Abstract
FREQUENCY (FRQ) is a crucial element of the circadian clock in Neurospora crassa. In the course of a circadian day FRQ is successively phosphorylated and degraded. Here we report that two PEST-like elements in FRQ, PEST-1 and PEST-2, are phosphorylated in vitro by recombinant CK-1a and CK-1b, two newly identified Neurospora homologs of casein kinase 1ε. CK-1a is localized in the cytosol and the nuclei of Neurospora and it is in a complex with FRQ in vivo. Deletion of PEST-1 results in hypophosphorylation of FRQ and causes significantly increased protein stability. A strain harboring the mutant frqΔPEST-1 gene shows no rhythmic conidiation. Despite the lack of overt rhythmicity, frqΔPEST-1 RNA and FRQΔPEST-1 protein are rhythmically expressed and oscillate in constant darkness with a circadian period of 28 h. Thus, by deletion of PEST-1 the circadian period is lengthened and overt rhythmicity is dissociated from molecular oscillations of clock components.
Keywords: casein kinase 1ε/circadian/frequency/Neurospora/PEST
Introduction
Most organisms use circadian clocks to regulate physiological and behavioral activities in a time-of-day specific manner. Circadian clocks are cell-autonomous and mediate rhythmic expression of a broad variety of clock-controlled genes with a periodicity of approximately one day even in the absence of environmental cues. Circadian clocks are entrainable by exogenous stimuli (zeitgebers) such as light, temperature and nutrition. Entrainment synchronizes the endogenous oscillations with the exogenous time and allows behavioral flexibility. On the molecular level circadian oscillations are supported by interconnected transcriptional–translational feedback loops of considerable complexity. The interconnected loops seem to function in a similar manner in fungi, flies and mammals (Dunlap et al., 1999; Young, 2000; Albrecht, 2001; Reppert and Weaver, 2001; Williams and Sehgal, 2001).
The circadian system of Drosophila melanogaster is well characterized on the molecular level. Drosophila CLOCK (CLK) and CYCLE (CYC) are Per-Arnt-Sim-like (PAS) domain-containing transcription factors that control transcription of period (per) and timeless (tim) RNA (Allada et al., 1998; Darlington et al., 1998; Rutila et al., 1998; McDonald et al., 2001; Wang et al., 2001). When PER and TIM proteins are synthesized they form a complex that enters the nucleus and represses transcription of their own genes by inactivating CLK–CYC (Hardin et al., 1990; Saez and Young, 1996; Lee et al., 1998, 1999). During the course of a day PER and TIM are degraded, which again allows synthesis of per and tim RNA by CLK–CYC (Young, 2000; Williams and Sehgal, 2001). The length of a circadian period is affected by the kinetics of nuclear entry and degradation of PER–TIM. TIM is phosphorylated by SHAGGY, which regulates nuclear translocation of the PER–TIM heterodimer (Martinek et al., 2001). The casein kinase 1ε (CK-1ε) homolog DOUBLETIME (DBT) triggers degradation of PER via phosphorylation and thereby regulates period length (Price et al., 1998; Rothenfluh et al., 2000; Kloss et al., 2001). PER and TIM not only repress synthesis of their own genes but also support the expression of CLK (Bae et al., 1998; Glossop et al., 1999). Furthermore, TIM increases per RNA levels through a post-transcriptional mechanism (Suri et al., 1999).
A central element of the circadian feedback loop of Neurospora crassa is frequency (frq). Expression of frq RNA depends on the oligomeric PAS domain containing transcription factors white collar (WC)-1 and WC-2 (Crosthwaite et al., 1997; Talora et al., 1999). On the other hand, FRQ controls the expression of wc-1 RNA (Merrow et al., 2001) and WC-1 (Lee et al., 2000; Cheng et al., 2001). As a consequence frq RNA and FRQ protein levels oscillate under free-running conditions (constant darkness) with a period of ∼22 h (Aronson et al., 1994; Garceau et al., 1997). frq RNA levels reach a maximum in the subjective late morning followed by a FRQ protein peak ∼4 h later (Garceau et al., 1997; Merrow et al., 1999). FRQ protein enters the nucleus and, when expressed at sufficiently high levels, represses synthesis of its own RNA (Aronson et al., 1994; Merrow et al., 1997; Luo et al., 1998).
In the course of the day, FRQ protein is progressively hyperphosphorylated and then degraded. When FRQ protein levels fall below a threshold frq RNA is synthesized again and a new circadian period begins (Garceau et al., 1997). The roles of the multiple phosphorylations of FRQ are not known. However, phosphorylation of FRQ at Ser513 by an unidentified kinase affects protein stability and the length of the circadian period (Liu et al., 2000).
FRQ contains two PEST-like sequences, PEST-1 and PEST-2 (Merrow and Dunlap, 1994). PEST sequences are rich in proline (P), aspartic and glutamic acid (E), serine (S), threonine (T) (Rogers et al., 1986). In many cases PEST sequences are implicated in the regulation of protein turnover (Rechsteiner and Rogers, 1996; Marchal et al., 2000; Roth and Davis, 2000).
Here we show that PEST-1 and PEST-2 of FRQ are targets for phosphorylation in vitro by CK-1a and CK-1b, two newly identified casein kinases 1 of N.crassa. Deletion of PEST-1 results in loss of the circadian rhythm of conidiation. FRQΔPEST-1 is expressed at high levels; it is hypophosphorylated and significantly more stable than authentic FRQ. Unexpectedly, frqΔPEST-1 RNA and FRQΔPEST-1 protein as well as WC-1 protein oscillate under constant conditions with a period of ∼28 h. The deletion of the PEST-1 sequence in FRQ dissociates the overt conidiation rhythm of Neurospora from molecular oscillations of critical clock components.
Results
Phosphorylation of PEST sequences in FRQ by casein kinases 1
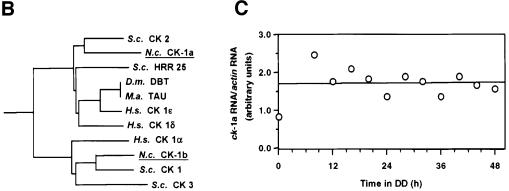
The length of a circadian period in Drosophila is regulated via phosphorylation of the clock protein PER by DBT, a homolog of human casein kinase 1ε (CK-1ε) (Kloss et al., 1998). Searching the Neurospora genome we found two CK-1 homologs, CK-1a, which is closely related to DBT, and CK-1b (Figure 1A and B). Attempts to inactivate CK-1a by repeat-induced point mutations (RIPs) (Selker and Garrett, 1988) were unsuccessful (data not shown), suggesting that the gene may be essential for cell viability. To characterize expression of CK-1a Neurospora was grown in the dark. Samples were harvested at 4 h intervals over a time course of 48 h and ck-1a RNA was analyzed by reverse transcription and real time quantitative polymerase chain reaction (RT–PCR) (Figure 1C) and northern blotting (not shown). ck-1a RNA was expressed at constant levels throughout the 48 h period.
Fig. 1. Identification of N.crassa casein kinases 1, N.c. CK-1a and N.c. CK-1b. (A) Sequence alignment of amino acid residues 1–298 of N.c. CK-1a and residues 1–302 of N.c. CK-1b with the N-terminal kinase domains of selected members of the casein kinase 1 family. The amino acid sequence of N.c. CK-1a is shown in the upper lane. Residues identical to N.c. CK-1a are indicated by a dot. (B) Phylogenetic relationship of members of the casein kinases 1 family. D.m. DBT: D.melanogaster DOUBLETIME; M.a. TAU: M.auratus casein kinase 1 TAU; S.c. HRR25, S.c. CK 1, S.c. CK 2 and S.c. CK 3: S.cerevisiae casein kinases 1; H.s. CK1α, H.s. CK1δ and H.s. CK1ε: Homo sapiens casein kinases 1α, 1δ and 1ε. (C) ck-1a RNA levels are constant throughout a circadian day. Light-grown Neurospora cultures were transferred to constant darkness (DD) and samples were harvested after the indicated time periods. From each sample total RNA was isolated and cDNA was prepared by using random hexamers for priming. Relative levels of ck-1 RNA and actin RNA were determined by reverse transcription and quantitative RT–PCR using the 5′ nuclease assay detection system (TaqManTM) as described in Materials and methods. For calibration, cDNA was prepared from serial dilutions of RNA from light-grown tissue. The relative ratios of ck-1/actin were plotted versus the corresponding time in DD.
