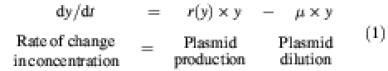Abstract
In one family of bacterial plasmids, multiple initiator binding sites, called iterons, are used for initiation of plasmid replication as well as for the control of plasmid copy number. Iterons can also pair in vitro via the bound initiators. This pairing, called handcuffing, has been suggested to cause steric hindrance to initiation and thereby control the copy number. To test this hypothesis, we have compared copy numbers of isogenic miniP1 plasmid monomer and dimer. The dimer copy number was only one-quarter that of the monomer, suggesting that the higher local concentration of origins in the dimer facilitated their pairing. Physical evidence consistent with iteron-mediated pairing of origins preferentially in the dimer was obtained in vivo. Thus, origin handcuffing can be a mechanism to control P1 plasmid replication.
Keywords: DNA looping/DNA replication/DNA supercoiling/plasmid/replication control
Introduction
All plasmids of bacterial origin studied to date have characteristic copy numbers. However, intrinsically random processes of initiating replication or partitioning copies between daughter cells generate spontaneous deviations. Without the capacity to correct for deviations, cells of a population can have large copy number variations (Lobner-Olesen, 1999; Paulsson and Ehrenberg, 2001). This can decrease segregational stability since cells with too few plasmids are at a higher risk of generating plasmid-free cells. At the other extreme, cells overloaded with plasmid copies are likely to be out-competed by cells with less of a metabolic burden. For stable maintenance, therefore, a plasmid must adjust for fluctuations of its copy number. At lower than average copy numbers, each replicon on average replicates more than once per cell cycle and, at higher copy numbers, they replicate less. This narrowing of copy number distribution by controlling DNA replication is the tenet of plasmid copy number control.
Many well studied plasmids, such as ColE1 and R1, control replication using plasmid-encoded RNA that serves as a replication inhibitor (Helinski et al., 1996; Summers, 1996). The RNA is made such that its concentration remains proportional to the plasmid copy number. An increase in plasmid concentration then leads to a corresponding increase in inhibitor concentration and lower replication frequency per plasmid copy. Such negative feedback control provides a mechanism to adjust fluctuations in copy numbers. The tighter the control, i.e. the higher the sensitivity amplification in the negative feedback loop, the greater the capacity to narrow the copy number distribution. Sensitivity amplification can be defined as the percentage change in the response (replication frequency) for a percentage change in the signal (plasmid or inhibitor concentrations). Traditional ways of obtaining high sensitivity include cooperative binding or multimerization of regulatory molecules as well as multistep control. Such mechanisms are referred to as ultrasensitive, compared with the standard hyperbolic curve (Koshland et al., 1982). Hyperbolic replication control can, at best, result in an inverse relationship between the replication frequency per copy and plasmid concentration. It is consistent with some proposed single-step kinetic models of inhibition, but is predicted to be rather inefficient in suppressing random copy number fluctuations. For plasmid ColE1, it has been suggested that a multistep inhibition process or the use of two different plasmid-encoded inhibitors, RNA I and the Rom protein, can give higher sensitivity and narrower distributions (Paulsson and Ehrenberg, 2001). Copy numbers in single cells are not easily measured, but experiments dealing with averages over large populations can sometimes be at least as instructive for inferring information about the control mechanism. One example is the comparison of average copy numbers in cells carrying either plasmid monomers or isogenic dimers. When both origins are active and non-interfering, dimerization means that replication can be triggered at either origin and that each plasmid copy codes for double sets of regulatory elements. The combined effect of these changes on the relative copy number of monomers and dimers then turns out to be a signature of replication control. In a theoretical analysis of plasmid ColE1, it was shown that if the plasmid uses a well-working hyperbolic mechanism, monomer and dimer copy numbers should be identical, whereas for highly sensitive mechanisms, the dimer copy number could be as much as 2-fold lower (Paulsson et al., 1998). Experimental results clearly support the latter for ColE1, indicating highly sensitive control.
