Abstract
RNases III are a family of double-stranded RNA (dsRNA) endoribonucleases involved in the processing and decay of a large number of cellular RNAs as well as in RNA interference. The dsRNA substrates of Saccharomyces cerevisiae RNase III (Rnt1p) are capped by tetraloops with the consensus sequence AGNN, which act as the primary docking site for the RNase. We have solved the solution structures of two RNA hairpins capped by AGNN tetraloops, AGAA and AGUU, using NMR spectroscopy. Both tetraloops have the same overall structure, in which the backbone turn occurs on the 3′ side of the syn G residue in the loop, with the first A and G in a 5′ stack and the last two residues in a 3′ stack. A non-bridging phosphate oxygen and the universal G which are essential for Rnt1p binding are strongly exposed. The compared biochemical and structural analysis of various tetraloop sequences defines a novel family of RNA tetraloop fold with the consensus (U/A)GNN and implicates this conserved structure as the primary determinant for specific recognition of Rnt1p substrates.
Keywords: dsRNA/NMR/ribonuclease/RNA processing/Rnt1p
Introduction
RNase III enzymes are double-stranded RNA (dsRNA) endoribonucleases involved in the processing and decay of a large number of prokaryotic and eukaryotic cellular RNAs. Besides these multiple cellular functions, it has been shown recently that Dicer, a Drosophila member of the RNase III family, is one of the nucleases involved in the RNA interference process, a potent method to inactivate gene expression (Bernstein et al., 2001). Yeast (Saccharomyces cerevisiae) RNase III (Rnt1p) was identified on the basis of sequence similarities with Escherichia coli RNase III (Abou Elela et al., 1996). Rnt1p processes the precursors of stable RNAs such as rRNAs (Abou Elela et al., 1996; Kufel et al., 1999), small nuclear RNAs (snRNAs) (Chanfreau et al., 1997; Abou Elela and Ares, 1998; Seipelt et al., 1999) and small nucleolar RNAs (snoRNAs) (Chanfreau et al., 1998a,b; Qu et al., 1999). Inspection of the Rnt1p cleavage sites revealed the presence of terminal tetraloops with the consensus sequence ‘AGNN’ located 13–16 bp from the cleavage sites of the substrates (Chanfreau et al., 2000). The first two residues of the loop are almost universally conserved as A and G, with only one exception in the U1 snRNA 3′ extension, which is capped by a UGGU tetraloop (Seipelt et al., 1999). The last two residues are not conserved, except that no cytosine has been found at the third position of the loop. Rnt1p specifically cleaves its dsRNA substrate within a fixed window of 13–16 bp from the AGNN tetraloop. The enzyme has been proposed to interact directly with the tetraloop in a way that positions the active site of the enzyme at the correct cleavage site on the dsRNA. Alteration of the loop to GNRA sequences dramatically reduces the rate of Rnt1p cleavage and decreases Rnt1p binding by 5-fold (Chanfreau et al., 2000; Nagel and Ares, 2000). AGNN tetraloops also seem to act as independent folding units, since some Rnt1p substrates show a configuration where a short hairpin capped by an AGNN tetraloop coaxially stacks against a long helix where the cleavage occurs (Chanfreau et al., 1998b, 2000).
Rnt1p contains an RNase III domain and a dsRNA-binding domain (dsRBD) at its C-terminus like E.coli RNase III, as well as a eukaryotic-specific domain at the N-terminus (Filippov et al., 2000). dsRBDs are common RNA-binding motifs found in many proteins that bind to dsRNA (March et al., 1985; St Johnston et al., 1992). Surprisingly, it was found that the binding specificity of Rnt1p for AGNN tetraloops resides in its dsRBD (Nagel and Ares, 2000), suggesting that the dsRBD of Rnt1p interacts directly with the AGNN tetraloop in the RNA–protein complex.
The requirement for the presence of an AGNN tetraloop in the Rnt1p substrates could be rationalized by two models. In the first model, the AGNN tetraloops adopt a particular, predetermined conformation, and it is this particular conformation that is recognized by Rnt1p in its initial phase of recognition of the substrate. In the second model, the protein recognizes exclusively the first two conserved bases (AG) of the loop independently of a specific structural context, when these two bases are exposed in a single-stranded conformation. In this case, one might not expect the AGNN tetraloops to be strongly structured, or their structure might be induced by binding of the protein. To differentiate between these two models and to investigate the RNA structural requirements for recognition of the substrate by Rnt1p, we have used a combination of structural and biochemical approaches. The solution structures of two RNA hairpins containing the tetraloops AGAA and AGUU which are recognized by Rnt1p were determined using NMR spectroscopy. These structures revealed that both tetraloops have a conserved fold with a syn G after which the backbone turns, which is also similar to the previously described UGAA tetraloop (Butcher et al., 1997). Furthermore, hairpins capped by a UGAA tetraloop are also recognized and cleaved by Rnt1p, while hairpins capped by an ACAA tetraloop are not cleaved efficiently. We show that this effect is due to a change in the loop conformation, since the ACAA tetraloop conformation differs significantly from the (U/A)GNN fold. These results demonstrate that the AGNN tetraloops adopt a predetermined conformation that may be one of the major determinants in the recognition of the dsRNA substrates by Rnt1p. In addition, these results identify a novel family of RNA tetraloop fold, defined by surprisingly simple sequence requirements.
Results
NMR spectra of AGAA and AGUU RNA hairpins
In order to determine the topology of AGNN tetraloops, two RNA hairpins capped by the AGAA and AGUU sequences were constructed. These sequences were chosen for study because (i) AGUU is the most common sequence in the list of Rnt1p substrates (Chanfreau et al., 2000) and (ii) AGAA is the sequence of the substrate that was used in our previous study (Chanfreau et al., 2000). In the 300 ms NOESY spectra of both molecules, sequential base H8/6– sugar H1′ connectivities are observed throughout the stems, consistent with the formation of a right-handed helix. An unusually intense intraresidue nuclear Overhauser effect (NOE) is observed between the loop G7 H8 and its own H1′, even at a short mixing time (50 ms), and is comparable in intensity with the pyrimidine H5–H6 cross-peaks, indicating that the G7 glycosidic angle is in the syn range. All other intranucleotide base–H1′ NOEs are weak and are only observable at long mixing times, as expected for nucleotides with anti glycosidic angles.