CK-1a does not contain a canonical nuclear import signal. Yet, when Neurospora cells were fractionated CK-1a was found in the cytosolic and in the nuclear fraction, similar to DBT in Drosophila (Figure 2A). To investigate whether CK-1a interacts with FRQ in vivo, total protein extracts were prepared from light-grown Neurospora and subjected to immunoprecipitation with affinity-purified antibodies against CK-1a (Figure 2B). CK-1a antibodies coprecipitated specifically FRQ while WC-1 was not precipitated (not shown). Neurospora cells were transferred to darkness and samples were harvested 8, 16 and 36 h after the light–dark (LD) transfer. Total cell lysates were prepared and subjected to immunoprecipitation with affinity-purified antibodies against CK-1a (Figure 2C). FRQ coimmunoprecipitated with CK-1a using extracts prepared at DD8, DD16 and DD36, suggesting that it was in a complex with FRQ throughout a circadian day. However, separation of the immunoprecipitates on long SDS gels revealed that forms of FRQ with low electrophoretic mobility were preferentially coprecipitated with CK-1a, suggesting that CK-1a is in a complex with highly phosphorylated FRQ. Thus, CK-1a either associates specifically with previously phosphorylated forms of FRQ or binds and rapidly phosphorylates FRQ such that complexes between CK-1a and non-phosphorylated FRQ may not accumulate.
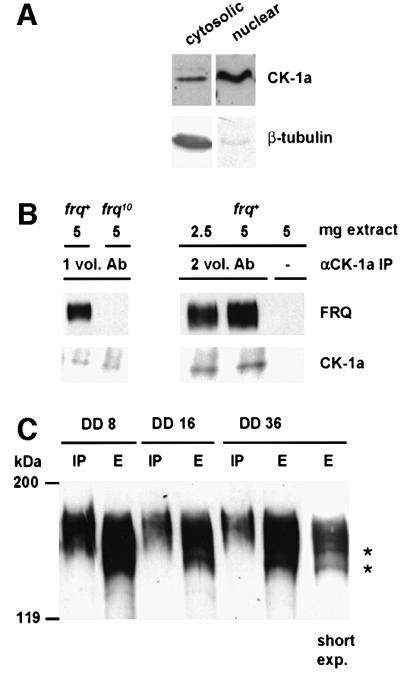
Fig. 2. CK-1a is in a complex with FRQ. (A) Subcellular localization of CK-1a. Light-grown Neurospora cells were fractionated as described (Luo et al., 1998). Cytosolic and nuclear fractions (200 µg each) were analyzed by SDS–PAGE and western blotting with affinity-purified antibodies against CK-1a (αCK-1a) (upper panels) and monoclonal antibodies against Neurospora tubulin (lower panels). (B) Co-immunoprecipitation of FRQ with CK-1a. Total cell extracts from the indicated strains were subjected to immunoprecipitation with affinity-purified CK-1a antibodies. 1 vol. and 2 vol. Ab correspond to the amounts of αCK-1a purified from 0.2 ml and 0.4 ml serum, respectively. Samples (1/3 of total) were analyzed by SDS–PAGE and western blotting with the monoclonal antibody FRQm3G11 (upper panels) and αCK-1a (lower panels). (C) CK-1a is in a complex with hyperphosphorylated FRQ. Total cell extracts were prepared from cells that were grown for the indicated time periods in DD. Samples were subjected to immunoprecipitation with αCK-1a (2 vol. Ab). Total cell extracts (E) and immunoprecipitates (IP) were analyzed by SDS–PAGE and western blotting with FRQm3G11. Right lane: short exposure of the FRQ signal in DD36 extract. Low phosphorylated forms of FRQ are indicated by asterisks.
We cloned the cDNAs encoding CK-1a and CK-1b, expressed His-tagged versions of both kinases in Escherichia coli and purified the recombinant proteins by Ni–NTA chromatography (see Materials and methods). The recombinant casein kinases were active in autophosphorylation assays and efficiently phosphorylated β-casein (not shown).
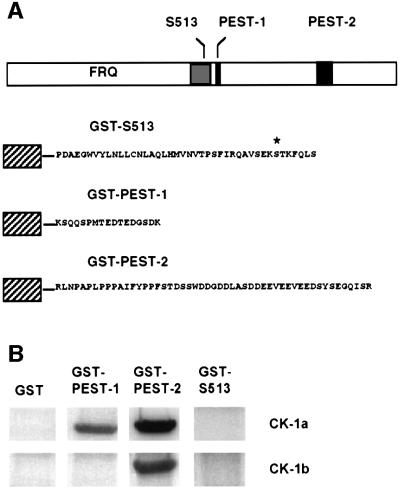
In Saccharomyces cerevisiae, casein kinases 1 phosphorylate uracil permease within a PEST sequence and, thereby trigger turnover of the protein (Marchal et al., 2000). FRQ contains two PEST-like elements, PEST-1 and PEST-2 (Figure 3A, upper portion), which are conserved in FRQ proteins from other fungi (Merrow and Dunlap, 1994). The PEST-find scores of 16.4 and 11.3, respectively, are significantly above the threshold level (5.0) (Rogers et al., 1986). PEST-1 is located in the central portion of FRQ and comprises a short confined segment of 17 amino acid residues. PEST-2 is located in the C-terminal portion and is 50 amino acid residues in length. We investigated whether the PEST sequences of FRQ are targets for phosphorylation by CK-1a and CK-1b. Fusion proteins between glutathione S-transferase (GST) and PEST sequences were constructed, expressed and purified. GST–PEST-1 and GST–PEST-2 contain the predicted PEST-like sequences comprising residues 544–560 and 843–893 of FRQ, respectively (Figure 3A, lower portion). Neither CK-1a nor CK-1b phosphorylated GST, which was used as a control (Figure 3B). Similarly, purified maltose-binding protein was not phosphorylated by the recombinant casein kinases (data not shown). GST–PEST-1 was specifically phosphorylated by CK-1a but not by CK-1b (Figure 3B). GST–PEST-2 was efficiently phosphorylated by CK-1a and with lower efficiency by CK-1b. Thus, PEST-2 appears to be a target for both casein kinases 1, while PEST-1 is a candidate sequence for specific phosphorylation of FRQ by CK-1a, the Neurospora homolog of Drosophila DBT.
Fig. 3. Phosphorylation of PEST-like elements in FRQ by recombinant CK-1a and CK-1b. (A) Construction of GST fusion proteins. FRQ is schematically outlined in the upper portion. Two PEST-like elements were identified with ‘PEST-find’ (http://www.at.embnet.org/embnet/tools/bio/PESTfind). PEST-1 (score 16.4) and PEST-2 (score 11.3) are indicated with black boxes. The gray box corresponds to residues from 478–519 of FRQ. Ser513 is indicated by an asterisk. FRQ-derived sequences that were fused to GST (hatched box) are shown in the lower part. (B) Specific phosphorylation of GST–PEST-1 by CK-1a and of GST–PEST-2 by CK-1a and CK-1b. The indicated GST fusion proteins (500 ng) were incubated with 50 ng recombinant His-tagged CK-1a (upper panel) and CK-1b (lower panel) for 60 min at 37°C in the presence of [32P]γ-ATP (2 Ci/mmol). Samples were analyzed by SDS–PAGE and autoradiography.
FRQ is phosphorylated at Thr501, Ser513 and Ser519 (Liu et al., 2000). To test whether these sites are phosphorylated by the Neurospora casein kinases 1, we constructed GST–S513, a fusion protein containing residues 478–519 of FRQ. Recombinant GST–S513 was neither phosphorylated by CK-1a nor by CK-1b (Figure 3B). Based on the evidence from in vitro phosphorylation assays, which must be interpreted with caution, FRQ might be phosphorylated by multiple kinases. However, a bona fide role of different kinases in the circadian system of Neurospora remains to be established.
