Abstract
Factors affecting competition between termination and elongation in vivo during translation of the fdhF selenocysteine recoding site (UGA) were studied with wild-type and modified fdhF sequences. Altering sequences surrounding the recoding site UGA without affecting RNA secondary structure indicated that the kinetics of stop signal decoding have a significant influence on selenocysteine incorporation efficiency. The UGA in the wild-type fdhF sequence remains ‘visible’ to the factor and forms a site-directed cross-link when mRNA stem–loop secondary structure is absent, but not when it is present. The timing of the secondary structure unfolding during translation may be a critical feature of competition between release factor 2 and tRNASec for decoding UGA. Increasing the cellular concentration of either of these decoding molecules for termination or selenocysteine incorporation showed that they were able to compete for UGA by a kinetic competition that is dynamic and dependent on the Escherichia coli growth rate. The tRNASec-mediated decoding can compete more effectively for the UGA recoding site at lower growth rates, consistent with anaerobic induction of fdhF expression.
Keywords: recoding/ribosome/selenocysteine/translation efficiency/translation termination
Introduction
During synthesis of formate dehydrogenase H (FDHH) in Escherichia coli, a cis-acting mRNA element, the selenocysteine incorporation sequence (SECIS), programmes recoding of an internal UGA as selenocysteine (Heider et al., 1992). The co-translational incorporation of selenocysteine requires tRNASec, a tRNA capable of decoding the fdhF recoding site (rsUGA) and SELB, a selenocysteyl-tRNASec-specific elongation factor. While the genetic requirements for selenocysteine incorporation in E.coli are well characterized (Böck et al., 1991), the interaction of the SELB quaternary complex with the ribosome is still not well understood. A variety of approaches indicates that SELB must be complexed with the SECIS element for a productive interaction with the ribosome. This suggests that SECIS element binding induces a conformational switch in SELB that facilitates formation of an anticodon–codon interaction between the Sec-tRNASec and the UGA codon. The SECIS element appears to act as a safety switch, preventing normal UGA termination codons being decoded as selenocysteine by SELB–GTP–Sec-tRNASec complexes (Klug et al., 1997; Hüttenhofer and Böck, 1998).
Incorporation of selenocysteine also necessitates that the canonical decoding of UGA as termination be subverted. It has been found previously that C following a UGA codon, as is found at the fdhF selenocysteine insertion site, induces a much lower termination efficiency in vivo than if any of the other three bases are in that position (Poole et al., 1995). There is now considerable evidence that the nucleotides surrounding the stop codon affect termination efficiency (Salser et al., 1969; Bossi and Roth, 1980; Engelberg-Kulka, 1981; Kopelowitz et al., 1992). Indeed, an emerging picture of the protein synthesis termination signal in prokaryotes suggests that the stop codon forms the core of a larger element containing both upstream and downstream sequences (Tate et al., 1996). For example, in vitro cross-linking experiments show that the release factor (RF) is in intimate contact with not only the stop codon but also the following three bases (Poole et al., 1998), and in vivo readthrough studies indicate that the two C-terminal amino acids of the nascent polypeptide (and thereby the two codons preceding the stop codon in the mRNA) also affect termination efficiency (Mottagui-Tabar et al., 1994; Björnsson et al., 1996). The kinetics of termination at the stop signal are due largely to the composition of these different elements that are presented to the RF. Furthermore, stop signals of differing efficacies appear to offer another level of control in regulating gene expression, particularly at recoding sites (Crawford et al., 1999).
Following the postulate that a consequence of having one signal to support two distinct decoding events is that there is competition for the signal, this work investigates how competition at the fdhF rsUGA is mediated in vivo. We show that the relative efficiencies of competing decoding events determine the translational fate of the fdhF rsUGA.
Results
Is the rsUGA decoded by RF2 like a typical UGA stop signal?
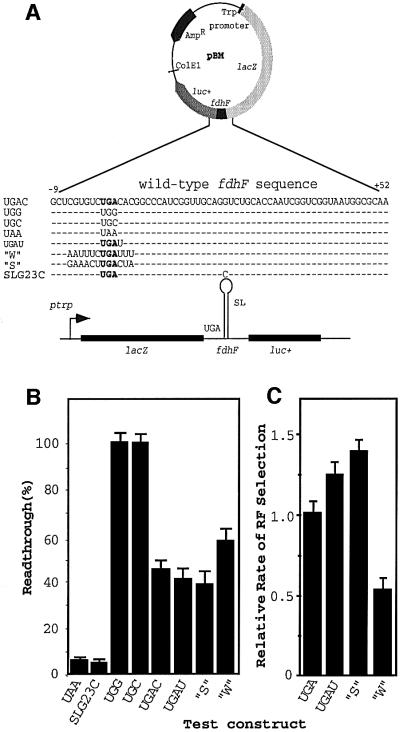
Vectors were constructed with the rsUGA (+1 to +3 bases) within the fdhF sequence (–9 to +52) cloned between two reporter genes (Figure 1A) to test whether changing the sequence surrounding the rsUGA affected the overall translational fate. Changes were made that were known to affect the kinetics of decoding UGA termination signals by RF2. The selenocysteine incorporation readthrough event is measured by the luciferase activity derived from expression of the luc+ downstream reporter, whereas a measure of both readthrough and termination events is determined from the activity of β-galactosidase expressed from the lacZ upstream reporter.
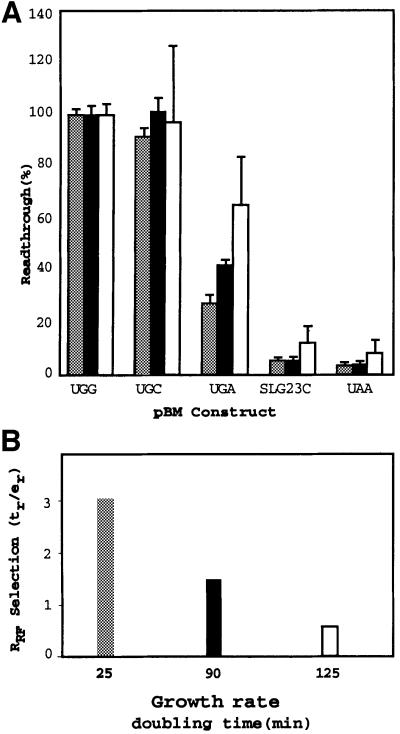
Fig. 1. An experimental system to investigate translational fate when the selenocysteine incorporation site of fdhF is altered. (A) Variations to the wild-type fdhF sequence inserted into pBM. (B) The mean readthrough efficiency (and standard errors) for each sequence expressed as a percentage. (C) The relative RF selection rate for different constructs. The SEM are shown.
Multiple isolates of clones containing the same construct, or containing different constructs, gave very similar β-galactosidase activities [within the standard error of the mean (SEM)], indicating similar expressions independently of the construct or clone (data not shown). The construct pBMUAA, with the rsUGA changed to UAA, was used as a control as it is unable to support translational elongation since the tRNASec anticodon is not complementary to this codon (Figure 1B). Nevertheless, a small amount of luciferase activity was observed with pBMUAA. Modification of the rsUGA context (SLG23C), where there is a base change in the apical loop of the fdhF hairpin known to abolish selenocysteine incorporation in vivo (Heider et al., 1992), gave a similar level of ‘apparent readthrough’. Therefore, both measurements are likely to reflect near-cognate readthrough and/or reinitiation downstream of the rsUGA. The rsUGA was replaced by two near-cognate sense codons, UGG (Trp) and UGC (Cys), to establish luciferase activity values when termination was precluded (100% readthrough). Both constructs gave very similar values (Figure 1B).