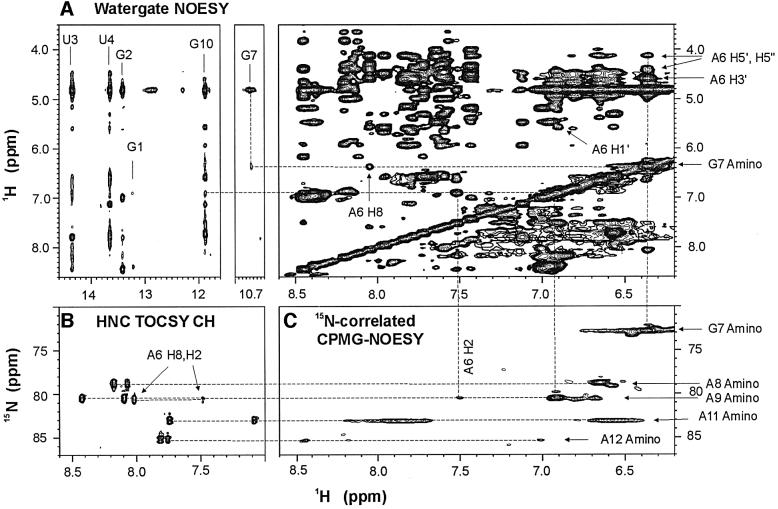
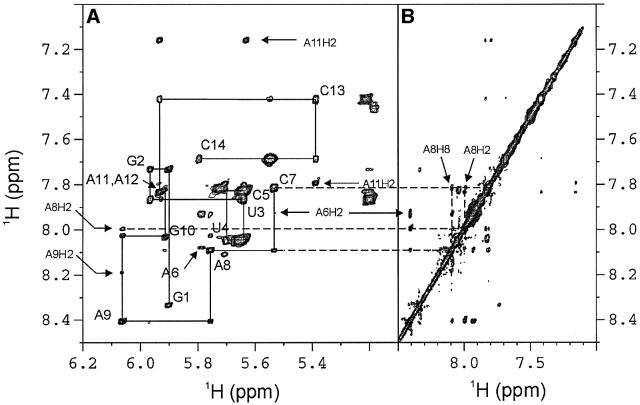
Five imino protons were assigned to the base pairs in the stem of the molecules based on sequential NOE connectivities observed in two-dimensional H2O NOESY spectra (Figure 1A). An additional upfield shifted imino proton resonating at ∼10.7 p.p.m. with an NOE cross-peak to an amino resonance at 6.4 p.p.m. was observed in the spectra of both the AGAA and the AGUU molecules. These were assigned to the imino and amino protons of G7, respectively (Figure 1A). The assignment of the G7 amino resonance was confirmed by its 1H–15N correlation in the CPMG-NOESY experiment (Figure 1C). NOEs between the G7 amino protons and the aromatic and sugar protons of A6 were observed in H2O NOESY spectra (Figure 1A), which indicate stacking interactions between A6 and G7.
Fig. 1. Unusual NOEs of the AGAA hairpin, 5′-GGUUC[AGAA]GAACC. (A) Portions of the NOESY spectrum of the AGAA hairpin recorded at 293 K on a 2 mM unlabeled sample in H2O. Note the cross-peak between the imino proton (10.7 p.p.m.) and the amino proton (6.4 p.p.m.) resonances. The NOE cross-peaks between the G7 amino proton and the A6 ribose and H8 protons are indicated on the spectral region on the right. (B) Portion of the spectrum of an HNC-TOCSY-CH experiment recorded at 293 K on a 1.0 mM 13C,15N-labeled sample. The amino nitrogen resonances of the adenines are correlated with both the H2 and H8 resonances within the same base. (C) Portion of the 15N-correlated CPMG-NOESY spectrum at 293 K on a 1.0 mM 13C,15N-labeled sample. The intense cross-peaks are between intraresidue amino proton and amino nitrogen resonances.
In order to assign the adenine amino 1H and 15N reson ances of the AGAA molecule unambiguously, a combination of long-range 1H–15N HMQC, HNC-TOCSY-CH and CPMG-NOESY experiments was used (Figure 1B and C). The NOE cross-peaks from the A9 amino to A6 H2, A6 H1′ and G10 H1 (Figure 1A) suggest that A9 stacks on G10 at the top of the stem and that there are possible hydrogen bonds between A6 and A9. Coupling constants estimated from TOCSY spectra indicated that all the loop nucleotides except A6 had S-type sugar puckers.
Solution structures of the AGAA and AGUU hairpins
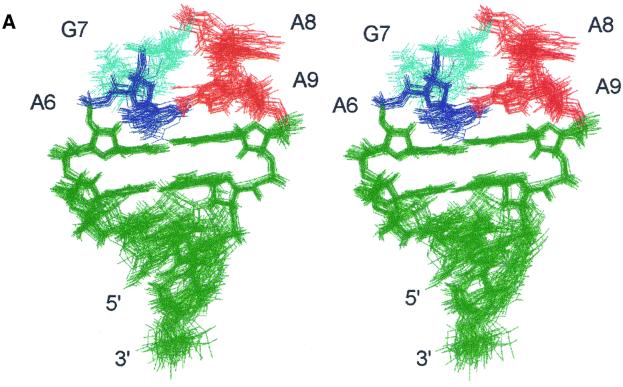
Both the AGAA and AGUU hairpin structures were determined from the NMR data (Figure 2). The stems of both molecules form an A-RNA helix, as expected. The 20 lowest energy structures of the AGAA hairpin (Figure 2A) are well defined, with an overall r.m.s.d. relative to the mean of 0.69 ± 0.18 Å and a local r.m.s.d. of 0.53 ± 0.15 Å within the loop. In the AGUU hairpin, U8 and U9 are not well defined in the 20 lowest energy structures (Figure 2B), which results in an overall r.m.s.d. of 1.41 ± 0.30 Å. This result is probably due to an insufficient number of NOE restraints for these two residues (only 8–9 NOE restraints per residue). However, we cannot rule out the possibility of local dynamics of the two uridine residues. Excluding U7 and U8, the structures have an overall r.m.s.d. value of 0.63 ± 0.20 Å.
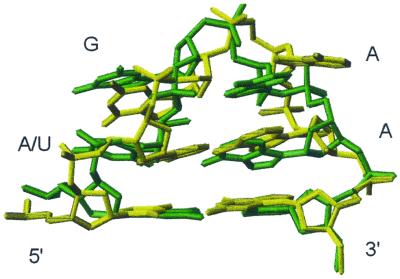
Fig. 2. Stereo views of superpositions of all heavy atoms of the 20 lowest energy solution structures of (A) AGAA (5′-GGUUC[AGAA]GAACC) and (B) AGUU (5′-GGUUC[AGUU]GAACC) RNA hairpins. Views are into the minor groove side of the loop.
