epstein et al., 2001b). Cy3- and Cy5-labeled cDNAs were prepared and hybridized to a microarray of 6219 yeast genes. Replica experiments were carried out using independent liquid cultures and with the opposite configuration of Cy3 and Cy5. The data were analyzed as described (Epstein et al., 2001a,b) using a cut-off of value of an average of 2.5-fold change in replica hybridizations.
Acknowledgments
Acknowledgements
We thank, G.Fink, C.Kaiser, P.O.Ljungdahl and M.C.Kielland-Brandt for yeast strains and plasmids, Tsan Xiao for technical support, and members of the Butow laboratory for helpful discussions. This work was supported by grants GM22525 and CA77811 from the National Institutes of Health, and I-0642 from The Robert A.Welch Foundation.
References
- Beck T. and Hall,M.N. (1999) The TOR signaling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature, 402, 689–692. [DOI] [PubMed] [Google Scholar]
- Berben G., Dumont,J., Gilliquet,V., Bolle,P.A. and Hilger,F. (1991) The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast, 7, 475–477. [DOI] [PubMed] [Google Scholar]
- Bork P., Sander,C. and Valencia,A. (1992) An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin and hsp70 heat shock proteins. Proc. Natl Acad. Sci. USA, 89, 7290–7294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd C., Turner,G.C. and Varshavsky,A. (1998) The N-end rule pathway controls the import of peptides through degradation of a transcriptional repressor. EMBO J., 17, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas M.E., Cutler,N.S., Lorenz,M.C., Di Como,C.J. and Heitman,J. (1999) The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev., 13, 3271–3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chelstowska A. and Butow,R.A. (1995) RTG genes in yeast that function in communication between mitochondria and the nucleus are also required for expression of genes encoding peroxisomal proteins. J. Biol. Chem., 270, 18141–18146. [DOI] [PubMed] [Google Scholar]
- Chelstowska A., Liu,Z., Jia,Y., Amberg,D. and Butow,R.A. (1999) Signaling between mitochondria and the nucleus regulates the expression of a new d-lactate dehydrogenase activity in yeast. Yeast, 15, 1377–1391. [DOI] [PubMed] [Google Scholar]
- Didion T., Regenberg,B., Jorgensen,M.U., Kielland-Brandt,M.C. and Andersen,H.A. (1998) The permease homologue Ssy1p controls the expression of amino acid and peptide transporter genes in Saccharomyces cerevisiae. Mol. Microbiol., 27, 643–650. [DOI] [PubMed] [Google Scholar]
- Epstein C.B., Hale,W.,IV and Butow,R.A. (2001a) Numerical methods for handling uncertainty in microarray data—an example analyzing perturbed mitochondrial function in yeast. Methods Cell Biol., 65, 439–482. [DOI] [PubMed] [Google Scholar]
- Epstein C.B., Waddle,J.A., Hale IV,W., Davé,V., Thornton,J., Macatee,T.L., Garner,H.R. and Butow,H.R. (2001b) Genome-wide responses to mitochondrial dysfunctions. Mol. Biol. Cell, 12, 297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph., 15, 133–138. [DOI] [PubMed] [Google Scholar]
- Fong H.K., Hurley,J.B., Hopkins,R.S., Miake-Lye,R., Johnson,M.S., Doolittle,R.F. and Simon,M.I. (1986) Repetitive segmental structure of the transducin β subunit: homology with the CDC4 gene and identification of related mRNAs. Proc. Natl Acad. Sci. USA, 83, 2162–2166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsburg S.L. and Guarente,L. (1989) Communication between mitochondria and the nucleus in regulation of cytochrome genes in the yeast Saccharomyces cerevisiae. Annu. Rev. Cell Biol., 5, 153–180. [DOI] [PubMed] [Google Scholar]
- Gangloff S.P., Marguet,D. and Lauquin,G.J.M. (1990) Molecular cloning of the yeast mitochondrial aconitase gene (Aco1) and evidence of a synergistic regulation of expression by glucose plus glutamate. Mol. Cell. Biol., 10, 3551–3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman O.B., Krupnick,J.G., Gurevich,V.V., Benovic,J.L. and Keen,J.H. (1997) Arrestin/clathrin interaction. Localization of the arrestin binding locus to the clathrin terminal domain. J. Biol. Chem., 272, 15017–15022. [DOI] [PubMed] [Google Scholar]
- Hardwick J.S., Kuruvilla,F.G., Tong,J.K., Shamji,A.F. and Schreiber,S.L. (1999) Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl Acad. Sci. USA, 96, 14866–14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herskowitz I. and Jensen,R.E. (1991) Putting the HO gene to work: practical uses for mating-type switching. Methods Enzymol., 194, 132–146. [DOI] [PubMed] [Google Scholar]