These modifications to the rsUGA (UAA, UGG or UGC, and SLG23C) set values for upper and lower limits of potential competition for decoding at the recoding site. The UGA and its natural surrounding context allowed almost 50% readthrough, reflecting near equal competition between the termination and elongation events under the growth conditions used for these experiments (Figure 1B, UGAC). This competition was affected by changes to the sequences surrounding the rsUGA to reflect a stronger or weaker stop signal. We have established that the native rsUGA site has an upstream sequence that supports efficient termination while the downstream sequence is relatively weak (Major, 2001). We predicted that there was potential to strengthen the termination signal modestly as well as to weaken it. The vectors UGAU and ‘S’ (strong upstream and downstream contexts) (Figure 1A) contain modified fdhF and have sequence elements predicted to increase the decoding rate of a UGA stop signal, while construct ‘W’ (weak upstream and downstream contexts) should decrease the decoding rate. Simply by changing the +4 base from C to U, a consistent decrease in luciferase activity occurred (reflecting a drop from ∼9 to 8 ribosomal passages in 20) and this was also the case with the strong context,‘S’. In contrast, the number of ribosomal passages supporting selenocysteine incorporation increased from 8 to 12 in 20 with the weak termination signal ‘W’. These data indicate that selenocysteine incorporation efficiency at rsUGA responds to parameters known to be important at a more typical UGA termination site.
The relative termination efficiencies supported by rsUGA present in each of these constructs can be analysed further by calculating the rate of RF selection at each site (Pedersen and Curran, 1991). The rate of RF selection (RRF) indicates the rate of stop signal recognition by the RF and, in this case, is the rate of termination (tr) relative to the rate of readthrough (er), i.e. RRF = tr/er. RRF for each of the constructs is shown in Figure 1C. These selection rates reflect the likelihood that a termination event will occur before selenocysteine incorporation when the rsUGA is at the A site during a given ribosomal passage. For example, a decrease in termination value observed with the weak stop signal (‘W’) (Figure 1B) reflected a 2-fold decrease in the rate of productive RF selection into the A site (Figure 1C), allowing for a relative increase in the competitiveness of Sec-tRNASec and increasing the likelihood that it would be the successful decoding molecule. In contrast, the two stronger stop signals have a 1.3- to 1.4-fold increase in RRF, decreasing the competitiveness of the Sec-tRNASec. At the fdhF selenocysteine incorporation site, just as at other sites of UGA stop signals, if the efficiency with which the signal is decoded by RF2 is increased, so is its competitiveness with cognate or near-cognate events (Major, 2001).
Is the fdhF rsUGA ‘visible’ to RF2 in the decoding site?
The in vivo studies suggested the Sec-tRNASec was able to compete effectively with premature chain termination under the conditions of our experiments. How might this be mediated? Data from structural predictions (Zinoni et al., 1990) and from use of chemical and enzymatic probes (Hüttenhofer et al., 1996a) suggest that the fdhF rsUGA is in a stem–loop, and this is a critical feature that allows the Sec-tRNASec to be ultimately competitive. However, during translation, the stem–loop must be unfolded, as decoding rsUGA by either cognate decoding molecule requires the codon to be presented as a single-stranded structure. Despite the fact that the signal can be strengthened or weakened, competition could be affected by an intrinsically poor rate of RF2 selection when the rsUGA (compared with a typical UGA signal) is occupying the A site. The specific sequences of the fdhF decoding site may make the rsUGA less accessible to the factor within the ribosomal active centre. A site-directed cross-linking strategy between the rsUGA and the factor was used to determine whether the rsUGA in the fdhF sequence context was accessible to RF2. We have shown previously that RF2 is in direct contact with a UGA termination codon positioned in the ribosomal A site. A zero-length cross-link can be formed between RF2 and a radioactively labelled mRNA analogue that contains a thioU at the +1 position of the stop signal (Brown and Tate, 1994).
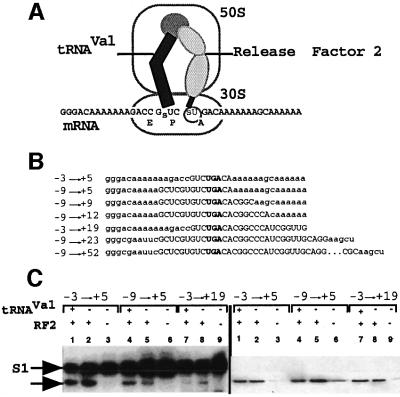
Figure 2A shows the site-directed cross-linking strategy with mRNA analogues containing fdhF sequences bound to the ribosome, with the rsUGA fixed in the A site by tRNAVal recognizing the previous codon in the P site. The designed mRNAs (used in the studies described in Figures 2 and 3) are shown in Figure 2B. They contain various lengths of fdhF sequence (–N to +N where +1 is the U of the rsUGA). For example, the –9 to +52 mRNA contains three upstream codons and extends downstream to include the stem–loop. Following the cross-linking reaction and before gel analysis, the products are digested with RNase T1, which leaves the factor cross-linked to a radiolabelled tetranucleotide derived from the mRNA. This complex is found at the same position as native RF2 on an SDS–polyacrylamide gel.
Fig. 2. Site-directed cross-linking studies to investigate whether RF2 can interact with the rsUGA at the selenocysteine incorporation site. (A) The experimental strategy. A designed mRNA is bound to the ribosome by tRNAVal located in the P site, allowing a thioU in the +1 position of the stop codon to form cross-links with the decoding RF in the A site. (B) The mRNA sequences used for the cross-linking studies (Figures 2 and 3). (C) Polyacrylamide gel separation of cross-linked complexes after RNase T1 digestion. The complexes were transferred to a nitrocellulose membrane and then detected by autoradiography (left panel) and immunodetection of RF2 (right panel). Lanes 1, 4 and 7, and lanes 3, 6 and 9 show cross-links formed in the presence or absence of RF2 and tRNAVal, respectively. Lanes 2, 5 and 8 show the cross-links formed in the presence of RF2 only. Cross-links between mRNA and RF2 (arrow) and S1 ribosomal protein are shown.
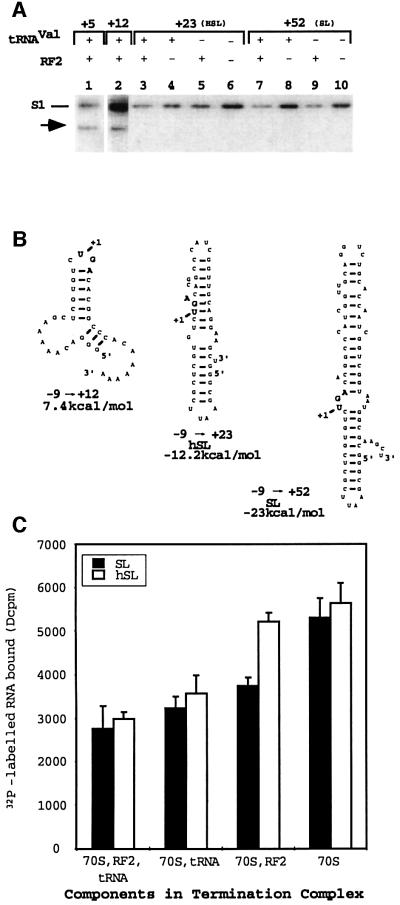
Fig. 3. Cross-linking and ribosomal binding studies with the quarter (+12), half (+23, hSL) and full (+52, SL) stem–loops. (A) Polyacrylamide gel separation of cross-linked complexes digested with RNase T1 then transferred to a nitrocellulose membrane, followed by autoradiographic detection. RF2- (arrow) and S1-specific cross-links are shown. RF2 and tRNAVal were present or absent from the reactions as indicated. (B) Secondary structure predictions for the quarter (–9 to +12), half (–9 to +23, hSL) and full (–9 to +52, SL) stem–loops determined by the program mFold. The free energy predictions for each structure are given. (C) The results of ribosomal binding assays with the hSL and SL stem–loops when different components of the termination complex are present.