Deletion of PEST-1 in FRQ results in loss of overt rhythmicity
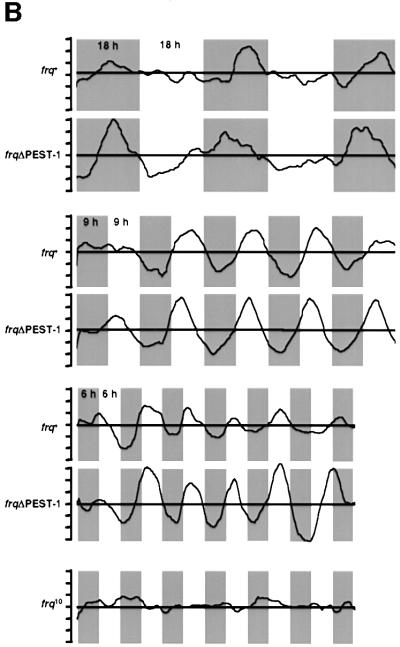
Since PEST-1 was specifically phosphorylated by CK-1a, we deleted the region encoding PEST-1 (17 amino acid residues) from frq and expressed the mutant gene under control of its own promoter in a frq-less Neurospora strain (frq10) (Aronson et al., 1994). When the resulting strain, frqΔPEST-1, was grown on race tubes in DD, no rhythmic conidiation (banding) was observed (Figure 4A). However, frqΔPEST-1 was growing with unregulated conidiation, while frq10 did not form conidia under these conditions. A control strain transformed with the wild-type frq gene showed a normal banding pattern with a periodicity of ∼22 h (not shown). Thus, deletion of the PEST-1 region of FRQ results in the loss of overt rhythmicity.
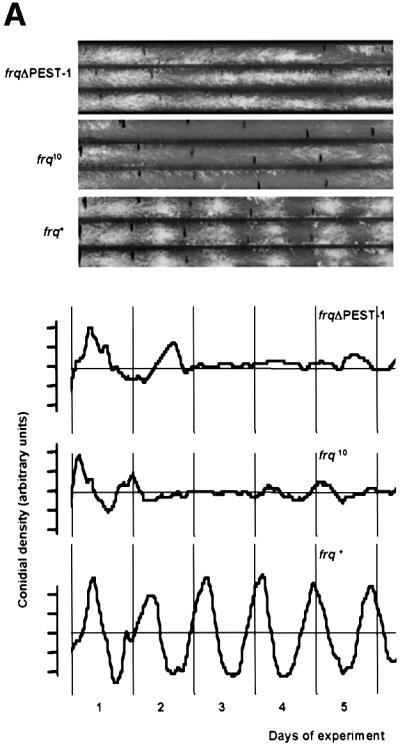
Fig. 4. frqΔPEST-1 has lost the overt conidiation rhythm. (A) Race tubes were inoculated with frqΔPEST-1, wild type (frq+) and frq null (frq10) and incubated at 25°C in DD. Upper panel: three replicate race tubes from each strain are shown. Lower panel: average conidial densities of triplicate race tubes. (B) frq+ and frqΔPEST-1 show driven conidial banding in 18 h/18 h, 9 h/9 h and 6 h/6 h LD cycles. Race tubes inoculated with the indicated strains were exposed to symmetrical LD cycles. The conidial densities of race tubes were determined and the averages of triplicates were plotted versus the time. The durations of the LD cycles are indicated. The gray background bars correspond to dark periods. Data of frq10 are shown only from the 6 h/6 h LD cycle; frq10 displayed no conidiation rhythm under all tested conditions.
Why is conidiation of frqΔPEST-1 arrhythmic in DD? To assess whether frqΔPEST-1 had generally lost the ability to form conidial bands, race tubes were inoculated with frq+, frqΔPEST-1 and frq10 and then exposed to symmetric LD cycles of different length (Figure 4B). frq+ showed banding in LD cycles while frq10 does not band under these conditions (Figure 4B, bottom panel and data not shown), confirming that light-regulated conidial banding requires functional FRQ (Chang and Nakashima, 1997; Merrow et al., 1999; Lakin-Thomas and Brody, 2000). Interestingly, frqΔPEST-1 banded in LD cycles that varied in length between 18 h/18 h and 6 h/6 h. In LD cycles conidiation is driven by light. In 12 h/12 h and longer cycles conidial bands are formed towards the end of the dark phase, while in shorter cycles conidiation, though initiated in the dark, is pushed into the light phase of the following day. This indicates that frqΔPEST-1 has the capacity to form bands when conidiation is driven by light. However, since frqΔPEST-1 does not band in DD it has apparently lost the ability to coordinate conidiation in a temporal fashion under constant conditions.
Physical and functional properties of FRQΔPEST-1
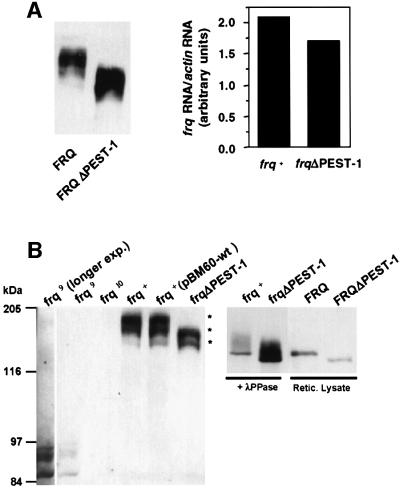
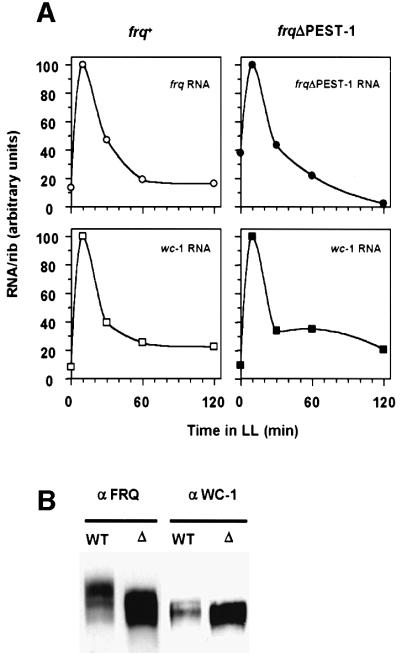
To characterize expression of the mutant FRQΔPEST-1 protein, total cell extracts were analyzed from wild-type and mutant strains that were grown in constant light (LL) (Figure 5A, left panel). As estimated by densitometry FRQΔPEST-1 was expressed at ∼3-fold higher levels than FRQ (not shown). Levels of frqΔPEST-1 RNA were slightly lower than levels of frq+ RNA (Figure 5A, right panel), suggesting that the elevated levels of FRQΔPEST-1 were due to an increased stability of the mutant protein (see below).
Fig. 5. Expression and phosphorylation profile of FRQΔPEST-1. (A) Left panel: protein extracts (200 µg) from light-grown frq+ and frqΔPEST-1 were analyzed by SDS–PAGE and western blotting with the monoclonal anti-FRQm3G11 antibody (αFRQ) (Merrow et al., 2001). Right panel: total RNA was prepared from light-grown strains. frq+ and frqΔPEST-1RNA were quantified by RT–PCR (as described in the legend to Figure 1C). (B) FRQΔPEST-1 is hypophosphorylated. Left panel: total protein extracts from the indicated light-grown strains. Major forms of FRQ that differ in electrophoretic mobility are indicated by asterisks. frq9: frameshift mutant with truncated FRQ; frq10: frq null mutant; frq+: wild-type frq; frq+ (pBM60-wt): frq10 transformed with pBM60 harboring wild-type frq; frqΔPEST-1: frq10 transformed with pBM60 harboring frqΔPEST-1. Central panel: protein extracts were subjected to immunoprecipitation with αFRQ and the precipitates were treated with 1000 U of λ-phosphatase (λ-PPase). Right panel: FRQ and FRQΔPEST-1 were synthesized by coupled in vitro transcription/translation in reticulocyte lysate (Retic. lysate). Samples were analyzed by SDS–PAGE and immunoblotting with αFRQ.
The phosphorylation pattern of FRQΔPEST-1 differed, however, significantly from that of FRQ. Light-grown control cells accumulated three major forms of FRQ that differed in electrophoretic mobility (Figure 5B). In contrast, only two major forms of FRQΔPEST-1 were observed. The mobility shifts were due to phosphorylation, since treatment of FRQ with phosphatase prior to SDS–PAGE resulted predominantly in a form of FRQ that co-migrated with in vitro synthesized, non-phosphorylated FRQ. Similarly, phosphatase treatment of FRQΔPEST-1 resulted in a species that co-migrated with in vitro synthesized FRQΔPEST-1 (Figure 5B). Due to the deletion of the PEST-1 region non-phosphorylated FRQΔPEST-1 migrated with a slightly higher mobility than non-phosphorylated FRQ.