In bacterial plasmids of a different type, the replication inhibitors are plasmid-specific repeated DNA (∼20 bp) sequences, called iterons (Helinski et al., 1996; Chattoraj, 2000). Iterons are found in the origin, but in many cases additional copies exist outside the origin, as in plasmids P1 and F (Tsutsui et al., 1983; Pal et al., 1986). Irrespective of their provenance, when cloned in trans, they invariably reduce plasmid copy number, the magnitude of the reduction being dependent upon the number of iterons cloned. Conversely, plasmid copy number is increased by deleting iterons that exist outside of the origin (Tsutsui et al., 1983; Pal et al., 1986). These results suggest that iterons serve the same purpose as inhibitors and that their concentration determines the replication rate.
Iterons bind plasmid-encoded initiator proteins both in vivo and in vitro. Iterons can also pair via the bound initiators in vitro (Mukherjee et al., 1985, 1988a,b; Chattoraj et al., 1988; McEachern et al., 1989). Based on these two properties of iterons, two models have been proposed for iteron-mediated inhibition of plasmid replication. Model I, initiator titration, assumes initiators to be limiting (Tsutsui et al., 1983; Chattoraj et al., 1984), with replication stopping when the iteron concentration becomes so high as to reduce initiator availability below the threshold needed for replication initiation. Model II assumes that a pairing of origin iterons (‘handcuffing’) results in a steric hindrance of replication initiation, with replication stopping when every origin is engaged in iteron-mediated pairing (Pal and Chattoraj, 1988; McEachern et al., 1989). Model I is disfavored currently because increasing initiator concentration beyond the physiological concentration does not increase copy number significantly, indicating that there must be a mechanism other than initiator limitation to prevent an uncontrolled increase in plasmid copy number (Pal and Chattoraj, 1988; Durland and Helinski, 1990; Uga et al., 1999). This other mechanism is now believed to be handcuffing.
The best support for the handcuffing model has come from characterization of high-copy initiator mutants that show a pairing defect in vitro (Mukhopadhyay et al., 1994; Miron et al., 1994; Blasina et al., 1996; Uga et al., 1999). However, only a handful of mutants have been tested and, in one study, one out of the three mutants tested was not defective for handcuffing (Miron et al., 1994). There can also be other modes of control. In several members of the iteron family of plasmids, when initiator expression is increased a few fold above the normal, the plasmid copy number decreases (Filutowicz et al., 1986; Muraiso et al., 1990; Ingmer and Cohen, 1993). The mechanism of this mode of control is not understood. In addition, replication in vitro of miniP1 plasmids carrying origin iterons only is not inhibited by increasing the plasmid concentration (Abeles and Austin, 1991). This result is troubling since it has been assumed that origin iterons control copy number when they are the only iterons, as in P1 and F deleted for the non-origin iterons or in plasmids that lack those iterons naturally, e.g. R6K and pSC101 (Chattoraj, 2000). This study was initiated to address the question of how copy number is controlled when origins are the only source of iterons.
In a classic experiment, Tsutsui and Matsubara showed that a hybrid of ColE1 + miniF plasmids replicated at a copy number (16–20) characteristic for the ColE1 plasmid but, when initiation from the ColE1 origin was blocked, the miniF origin was not activated until the copy number dropped to a value characteristic of miniF (1–2). A small deviation in copy number above the characteristic limit thus led to efficient replication switch-off. When chromosomes have an integrated copy of plasmid P1, a cytoplasmic copy cannot be maintained even under selection (Pal and Chattoraj, 1988; McEachern et al., 1989). These results can be taken as evidence for a sensitive inhibition mode, but do not distinguish between initiator titration and handcuffing. To address the issue, we have measured instead the average copy numbers of monomers and isogenic dimers and, in analogy with ColE1, use relative copy numbers to infer information about the control mechanism. However, the theoretical analysis of ColE1 (Paulsson et al., 1998) was specific to that particular situation and did not suggest how to distinguish between initiator titration and handcuffing. We have therefore generalized the analysis only assuming that replication at an origin depends on the total number of regulatory genetic elements, such as iterons, replication initiators or even chromosome-encoded molecules. All molecular details about kinetic interactions between these elements can be left unspecified. A direct conclusion from the model is that dimers cannot have less than half the average copy number of monomers. If dimer copy numbers still turn out to be lower than this limit, then dimerization must play a more involved role in replication control. Such a dependency is inconsistent with the initiator titration model but follows naturally from the handcuffing model. Origin pairing is expected to be favored in the dimer because the proximity of the two origins should increase the on-rate. From studies of site-specific recombination, it has been well established that communication between distant sites is facilitated when they are linked (Oram et al., 1997). It is also possible that the paired complexes would be more stable in the dimer because of a lower off-rate; the supercoiling of DNA may help to stabilize origin pairs by reinforcing the protein bridge between sites in cis only (‘spring clip’ stabilization) (Summers, 1998).