Figure 2C (left panel) indicates that RF2 can cross-link to the +1 thioU of UGA in mRNAs (arrowed) where just the upstream valine codon, UGA, and two more nucleotides of the fdhF sequence are present (–3 to +5, lanes 1 and 2). Increasing the upstream sequence from one to three codons of fdhF sequence (–9 to +5, lanes 4 and 5), or the downstream sequence to +19 (lanes 7 and 8) still supported the cross-link but with decreased intensity. In the absence of tRNAVal, cross-links formed with RF2 (lanes 2, 5 and 8), consistent with previous experiments, demonstrating that RF2 contributes to termination complex alignment (E.S.Poole and W.P.Tate, unpublished data). No cross-links were found when both tRNAVal and RF2 were excluded from the reaction (lanes 3, 6 and 9). A strong cross-link to ribosomal protein S1 could be seen in all lanes and appeared more intense when tRNAVal was absent, consistent with an unoccupied P site allowing the –2 thioU of the GUC codon to contribute to the cross-link reaction in addition to the +1 thioU. The right panel shows an immunoanalysis of a duplicate gel after western transfer, showing RF2 at the same position as the 32P-labelled mRNA fragment. Faint bands in lanes 3, 6 and 9 reflect a small amount of RF2 remaining in the purified ribosome preparations.
Cross-linking reactions using mRNA analogues containing the three codons of upstream sequence and a quarter (–9 to +12), half (–9 to +23) or the full stem–loop (–9 to +52) are shown in Figure 3A. In these cases with a static termination complex, inclusion of fdhF sequences supported cross-links to RF2 with the control (–3 to +5, lane 1) and quarter stem–loop (lane 2) but not with the half stem–loop sequence (lanes 3–6) or when the full-length stem–loop is present (lanes 7–10). Despite this, these mRNAs formed cross-links to ribosomal protein S1 (Figure 3A, all lanes), indicating they were able to bind to the ribosome. The S1 cross-link intensity can vary with the mRNA sequence and length, as well as whether other components of the termination complex are present, as observed in Figure 3A. However, even at very long exposures (data not shown), no cross-links to RF2 were observed with the half and full stem–loop mRNAs.
Putative secondary structures formed by these mRNAs containing fdhF sequences and surrounding reporter sequences are shown in Figure 3B. The rsUGA (bold) and possible extra secondary structure contributed by the sequences are shown. Although the –9 to +23 sequence is unable to form the canonical stem–loop structure, the RNA folding program mFold (Zucker, 1989) indicates that it has the potential to form a stable secondary structure. On the other hand, the putative secondary structure of the shorter –9 to +12 mRNA is much weaker. The mRNA secondary structures in these static experimental complexes may contribute to their inability to be positioned on the ribosome in the right orientation for optimal RF2 UGA decoding.
We then used ribosomal binding assays to measure relative binding of the half stem–loop (hSL, –9 to +23) and full stem–loop (SL, –9 to +52) mRNAs to the ribosome in the presence or absence of the other components (Figure 3C). While the two mRNAs bound to the ribosome in a complete termination complex, omission of either RF2 or tRNA improved binding and there was a 2-fold increase in bound mRNA when both RF2 and tRNAVal were omitted. The half stem–loop showed significantly increased ribosomal binding when only tRNAVal was omitted, whereas the full stem–loop mRNA required the absence of both RF2 and tRNAVal before a significant change was observed. Consistent with the observations of Hüttenhofer et al. (1996b), the percentage of the mRNA bound to the ribosome decreased as the mRNA secondary structure increased (∼10% hSL, ∼7.5% SL) and decreased further when all the termination complex components were present.
A cross-link between the +1 U of the UGA and RF2 is a measure of a productive orientation of the stop codon and factor at the decoding site. For example, we have shown previously that the cross-link intensity can be affected significantly by the identity of the P site tRNA (McCaughan et al., 1998), and the results shown in Figure 3A and C indicate that the orientation of the rsUGA mRNAs with respect to RF2 is seriously disturbed in these static termination complexes. Heating the mRNAs to relax secondary structures immediately before complex formation did not restore the cross-linked product, although that with ribosomal protein S1 was somewhat enhanced (data not shown). However, rapid reformation of secondary structure is possible under the conditions used for these experiments. In vivo, the critical moment would be when the translating ribosome disrupts the secondary structure of the stem–loop to allow RF2 and Sec-tRNASec access to the rsUGA as it reaches the A site in a single-stranded conformation. This scenario is distinct from the static experiments with the rsRNA analogues used here.
Can competition for decoding the rsUGA be influenced by changes in the concentrations of the decoding molecules, RF2 or tRNASec?
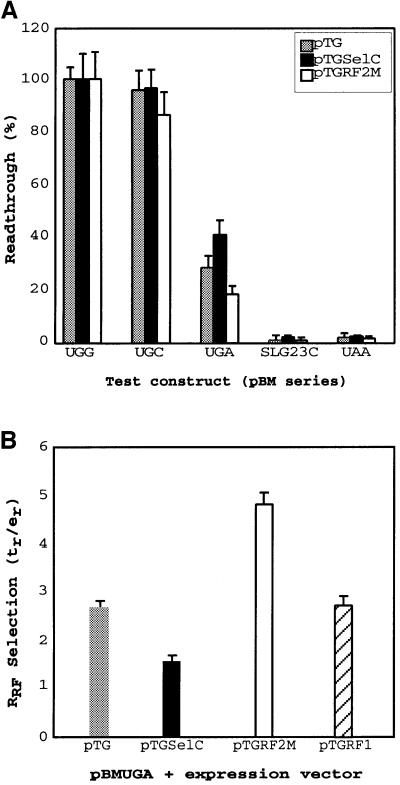
Expression vectors were constructed containing prfB (pTGRF2M) encoding RF2T246S, selC (pTGSelC) encoding tRNASec, or prfA (pTGRF1) encoding RF1, and were introduced into bacteria containing the test fdhF constructs. Expression of RF2T246S, where the threonine residue at position 246 is substituted with serine, allows more reliable production of a functionally active factor than when native RF2 is expressed, and has been used in these experiments. The host vector, pTG, was included as a control to enable any effects of the overproduction of tRNASec, RF1 or RF2 to be separated from general effects resulting from the presence of the additional vector. Overexpression allowed selection rates of the decoding factors (RF1 or RF2) and Sec-tRNASec to be influenced directly through changes in their cellular concentrations. No differential effects on the activity of the downstream product, luciferase, in the control UGG- or UGC-modified test vectors were observed with any of the pTG recombinant series (Figure 4A). They also had little effect on UAA and SLG23C (altered stem–loop) controls where decoding was near 100% termination (Figure 4A). However, at the native rsUGA, overexpression of RF2 decreased readthrough, but overexpression of tRNASec increased readthrough. When the data were analysed for relative rate of RF selection at rsUGA, there was a nearly 2-fold decrease as tRNASec was overexpressed, which contrasted with a 2-fold increase as RF2 was overexpressed (Figure 4B). Overexpression of RF2 was measured by immunoanalysis and gave a 3- to 5-fold change in cellular RF2 concentration. Significantly, overexpression of the non-cognate factor RF1 (recognizing UAG and UAA) did not influence competition.