Thus, the absence of one of the three phosphorylated species in FRQΔPEST-1 indicates that the PEST-1 region is either a direct target for phosphorylation and/or required to direct phosphorylation at other sites.
We then analyzed physical properties of FRQΔPEST-1. Deletion of PEST-1 did not alter the subcellular localization of the mutant protein as determined by subcellular fractionation and western blotting (not shown). Sedimentation analysis in sucrose density gradient suggested that the oligomeric state of FRQ (Denault et al., 2001) was not affected by deletion of the PEST-1 sequence (not shown).
When Neurospora is transferred from dark to light, synthesis of frq and wc-1 RNA are induced (Crosthwaite et al., 1997; Linden and Macino, 1997). The levels of light-induced RNAs are dependent on FRQ (Heintzen et al., 2001; Merrow et al., 2001). To assess functional properties of FRQΔPEST-1, we measured light inducibility of frq RNA and wc-1 RNA. In frq+ cells both RNAs were rapidly induced by light (Figure 6A, left panels). As expected, RNA levels reached a maximum after ∼10 min and adapted to low levels after longer light exposure of the cells (Arpaia et al., 1999; Heintzen et al., 2001). Similarly, frqΔPEST-1 RNA and wc-1 RNA were rapidly induced by light in frqΔPEST-1 cells and adapted subsequently to lower levels (Figure 6A, right panels). Thus, FRQΔPEST-1 supports inducibility by light as well as adaptation to light of wc-1 and frqΔPEST-1 RNA.
Fig. 6. Properties of frqΔPEST-1. (A) FRQΔPEST-1 supports light induction of frq and wc-1 RNA. frq+ and frqΔPEST-1 were grown in the dark for 28 h and then exposed to saturating light. Samples were harvested after the indicated time periods and RNA were prepared. frq RNA, frqΔPEST-1 RNA, wc-1 RNA and ribosomal RNA were detected by northern blot analysis and signals were quantified as described (Merrow et al., 2001). The data were normalized to the amount of ribosomal RNA (RNA/rib.RNA). The maximal signal was set to 100. Data are the average of duplicates. (B) Expression levels in LL of FRQΔPEST-1 and WC-1 are elevated in frqΔPEST-1. Protein extracts (200 µg) from frq+ (WT) and frqΔPEST-1 (Δ) were analyzed by SDS–PAGE and immunoblotting with the monoclonal antibodies αFRQ and anti-WC-1m4H4 (αWC-1). (C) FRQΔPEST-1 supports negative feedback after LD transfer. Light-grown frq+ and frqΔPEST-1 cells were transferred to darkness. After the indicated time periods mycelial pads were harvested and RNA was prepared. frq RNA (left panel) and frqΔPEST-1 RNA (right panel) were analyzed by northern blotting and quantified. The maximal signals were set equal to 100. Data are the average of duplicate experiments. (D) Turnover of FRQ and FRQΔPEST-1 after LD transition. Light-grown frq+ and frqΔPEST-1 cells were transferred to the dark and protein extracts were prepared after the indicated time periods. Samples were analyzed by SDS–PAGE and western blotting (upper panels). FRQ was detected by ECL and quantified by densitometry of X-ray films (lower panels). The solid lines represent exponential curve fits (data from t = 0 h were not included). One representative experiment out of three is shown. (E) Stability of FRQΔPEST-1 after the addition of CHX. Cultures were grown in LL for 2 days prior to addition of CHX at time point 0 h. Samples were harvested after the indicated time periods, analyzed by western blotting with αFRQ antibodies and quantified.
FRQ facilitates expression of WC-1 in a post-transcriptional manner (Lee et al., 2000; Cheng et al., 2001). Since FRQΔPEST-1 was expressed at high levels, we compared expression levels of WC-1 in light-grown frqΔPEST-1 and frq+. Equal amounts of total cell extracts were analyzed by SDS–PAGE and western blotting (Figure 6B). WC-1 was expressed in frqΔPEST-1 at significantly higher levels than in frq+. wc-1 RNA levels were similar in both strains (not shown), indicating that FRQΔPEST-1 supports the expression of WC-1 in a manner similar to that proposed for FRQ (Lee et al., 2000).
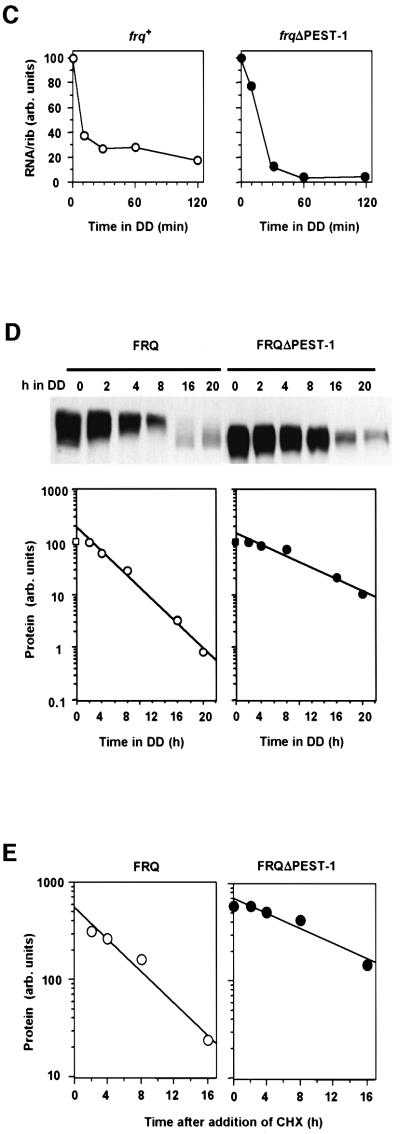
Synthesis of frq RNA is repressed when Neurospora cells are transferred from light to darkness. We measured the kinetics of RNA repression in frq+ and frqΔPEST-1 (Figure 6C). In both strains frq RNA levels decreased within 30 min after LD transfer. When frq9 was transferred from light to dark, no significant reduction in the amount of frq9 RNA was observed (not shown and Merrow et al., 1999). This demonstrates that deletion of PEST-1 does not compromise the ability of the cells to negatively feed back on frq transcription.
We then measured the kinetics of turnover of FRQ and FRQΔPEST-1 protein. Light-grown cells were transferred to dark to repress new synthesis of frq and frqΔPEST-1 RNA. Subsequently samples were analyzed over a time course of 20 h. Following the LD transfer FRQ and FRQΔPEST-1 were hyperphosphorylated. Due to changes in the phosphorylation pattern it was difficult to compare protein levels between 0 and 2 h (Figure 6D, upper panel). Yet, levels did not appear to change significantly. Subsequently FRQ was rapidly degraded with a half-life of 2.7 ± 0.2 h (Figure 6D, lower panels). In comparison, degradation of FRQΔPEST-1 was slow. The half-life for turnover was 5.6 ± 0.3 h, indicating that the mutant protein was significantly more stable than authentic FRQ. After 16 h in DD hyperphosphorylated FRQ was completely degraded and new synthesis of low phosphorylated forms of FRQ was observed, demonstrating that a new circadian oscillation had commenced. No new synthesis of FRQΔPEST-1 occurred in the observed 20 h time period.
To confirm the differences in protein stability, frq and frqΔPEST-1 cells were treated with cycloheximide (CHX) to inhibit protein synthesis, and samples were analyzed over a 16 h period. Again, turnover of FRQ was faster than the turnover of FRQΔPEST-1 (Figure 6E). The overall kinetics of degradation of both proteins were, however, slower in the presence of CHX than in the absence of the drug. FRQ was degraded with a half-life of 3.7 h, while the half-life of degradation of FRQΔPEST-1 was 7.7 h. The slower degradation kinetics may be due to pleiotropic effects of CHX and indicate that ongoing protein synthesis is required for the turnover of FRQ at physiological rates.
In summary, the data show that deletion of PEST-1 results in a pronounced stabilization of FRQΔPEST-1 protein.