We show the copy number of isogenic dimers to be about one-quarter of that of the monomer. By controlling the amount of the initiator protein (called RepA), we have ruled out that this effect is due to insufficient RepA. We also provide evidence consistent with physical interactions between origins in vivo. These results are significant in two respects: they show that intact origins are used for negative control of plasmid copy number, and they uphold handcuffing as a mechanism for the negative control.
Results
Copy number of monomeric and dimeric forms of a miniP1 plasmid
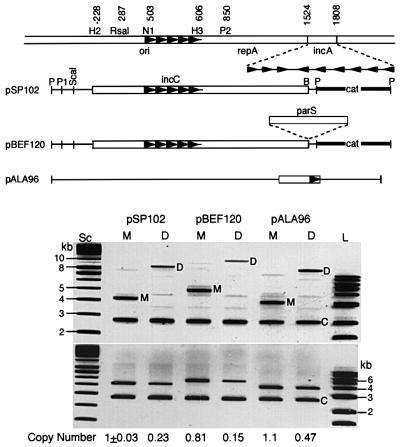
When monomeric and dimeric forms of a miniP1 were established in recA bacteria, the dimer copy number was one-quarter that of the monomer (pSP102, Figure 1). The results were the same with a second miniP1 plasmid that additionally contained a parS site (centromere analog), and the cognate Par proteins, ParA and ParB, were supplied in trans (pBEF120). In a pBR322-based replicon, the ratio of monomer to dimer copy number was roughly half, as was found by others (pALA96) (Chiang and Bremer, 1988; Summers, 1998). In the P1 system, origins in cis were more effective in lowering copy number, apparently because the arrangement facilitated their physical interaction.
Fig. 1. Plasmid copy number in cells carrying either a plasmid monomer (M) or an isogenic plasmid dimer (D). Top: schematic map of plasmids. Open bars represent P1 DNA, thin lines pBR322 DNA, and thick lines a drug marker DNA. Coordinates in the top line are from Abeles et al. (1984). The enzyme names have been abbreviated in some cases as follows: HincII (H2), NruI (N1), HindIII (H3), PvuII (P2), PstI (P), PvuI (P1) and BamHI (B). Arrowheads represent iterons. The three genetic elements that comprise the P1 plasmid replicon are ori (387–611), initiator gene repA (664–1521) and the control locus, incA (1524–1808). pSP102 is a miniP1 plasmid. pBEF120 is identical to pSP102 except for the presence of parS. pALA96 is derived from pBR322 (Chattoraj et al., 1984). Bottom: lanes Sc and L represent supercoiled and linear DNAs that served as size markers in kb units. Plasmid DNA was isolated from cells carrying the two (M and D) forms. Before DNA isolation, a control culture containing pNEB193 (C) was added to each of the experimental cultures to account for loss during DNA isolation. The upper and lower panels are identical except that the DNAs were digested with HindIII in the lower panel for ease of measuring total plasmid DNA. The band intensities were normalized with respect to the pSP102 monomer band and adjusted further for plasmid size to determine relative copy numbers.