Fig. 4. The effect of increased cellular concentration of RF2 and tRNASec on selenocysteine incorporation at different signals. (A) Selenocysteine incorporation efficiency on overexpression of the control vector (pTG), tRNASec (pTGSelC) and RF2 (pTGRF2M) at variant sequences of rsUGA as indicated, and the stem–loop variant SLG23C. (B) The relative RF selection rate at the natural selenocysteine incorporation site under conditions of overexpression of the control vector (pTG, shaded bar), tRNASec (pTGSelC, closed bar), RF2 (pTGRF2M, open bar) and non-cognate RF1 (pTGRF1, hatched bar). The standard errors of the mean are shown.
Growth conditions modulate competition at the fdhF rsUGA
To determine whether the observed competition for rsUGA decoding is regulated according to the physiological state of the bacterial cell, the effect of growth rate was investigated (Figure 5). To achieve divergent growth rates, parallel cultures were grown from the same inocula, but in rich or two different minimal media. As growth rate decreased, luciferase activity increased, indicating that readthrough at the natural rsUGA site was significantly enhanced (Figure 5A). Experiments with two different readthrough constructs (pBMUGG and pBMUGC) and a termination construct (pBMUAA) were used to assess whether background readthrough or termination rates changed under different growth conditions. No significant differences were observed. At the highest growth rate, <30% of ribosomal passages through the rsUGA resulted in selenocysteine incorporation while, at the slowest growth rates, this increased to 60% of ribosomal passages. The very slow growth rate of the E.coli FJU112 strain in this latter case results in an elevation of the background level of luciferase activity with the control plasmids pBMG23C and pBMUAA. This is likely to result from increased frequency of events that are independent of the recoding event. The RRF for the different media conditions is shown in Figure 5B.
Fig. 5. The effect of growth rate on selenocysteine incorporation. (A) The effect of growth rate on selenocysteine insertion at different recoding site signals. Readthrough (%) was determined under high (Rich, doubling time = 25 min, shaded bars), medium (Min C, doubling time = 90 min, closed bars) and slow (Min G, doubling time = 125 min, open bars) bacterial growth conditions with constructs for the different fdhF recoding sites as indicated. The SEM are shown. (B) RF2 selection rates at the natural rsUGA under the different growth conditions.
Discussion
Termination efficiency is a determinant of selenocysteine incorporation efficiency at the fdhF rsUGA
Sequence context around the fdhF rsUGA could modulate selenocysteine incorporation efficiency through a direct influence on the intrinsic selection rate of either of the two competing decoding molecules, RF2 and Sec-tRNASec. Unless higher level structures in the mRNA affect binding of one or both of these trans factors into the ribosomal A site so that they are selected at markedly different rates, increasing or decreasing the selection rate of one of the decoding molecules would be predicted to lead to a converse effect on the selection rate of the other molecule. Indeed, this simple competition seems to be operating at the fdhF selenocysteine insertion site.
The sequence modification to the recoding site present in pBMUGAU favoured termination according to prediction. The substitution of U for C at the position following the stop codon has been shown previously to increase termination efficiency at UGA signals in vivo (Poole et al., 1995). The increase in termination efficiency observed for pBMUGAU is not as dramatic as the effect of C→U substitution on the efficiency of termination at the RF2 frameshift rsUGA (Poole et al., 1995), but the mechanisms of the competing events and the kinetic consequences are distinct in the two cases. Studies have demonstrated that the recruitment for suppressor tRNAs is also influenced by the base following a stop codon, although this effect was shown to be mediated through the enhanced ability of purines 3′ to the codon to form stacking interactions with the suppressor tRNA (Kopelowitz et al., 1992). The C→U transition at the base 3′ to the rsUGA in pBMUGAU would not be predicted to affect the stability of the anticodon– codon interaction between tRNASec and rsUGA.
The sequence elements chosen for upstream and downstream contexts of the recoding site UGA in pBM‘S’ and pBM‘W’ have been shown to increase and decrease termination efficiency, respectively, as predicted (Major, 2001). It is most likely that the changes in selenocysteine incorporation efficiency observed here are also mediated through changes to the intrinsic rate of RF selection. The increase in termination with pBM‘S’ was modest, but the natural sequence context upstream of the wild-type fdhF rsUGA favours relatively efficient termination. A strong upstream termination context overcoming the effect of the weaker downstream context is consistent with observations that this can occur in the natural sequence of a poorly expressed E.coli gene (Mottagui-Tabar and Isaksson, 1997).
The changes in selenocysteine incorporation efficiency in response to modifications to the sequence context are unlikely to be due to perturbations of the mRNA stem–loop that programmes UGA recoding. The stem– loops encoded by pBM‘S’ and pBM‘W’ are similar in terms of their predicted secondary structure and stabilities (data not shown), yet the sequence modifications have opposing effects on selenocysteine incorporation. This suggests that neither sequestering the rsUGA codon to prevent recognition by RF2, nor an increased ribosomal pause at the site provided by the additional stability conferred on the entire stem–loop from the lower helical region are essential for efficient selenocysteine incorporation. This is consistent with results of a mutational analysis of the structural requirements of the fdhF hairpin for selenocysteine incorporation (Liu et al., 1998).
In an innovative approach, Sandman and Noren (2000) have used phage display to show that the nucleotide following the internal TGA in the fdhF gene determines whether suppression with a near-cognate tRNA competes with co-translational selenocysteine incorporation: a purine nucleotide in this position supports competition whereas a pyrimidine, such as the naturally occurring C, does not. Liu et al. (1999) propose an extended selenocysteine element in the mRNA that includes the nucleotide following the rsUGA and the two preceding codons, preventing near-cognate readthrough of the UGA when selenium is limiting. We interpret this to reflect elements that are important for the termination signal and strengthen or weaken termination efficiency (Figure 1B and C; Major, 2001). Selenocysteine occupies the active site of FDHH. Therefore, the codons immediately upstream and downstream of the rsUGA encode residues that influence the local environment of selenocysteine and, consequently, the activity of the site. Furthermore, the codon immediately downstream of the rsUGA encodes His141, a residue critical for catalytic activity of the enzyme (Boyington et al., 1997). This suggests that there has been an evolutionary optimization at the fdhF rsUGA to ensure a functional active site and to be certain that premature termination during synthesis is not too competitive so that production of a full-length protein is possible.