Rhythmicity of frqΔPEST-1 on the molecular level
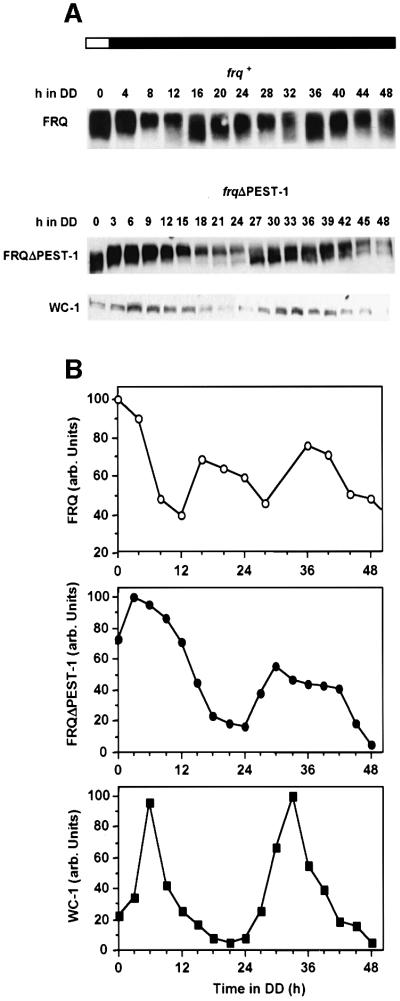
The deletion of PEST-1 does not interfere with crucial FRQ-dependent properties such as regulation of conidiation by light, adaptation of RNA expression levels to light and negative feedback on frq RNA synthesis after LD transfer of cells (see above). We therefore tested whether components of the circadian clock are rhythmically expressed in frqΔPEST-1 cells despite the lack of an overt conidiation rhythm. frqΔPEST-1 cells were grown in liquid culture and harvested at 3 h intervals spanning a time period of 48 h. As a control, frq+ cells were harvested at 4 h intervals over the same period. Total cell lysates of each time point were subjected to western blot analysis. As expected, FRQ was hyperphosphorylated and rapidly turned over (Figure 7A and B, upper panels). New synthesis of unphosphorylated FRQ was observed after 16 h in DD. Surprisingly FRQΔPEST-1 was also rhythmically expressed (Figure 7A and B, central panels). After LD transfer FRQΔPEST-1 protein showed the characteristic mobility shifts due to progressive phosphorylation. The phosphorylated protein was slowly degraded over an extended time period. After 24 h in DD low phosphorylated FRQΔPEST-1 protein was newly synthesized. This indicates that the negative feedback was relieved and a new circadian cycle had started. However, compared to synthesis of FRQ in control cells, the phase of FRQΔPEST-1 was delayed by ∼8 h. This is presumably due to the high expression levels of FRQΔPEST-1 in LL and its increased stability. The newly synthesized FRQΔPEST-1 was then gradually phosphorylated and slowly degraded. It was still detectable after 48 h, indicating that the circadian period of frqΔPEST-1 was longer than 24 h.
Fig. 7. Circadian oscillation of FRQΔPEST-1 and WC-1 in frqΔPEST-1. (A) Light-grown frq+ and frqΔPEST-1 were transferred to darkness (black bar). Samples were harvested at 4 h intervals (frq+) and 3 h intervals (frqΔPEST-1) over a time course of 48 h. Protein extracts were prepared and analyzed by SDS–PAGE, western blotting and ECL. FRQ (upper panel) and FRQΔPEST-1 (central panel) were detected with αFRQ antibodies and WC-1 was detected with αWC-1 antibodies (lower panel). (B) The indicated antigens were quantified by densitometry of ECL signals on selected X-ray films. The maximal signals were set to 100.
WC-1 was also expressed in a rhythmic fashion in frqΔPEST-1 cells (Figure 7A and B, lower panels). WC-1 reached a first maximum 6 h after LD transfer and a second maximum after 33 h in DD. WC-1 levels were minimal at DD21 and DD48. This suggests that the circadian clock was oscillating with a period of ∼27 h in frqΔPEST-1 cells.
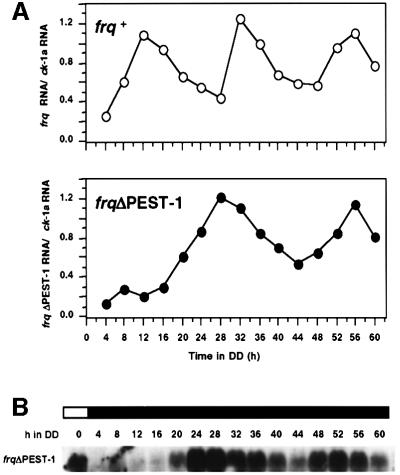
To analyze circadian rhythmicity on the transcriptional level, RNA from frq+ and frqΔPEST-1 cells was prepared at 4 h intervals over a time course of 60 h. frq+ and frqΔPEST-1 RNA was quantified by RT–PCR (Figure 8A, upper panel). ck-1a RNA, which is not rhythmically expressed (see above), was also quantified and data were used for normalization. Within the observed time course frq+ RNA levels peaked three times, around DD12, DD32 and DD56, in agreement with a circadian period of 22 h. frqΔPEST-1 RNA was also rhythmically expressed (Figure 8A, lower panel and panel B). However, frqΔPEST-1 RNA peaked only twice, at DD28 and DD56. This indicates that the circadian period was lengthened to ∼28 h. The appearance of the first peak of frqΔPEST-1 RNA at DD28 indicates that the phase was substantially delayed following the light-to-dark transfer.
Fig. 8. Rhythmic expression of frqΔPEST-1 RNA. Light-grown frq+ and frqΔPEST-1 were transferred to the dark. Samples were harvested at 4 h intervals over a time course of 60 h and total RNA was prepared. (A) frq, frqΔPEST-1 and ck-1 RNA was quantified by RT–PCR using the 5′ nuclease assay detection system (TaqManTM). For calibration, cDNA was prepared from serial dilutions of RNA from light-grown tissue. The relative ratios of frq/ck-1 and frqΔPEST-1/ck-1 were plotted versus the corresponding time in DD. (B) frqΔPEST-1 RNA was analyzed by northern blotting.
Discussion
We show that CK-1a, a Neurospora homolog of Drosophila DBT interacts with FRQ in vivo and that CK-1a phosphorylates two PEST regions of FRQ in vitro. Deletion of the PEST-1 region in FRQ results in an arrhythmic conidiation phenotype. Yet, on the molecular level, frq RNA, as well as FRQ and WC-1 protein, oscillate with a circadian period of ∼28 h. What causes the loss of overt rhythmicity and the lengthening of the period in frqΔPEST-1? FRQ-dependent functions, such as light-dependent conidiation, repression of frq RNA synthesis upon LD transfer and adaptation of light-induced frq and wc-1 RNA levels, are functional in frqΔPEST-1. As with frq+, the lag between frqΔPEST-1 RNA transcription and FRQΔPEST-1 protein synthesis comprises ∼4 h and thus does not account for the observed period lengthening in frqΔPEST-1. However, FRQΔPEST-1 is significantly more stable than FRQ and the mutant protein accumulates at high levels in light-grown cells. The circadian expression profile of FRQΔPEST-1 is broadened in a manner that corresponds to the observed period length in frqΔPEST-1. The persistence of high levels of stable FRQΔPEST-1 leads apparently to a prolonged negative feedback. This may cause the pronounced phase delay in frqΔPEST-1 RNA transcription and the lengthening of the circadian period.
Recently it was shown that the FRQ feedback loop is interconnected with WC-1 expression (Lee et al., 2000). Although the molecular mechanism is not yet fully understood, expression and rhythmicity of WC-1 are controlled by FRQ in a post-transcriptional manner (Lee et al., 2000; Cheng et al., 2001). WC-1 on the other hand is required for adequate expression levels of frq RNA (Crosthwaite et al., 1997; Cheng et al., 2001). The interconnected loops contribute to the robustness of the circadian oscillation (Cheng et al., 2001). WC-1 levels are elevated in light-grown frqΔPEST-1 cells and circadian oscillation of WC-1 is supported by FRQΔPEST-1 in DD. This suggests that the interconnected feedback loop is functional in frqΔPEST-1.