Monomer–dimer copy number difference is not eliminated by initiator oversupply
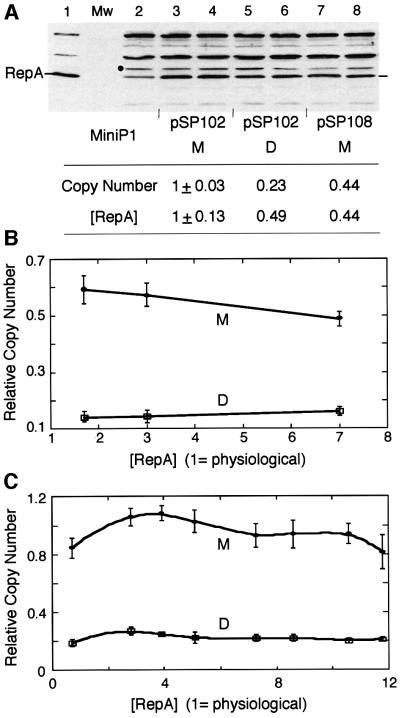
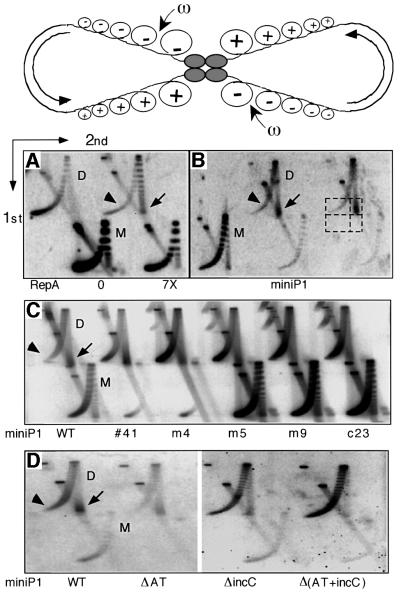
The copy number of miniP1 plasmids with only the origin iterons depends upon the concentration of initiator protein, and optimal replication requires ∼3-fold more RepA than the level made by the wild-type plasmid (physiological level) (Pal and Chattoraj, 1988). One reason for the lower copy number observed for dimers could be that not enough RepA was made from dimers. RepA concentration was indeed lower in dimer-carrying cells compared with monomer-carrying cells (Figure 2A). However, 6- to 12-fold additional RepA, supplied in trans from two different sources, did not significantly alter the monomer– dimer copy number difference (Figure 2B and C). We conclude that RepA concentration was adequate in the dimer and it was not the cause of the lower dimer copy number.
Fig. 2. Relationship of [RepA] and [origin] of miniP1 (pSP102) M and D. (A) [RepA] was determined by immunoblotting. In lane 1, cells with an overproducer of RepA source were used to locate RepA. In the next lane, protein molecular weight (Mw) standards were used that did not react with RepA antibody. In lane 2, cells do not have a RepA source but show a cross-reacting band of the size of RepA. The band was also seen with a different RepA antibody (gift of S.Wickner). The intensity of this band was subtracted from the bands in corresponding positions of lanes 3–8 for determination of relative [RepA]. A lower copy number derivative of pSP102 (pSP108, lanes 7–8; Pal et al., 1986) was used to test the effect of lowering the copy number on RepA production from a monomeric source. The black dot represents the band chosen for normalizing RepA content among different lanes. Duplicate samples were used in lanes 3–8 and their average RepA content is shown in the table below the figure. (B and C) Copy number of miniP1 M and D at increasing [RepA] in trans. In (B), the miniP1 plasmid was pVM11 which is deleted for its own repA gene. RepA was provided from compatible plasmids pALA69, pALA177 and pALA176 that constitutively produced 1.7×, 3× and 7× protein relative to the amount produced by wild-type P1 plasmid (physiological amount = 1) (Swack et al., 1987). In (C), the miniP1 plasmid was pSP302, which is identical to pSP102 except for the presence of a repA103(Am) mutation (Austin et al., 1985), and the host (MC4100) was non-suppressing (Sozhamannan and Chattoraj, 1993). RepA was supplied at varying concentrations using different inducer concentrations from pSP206, which contains repA under the control of the lac promoter (Pal and Chattoraj, 1988). Note that in both (B) and (C), the copy number difference is largely maintained irrespective of [RepA].