The sequence context surrounding the rsUGA does not preclude recognition by RF2
RF2 was able to cross-link to the +1 thioU in the rsUGA in a variety of test mRNA analogues containing different sequence elements around the fdhF rsUGA. This indicated that neither the primary sequence context adjacent to the rsUGA nor the presence of tRNAVal in the ribosomal P site prevented stop codon recognition by RF2. In contrast, RF2 did not cross-link to mRNA with the fdhF stem–loop and failed to cross-link to mRNA capable of forming a stable secondary structure different from the canonical stem– loop. mRNAs with predicted secondary structures of lower stability supported cross-links to RF2, but in lower yields than comparable mRNAs less likely to form any stable secondary structures.
This suggests that any significant downstream secondary structures, such as the fdhF stem–loop, have the potential to prevent, or greatly decrease, formation of an mRNA–RF2 interaction. The stability of such structures during in vivo translation and at the transient stage of their melting may contribute to a weaker RF2 competition at the rsUGA in fdhF than would normally occur at other UGA termination sites. However, our data indicate that RF2 is still able to decode rsUGA efficiently in vivo, and is affected by the same parameters influential at a typical UGA termination site, reinforcing expectations that the secondary structure must be melted before entry of the rsUGA codon into the ribosomal A site.
RF2 and tRNASec compete for rsUGA
The 2-fold elevation in the determined rate of Sec-tRNASec selection relative to termination brought about by increasing the pool of tRNASec (see Figure 4) is in accord with the previous observation that a similar elevation of readthrough of a UGA-containing fdhF–lacZ fusion was mediated by overexpression of tRNASec (Tormay et al., 1996). From this observation, the authors concluded that Sec-tRNASec is the limiting component of the selenocysteine incorporation machinery. Indeed, while estimates of tRNASec have varied between ∼220 (Dong et al., 1996) and 350 molecules per cell (Tormay et al., 1996), both estimates are several fold lower than that deduced for the specific elongation factor, SELB (1100 molecules per cell) (Forchhammer et al., 1990). This suggests that a number of selenocysteine incorporation complexes (in the form of mRNA–SELB–GTP ternary complexes) lack Sec-tRNASec. The increased efficiency of selenocysteine incorporation observed on expression of pTGSelC (see Figure 4) was most probably the result of an increase in the amount of Sec-tRNASec available to interact with the mRNA–SELB–GTP ternary complex, which led to increased levels of the competent quaternary complex, mRNA–SELB–GTP–Sec-tRNASec.
Previously, it has been shown that increasing decoding RF concentration at the RF2 frameshift rsUGA leads to an increase in RF selection rate at stop signals introduced at the site (Crawford et al., 1999). The results at the fdhF rsUGA with overexpression of RF2 (see Figure 4) indicate that there was an improved RF2 competitiveness specifically with the SELB quaternary complex for decoding wild-type rsUGA, and suggest that even in the presence of the SELB quaternary complex, the rsUGA in the A site is ‘visible’ to RF2, consistent with previous observations (Tormay and Böck, 1997; Suppmann et al., 1999).
Selenocysteine incorporation is likely to be an efficient process during anaerobically induced expression of fdhF
We have conducted many studies at high growth rates in rich media where termination efficiency at the wild-type fdhF rsUGA allows only residual incorporation of 75Se (Mansell, 1999). Suppmann et al. (1999) also found a low absolute efficiency of selenocysteine incorporation in rapidly growing cells. Increased selenocysteine incorporation efficiency occurs relative to termination at the rsUGA as the growth rate of the host bacterium decreases (see Figure 5). Critical differences in relative concentrations of each key component of the quaternary complex (mRNA–SELB–GTP–Sec-tRNASec) at different bacterial growth rates may result in complete or incomplete complex assembly on the mRNA. This may be responsible for higher or lower competitiveness of the complex with RF at different E.coli growth rates (see Figure 5). However, the expression levels of the upstream reporter, lacZ, from pBMUGA at all growth rates were comparable (data not shown), suggesting that the decreased rate of Sec-tRNASec selection relative to termination at high growth rates does not result simply from an increased rate of ribosomal passage on lacZ–fdhF–luc+ mRNAs that have no associated complex.
It must be considered that fdhF is expressed only during anaerobic growth, when growth rates are likely to be significantly slower than the high rates achieved by cells growing aerobically in relatively rich media. As growth rate decreases, the proportion of RF2 associated with ribosomes is lowered and the overall number of RF2 molecules is reduced in the cell, although not apparently their free concentration (Adamski et al., 1994). The results presented here suggest that at very slow growth rates, the selection rate of Sec-tRNASec may exceed the RF2 selection rate (see Figure 5A). Parallel experiments performed in the same growth media under anaerobic conditions (data not shown) showed that the dominant factor giving differences in competition between selenocysteine incorporation and termination at the rsUGA was the nature of the medium rather than the aerobic or anaerobic condition, even though anaerobic growth rates were slower for each medium. Consequently, the parameters affecting competition may be more involved than simply the growth rate.
How is competition between the RF2 and Sec-tRNASec decoding molecules mediated?
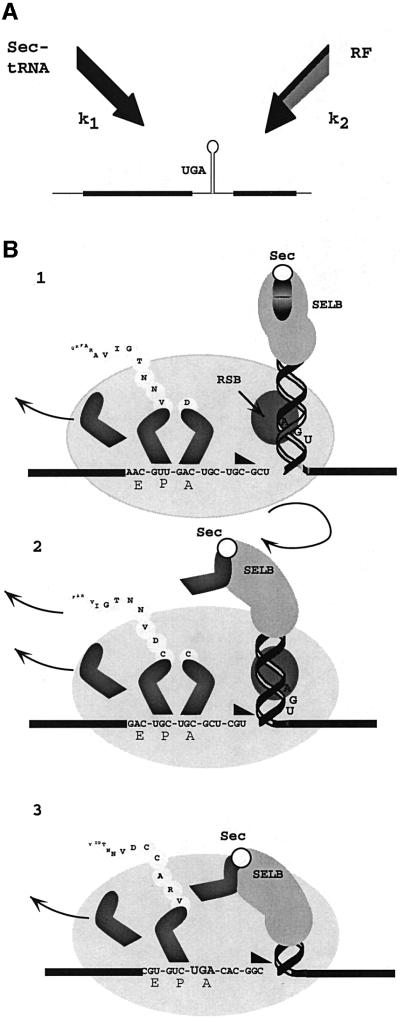
The results demonstrate that there is a reciprocal relationship between the termination and selenocysteine incorporation efficiencies at the fdhF rsUGA and provide compelling evidence for competition between the canonical and non-canonical decoding events (Figure 6A). The successful event will depend on the selection rates of the two decoding molecules, Sec-tRNASec (k1) and RF2 (k2). The competitiveness of a decoding molecule can change according to the relative concentrations of the participating molecules. However, SELB-mediated decoding is unusual in that the complex must be pre-bound to the fdhF SECIS in order to interact efficiently with the ribosome and mediate rsUGA decoding. Under non-limiting conditions with a pre-bound complex, k1 will be invariant. In contrast, the rate at which the rsUGA codon can recruit RF2 will depend on factor concentration, and k2 will alter. Under these circumstances, k2 will determine changes in competition outcome. When the SELB complex is limiting, changes in the concentration of the decoding Sec-tRNASec and, thereby, the proportion of fdhF mRNA molecules loaded with complex will critically affect competition.