- Iraqui I., Vissers,S., Bernard,F., DeCraene,J.O., Boles,E., Urrestarazu,A. and Andre,B. (1999) Amino acid signaling in Saccharomyces cerevisiae: a permease-like sensor of external amino acids and F-box protein Grr1p are required for transcriptional induction of the AGP1 gene, which encodes a broad-specificity amino acid permease. Mol. Cell. Biol., 19, 989–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Y., Rothermel,B., Thornton,J. and Butow,R.A. (1997) A basic helix–loop–helix zipper transcription complex functions in a signaling pathway from mitochondria to the nucleus. Mol. Cell. Biol., 17, 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klasson H., Fink,G.R. and Ljungdahl,P.O. (1999) Ssy1p and Ptr3p are plasma membrane components of a yeast system that senses extracellular amino acids. Mol. Cell. Biol., 19, 5405–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A., Wedaman,K.P., O’Shea,E.K. and Powers,T. (2000) Mechanism of metabolic control: target of rapamycin signaling links nitrogen quality to the activity of the Rtg1 and Rtg3 transcription factors. J. Cell Biol., 151, 863–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraulis P.J. (1991) MOLSCRIPT: a program to produce both detailed and schematic plots of proein structures. J. Appl. Crystallogr., 24, 946–950. [Google Scholar]
- Liao X. and Butow,R.A. (1993) RTG1 and RTG2: two yeast genes required for a novel path of communication from mitochondria to the nucleus. Cell, 72, 61–71. [DOI] [PubMed] [Google Scholar]
- Liao X.S., Small,W.C., Srere,P.A. and Butow,R.A. (1991) Intramitochondrial functions regulate nonmitochondrial citrate synthase (CIT2) expression in Saccharomyces cerevisiae. Mol. Cell. Biol., 11, 38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z. and Butow,R.A. (1999) A transcriptional switch in the expression of yeast tricarboxylic acid cycle genes in response to a reduction or loss of respiratory function. Mol. Cell. Biol., 19, 6720–6728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljungdahl P.O., Gimeno,C.J., Styles,C.A. and Fink,G.R. (1992) SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast. Cell, 71, 463–478. [DOI] [PubMed] [Google Scholar]
- Meritt E.A. (1997) Raster3D: photorealistic molecular graphics. Methods Enzymol., 277, 505–524. [DOI] [PubMed] [Google Scholar]
- Mumberg D., Muller,R. and Funk,M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene, 156, 119–122. [DOI] [PubMed] [Google Scholar]
- Peitsch M.C. (1996) ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans., 24, 274–279. [DOI] [PubMed] [Google Scholar]
- Regenberg B., Holmberg,S., Olsen,L.D. and Kielland-Brandt,M.C. (1998) Dip5p mediates high-affinity and high capacity transport of l-glutamate and l-aspartate in Saccharomyces cerevisiae. Curr. Genet., 33, 171–177. [DOI] [PubMed] [Google Scholar]
- Regenberg B., During-Olsen,L., Kielland-Brandt,M.C. and Holmberg,S. (1999) Substrate specificity and gene expression of the amino-acid permeases in Saccharomyces cerevisiae. Curr. Genet., 36, 317–328. [DOI] [PubMed] [Google Scholar]
- Roberg K.J., Bickel,S., Rowley,N. and Kaiser,C.A. (1997a) Control of amino acid permease sorting in the late secretory pathway of Saccharomyces cerevisiae by SEC13, LST4, LST7 and LST8. Genetics, 147, 1569–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberg K.J., Rowley,N. and Kaiser,C.A. (1997b) Physiological regulation of membrane protein sorting late in the secretory pathway of Saccharomyces cerevisiae. J. Cell Biol., 137, 1469–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M.D., Winston,F. and Heiter,P. (1990) Methods in Yeast Genetics: A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Rosenkrantz M., Kell,C.S., Pennell,E.A. and Devenish,L.J. (1994) The HAP2,3,4 transcriptional activator is required for derepression of the yeast citrate synthase gene, CIT1. Mol. Microbiol., 13, 119–131. [DOI] [PubMed] [Google Scholar]
- Sekito T., Thornton,J. and Butow,R.A. (2000) Mitochondria-to-nuclear signaling is regulated by the subcellular localization of the transcription factors Rtg1p and Rtg3p. Mol. Biol. Cell, 11, 2103–2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamji A.F., Kuruvilla,F.G. and Schreiber,S.L. (2000) Partitioning the transcriptional program induced by rapamycin among the effectors of the TOR proteins. Curr. Biol., 10, 1574–1581. [DOI] [PubMed] [Google Scholar]
- Sondek J., Bohm,A., Lambright,D.G., Hamm,H.E. and Sigler,P.B. (1996) Crystal structure of a G-protein βγ dimer at 2.1 Å resolution. Nature, 379, 369–374. [DOI] [PubMed] [Google Scholar]
- terHaar E., Musacchio,A., Harrison,S.C. and Kirchhausen,T. (1998) Atomic structure of clathrin: a β propeller terminal domain joins an α zigzag linker. Cell, 95, 563–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traven A., Wong,J.M., Xu,D., Sopta,M. and Ingles,C.J. (2001) Inter-organellar communication: altered nuclear gene expression profiles in a yeast mitochondrial DNA mutant. J. Biol. Chem., 276, 4020–4027. [DOI] [PubMed] [Google Scholar]