Control-defective RepA mutants reduce monomer/dimer copy number ratio
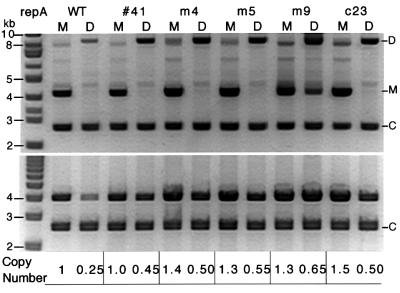
If the lower copy number of dimers is due to more effective control than normally operates on monomeric P1 plasmids, we can expect that the monomer–dimer copy number difference would be reduced in mutant plasmids with control-defective (high-copy) mutations in repA. Plasmids with three previously characterized (m4, m5 or m9; Mukhopadhyay et al., 1994) or two uncharacterized (41 or c23) high-copy mutations in repA were analyzed in the present system. Wild-type RepA requires chaperones to activate the protein for binding to iterons. The mutant RepAs (corresponding to mutations m4, m5 and m9) were selected for chaperone-independent iteron binding. They also confer high-copy phenotype and exhibit reduced handcuffing in vitro. RepA41 differs from wild-type protein in that it does not inhibit miniP1 replication when overproduced in trans (data not shown). RepAc23 allows replication of miniP1 plasmids in the presence of an excess of iterons that would normally inhibit replication initiation by wild-type RepA. All five mutant RepAs reduced the copy number difference between monomer and dimer (Figure 3). However, the dimer copy number was at best half that of the isogenic monomer (i.e. m9). This suggests that the mutants are only partially affected in control, as is also evident from the modest increase of the copy number of the mutant monomers compared with the wild-type. The results also suggest that the control system that operates on dimers is identical to that normally operating on monomers.
Fig. 3. Comparison of copy number of miniP1 M and D carrying initiator gene (repA) mutations that increase the plasmid copy number. Copy number was determined as in Figure 1. All five mutant plasmids show a reduced difference in copy number between M and D (table below the gel pictures).
Dimer copy number with one origin mutated
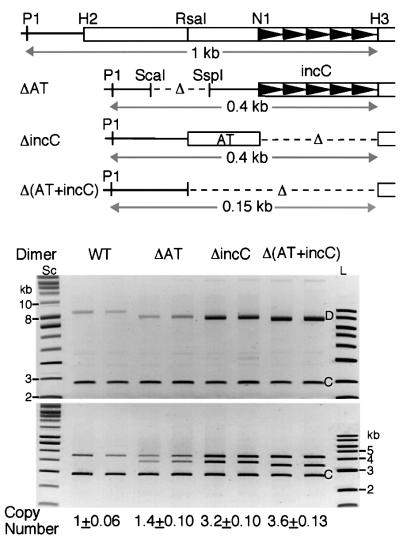
As a first step towards defining the elements of the origin involved in its negative control, we deleted one of the origins in the dimer either partially or completely (Figure 4). The partially deleted origins either deleted the AT-rich half of the origin (as in plasmid ΔAT) or the iterons (as in plasmid ΔincC). Dimer copy number remained low when the iterons were intact (lane ΔAT). The copy number increased when the iterons were deleted from one of the origins [lanes ΔincC and Δ(AT+incC)]. These results indicate that the iterons of the origin are primarily responsible for the negative control of the copy number.
Fig. 4. D copy number with one mutated origin. The maps show an intact miniP1 origin and the regions deleted from one of the origins in mutant plasmid dimers. Open bars represent P1 DNA and arrowheads the iterons as in Figure 1. In the ΔAT mutant plasmid, the H2–N1 region is deleted. The plasmid is also deleted for the ScaI–SspI region of pBR322 DNA containing a bla promoter. In the ΔincC mutant plasmid, the N1–H3 region carrying the incC iterons is deleted. In the Δ(AT+incC) mutant plasmid, the RsaI–H3 region is deleted. Two independent cultures were used for each plasmid. Deletion of the iterons from one of the origins had the most dramatic effect on dimer copy numbers [lanes ΔincC and Δ(AT+incC)].