Fig. 6. A possible mechanism for regulation of competition between the RF2 and Sec-tRNASec decoding molecules at the fdhF selenocysteine insertion site. (A) A kinetic scheme of the competition between the decoding molecules, RF2 and Sec-tRNASec, at the rsUGA. k1 and k2 represent the rate of Sec-tRNASec and RF2 association, respectively. (B) The ‘helical approach’ model for competitive delivery of Sec-tRNASec to the rsUGA. (1) The SELB complex carrying the Sec-tRNASec is bound to the apical tip of the fdhF stem–loop as the sequence approaches the ribosomal decoding site. (2) The ribosomal melting impetus (triangle) encounters the stem–loop and induces unwinding. (3) The SELB complex is positioned towards the ribosomal SELB-binding site (RSB) allowing proximity and delivery of Sec-tRNASec to the ribosomal A site in competition with RF2.
Mansell (1999) has proposed a ‘helical approach’ model as a mechanism for how competition is mediated at the A site of the ribosome (Figure 6B). The SELB complex carrying the Sec-tRNASec is bound to the apical loop of the fdhF stem–loop as the sequence approaches the ribosomal decoding site (1). If the complex is to remain bound, it must rotate about the axis of the helical stem as the secondary structure unwinds. There is likely to be a ribosomal pause or translational ‘slow-down’ because of the increased torsional load imposed by the unwinding hairpin (2). Indeed, Suppmann et al. (1999) observed such a ribosomal pause at the rsUGA. At this point, residual secondary structure would spatially constrain the quaternary complex so that it is ideally positioned to deliver Sec-tRNASec just as the rsUGA reaches the A site (3). Thanbichler et al. (2000) show that the affinity of SELB for its binding site would decrease when Sec-tRNASec is released. Under conditions of low RF2 concentration (k2 low), this would give the Sec-tRNASec a significant advantage over the decoding RF2 to be the first to reach the inner cavern of the ribosomal active centre. Indeed, this may be why the relatively strong termination context of the rsUGA performs poorly against this competition. At high RF2 concentrations (k2 high), the creation of a translational ‘slow-down’ by the ‘helical approach’ of the quaternary complex may be just sufficient to provide the opportunity for RF2 to be successful for decoding rsUGA.
Recent structural determinations of the ribosome suggest that the mRNA threads through a hole formed at the decoding site and that several bases downstream from the A site codon would be covered (Yusupova et al., 2001). How does this affect the ‘helical approach’ model? The SELB complex binds to the apex of the stem–loop of the SECIS element, with a minihelix containing 17 bases, sufficient to support selenocysteine incorporation (Thanbichler et al., 2000). This implies that it should be possible to unwind the helix, including the UGA and up to 10 further bases, to thread through the mRNA track without the complex being affected. By the time the UGA moves into the A site, it should still be possible for the Sec-tRNASec to be released from the complex and for SELB to dissociate without major disruption to the structure of the ribosomal mRNA channel.
Materials and methods
Materials
Plasmids were purified using a QIAprep® Spin Miniprep Kit (Qiagen) and were introduced into bacterial cells by electroporation using an Electro Cell Manipulator® 600 (BTX). β-galactosidase activity was analysed using a EL340 (BIO-TEK) or EAR340 (SLT) microplate reader, and luciferase activity was measured in an AutoLumat LB953 luminometer (EG&G Berthold). [α-32P]GTP (3000 Ci/mmol) and the ECL western blotting analysis system were purchased from Amersham. In vitro transcription reactions were carried out using a RiboMAX™ Large Scale RNA Production System (Promega). tRNAVal was purchased from Sigma and restriction endonucleases and RNase T1 were purchased from Roche. UV irradiation was performed using a 15 W National Matsushita black light (FL 15 BL) in a Griffin UV light box through a glass filter that was impermeable to light of wavelength <300 nm. A Mini-PROTEAN II electrophoresis cell (Bio-Rad) and Mini Trans-Blot electrophoretic transfer cell (Bio-Rad) were used for gel electrophoresis and protein transfer, respectively. For the ribosome binding assays, GF/C glass fibre filters (Whatman) were used and the radiation was measured in OPTI-FLOUR® scintillant (Packard) using a LKB 1211 RACKBETA liquid scintillation counter.
Bacterial strains and growth media
Expression studies were carried out in the E.coli strain FJU112 [Δ(lac-pro) gyrA ara recA56 Tn10, F′ lacIQ1] (Jørgensen and Kurland, 1990). This strain has wild-type ribosomes, and no suppressor tRNAs that could compete with termination or selenocysteine incorporation events. Bacteria were grown aerobically in M9 medium supplemented with 0.2% glucose, 10 µg/ml thiamine, 1 mM MgSO4, 0.1 mM CaCl2, 1 µM Na2SeO3 and the trace elements according to Neidhardt et al. (1974). To achieve divergent growth rates, bacteria were grown in: (i) rich medium (1% tryptone, 0.5% yeast extract, 0.5% glycerol, 0.1 M potassium phosphate pH 7.0, 1 mM MgSO4, 0.1 mM CaCl2, 1 µM Na2SeO3 and the trace elements as above); (ii) minimal C medium (M9 supplemented with 0.3% glucose, 0.3% casamino acids, 1 µM Na2SeO3 and the trace elements as above); or (iii) minimal G medium (M9 supplemented with 0.43% glucose, 1 µM Na2SeO3 and the trace elements as above). Antibiotics ampicillin and gentamicin were added to final concentrations of 100 and 10 µg/ml, respectively.
Plasmid construction
Standard recombination techniques were used in the construction of test plasmids. Plasmid pTG contains the p15A origin of replication from pACYC184 (New England Biolabs), allowing co-existence with plasmids with a ColE1 origin of replication. A synthetic trpE promoter and minimal polylinker fragment were inserted by annealing complementary oligonucleotides prior to ligation with a fragment derived from pACYC184 containing both the p15A origin of replication and the gene conferring tetracycline resistance. Next, the tetracycline resistance gene was replaced with a fragment conferring gentamicin resistance from plasmid pOT182. Plasmid pTGRF1 contains the prfA gene encoding RF1 obtained from plasmid pRF1-2 (Donly et al., 1990). Plasmid pTGRF2M was constructed by D.Wilson, and contains the prfB gene with a Thr246Ser mutation, which alleviates the decrease in specific activity observed when RF2 is overexpressed (Wilson et al., 2000). Plasmid pTGSelC was constructed by ligation of a fragment containing the selC gene from pMN81 (Leinfelder et al., 1988) into pTG.
Construction of the pBM series of plasmids has been described in detail elsewhere (Mansell, 1999). Briefly, pBM contains a ColE1 origin of replication, ampicillin resistance and the lacZ–fdhF–luc+ fusion under the control of an inducible trpE promoter. Test fdhF sequence elements were introduced by ligation after either annealing synthetic oligonucleotides or using PCR primers to amplify target sequences in plasmids in which the rsUGA codon had been replaced. The likelihood of translation reinitiation giving β-galactosidase-independent luciferase activity was reduced by removing the initiation codon at the start of the luc+ gene by site-directed mutagenesis. Plasmid pBMG23C contains the G23→C mutation shown previously to abolish selenocysteine incorporation in vivo (Heider et al., 1992).
The pGhSL and pGSL vectors were designed so that in vitro transcription reactions from a T7 promotor would provide mRNA for cross-linking studies (Mansell, 1999). In brief, the pGem3Zf(+) vector (Promega) was ligated with annealed oligonucleotides that comprised the half stem–loop sequence (pGhSL) or ligated with an excised fragment that contained the complete stem–loop sequence (pGSL).