It is surprising that frqΔPEST-1 does not form conidial bands on race tubes although frqΔPEST-1 RNA and FRQΔPEST-1 protein oscillate in DD. Thus, the endogenous molecular rhythm of frqΔPEST-1/FRQΔPEST-1 is not accompanied by an overt conidiation rhythm, a phenomenon hitherto not described. The length of the circadian period of frqΔPEST-1 inferred from the molecular oscillations (28 h) is similar to that of the long period mutant frq7 (29 h) (Feldman and Hoyle, 1973). Yet, frq7 bands on race tubes in DD. We have no explanation of why frqΔPEST-1 has lost the ability to band in DD. However, the circadian expression profiles of clock proteins in frqΔPEST-1 differ significantly from those in frq+ and frq7. Circadian expression profiles of FRQ and WC-1 are separated in frq+ and in frq7 (Lee et al., 2000). In contrast, the temporal expression profile of FRQΔPEST-1 is broad due to its increased stability and it overlaps temporally with the peak of WC-1 expression. FRQΔPEST-1 levels reach a maximum at DD30 and remain high for ∼12 h. Thus, it resembles for most of a circadian day the expression of FRQ in light-grown frq+ cells, which could explain why conidial banding is disturbed in frqΔPEST-1.
Although frqΔPEST-1 does not display an overt conidiation rhythm in DD, it forms conidial bands in LD cycles. In this respect, the strain resembles frq10 qa-2p-FRQ, where FRQ is constantly expressed at high levels (Merrow et al., 2001). frq10 qa-2p-FRQ is arrhythmic in DD but bands in LD cycles. Rhythmic expression of WC-1 and of light-induced genes should be driven in LD cycles and may allow banding of strains which maintain high levels of FRQ throughout a circadian day.
What is the function of the PEST-1 element of FRQ? During the course of a circadian period FRQ is progressively phosphorylated and then degraded (Garceau et al., 1997). Under continuous illumination three major forms of FRQ accumulate to a steady state, which correspond to a species that differ in their extent of phosphorylation. One of these forms is absent when the PEST-1 segment is deleted, suggesting that PEST-1 is a target for phosphorylation in vivo. FRQΔPEST-1 is significantly more stable than FRQ, demonstrating that PEST-1 affects protein turnover. No other function of FRQ is apparently affected by the deletion of PEST-1. The PEST-1 sequence therefore appears to be a functional element that regulates turnover of FRQ in response to phosphorylation. Neurospora CK-1a in vitro phosphorylates PEST-1. PEST sequences are often targets for phosphorylation and implicated in regulated protein turnover. In S.cerevisiae, uracil permease is phosphorylated within a PEST-like sequence by casein kinases 1, triggering ubiquitylation, endocytosis and degradation of the plasma membrane transporter (Marchal et al., 2000). Casein kinases phosphorylate serine and threonine residues in the environment of aspartic and glutamic acids residues (Flotow and Roach, 1991; Pulgar et al., 1999). Since these residues are enriched in PEST sequences, they may in many cases be targets for phosphorylation by casein kinases.
Phosphorylation-dependent shifts in electrophoretic mobility of FRQ suggest that the protein is phosphorylated at multiple sites. FRQ contains a second PEST-like sequence that is phosphorylated by CK-1a and CK-1b in vitro. Whether the PEST-2 sequence has a function in protein turnover is currently under investigation. Recently Liu et al. (2000) reported phosphorylation of FRQ at three sites outside of the PEST sequences, Thr501, Ser513 and Ser519. Neurospora Ca/CaM-kinases were suggested to be involved in these phosphorylations (Yang et al., 2001). These sites were not phosphorylated by CK-1a or CK-1b in vitro and the sequence contexts have no similarity to casein kinase 1 consensus sites. This raises the question whether FRQ is phosphorylated by more than one kinase in vivo. An exchange of Ser513 affected the stability of FRQ and the length of the circadian period in a similar manner as deletion of PEST-1 (Liu et al., 2000). Whether the different phosphorylations of FRQ directly affect the kinetics of protein turnover is not known. Various steps in the lifetime of FRQ might be regulated by phosphorylation. If one of these steps is slowed down, this may indirectly result in slower kinetics of FRQ turnover.
Interconnected feedback loops are a key feature of circadian systems in all species (Loros and Dunlap, 2001; Reppert and Weaver, 2001; Williams and Sehgal, 2001). Casein kinases 1 might be conserved components, which are recruited by the circadian clocks to regulate period length via phosphorylation of components of the negative feedback loops.
Materials and methods
Strains, media and growth conditions
Neurospora standard laboratory strains used in this study were frq+ (frq wild type), frq7 (long period mutant strain with point mutation in FRQ), frq9 (frameshift mutant with truncated FRQ) and frq10 (frq null mutant). All strains carry the bd mutation (Loros et al., 1986; Aronson et al., 1994; Merrow et al., 1999). The Δwc-1;bd strain was generated by RIP (Talora et al., 1999), and crossed for bd, as previously described (Merrow et al., 2001). Neither wc-1 RNA nor WC-1 protein is detectable in this strain. For transformations the bd;frq10;his-3 strain was used (Bell-Pedersen et al., 1996).
Race tube medium contained 0.3% glucose, 1× Vogel’s medium (Vogel, 1964), 0.5% l-arginine, 10 ng/ml biotin and 2% agar. For all other experiments, the standard medium was 2% glucose, 1× Vogel’s, 0.5% l-arginine and 10 ng/ml biotin. For inoculation 8- to 10-day-old conidia from solid standard medium were harvested and for each circadian time point 25 ml of media in 100 ml flasks were inoculated with 3.5 × 106 conidia.
All liquid cultures were incubated in LL at 30°C for 1–2 days prior to transfer to DD at 25°C. Light induction was performed after cultivation of cells for 28 h in DD. For circadian time courses the cultures were pre-incubated with shaking in LL until a staggered light-to-dark transfer of the cultures was performed. Age-matched cultures were harvested by filtration and immediately frozen in liquid nitrogen.
Race tube analyses and circadian time courses
Race tubes were inoculated, germinated in LL for ∼24 h and transferred either to DD or different light–dark cycles (LD) at 25°C. The fluence rate was 20 µE delivered by cool white fluorescent bulbs. Race tubes were scanned and the conidial densities were quantified using the Chrono program (Roenneberg and Taylor, 2000). Circadian time courses were performed essentially as described (Aronson et al., 1994).
Plasmid constructs and Neurospora transformations
Cloning and expression of CK-1a and CK-1b. The open reading frame (ORF) of CK-1a was amplified from Neurospora cDNA using the oligo nucleotide primer pair: CCCCGTCGACAAGGTACCATGACTACC ATGGATCTC and CCCCAAGCTTGCGGCCGCTTATCGCAAAAT CCTGTC. A BamHI–HindIII fragment was cloned into pQE30. CK-1b was amplified with the primers CCCCGTCGACATGGCTTCC TCGACGTCCTCCAAC and CCCCAAGCTTCAGCTGCAGCACA GGG and a SalI–HindIII fragment was cloned into pQE30. The cloned cDNAs were sequenced and His-tagged versions of the recombinant kinases were expressed in E.coli and purified by Ni–NTA chromatography.
Construction of GST-fusion proteins: fragments encoding residues 478–519 (S513), 544–560 (PEST-1) and 843–893 (PEST-2) of FRQ were amplified by PCR. A BamHI site was introduced in frame at the 5′ end and a stop codon and NotI site at the 3′ end. The fragments were cloned into pGEX4T2.
The ORF of the large form of FRQ was cloned into pGEM4 resulting in pGEM4-ORF2. In pGEM4ORF2ΔPEST-1 the region encoding PEST-1 (1630–1680) was replaced with TGCGGCCGC.
pBM60-wt contains the ClaI-fragment (8.67 kbp) that is necessary for circadian rhythmicity (McClung et al., 1989). An exchange of the AflII–SfiI fragment with the deleted PEST-1 region resulted in pBM60frqΔPEST-1. Spheroplasts of frq10;bd;his-3 were generated as described, targeted transformations at the his-3 locus were performed with pBM60frqΔPEST1 and pBM60-wt (wild-type control) as described (Madi et al., 1994). Several mutant clones were selected and microconidia were generated for homogenous strains.
RNA analysis
RNA was prepared essentially as described except that only a single phenol extraction was performed (Crosthwaite et al., 1995). RNA was run on 1.2% agarose formaldehyde gels and blotted onto Nylon membranes (HybondN, Amersham Corp.) (Merrow et al., 1997). Riboprobes were used to probe the blots for frq, wc-1 and ribosomal RNA as described (Crosthwaite et al., 1995; Merrow et al. 2001).