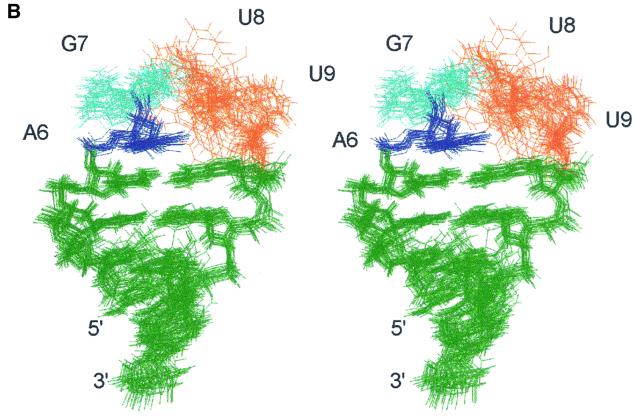
In the 20 lowest energy structures of the two hairpins (Figure 2), the universal G in the second position of the loop has a syn glycosidic torsion angle and its base stacks nicely on the preceding base, which results in the backbone turn at the GpN step. The bases in the tetraloop are distributed equally in a 5′ and 3′ stacking arrangement. A6 and G7 are stacked over the preceding C5 at the top of the stem. In the AGAA tetraloop, A8 and A9 are stacked over the stem G10 (Figure 3B). This arrangement exposes the non-bridging phosphate oxygen atoms between G7 and A8 at the top of the loop. The exposure of these oxygen atoms may have important functional consequences. Substitution of the Rp non-bridging phosphate oxygen at the 3′ side of the universal G with a phosphorothioate dramatically reduces the binding of Rnt1p to its dsRNA substrate, snR47 (Chanfreau et al., 2000). A likely explanation would be that this non-bridging phosphate oxygen is in direct contact with Rnt1p in the RNA–protein complex. The loop conformation favors the presentation of this phosphate oxygen to the protein. However, we cannot rule out that incorporation of phosphorothioate groups may alter the conformation of the loop, as demonstrated in the case of an RNA hairpin containing the binding site for bacteriophage MS2 capsid protein (Smith and Nikonowicz, 2000).
Fig. 3. (A) Stereo view of the lowest energy structure of the AGAA tetraloop. The view is into the minor groove side of the loop. Two potential hydrogen bonds in the tetraloop are indicated with dashed lines, G7H22–G7 O2P and A9 H62–A6 N3. The arrow points to the ‘turning’ phosphate. (B) Top view of the sheared A–A base pair stacked on the closing C·G base pair. (C) Top view showing the stacking interactions of the four bases of the AGAA tetraloop.
In the 20 lowest energy structures of the AGAA tetraloop, there are two potential hydrogen bonds which stabilize the tetraloop (Figure 3). The amino group of A9 is positioned to form a hydrogen bond to the N3 of A6, and the bases form a classical sheared A–A base pair. The amino proton of G7 is within hydrogen bonding distance of one of the non-bridging oxygens on its 5′ phosphate group due to the syn conformation of G7. This hydrogen bond could not form if there was an A at position 7 instead of a G. The observed G7 and A9 amino proton resonances, indicating slow exchange with water, and their NOE cross-peaks are consistent with hydrogen bond formation.
AGNN tetraloops have the same fold as the UGAA tetraloop
Unexpectedly, the AGAA and AGUU tetraloops have a structure which is very similar to the previously described structure of an unmodified UGAA tetraloop (Butcher et al., 1997). This tetraloop sequence is highly conserved at the 3′ end of the eukaryotic 18S rRNA (position 1777–1780 in S.cerevisiae), but both adenine bases of the UGAA loop are N6-dimethylated in vivo. The biological function of adenine dimethylation at these positions is unknown (Lafontaine et al., 1995). However, adenine N6-dimethylation would disrupt the structure of the unmodified tetraloop (Butcher et al., 1997).
An overlay of the lowest energy structures of AGAA and UGAA tetraloops is presented in Figure 4. The AGAA and AGUU tetraloops share many structural features with the UGAA tetraloop. First, the guanosine residue is syn and stacks on its preceding base. Secondly, the backbone makes an almost 180° turn at the GpN step, which exposes the non-bridging phosphate oxygen atoms following the guanosine residue. Finally, the four residues of the loop are distributed equally in a 5′ and 3′ stacking arrangement. Both the AGAA and UGAA tetraloops are stabilized by formation of a non-Watson–Crick pair between the first and last nucleotides. However, since the U–A pair found in the UGAA tetraloop must have a different geometry from that of the sheared A–A base pair of the AGAA tetraloop, the stacking of the residues in the two tetraloops is not identical. The A6 is stacked more into the interior of the loop than the U6. The G7 in the UGAA also has a hydrogen bond from the amino proton to its 5′ phosphate group but, in contrast to the AGAA tetraloop, this hydrogen bond is to the O5′ rather than the non-bridging oxygen.
Fig. 4. An overlay of the lowest energy structures of the G[UGAA]C (green) and C[AGAA]G (yellow) tetraloops with their closing base pairs.
Rnt1p cleaves RNA substrates with UGAA and AGCA but not ACAA tetraloops
To investigate further the tetraloop sequence requirements for Rnt1p cleavage, we constructed three mutant substrates and compared their ability to be recognized and cleaved by Rnt1p with a wild-type substrate containing an AGAA tetraloop. The first mutant substrate contains an ACAA tetraloop, a mutation in the strongly conserved second nucleotide position of the loop. The second mutant contains a UGAA tetraloop, a mutation in the strongly conserved first nucleotide of the loop. Only one Rnt1p substrate, the 3′ processing site in the 3′ extension of the U1 snRNA, has a U in the first position of the tetraloop. This substrate was chosen because, as shown above, the UGAA tetraloop has a structure very similar to that of the AGNN tetraloops. Thus, one predicts that if the conformation of the loop dictates efficient cleavage, this substrate should be cleaved efficiently. The third mutant substrate has an AGCA tetraloop. A cytosine was introduced into the third position because none of the Rnt1p substrates discovered so far contains a C in the third position of the tetraloop, which suggested that a cytosine at this position could be an antideterminant of cleavage.