Replication fork progression is not hindered in a dimer
Here we have considered whether the lowering of the dimer copy number is due to stalling of replication fork elongation as it encounters an origin (Brewer and Fangman, 1993; Viguera et al., 1996). At the time of initiation at one of the dimer origins, the replisome assembly at the other origin may be incomplete but substantial enough to hinder fork progression. This possibility is considered unlikely due to the following results. First, increasing RepA does not decrease dimer copy number (Figure 2B and C) as might be expected if RepA–iteron complexes form the core of the road-block. Secondly, in the case of high-copy mutants, the reduced difference in monomer–dimer copy numbers may mean that fork stalling is not a necessary feature of a replicating dimer (Figure 3). Thirdly, miniP1 monomers replicate bidirectionally in vivo but the two forks progress asymmetrically (Park and Chattoraj, 2001). One of the forks is impeded due to converging transcription from the cat gene of the plasmid. This apparently is not a problem since the fork moving in the opposite direction can complete replication.
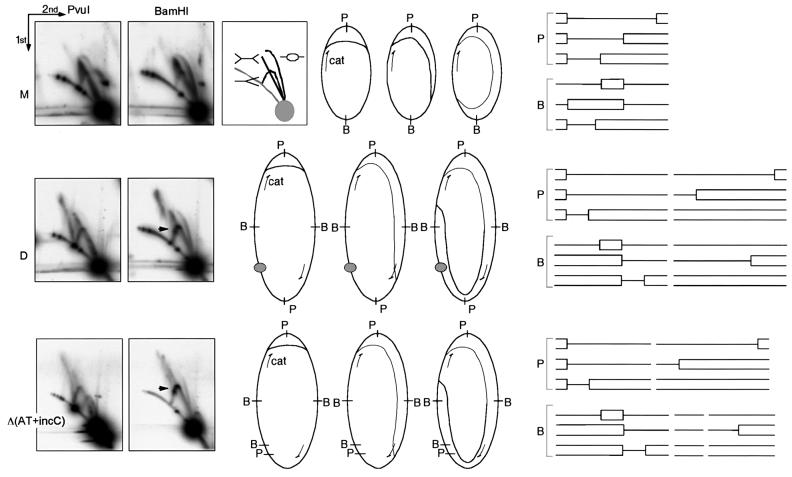
The pausing at an origin of a dimer was tested by analyzing replication intermediates (RIs) by two-dimensional gel electrophoresis (Friedman and Brewer, 1995) (Figure 5). As shown earlier, RIs of pSP102 monomer, linearized by PvuI, migrate as if they are of double-Y pattern, indicating replication initiation close to the PvuI site (Figure 5, M) (Park and Chattoraj, 2001). RIs linearized at the BamHI site, which is diametrically opposite to the PvuI site, migrate in a bubble arc initially and then switch to the double-Y line. When plasmid dimers were examined, the most conspicuous change was the appearance of the simple-Y pattern (Figure 5, D). Studies with ColE1 multimers have shown that initiation at one of the origins in a multimer suffices to complete replication, and simultaneous use of more than one origin is rare (Martin-Parras et al., 1992). If the dimer is replicated from one of the origins, the intermediates generated from the plasmid half that replicates passively are predicted to be of simple-Y form.
Fig. 5. Two-dimensional gel electrophoresis of replication intermediates (RIs) of pSP102 M, D and Δ(AT+incC). Top row: the box next to the gel pictures shows the expected gel running patterns of RIs with bubble, simple-Y and double-Y forms. The gray circle and the gray arc represent linear DNA of 4.3 kb (length of the pSP102 monomer) and longer lengths, respectively. The gel figures show the RIs after linearization with PvuI (P) and BamHI (B). The RIs expected from asymmetric bidirectional replication, initiated near the PvuI (P) site and migrated to various lengths, are also shown schematically before and after linearization. The asymmetry in fork migration is attributed to converging cat transcription (arrow). The arrowhead in BamHI-digested samples points to stalled forks in dimeric plasmids [D and Δ(AT+incC)]. The stalling is attributed to converging cat transcription and not to road-block mediated by RepA–iteron complexes (represented by a filled circle) since identical stalling is seen in dimers with one origin [Δ(AT+incC)]. Note that the spot for linear DNA splits into two in Δ(AT+incC) when digested by PvuI and into three when digested by BamHI (the two smaller spots are not seen as they have migrated beyond the gel limit).