Reverse transcription and quantitative PCR (quantitative RT–PCR): cDNA was synthesized from 2 µg total RNA using random hexamers and treated with DNase. Duplicate reactions (50 µl) containing cDNA equivalent to 0.1 µg RNA were performed in 96-well plates with the GeneAmp® 5700 Sequence Detection System (Applied Biosystems) using the following primer pairs (900 nM) and TaqMan probes (250 nM):
Frequency:
5′-TTGTAATGAAGGTGTCCGAAGGT-3′ (frq forw-primer)
5′-GGAGGAAGAAGCGGAAAACG-3′ (frq rev-primer)
5′-FAM—ACCTCCCAATCTCCGAACTCGCCTG-3′ (frq TaqMan)
Casein kinase 1a:
5′-TTACTGGCACTGCCCGTTATG-3′ (ck-1 forw-primer)
5′-CGTAGCCAAGGGACTCCATGT-3′ (ck-1 rev-primer)
5′-FAM—CTCCATCAACACGATCTCGGCG-3′ (ck-1 TaqMan)
Actin:
5′-AATGGGTCGGGTATGTGCAA-3′ (actin forw-primer)
5′-CTTCTGGCCCATACCGATCAT-3′ (actin rev-primer)
5′-FAM—CAGAGCTGTTTTCCCTTCCATCGTTGGT-3′ (actin Taq Man)
RNA from light-grown cells was stepwise diluted (2, 0.2, 0.02, 0.002, 0.0002 µg) and used to generate quantitative RT–PCR calibration curves for each primer system.
Protein analyses
Protein was extracted from mycelia by previously described methods (Garceau et al., 1997). Total protein (300 µg) per lane was subjected to SDS–PAGE on 7.5% gels for FRQ and WC-1, and 16% gels for CK-1a.
Western blots were decorated with monoclonal antibodies against FRQ (αFRQm3G11) or against WC-1 (αWC-1m4H4) or affinity-purified rabbit antibodies against CK-1a. β-tubulin antibodies were a generous gift of Dr Manfred Schliwa. The FRQ antibody recognizes an epitope between residues 64 and 100 of FRQ and thus specifically detects the large form of FRQ. For αWC-1 a purified GST–WC-1 fusion (970 – 1071 amino acid residues) was used for immunization of mice. Western blots were performed using enhanced chemilumininescence (ECL) and ECL plus (Boehringer Mannheim). To control uniform loading of the gels, nitrocellulose filters were stained with Ponceau S. In most cases cross-reacting bands appeared after longer exposures of the ECL blots, which were used to estimate equal loading of the gels. Similarly, the reactivities to β-tubulin and CK-1a, which are expressed at constant levels, were used for normalization.
Immunoprecipitation and phosphatase treatment
For immunoprecipitation 1 ml of αFRQm3G11 or the indicated amounts of affinity-purified antibodies against CK-1a were incubated with 20 µl protein A–Sepharose beads (Pharmacia) in 1× TBS buffer for 60 min at room temperature. The beads were washed with TBS and 5 mg protein extract were added in a total volume of 500 µl extraction buffer (50 mM HEPES pH 7.4, 10% glycerol, 137 mM NaCl, 5 mM EDTA, 1 mM phenylmethyl sulfonyl fluoride, 1 µg/ml leupeptin, 1 µg/ml pepstatin A). Samples were incubated under agitation for 2 h to 10 h at 4°C. Protein A–Sepharose was sedimented and washed twice for 5 min with 1 ml extraction buffer. For dephosphorylation of FRQ the immunoprecipitate was incubated with λ–protein phosphatase (1000 U, New England Biolabs) for 30 min at 30°C in a total volume of 25 µl. The reaction was stopped with SDS–sample buffer.
Acknowledgments
Acknowledgements
We would like to thank Christiane Kotthoff for excellent technical assistance and Ina Contag for her help in producing monoclonal antibodies. This work was supported by grants from the Deutsche Forschungs-Gemeinschaft.
References
- Albrecht U. (2001) Circadian rhythms: a fine c(l)ocktail! Curr. Biol., 11, 517–519. [DOI] [PubMed] [Google Scholar]
- Allada R., White,N.E., So,W.V., Hall,J.C. and Rosbash,M. (1998) A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell, 93, 791–804. [DOI] [PubMed] [Google Scholar]
- Aronson B.D., Johnson,K.A., Loros,J.J. and Dunlap,J.C. (1994) Negative feedback defining a circadian clock: autoregulation of the clock gene frequency. Science, 263, 1578–1584. [DOI] [PubMed] [Google Scholar]
- Arpaia G., Cerri,F., Baima,S. and Macino,G. (1999) Involvement of protein kinase C in the response of Neurospora crassa to blue light. Mol. Gen. Genet., 262, 314–322. [DOI] [PubMed] [Google Scholar]
- Bae K., Lee,C., Sidote,D., Chuang,K.Y. and Edery,I. (1998) Circadian regulation of a Drosophila homolog of the mammalian Clock gene: PER and TIM function as positive regulators. Mol. Cell. Biol., 18, 6142–6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell-Pedersen D., Loros,J.J. and Dunlap,J.C. (1996) Distinct cis-acting elements mediate clock, light and developmental regulation of the Neurospora crassa eas (ccg-2) gene. Mol. Cell. Biol., 16, 513–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang B. and Nakashima,H. (1997) Effects of light–dark cycles on the circadian conidiation rhythm in Neurospora crassa. J. Plant Res., 110, 449–453. [Google Scholar]
- Cheng P., Yang,Y. and Liu,Y. (2001) Interlocked feedback loops contribute to the robustness of the Neurospora circadian clock. Proc. Natl Acad. Sci. USA, 98, 7408–7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crosthwaite S.K., Loros,J.J. and Dunlap,J.C. (1995) Light-induced resetting of a circadian clock is mediated by a rapid increase in frequency transcript. Cell, 81, 1003–1012. [DOI] [PubMed] [Google Scholar]
- Crosthwaite S.K., Dunlap,J.C. and Loros,J.J. (1997) Neurospora wc-1 and wc-2: transcription, photoresponses and the origin of circadian rhythmicity. Science, 276, 763–769. [DOI] [PubMed] [Google Scholar]
- Darlington T.K, Wager-Smith,K., Ceriani,M.F., Staknis,D., Gekakis,N., Steeves,T.D., Weitz,C.J., Takahashi,J.S. and Kay,S.A. (1998) Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science, 280, 1548–1549. [DOI] [PubMed] [Google Scholar]
- Denault D.L., Loros,J.J. and Dunlap,J.C. (2001) WC-2 mediates WC-1–FRQ interaction within the PAS protein-linked circadian feedback loop of Neurospora. EMBO J., 20, 109–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap J.C., Loros,J.J., Liu,Y. and Crosthwaite,S.K. (1999) Eukaryotic circadian systems: cycles in common. Genes Cells, 4, 1–10. [DOI] [PubMed] [Google Scholar]
- Feldman J.F. and Hoyle,M.N. (1973) Isolation of circadian clock mutants of Neurospora crassa. Genetics, 75, 605–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotow H. and Roach,P.J. (1991) Role of acidic residues as substrate determinants for casein kinase I. J. Biol. Chem., 266, 3724–3727. [PubMed] [Google Scholar]
- Garceau N.Y., Liu,Y., Loros,J.J. and Dunlap,J. (1997) Alternative initiation of translation and time specific phosphorylation yield multiple forms of essential clock protein FREQUENCY. Cell, 89, 469–476. [DOI] [PubMed] [Google Scholar]
- Glossop N.R., Lyons,L.C. and Hardin,P.E. (1999) Interlocked feedback loops within the Drosophila circadian oscillator. Science, 286, 766–768. [DOI] [PubMed] [Google Scholar]
- Hardin P.E., Hall,J.C. and Rosbash,M. (1990) Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature, 343, 536–540. [DOI] [PubMed] [Google Scholar]
- Heintzen C., Loros,J.J. and Dunlap,J.C. (2001) The PAS protein VIVID defines a clock-associated feedback loop that represses light input, modulates gating and regulates clock resetting. Cell, 104, 453–464. [DOI] [PubMed] [Google Scholar]
- Kloss B., Price,J.L, Saez,L., Blau,J., Rothenfluh,A., Wesley,C.S. and Young,M.W. (1998) The Drosophila clock gene double-time encodes a protein closely related to human casein kinase Iε. Cell, 94, 97–107. [DOI] [PubMed] [Google Scholar]
- Kloss B., Rothenfluh,A., Young,M.W. and Saez,L. (2001) Phosphorylation of period is influenced by cycling physical associations of double-time, period and timeless in the Drosophila clock. Neuron, 30, 699–706. [DOI] [PubMed] [Google Scholar]