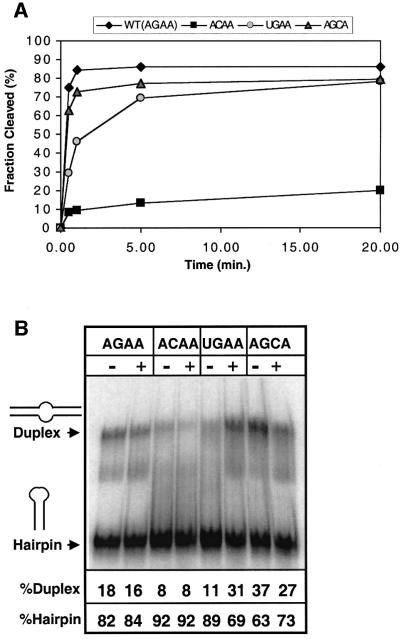
The wild-type and mutant substrates were submitted to cleavage by Rnt1p under single-turnover conditions (Figure 5A). Native gel shift analysis of the substrates in the absence of protein indicated that a fraction of each of the RNAs was in a duplex rather than a hairpin conformation (Figure 5B). This duplex conformation would have an internal loop rather than a hairpin tetraloop and would not be expected to be a substrate for cleavage by Rnt1p. Thus, in order to compare the cleavage results on the substrate (hairpin) conformation only, the data were normalized to the total fraction of hairpin for each substrate (Materials and methods). Results for AGAA (wild-type), ACAA, UGAA and AGCA substrates are shown plotted in Figure 5A. Upon incubation with Rnt1p, almost 90% of the AGAA hairpin was cleaved, with most of the reaction occurring in <1 min. The UGAA hairpin was cleaved somewhat less efficiently than the wild-type AGAA hairpin, with a slower initial rate and a plateau after ∼75% of the substrates are cleaved. Contrary to our expectations and to the lack of Cs at the third position of the AGNN tetraloops, the AGCA hairpin was also cleaved efficiently. The initial rate is almost the same as for the wild-type, and the plateau is also ∼75%.
Fig. 5. (A) Single turnover cleavage kinetics of the wild-type and mutant substrates by recombinant Rnt1p. Shown is the fraction of RNA cleaved as a function of time. The values indicated are the average of 10 independent experiments, five with and five without the denaturation–renaturation procedure. (B) Native gel electrophoresis analysis of the wild-type and mutant substrates. RNAs were loaded on a native gel with (+) or without (–) denaturation–renaturation treatment. The fraction of RNA in the hairpin and in the duplex conformation is indicated for each substrate, and the values indicated at the bottom are the average of two independent experiments. The faint extra bands on the gel may be due to another alternative conformation of the substrate or to degradation.
The mutation of the universally conserved G to a C in the ACAA hairpin resulted in a dramatic decrease in cleavage efficiency. The initial rate of cleavage was slower than for the wild-type and for the other mutant substrates, and <10% of the ACAA is cleaved after 1 min of incubation with the enzyme and <20% after 20 min. These results indicate that the ACAA tetraloop is a very poor Rnt1p substrate.
The enzyme cleavage experiments indicate that tetraloops containing the universally conserved G at position 2 are recognized as substrates by Rnt1p. Tetraloops such as UGAA which contain a G at the second position and have a sequence that allows a similar loop conformation with a syn G and a backbone turn between the second and third nucleotides are still recognized by Rnt1p, albeit with lower efficiency. On the other hand, substitution of the universal G for a C dramatically lowers the cleavage efficiency in vitro and would be expected to abolish cleavage completely in vivo. This observation suggested that this single mutation results in a dramatic change in the loop conformation, that could no longer be recognized by Rnt1p.
The ACAA sequence adopts a conformation that differs from the AGNN family
To test the hypothesis that the ACAA sequence induced a loop conformation incompatible with recognition of the dsRNA by Rnt1p and different from the AGNN family, we examined the structure of the ACAA tetraloop by NMR. A weak intraresidual NOE between C7 H6 and H1′ in the NOESY spectrum at short mixing time indicates that C7 is in the anti glycosidic conformation (Figure 6). This is consistent with the general rule that the syn conformation is sterically hindered for pyrimidines. No sequential NOEs are observed from C5 to A6 and A6 to C7, which suggests that the backbone is stretched out around A6, where the backbone most probably makes its turn. NOEs from C7 H6 to A8 H8 and A8 H2 suggest that the bases of C7 and A8 stack on each other, unlike the AGAA tetraloop in which G7 and A8 are located on opposite sides of the backbone and G7 stacks on the preceding base. Thus, substitution of the universal G with a C causes the tetraloop to adopt a different conformation where the second nucleotide is no longer syn and where the backbone turn occurs after the first nucleotide instead of after the second. It is likely that this conformation is therefore no longer recognized by the nuclease, which would explain the profound cleavage defect exhibited by the ACAA substrate.
Fig. 6. Portions of a NOESY spectrum (τm = 300 ms) of the ACAA hairpin recorded at 293 K in D2O. (A) H1′–base H8/6 region of the NOESY spectrum. Solid lines show the base–H1′ sequential connectivities. The H1′–H8/6 intraresidue NOE cross-peaks are labeled. Note the missing sequential connectivities from C5 to C6 to C7. (B) Aromatic region of the NOESY spectrum. NOE cross-peaks between C7 H6 and A8 H8 are indicated with arrows.
Discussion
Comparison with other tetraloops
Tetraloops are common RNA motifs that play important roles in RNA folding as well as in their biological functions. AGNN tetraloops adopt a distinct folding topology as compared with the three classes of hyperabundant tetraloops first identified in rRNAs, UNCG, GNRA and CUYG (Tuerk et al., 1988; Woese et al., 1990). All of these tetraloops are closed by a non-Watson–Crick or buckled base pair which shortens the phosphate–phosphate distance across the top of the helix so that the remaining two nucleotides can span the distance to close the loop, and this is also the case for the AGAA tetraloop. The most striking features that distinguish AGNN tetraloops from these other tetraloops are the stacking of the first and last two residues on the 5′ and 3′ sides of the tetraloop, respectively, and the reversal of the backbone on the 3′ side of the guanosine residue which exposes its non-bridging phosphate oxygen atoms.