If the passive origin served as a road-block in the miniP1 dimer, we would have seen a spot representing stalled RIs on the simple-Y pattern near the origin (Martin-Parras et al., 1992). In fact, a spot was seen in the BamHI-digested sample (arrow, Figure 5, D). However, a similarly located spot was also seen when the second origin was deleted entirely from the dimer [Figure 5, Δ(AT+incC)]. In this sample, the passively replicated (origin-deleted) half of the dimer was split into two smaller parts because of the presence of an extra BamHI site. These run separately from the full-length RIs of the origin containing half of the dimer. Persistence of the spot indicates that stalling must have been on the dimer half with the active origin, and was most likely to be due to the converging cat transcription. We conclude that origins do not interfere with fork migration in a dimer.
A topological assay for origin interactions in a dimer
The results described so far support the view that origins interact physically. More direct evidence for such interactions was obtained by taking advantage of their effect on the diffusion of superhelical tension. It has been shown that simultaneous binding of a lac repressor, which is a stable tetramer, to two lac operators at diametrically opposite positions in a dimer can prevent diffusion of transcription-generated superhelical tension across the operators (Wu and Liu, 1992; Figure 6, top panel). Repressor binding to a single operator in a plasmid monomer (or dimer) cannot create such a barrier. Simultaneous binding in a dimer (or two monomers in trans) can lead to accumulation of positive supercoils on one side of the barrier and negative supercoils on the other. When enzymes that can remove positive supercoils (gyrase and topo IV; Zechiedrich et al., 1997) are inactivated but not the one that removes negative supercoils (ω protein), dimeric or higher oligomeric plasmids are found to be more positively supercoiled.
Fig. 6. Topological assay of physical interactions between miniP1 iterons in dimeric plasmids. Top: schematic showing simultaneous binding of a tetrameric protein to two diametrically opposite sites in a homodimer that blocks diffusion of + and – supercoils (Sc), generated by transcription (semicircular arrows), across the sites. Relaxation specifically of –Sc by a topoisomerase (ω) could result in accumulation of +Sc in the dimer. Protein binding to a single site cannot block Sc diffusion, hence no +Sc can accumulate (adapted from Wu and Liu, 1992). Topoisomers of plasmid M and D are separated by two-dimensional agarose gel electrophoresis. In the first dimension, plasmids with more Sc, + or –, migrate faster. In the second dimension, run in the presence of chloroquine, plasmids with +Sc run ahead of those with –Sc. The leading and trailing edges of the plasmid topoisomer distribution contain plasmids with most +Sc (arrow) and most –Sc (arrowhead), respectively. (A) Distribution of a pBR322-derived plasmid (pAO; Wu and Liu, 1992) to which the incC iterons have been added. The 0 and 7× RepA were supplied from compatible plasmids pST52 and pALA198, respectively (Park et al., 1998) (1× is physiological RepA). Plasmids with +Sc are seen only when RepA was supplied. (B) Topoisomer distribution of a miniP1 plasmid, pSP102. The dashed boxes show the region used to quantify the +Sc/–Sc ratio. The intensity of the lower two (empty) boxes was used as background and subtracted from the corresponding upper boxes before calculating the ratio. Plasmids with +Sc predominate in the D but not in the M population. (C) Relative levels of +Sc in pSP102 and its five high-copy derivatives (plasmids are the same as in Figure 3). From the comparison of band intensities of plasmids with +Sc and –Sc, from the two edges of the topoisomer distribution (arrow and arrowhead, respectively), it is apparent that there are relatively fewer plasmids with +Sc in mutants than in the wild-type (WT). (D) Participation of iterons in the generation of +Sc in plasmid dimers. One of the two origins is defective in mutant dimers (plasmids are the same as in Figure 4). Plasmids with +Sc are reduced particularly when the incC iterons are deleted [ΔincC and Δ(AT+incC), last two lanes]. Comparison of ΔincC and Δ(AT+incC) plasmids also shows that the AT-rich region contributes somewhat to the accumulation of +Sc.