Induction of βgal–fdhF–luc+ fusion protein expression
Freshly transformed individual colonies were grown overnight in M9 medium supplemented as described above with the addition of 20 µg/ml tryptophan to repress expression. Aliquots (200 µl) of overnight cultures were inoculated into 4 ml of fresh medium without tryptophan, and bacteria were grown to an OD600 of 0.5–0.6 before the addition of 3-β-indoleacrylic acid to 50 µg/ml to induce expression of βgal–fdhF–luc+ fusion proteins. Bacteria were harvested 2–3 h post-induction.
β-galactosidase and luciferase activity assays
β-galactosidase activity was determined as described (Miller, 1972), with the following modifications. The OD600 of cultures was measured, then equal numbers of cells pelleted by centrifuging at 4000 g. Cells were resuspended in 200 µl of luciferase resuspension buffer [0.1 M Tricine pH 7.8, 2 mM dithiothreitol (DTT), 2 mM EDTA, 10% (v/v) glycerol, 1% (v/v) Triton X-100] (Tanguay and Gallie, 1996) and lysed by freezing on dry ice for 10 min followed by thawing at room temperature. Dilutions of cell lysates in microtitre plates were assayed for β-galactosidase activity.
For the luciferase activity assay, equal volumes of lysate typically 5–10 µl) were added to 50 µl of luciferase assay buffer [20 mM Tricine pH 7.8, 1.07 mM (MgCO3)4·Mg(OH)2·5H2O, 2.67 mM MgSO4, 0.1 mM EDTA, 33.3 mM DTT, 0.27 mM CoA, 0.5 mM luciferin, 0.5 mM ATP] (Tanguay and Gallie, 1996).
Calculation of readthrough values and relative rate of RF selection
The readthrough value was obtained from the ratio of luciferase activity to β-galactosidase activity. The mean readthrough control (RTC) value derived from clones containing the RTC plasmid pBMUGG was set to 100%. The luciferase:β-galactosidase ratio for each individual test clone was expressed as a percentage of the mean RTC value to give the readthrough value for that clone. The result of several experiments for a given construct allowed a mean readthrough value for each clone to be determined. The SEM of the readthrough values was determined for each clone from the multiple analyses.
The RF selection rate (Pedersen and Curran, 1991) is a measure of the efficiency of stop signal recognition by the RF. Pedersen and Curran determined the rates of RF selection (RRF) at different UAGN signals and their formula was adapted for these experiments. In our experiments, the rates of termination (T) and readthrough (I) were determined within a single experiment. Therefore, the RRF relative to readthrough was the ratio of termination to readthrough, RRF = T/I. A background level given by the apparent readthrough value of pBMUAA (background luciferase activity) was subtracted from the readthrough values of the remaining constructs. The mean RTC value determined with UGG was set as 100%. The values from test constructs then were expressed as a percentage. The error calculated in the corrected readthrough value for each construct was the sum of the SEM of pBMUAA and the SEM for that construct.
UV cross-linked studies
In vitro transcription reactions to incorporate thioUTP and [α-32P]GTP into designed RNAs, the formation of ribosomal complexes and site-directed cross-links, and gel analysis of cross-linked complexes were carried out as described (Poole et al., 1997). Transcription reactions were performed in a volume of 50 µl and ribosomal complexes contained 50 pmol RF2. ECL immunodetection of cross-linked RF2 after western transfer onto a nitrocellulose membrane was achieved using a 1:4000 dilution of primary antibody against RF2 followed by a 1:10 000 dilution of secondary antibody. In vitro transcription reactions to produce half stem–loop (hSL, –9 to +23) and full stem–loop (SL, –9 to +52) mRNA were performed as described except that ∼10 µg of linearized pGhSL and pGSL vectors were used as a template. Control ribosomal complexes lacked both tRNAVal and RF2, or tRNAVal only.
Ribosome binding assay
Assays to measure the ribosomal binding capacity of half stem–loop (hSL, –9 to +23) and full stem–loop (SL, –9 to +52) [α-32P]GTP-labelled mRNA transcripts used the method of Tate and Caskey (1990), except that reactions were incubated for 30 min and contained 50 pmol ribosomes, 100 pmol mRNA, 200 pmol tRNAVal and/or 100 pmol RF2, as appropriate, in binding buffer [50 mM Tris(OAc)2 pH 7.2, 20 mM Mg(OAc)2 and 100 mM NH4Cl]. Specific activities of hSL and SL mRNA transcripts were 720 and 1035 c.p.m./pmol, respectively. Bound mRNA was calculated from radioactivity retained on glass fibre filters measured by Cerenkov or liquid scintillant radiation.
Acknowledgments
Acknowledgements
The authors would like to thank C.Edgar, P.Flawn and Dr L.Major for their technical support, and K.Barwell for the graphic design in the ‘helical approach’ model. This work was supported by a grant awarded to W.P.T. and a postgraduate scholarship to J.B.M. from the Health Research Council of New Zealand, and a grant from the Human Frontier Science Program.
References
- Adamski F.M., McCaughan,K.K., Jørgensen,F., Kurland,C.G. and Tate, W.P. (1994) The concentration of polypeptide chain release factors 1 and 2 at different growth rates of Escherichia coli. J. Mol. Biol., 238, 302–308. [DOI] [PubMed] [Google Scholar]
- Björnsson A., Mottagui-Tabar,S. and Isaksson,L.A. (1996) Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J., 15, 1696–1704. [PMC free article] [PubMed] [Google Scholar]
- Böck A., Forchhammer,K., Heider,J. and Baron,C. (1991) Selenoprotein synthesis: an expansion of the genetic code. Trends Biochem. Sci., 16, 463–467. [DOI] [PubMed] [Google Scholar]
- Bossi L. and Roth,JR. (1980) The influence of codon context on genetic code translation. Nature, 286, 123–127. [DOI] [PubMed] [Google Scholar]
- Boyington J.C., Gladyshev,V.N., Khangulov,S.V., Stadtman,T.C. and Sun,P.D. (1997) Crystal structure of formate dehydrogenase H: catalysis involving Mo, molybdopterin, selenocysteine and an Fe4S4 cluster. Science, 275, 1305–1308. [DOI] [PubMed] [Google Scholar]
- Brown C.M. and Tate,W.P. (1994) Direct recognition of mRNA stop signals by Escherichia coli polypeptide chain release factor two. J. Biol. Chem., 269, 33164–33170. [PubMed] [Google Scholar]
- Crawford D.J., Ito,K., Nakamura,Y. and Tate,W.P. (1999) Indirect regulation of translational termination efficiency at highly expressed genes and recoding sites by the factor recycling function of Escherichia coli release factor RF3. EMBO J., 18, 727–732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H.J., Nilsson,L. and Kurland,C.G. (1996) Co-variation of tRNA abundance and codon usage in Escherichia coli at different growth rates. J. Mol. Biol., 260, 649–663. [DOI] [PubMed] [Google Scholar]
- Donly B.C., Edgar,C.D., Williams,J.M. and Tate,W.P. (1990) Tightly controlled expression systems for the production and purification of Escherichia coli release factor 1. Biochem. Int., 20, 437–443. [PubMed] [Google Scholar]
- Engelberg-Kulka H. (1981) UGA suppression by normal tRNATrp in Escherichia coli: codon context effects. Nucleic Acids Res., 9, 983–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K., Rucknagel,K.P. and Böck,A. (1990) Purification and biochemical characterization of SELB, a translation factor involved in selenoprotein synthesis. J. Biol. Chem., 265, 9346–9350. [PubMed] [Google Scholar]