The solution structure of a fragment of the first stem– loop of the SL1 RNA of Caenorhabditis elegans was determined previously by NMR (Greenbaum et al., 1996). In these structures, an AGUU tetraloop on top of a buckled A·U Watson–Crick base pair is located in the loop region. The guanosine (G16) of the tetraloop is also in the syn conformation, but the orientation of its base is in the opposite direction compared with our structures of the AGUU and AGAA tetraloops. In contrast to our structures, where the backbone turn is on the 3′ side of the conserved guanosine, reversal of the backbone direction is on the 5′ side of G16. While it is possible that the presence of an A·U base pair instead of a C·G base pair at the top of the stem might alter the conformation of the loop, we believe that the difference in the two structures is most likely to be due to the absence of critical NOE restraints in the calculations that define the structure of the SL1 tetraloop (Greenbaum et al., 1996). In our spectra as well as those of the SL1 RNA, there is an imino resonance at 10.7 p.p.m., which we assigned to the imino proton of the guanosine in the tetraloop. In the SL1 RNA, this imino proton resonance was incorrectly assigned to G21, a residue in the stem. The chemical shift of the guanosine imino proton and its NOE with a resonance of 6.4 p.p.m. are consistent with a syn conformation (Greenbaum et al., 1995). Since no restraints from the imino or amino protons of the G were used in the structure calculations for SL1, the misassignment had little effect on the calculated structures. However, our assigned NOEs between the amino protons of G7 and the sugar protons of the preceding residue were important for defining the conformation of the syn G in our tetraloop structures.
A novel class of RNA tetraloops defined by a syn guanosine at position 2 is recognized by Rnt1p
The fact that the AGAA and AGUU tetraloops adopt the same overall conformation which is similar to the UGAA structure suggests that the (A/U)GNN sequence defines a structural fold based on a surprisingly simple sequence requirement. The conserved G in position 2 plays an essential role in the tetraloop structure. The syn conformation of the G is required for the stacking on the first base and for hydrogen bond formation between the G7 amino proton and its phosphate group, and it also helps set up the backbone turn which occurs after the nucleotide. An A in this position could not form a hydrogen bond to the phosphate group, and therefore would be less likely to be stabilized in the syn conformation.
In the Rnt1p substrates, a guanosine at the second position in the tetraloop is absolutely required, and the first position is almost always an adenosine. A guanosine at the first position may result in a GNRA sequence, which adopts a different type of stable tetraloop structure; this probably explains the absence of G at the first position of natural substrates identified to date. Consistent with this, substrates capped by GNRA tetraloops are not cleaved by Rnt1p (Chanfreau et al., 2000; Nagel and Ares, 2000). Mutations such as UGAA or AGCA that make the sequence differ slightly from the optimal consensus are cleaved slightly less efficiently than substrates capped by an AGAA tetraloop (Figure 5). A cytosine at the first position moderately inhibits cleavage (Chanfreau et al., 2000), but we do not know if this suboptimal cleavage efficiency is due to a perturbation of the AGNN tetraloop conformation or to a change in the identity of one of the bases required for recognition by the protein. The AGNN structural fold is utilized by yeast RNase III to discriminate between dsRNA targets and non-targets, and it is likely that it could be utilized as a specific RNA recognition motif by other proteins in different species. It is remarkable that such a simple consensus sequence and structure can act as a determinant for the activity of this important class of enzymes.
How does Rnt1p recognize the AGNN tetraloop?
As reported, the primary determinants of tetraloop binding specificity are located in the C-terminal dsRBD of Rnt1p (Nagel and Ares, 2000). dsRBDs, also known as dsRBMs, are common sequence motifs found in many proteins that bind to dsRNA. The structures of several dsRBDs as well as their complexes with dsRNA have been determined using NMR and crystallography (Kharrat et al., 1995; Nanduri et al., 1998; Ryter and Schultz, 1998; Ramos et al., 2000). The dsRBD domains have an αβββα topology in which two terminal α-helices pack against one face of a three-stranded antiparallel β-sheet. As illustrated in the solution structures of the Staufen dsRBD3–dsRNA complex (Ramos et al., 2000), as well as in the crystal structure of the complex of Xlrbpa dsRBD with dsRNA (Ryter and Schultz, 1998), dsRBDs contact one side of the dsRNA using three regions of the protein: the N-terminal α-helix (α-helix 1) and the loop between β1 and β2 interact with the RNA minor groove and the β2 strand interacts with the intervening major groove of dsRNA. Most of the contacts involve the phosphodiester backbone or the ribose 2′-OH groups, which is consistent with the low sequence specificity of most dsRBDs. In contrast, the dsRBD of Rnt1p binds to dsRNAs containing AGNN tetraloops with a 5-fold higher affinity than to a dsRNA capped by a GNRA tetraloop (Nagel and Ares, 2000), suggesting that it binds the tetraloop directly. Also, the fact that the cleavage site is determined from the distance to the tetraloop (Chanfreau et al., 2000) makes it unlikely that there is no direct contact with the tetraloop. Such direct binding of a dsRBD to a structured tetraloop is not unprecedented (Ramos et al., 2000). It has been hypothesized that loop 2 of Rnt1p dsRBD, which lacks the GxxH motif found in many other dsRBDs, may contact the AGNN tetraloop directly (Nagel and Ares, 2000). In this view, residues in loop 2 would interact with the minor groove side of the tetraloop, where the backbone of the conserved A/U and G residues is located. However, in the Staufen–dsRNA complex, there is an interaction between the α-helix 1 and a UUCG tetraloop that connects the two strands of the RNA (Ramos et al., 2000). This ‘fortuitous’ interaction may be relevant to the specific recognition of the AGNN tetraloops by Rnt1p. If Rnt1p interacts with its substrate in a similar manner to that observed for Staufen, then it would be expected that α-helix 1 rather than loop 2 would interact with the minor groove of the tetraloop.
The conserved structures of the (A/U)GNN tetraloops along with the enzymatic cleavage results suggest that the primary requirement for specific binding of Rnt1p is a syn guanine at the second position of the tetraloop. As discussed, the positioning of the guanine exposes the non-bridging oxygen of its 3′ phosphate at the top of the loop, which is important for Rnt1p binding (Chanfreau et al., 2000). The results reported here are consistent with the hypothesis that a conserved structure is the primary determinant of Rnt1p substrate recognition. Base-specific contacts to the first two conserved bases are also possible, but the only hydrogen bond donors or acceptors exposed in the minor groove are A6 N3 (or U6 O2). The G7 ribose points down into the minor groove, but the base is entirely in the major groove. Thus, if there is specific base recognition, it would probably involve positioning part of the protein into the major groove, which is not consistent with currently proposed models for dsRBD binding, or a change in the tetraloop structure upon protein binding.