- Lakin-Thomas P.L. and Brody,S. (2000) Circadian rhythms in Neurospora crassa: lipid deficiencies restore robust rhythmicity to null frequency and white-collar mutants. Proc. Natl Acad. Sci. USA, 97, 256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C., Bae,K. and Edery,I. (1998) The Drosophila CLOCK protein undergoes daily rhythms in abundance, phosphorylation and interactions with PER–TIM complex. Neuron, 21, 857–867. [DOI] [PubMed] [Google Scholar]
- Lee C., Bae,K. and Edery,I. (1999) PER and TIM inhibit the DNA binding activity of a Drosophila CLOCK-CYC/dBMAL1 heterodimer without disrupting formation of the heterodimer: a basis for circadian transcription. Mol. Cell. Biol., 19, 5316–5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Loros,J.J. and Dunlap,J.C. (2000) Interconnected feedback loops in the Neurospora circadian system. Science, 289, 107–110. [DOI] [PubMed] [Google Scholar]
- Linden H. and Macino,G. (1997) White collar 2, a partner in blue-light signal transduction controlling expression of light-regulated genes in Neurospora crassa. EMBO J., 16, 98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Loros,J.J. and Dunlap,J. (2000) Phosphorylation of the Neurospora clock protein FREQUENCY determines its degradation rate and strongly influences the period length of the circadian clock. Proc. Natl Acad. Sci. USA, 97, 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loros J.J. and Dunlap,J.C. (2001) Genetic and molecular analysis of circadian rhythms in Neurospora. Annu. Rev. Physiol., 63, 757–794. [DOI] [PubMed] [Google Scholar]
- Loros J.J., Richman,A. and Feldman,J.F. (1986) A recessive circadian clock mutation at the frq locus of Neurospora crassa. Genetics, 114, 1095–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C., Loros,J.J. and Dunlap,J.C. (1998) Nuclear localization is required for function of the essential clock protein FRQ. EMBO J., 17, 1228–1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madi L., Ebbole,D.J., White,B.T. and Yanofsky,C. (1994) Mutants of Neurospora crassa that alter gene expression and conidia development. Proc. Natl Acad. Sci. USA, 91, 6226–6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C., Haguenauer-Tsapis,R. and Urban-Grimal,D. (2000) Casein kinase I-dependent phosphorylation within a PEST sequence and ubiquitination at nearby lysines signal endocytosis of yeast uracil permease. J. Biol. Chem., 275, 23608–23614. [DOI] [PubMed] [Google Scholar]
- Martinek S., Inonog,S., Manoukian,A.S. and Young,M.W. (2001) A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell, 105, 769–779. [DOI] [PubMed] [Google Scholar]
- McClung, C.R., Fox,B.A. and Dunlap,J.C. (1989) The Neurospora clock gene frequency shares a sequence element with the Drosophila clock gene period. Nature, 339, 558–562. [DOI] [PubMed] [Google Scholar]
- McDonald M.J., Rosbash,M. and Emery,P. (2001) Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol. Cell. Biol., 21, 1207–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M.W. and Dunlap,J.C. (1994) Intergeneric complementation of a circadian rhythmicity defect: phylogenetic conservation of structure and function of the clock gene frequency. EMBO J., 13, 2257–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M.W., Garceau,N. and Dunlap,J. (1997) Dissection of a circadian oscillation into discrete domains. Proc. Natl Acad. Sci. USA, 94, 3877–3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrow M.W., Brunner,M. and Roenneberg,T. (1999) Assignment of circadian function for the Neurospora clock gene frequency. Nature, 399, 584–586. [DOI] [PubMed] [Google Scholar]
- Merrow M.W., Franchi,L., Dragovic,Z., Görl,M., Johnson,J., Brunner,M., Macino,G. and Roenneberg,T. (2001) Circadian regulation of the light input pathway in Neurospora crassa. EMBO J., 20, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price J.L., Blau,J., Rothenfluh,A., Abodeely,M., Kloss,B. and Young,M.W. (1998) double-time is a novel Drosophila clock gene that regulates PERIOD protein accumulation. Cell, 94, 83–95. [DOI] [PubMed] [Google Scholar]
- Pulgar V., Marin,O., Meggio,F., Allende,C.C., Allende,J.E. and Pinna,L.A. (1999) Optimal sequences for non-phosphate-directed phosphorylation by protein kinase CK1 (casein kinase-1): a re-evaluation. Eur. J. Biochem., 260, 520–526. [DOI] [PubMed] [Google Scholar]
- Rechsteiner M. and Rogers,S.W. (1996) PEST sequences and regulation by proteolysis. Trends Biochem. Sci., 7, 267–271. [PubMed] [Google Scholar]
- Reppert S.M. and Weaver,D.R. (2001) Molecular analysis of mammalian circadian rhythms. Annu. Rev. Physiol., 63, 647–676. [DOI] [PubMed] [Google Scholar]
- Roenneberg T. and Taylor,W. (2000) Automated recordings of bioluminescence with special reference to the analysis of the circadian rhythms. Methods Enzymol., 305, 104–119. [DOI] [PubMed] [Google Scholar]
- Rogers S., Wells,R. and Rechsteiner,M. (1986) Amino acid sequences common to rapidly degraded proteins: the PEST hypothesis. Science, 234, 364–368. [DOI] [PubMed] [Google Scholar]
- Roth A.F. and Davis,N.G. (2000) Ubiquitination of the PEST-like endocytosis signal of the yeast a-factor receptor. J. Biol. Chem., 275, 8143–8153. [DOI] [PubMed] [Google Scholar]
- Rothenfluh A., Abodeely,M. and Young,M.W. (2000) Short-period mutations of per affect a double-time-dependent step in the Drosophila circadian clock. Curr. Biol., 10, 1399–1402. [DOI] [PubMed] [Google Scholar]
- Rutila J.E., Suri,V., Le,M., So,W.V., Rosbash,M. and Hall,J.C. (1998) CYCLE is a second bHLH–PAS clock protein essential for circadian rhythmicity and transcription of Drosophila period and timeless. Cell, 93, 805–814. [DOI] [PubMed] [Google Scholar]
- Saez L. and Young,M.W. (1996) Regulation of nuclear entry of the Drosophila clock proteins period and timeless. Neuron, 17, 911–920. [DOI] [PubMed] [Google Scholar]
- Selker E.U. and Garrett,P.W. (1988) DNA sequence duplications trigger gene inactivation in Neurospora crassa. Proc. Natl Acad. Sci. USA, 85, 6870–6874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suri V., Lanjuin,A. and Rosbash,M. (1999) TIMELESS-dependent positive and negative autoregulation in the Drosophila circadian clock. EMBO J., 18, 675–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talora C., Franchi,L., Linden,H., Ballario,P. and Macino,G. (1999) Role of a white collar-1-white collar-2 complex in blue-light signal transduction. EMBO J., 18, 4961–4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel H.J. (1964) Distribution of lysine pathways among fungi: evolutionary implications. Am. Naturalist, 98, 435–446. [Google Scholar]
- Wang G.K., Ousley,A., Darlington,T.K., Chen,D., Chen,Y., Fu,W., Hickman,L.J., Kay,S.A. and Sehgal,A. (2001) Regulation of the cycling of timeless (tim) RNA. J. Neurobiol., 47, 161–175. [DOI] [PubMed] [Google Scholar]
- Williams J.A. and Sehgal,A. (2001) Molecular component of the circadian system in Drosophila. Annu. Rev. Physiol., 63, 729–755. [DOI] [PubMed] [Google Scholar]
- Yang Y., Cheng,P., Zhi,G. and Liu,Y. (2001) Identification of a calcium/calmodulin-dependent protein kinase that phosphorylates the Neurospora circadian clock protein FREQUENCY. J. Biol. Chem., 276, 41064–41072. [DOI] [PubMed] [Google Scholar]
- Young M.W. (2000) Life’s 24-hour clock: molecular control of circadian rhythms in animal cells. Trends Biochem. Sci., 25, 601–606. [DOI] [PubMed] [Google Scholar]