- Heider J., Baron,C. and Böck,A. (1992) Coding from a distance: dissection of the mRNA determinants required for the incorporation of selenocysteine into protein. EMBO J., 11, 3759–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A. and Böck,A. (1998) Selenocysteine inserting RNA elements modulate GTP hydrolysis of elongation factor SelB. Biochemistry, 37, 885–890. [DOI] [PubMed] [Google Scholar]
- Hüttenhofer A., Westhof.E. and Böck,A. (1996a) Solution structure of mRNA hairpins promoting selenocysteine incorporation in Escherichia coli and their base-specific interaction with special elongation factor SELB. RNA, 2, 354–366. [PMC free article] [PubMed] [Google Scholar]
- Hüttenhofer A., Heider,J. and Böck,A. (1996b) Interaction of the Escherichia coli fdhF mRNA hairpin promoting selenocysteine incorporation with the ribosome. Nucleic Acids Res., 24, 3903–3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen F. and Kurland,C.G. (1990) Processivity errors of gene expression in Escherichia coli.J. Mol. Biol., 215, 511–521. [DOI] [PubMed] [Google Scholar]
- Klug S.J., Hüttenhofer,A., Kromayer,M. and Famulok,M. (1997) In vitro and in vivo characterization of novel mRNA motifs that bind special elongation factor SelB. Proc. Natl Acad. Sci. USA, 94, 6676–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopelowitz J., Hampe,C., Goldman,R., Reches,M. and Engelberg-Kulka,H. (1992) Influence of codon context on UGA suppression and readthrough. J. Mol. Biol., 225, 261–269. [DOI] [PubMed] [Google Scholar]
- Leinfelder W., Zehelein,E., Mandrand-Berthelot,M.A. and Böck,A. (1988) Gene for a novel tRNA species that accepts l-serine and cotranslationally inserts selenocysteine. Nature, 331, 723–725. [DOI] [PubMed] [Google Scholar]
- Liu Z., Reches,M., Groisman,I. and Engelberg-Kulka,H. (1998) The nature of the minimal ‘selenocysteine insertion sequence’ (SECIS) in Escherichia coli. Nucleic Acids Res., 26, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z., Reches,M. and Engelberg-Kulka,H. (1999) A sequence in the Escherichia coli fdhF ‘selenocysteine insertion sequence’ (SECIS) operates in the absence of selenium. J. Mol. Biol., 294, 1073–1086. [DOI] [PubMed] [Google Scholar]
- Major L.L. (2001) Is the prokaryotic termination signal a simple triplet codon or an extended sequence element? PhD Thesis, University of Otago, Otago, New Zealand.
- Mansell J.B. (1999) UGA: selenocysteine or stop? PhD Thesis, University of Otago, Otago, New Zealand.
- McCaughan K.K., Poole,E.S., Pel,H.J., Mansell,J.B., Mannering,S.A. and Tate,W.P. (1998) Efficient in vitro translational termination in Escherichia coli is constrained by the orientations of the release factor, stop signal and peptidyl-tRNA within the termination complex. Biol. Chem., 379, 857–866. [DOI] [PubMed] [Google Scholar]
- Miller J.F. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Mottagui-Tabar S. and Isaksson,L.A. (1997) Only the last amino acids in the nascent peptide influence translation termination in Escherichia coli genes. FEBS Lett., 414, 165–170. [DOI] [PubMed] [Google Scholar]
- Mottagui-Tabar S., Björnsson,A. and Isaksson,L.A. (1994) The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J., 13, 249–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neidhardt F.C., Bloch,P.L. and Smith,D.F. (1974) Culture medium for enterobacteria. J. Bacteriol., 119, 736–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen W.T. and Curran,J.F. (1991) Effects of the nucleotide-3′ to an amber codon on ribosomal selection rates of suppressor transfer RNA and release factor-1. J. Mol. Biol., 219, 231–241. [DOI] [PubMed] [Google Scholar]
- Poole E.S., Brown,C.M. and Tate,W.P. (1995) The identity of the base following the stop codon determines the efficiency of in vivo translational termination in Escherichia coli. EMBO J., 14, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole E.S., Brimacombe,R. and Tate,W.P. (1997) Decoding the translational termination signal: the polypeptide chain release factor in Escherichia coli crosslinks to the base following the stop codon. RNA, 3, 974–982. [PMC free article] [PubMed] [Google Scholar]
- Poole E.S., Major,L.L., Mannering,S.A. and Tate,W.P. (1998) Translational termination in Escherichia coli: three bases following the stop codon crosslink to release factor 2 and affect the decoding efficiency of UGA-containing signals. Nucleic Acids Res., 26, 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salser W., Fluck,M. and Epstein,R. (1969) The influence of the reading context upon the suppression of nonsense codons, III. Cold Spring Harbor Symp. Quant. Biol., 34, 513–520. [DOI] [PubMed] [Google Scholar]
- Sandman K. and Noren,C.J. (2000) The efficiency of Escherichia coli selenocysteine insertion is influenced by the downstream nucleotide. Nucleic Acids Res., 28, 755–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suppmann S., Persson,B.C. and Böck,A. (1999) Dynamics and efficiency in vivo of UGA-directed selenocysteine insertion at the ribosomes. EMBO J., 18, 2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanguay R.L. and Gallie,D.R. (1996) Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol., 16, 146–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate W.P. and Caskey,C.T. (1990) Termination of protein synthesis. In Spedding,G. (ed.), Ribosomes and Protein Synthesis: A Practical Approach. IRL Press, Oxford, UK, pp. 81–100.
- Tate W.P., Poole,E.S., Dalphin,M.E., Major,L.L., Crawford,D.J.G. and Mannering,S.A. (1996) The translational stop signal: codon with a context, or extended factor recognition element? Biochimie, 78, 945–952. [DOI] [PubMed] [Google Scholar]
- Thanbichler M., Böck,A. and Goody,R.S. (2000) Kinetics of the interaction of translation factor SelB from Escherichia coli with guanine nucleotides and selenocysteine insertion sequence RNA. J. Biol. Chem., 275, 20458–20466. [DOI] [PubMed] [Google Scholar]
- Tormay P. and Böck,A. (1997) Barriers to heterologous expression of a selenoprotein gene in bacteria. J. Bacteriol., 179, 576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormay P., Sawers,A. and Böck,A. (1996) Role of stoichiometry between mRNA, translation factor SelB and selenocysteyl-tRNA in selenoprotein synthesis. Mol. Microbiol., 21, 1253–1259. [DOI] [PubMed] [Google Scholar]
- Wilson D.N., Guévremont,D. and Tate,W.P. (2000) The ribosomal binding and peptidyl-tRNA hydrolysis functions of Escherichia coli release factor 2 are linked through residue 246. RNA, 6, 1704–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupova G.Zh., Yusupov,M.M., Cate,J.H.D. and Noller,H.F. (2001) The path of the messenger RNA through the ribosome. Cell, 106, 233–241. [DOI] [PubMed] [Google Scholar]
- Zinoni F., Heider,J. and Böck,A. (1990) Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc. Natl Acad. Sci. USA, 87, 4660–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucker M. (1989) On finding all suboptimal foldings of an RNA molecule. Science, 244, 48–52. [DOI] [PubMed] [Google Scholar]