Materials and methods
NMR sample preparation
RNA samples for NMR studies were prepared from DNA templates using T7 RNA polymerase (Milligan et al., 1987). 13C,15N-labeled NTPs were isolated from Methylobacterium extorquens bacteria strain AM1 which had been grown in media containing [13C]methanol and [15N]ammonia as the sole carbon and nitrogen sources, purified, and converted to NTPs as described (Batey et al., 1992; Nikonowicz et al., 1992; Peterson et al., 1994). After transcription with either commercially purchased unlabeled NTPs or the enzymatically prepared 13C,15N-labeled NTPs, the RNA was ethanol precipitated and purified by HPLC on a PEI anion exchange column. The RNA was concentrated and then desalted by Sephadex G-25 gel filtration and lyophilized to dryness. NMR samples were dissolved in 90% H2O/10% 2H2O to final concentrations of 1.0–2.0 mM and the pH was adjusted to 6.2 with 3–5 µl of 1 M NaOH. 2H2O samples were prepared by lyophilizing the 90% H2O/10% 2H2O samples and resuspending in 99.999% 2H2O. The sequences of the RNA molecules used in these studies are: 5′-GGUUC[AGAA]GAACC (AGAA hairpin); 5′-GGUUC[AGUU]GAACC (AGUU hairpin) and 5′-GGUUC[ACAA] GAACC (ACAA hairpin) [loop residues are bracketed].
NMR spectroscopy and resonance assignments
All spectra were recorded on Bruker DRX 500 and 600 MHz spectrometers. Complete assignments of non-exchangeable protons and their bound carbons were obtained by analysis of a series of two-dimensional experiments [NOESY (Macura et al., 1980), DQF-COSY (Rance et al., 1983), CITY-TOCSY (Bax and Davis, 1985; Kadkhodaei et al., 1993), CT-HSQC and HCNCH (Sklenar et al., 1993)] and three-dimensional experiments [3D HCCH-TOCSY (Bax et al., 1990), 3D HCCH-COSY (Clore et al., 1990) and 3D NOESY-1H–13C-HMQC (Nikonowicz and Pardi, 1993)] acquired at 293 K following established protocols (Dieckmann and Feigon, 1997). An HCP-TOCSY experiment (Marino et al., 1995) was used to confirm the sequential assignments of 1H1′ and 31P resonances. Exchangeable 1H and 15N resonance were assigned through two-dimensional NOESY, long-range 1H–15N HMQC, 15N-correlated CPMG-NOESY (Mueller et al., 1995) and HNC-TOCSY-CH (Simorre et al., 1996) experiments in 90% 1H2O /10% 2H2O at 274 and/or 293 K. All NMR data were processed and analyzed with XWINNMR (Bruker) and Felix 2000 (MSI).
Structure calculations of AGAA and AGUU RNA hairpins
Most non-exchangeable inter-proton distance restraints were obtained from integration of cross-peaks in two-dimensional NOESY spectra acquired at 293 K with various mixing times of 50, 100, 150, 200 and 250 ms, using the average pyrimidine H5–H6 cross-peak intensity as a standard reference of 2.45 Å. The upper bounds of these distance restraints were set to be 120% of the NOE distances and the lower bounds equal to the sum of the van der Waals radii (1.8 Å). NOE distances of exchangeable protons and those obtained from three-dimensional NOESY-HMQC were classified semi-quantitatively as strong (1.8–3.5 Å), medium (1.8–5.0 Å) or weak (1.8–6.0 Å). Intranucleotide NOE restraints that may be affected by spin diffusion were restrained to 1.8–7.0 Å. The ribose conformations were analyzed with TOCSY experiments. Residues with absent H1′–H2′ cross-peaks (all stem residues except the first base pair and G10) were restrained to an N-type range (85 ± 30°), those with strong H1′–H2′ and H1′–H3′ cross-peaks were restrained to an S-type range (145 ± 30°) (G7, A8, A9 and U8, U9) and residues with intermediate cross-peaks intensities (G1, A6, G10, C14) were left unrestrained. A (31P, 1H) HETCOR (Sklenárõ et al., 1986) experiment was used to estimate the α backbone torsion angles around O3′-P and P-O5′, respectively. The α backbone dihedral angles were restrained to exclude the trans range (0 ± 120°) for stem residues except G10, since they had phosphate chemical shifts within the –4 to –5 p.p.m. range (Allain and Varani, 1995). β and ε dihedral angles were measured using 31P spin-echo difference CT-HSQC and spin-echo difference CH-HCCH correlation (Kolk et al., 1998; Legault et al., 1995). Both α and β dihedral angles were left unrestrained for the loop residues in the structure calculations.
The structures of AGAA and AGUU hairpins were calculated from an extended RNA conformation with a standard simulated annealing protocol using XPLOR 3.1 (Brünger, 1992). For AGAA, 300 NOE restraints and 54 dihedral angle restraints were obtained and used in the structure calculation. For AGUU, 261 NOE restraints and 54 dihedral restraints were obtained and used in structure calculation. An additional 13 hydrogen bond distance restraints were used to restrain the five base pairs in the stem to be Watson–Crick base pairs, which is consistent with the experiment data. The simulated annealing protocol consisted of 24 ps of dynamics at 2000 K (integration time 8000 steps of 3 fs) and 48 ps of slow cooling from 2000 to 100 K (25 K/cycle). It was then followed by 500 steps of energy minimization using Powell’s algorithm. For each molecule, 100 structures were calculated, of which 20 structures were selected for the final ensemble to be analyzed based on lowest total and NOE energy. The structures were analyzed with Insight II and MolMol (Koradi et al., 1996). Hydrogen bonds in the tetraloops were analyzed with Insight II (Biosym), using criteria in which the angle between proton donor and acceptor must be >120° and the distance <2.5 Å. The structure determination statistics are given in Table I.
Table I. Structure determination statistics for AGAA and AGUU hairpins.
| AGAA | AGUU | |
|---|---|---|
| NMR-derived distance and dihedral angle restraints | ||
| Loop intranucleotide | 23 | 20 |
| Loop internucleotide | 69 | 32 |
| Total intranucleotide | 53 | 60 |
| Total internucleotide | 247 | 201 |
| Dihedral angles | 54 | 54 |
| Structure statistics | ||
| NOE violations (average) | 0 >0.1 Å | 0 >0.5 Å, 1.0 >0.1 Å |
| Angle violations | 0 >5° | 0> 5° |
| Average residual angle violations | 0 | 0 |
| Mean deviation from ideal covalent geometry | ||
| Bond length (Å) | 0.0048 | 0.0043 |
| Bond angles (°) | 1.08 | 1.01 |
| Improper (°) | 0.38 | 0.37 |
| R.m.s.d. for all heavy atoms relative to mean structure | ||
| All residues (Å) | 0.69 ± 0.18 | 1.41 ± 0.30 |
| Stem residues (Å) | 0.49 ± 0.15 | 0.52 ± 0.17 |
| Loop residues (Å) | 0.53 ± 0.15 | |
| All residues except U8 and U9 of AGUU (Å) | 0.63 ± 0.20 | |
Rnt1p cleavage assays
The Rnt1p substrates were prepared as described (Chanfreau et al., 2000), except that transcripts were internally labeled with [α-32P]UTP. Two sets of cleavage assays were done for each substrate. In one, the gel-purified substrates were used directly in the cleavage assay. In the other set, the gel-purified substrates first were denatured at 85°C for 3 min in 50 mM Tris–HCl pH 7.6, 200 mM KCl, 0.1 mg/ml wheat germ tRNA, 5 mM MgCl2 and then cooled down to 55°C (0.2°C/s) and to 23°C (0.1°C/s) in a Peltier thermocycler. The renatured substrates were then used in the cleavage assay as described (Chanfreau et al., 2000). The potential of wild-type and mutant RNAs to form intermolecular duplexes was assayed by running these RNAs in the absence of protein on native gels, as described (Chanfreau et al., 2000). All quantitations were performed by PhosphorImager analysis. In order to compare the cleavage results on the substrate (hairpin) conformation only, the data were normalized to the total fraction of hairpin for each substrate, estimated by analysis of the native gels (Figure 5B). After this normalization, the cleavage kinetics with or without the denaturation–renaturation procedure showed no significant differences (data not shown). Thus, the data from the two conditions were pooled and the average of in vitro cleavage kinetics from 10 independent experiments were plotted and analyzed (Figure 5A).
Coordinate deposition
Coordinates for the 20 lowest energy structures of the AGAA and AGUU hairpins have been deposited in the RCSB PDB (accession Nos 1k4a and 1k4b, respectively.)
Acknowledgments
Acknowledgements
We thank Mr David Finger and Dr Robert Peterson for NMR assistance. This work was supported by NSF grant MCB 9808072 and NIH grant GM R01 37254 to J.F. and NIH grant GM R01 61518 to G.C.
References
- Abou Elela S. and Ares,M. (1998) Depletion of yeast RNase III blocks correct U2 3′ end formation and results in polyadenylated but functional U2 snRNA. EMBO J., 17, 3738–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Elela S., Igel,H. and Ares,M. (1996) RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell, 85, 115–124. [DOI] [PubMed] [Google Scholar]
- Allain F.H.-T. and Varani,G. (1995) Structure of the P1 helix from group I self-splicing introns. J. Mol. Biol., 250, 333–353. [DOI] [PubMed] [Google Scholar]
- Batey R.T., Inada,M., Kujawinski,E., Puglisi,J.D. and Williamson,J.R. (1992) Preparation of isotopically labeled ribonucleotides for multidimensional NMR spectroscopy of RNA. Nucleic Acids Res., 20, 4515–4523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bax A. and Davis,D.G. (1985) MLEV-17-based two-dimensional homonuclear magnetization transfer spectroscopy. J. Magn. Reson., 65, 355–360. [Google Scholar]
- Bax A., Clore,G.M., Driscoll,P.C., Gronenborn,A.M., Ikura,M. and Kay,L.E. (1990) Practical aspects of proton–carbon–carbon–proton three-dimensional correlation spectroscopy of 13C-labeled proteins. J. Magn. Reson., 87, 620–627. [Google Scholar]
- Bernstein E., Caudy,A.A., Hammond,S.M. and Hannon,G.J. (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature, 409, 363–366. [DOI] [PubMed] [Google Scholar]
- Brünger A.T. (1992) X-PLOR (Version 3.1) Manual. Yale University Press, New Haven, CT.
- Butcher S.E., Dieckmann,T. and Feigon,J. (1997) Solution structure of the conserved 16S-like ribosomal RNA UGAA tetraloop. J. Mol. Biol., 268, 348–358. [DOI] [PubMed] [Google Scholar]
- Chanfreau G., Abou Elela,S., Ares,M. and Guthrie,C. (1997) Alternative 3′-end processing of U5 snRNA by RNase III. Genes Dev., 11, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau G., Legrain,P. and Jacquier,A. (1998a) Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol., 284, 975–988. [DOI] [PubMed] [Google Scholar]
- Chanfreau G., Rotondo,G., Legrain,P. and Jacquier,A. (1998b) Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J., 17, 3726–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau C., Buckle,M. and Jacquier,A. (2000) Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl Acad. Sci. USA, 97, 3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G.M., Bax,A., Driscoll,P.C., Wingfield,P.T. and Gronenborn,A.M. (1990) Assignment of the side-chain 1H and 13C resonances of interleukin-1β using double- and triple-resonance heteronuclear three-dimensional NMR spectroscopy. Biochemistry, 29, 8172–8184. [DOI] [PubMed] [Google Scholar]
- Dieckmann T. and Feigon,J. (1997) Assignment methodology for larger oligonucleotides: application to an ATP-binding RNA aptamer. J. Biomol. NMR, 9, 259–272. [DOI] [PubMed] [Google Scholar]
- Filippov V., Solovyev,V., Filippova,M. and Gill,S.S. (2000) A novel type of RNase III family proteins in eukaryotes. Gene, 245, 213–221. [DOI] [PubMed] [Google Scholar]
- Greenbaum N.L., Radhakrishnan,I., Hirsh,D. and Patel,D.J. (1995) Determination of the folding topology of the SL1 RNA from Caenorhabditis elegans by multidimensional heteronuclear NMR. J. Mol. Biol., 252, 314–327. [DOI] [PubMed] [Google Scholar]
- Greenbaum N.L., Radhakrishnan,I., Patel,D.J. and Hirsh,D. (1996) Solution structure of the donor site of a trans-splicing RNA. Structure, 4, 725–733. [DOI] [PubMed] [Google Scholar]
- Kadkhodaei M., Hwang,T.L., Tang,J. and Shaka,A.J. (1993) A simple windowless mixing sequence to suppress cross relaxation in TOCSY experiments. J. Magn. Reson. A, 105, 104–107. [Google Scholar]
- Kharrat A., Macias,M.J., Gibson,T.J., Nilges,M. and Pastore,A. (1995) Structure of the dsRNA binding domain of E.coli RNase III. EMBO J., 14, 3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk M.H., Wijmenga,S.S., Heus,H.A. and Hilbers,C.W. (1998) On the NMR structure determination of a 44n RNA pseudoknot: assignment strategies and derivation of torsion angle restraints. J. Biomol. NMR, 12, 423–433. [Google Scholar]
- Koradi R., Billeter,M. and Wüthrich,K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph., 14, 51–55. [DOI] [PubMed] [Google Scholar]
- Kufel J., Dichtl,B. and Tollervey,D. (1999) Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA, 5, 909–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine D., Vadenhaute,J. and Tollervey,D. (1995) The 18S rRNA dimethylase Dim1p is required for pre-ribosomal RNA processing in yeast. Genes Dev., 9, 2470–2481. [DOI] [PubMed] [Google Scholar]
- Legault P., Jucker,F.M. and Pardi,A. (1995) Improved measurement of C-13, P-31 J coupling constants in isotopically labeled RNA. FEBS Lett., 362, 156–160. [DOI] [PubMed] [Google Scholar]
- Macura S., Huang,Y., Suter,D. and Ernst,R.R. (1980) Two-dimensional chemical exchange and cross-relaxation spectroscopy of coupled nuclear spins. J. Magn. Reson., 43, 259–281. [Google Scholar]
- March P.E., Ahnn,J. and Inouye,M. (1985) The DNA sequence of the gene (rnc) encoding ribonuclease III of Escherichia coli. Nucleic Acids Res., 13, 4677–4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marino J.P., Schwalbe,H., Anklin,C., Bermel,W., Crothers,D.M. and Griesinger,C. (1995) Sequential correlation of anomeric ribose protons and intervening phosphorus in RNA oligonucleotides by H-1, C-13, P-31 triple resonance experiment HCP-CCH-TOCSY. J. Biomol. NMR, 5, 87–92. [DOI] [PubMed] [Google Scholar]
- Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7-RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller L., Legault,P. and Pardi,A. (1995) Improved RNA structure determination by detection of NOE contacts to exchange-broadened amino protons. J. Am. Chem. Soc., 117, 11043–11048. [Google Scholar]
- Nagel R. and Ares,M. (2000) Substrate recognition by a eukaryotic RNase III: the double-stranded RNA-binding domain of Rnt1p selectively binds RNA containing a 5′-AGNN-3′ tetraloop. RNA, 6, 1142–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanduri S., Carpick,B.W., Yang,Y.W., Williams,B.R.G. and Qin,J. (1998) Structure of the double-stranded RNA-binding domain of the protein kinase PKR reveals the molecular basis of its dsRNA-mediated activation. EMBO J., 17, 5458–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikonowicz E.P. and Pardi,A. (1993) An efficient procedure for assignment of the proton, carbon and nitrogen resonances in 13C/15N labeled nucleic acids. J. Mol. Biol., 232, 1141–1156. [DOI] [PubMed] [Google Scholar]
- Nikonowicz E.P., Sirr,A., Legault,P., Jucker,F.M., Baer,L.M. and Pardi,A. (1992) Preparation of C-13 and N-15 labelled RNAs for heteronuclear multi-dimensional NMR studies. Nucleic Acids Res., 20, 4507–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R.D., Bartel,D.P., Szostak,J.W., Horvath,S.J. and Feigon,J. (1994) 1H NMR studies of the high-affinity Rev binding site of the Rev responsive element of HIV-1 mRNA: base pairing in the core binding element. Biochemistry, 33, 5357–5366. [DOI] [PubMed] [Google Scholar]
- Qu L.H. et al. (1999) Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol., 19, 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos A., Grunert,S., Adams,J., Micklem,D.R., Proctor,M.R., Freund,S., Bycroft,M., St Johnston,D. and Varani,G. (2000) RNA recognition by a Staufen double-stranded RNA-binding domain. EMBO J., 19, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rance M., Sørensen,M., Bodenhausen,G., Wagner,G., Ernst,R.R. and Wüthrich,K. (1983) Improved spectral resolution in COSY 1H NMR spectra of proteins via double quantum filtering. Biochem. Biophys. Res. Commun., 117, 479–485. [DOI] [PubMed] [Google Scholar]
- Ryter J.M. and Schultz,S.C. (1998) Molecular basis of double-stranded RNA–protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J., 17, 7505–7513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seipelt R.L., Zheng,B.H., Asuru,A. and Rymond,B.C. (1999) U1 snRNA is cleaved by RNase III and processed through an Sm site-dependent pathway. Nucleic Acids Res., 27, 587–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simorre J.P., Zimmermann,G.R., Mueller,L. and Pardi,A. (1996) Triple-resonance experiments for assignment of adenine base resonances in C-13/N-15-labeled RNA. J. Am. Chem. Soc., 118, 5316–5317. [Google Scholar]
- Sklenárõ V., Miyashiro,H., Zon,G., Miles,H.T. and Bax,A. (1986) Assignment of the 31P resonances in oligonucleotides by two-dimensional NMR spectroscopy. FEBS Lett., 208, 94–98. [DOI] [PubMed] [Google Scholar]
- Sklenárõ V., Peterson,R.D., Rejante,M.R., Wang,E. and Feigon,J. (1993) Two-dimensional triple-resonance HCNCH experiment for direct correlation of ribose H1′ and base H8, H6 protons in 13C, 15N-labeled RNA oligonucleotides. J. Am. Chem. Soc., 115, 12181–12182. [Google Scholar]
- Smith J.S. and Nikonowicz,E.P. (2000) Phosphorothioate substitution can substantially alter RNA conformation. Biochemistry, 39, 5642–5652. [DOI] [PubMed] [Google Scholar]
- St Johnston D., Brown,N.H., Gall,J.G. and Jantsch,M. (1992) A conserved double-stranded RNA-binding domain. Proc. Natl Acad. Sci. USA, 89, 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C. et al. (1988) CUUCGG hairpins: extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc. Natl Acad. Sci. USA, 85, 1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C.R., Winker,S. and Gutell,R.R. (1990) Architecture of ribosomal RNA: constraints on the sequence of ‘tetra-loops’. Proc. Natl Acad. Sci. USA, 87, 8467–8471. [DOI] [PMC free article] [PubMed] [Google Scholar]