We initially tested a pBR322-derived plasmid that was used to show interactions between lac operators (Wu and Liu, 1992), except that the operators were replaced with incC iterons. When RepA was provided in trans, dimeric plasmids were enriched for positive supercoils (Figure 6A). The appearance of positive supercoils depended on the presence of both iterons and RepA (data not shown). The results were similar for dimers of pSP102, the miniP1 plasmid with which most of the results were obtained in this study (Figure 6B). The plasmid has two genes, cat and repA, both directed the same way, and their transcription most probably provided the force to generate the positive supercoils. The ratio of positive to negative supercoils from the two extremities of the topoisomer distribution (arrow and arrowhead/dashed box area, Figure 6B) was 2.1 ± 0.84 for dimers and 0.34 ± 0.045 for monomers. In other words, positive supercoils were enriched in the dimer by ∼6-fold. When pSP102 derivatives with high-copy initiator mutations (same as in Figure 3) were used, the ratio of the intensities of positive and negative supercoiled dimer DNAs was 0.88 ± 0.045, 0.29 ± 0.06, 0.45 ± 0.06, 0.32 ± 0.06 and 0.28 ± 0.09 for the mutant 41, m4, m5, m9 and c23, respectively (Figure 6C). Thus, there has been a 2- to 8-fold reduction in the accumulation of positive supercoils in the mutants. These results provide supportive evidence for physical interactions between miniP1 origins in vivo, and reduction of such interactions in control-defective initiator mutants.
To test the role of iterons in the accumulation of the positive supercoils, dimeric plasmids of Figure 4 with various deletions in one of the origins were used (Figure 6D). The positive to negative supercoil ratio was 2.0 ± 0.81, 1.8 ± 0.67, 0.58 ± 0.10 and 0.19 ± 0.04 for the wild-type, ΔAT, ΔincC and Δ(AT+incC) plasmids, respectively. The absence of the incC iterons from the second origin significantly reduced accumulation of positive supercoils. However, some positive supercoils were still seen when only iterons were deleted from the second origin (Figure 6D, lanes ΔincC). It is possible that origin-binding proteins, such as DnaA and SeqA, were responsible for the residual positive supercoils since both these proteins can oligomerize (Krause et al., 1997; Brendler et al., 2000; Skarstad et al., 2000). The consequence for copy number control of this non-iteron-mediated pairing activity must be nominal, as evident from copy number measurements (Figure 4). It should be noted that the topological assay used here is best applied to cases where isogenic monomer and dimer plasmids can be compared from the same cell culture. This obviates the need to know the absolute levels of transcriptional and DNA relaxation activities. Although we have used heterodimers in experiments of Figure 6D, it is reassuring that dimers with one origin [Δ(AT+incC)] behaved like monomers both in copy number measurements [M/D = 4, Figure 1 versus Δ(AT+incC)/D = 3.6, Figure 4] and in supercoiling assays (Figure 6D).
A theoretical analysis of monomer–dimer copy number ratio
Changes in plasmid concentration can be described by the rate equation:
Plasmids replicate with rate r(y) per copy and are diluted due to exponential cell growth with rate µ. A growth rate µ corresponds to a generation time ln2/µ. To normalize the analysis, we chose our time unit such that µ = 1 and, correspondingly, the generation time is ln2 time units. Equation (1) then becomes parameter free [dy/dt = r(y) × y – y), which simplifies analysis. Because replication control is negative, the replication frequency per copy is lower at higher plasmid concentrations, i.e. r(y) decreases monotonically with y. At the steady-state concentration ȳ, production balances dilution, r(ȳ) = 1, so that:
ȳ = r–1 (1)(2)
where r–1 is the inverse function, not the inverse value 1/r. For instance, if r = e–y, then the inverse function is r–1 = –lny, not 1/e–y = ey.
If the replication frequency at an origin only depends on the total number of basic replicons (iterons, genes for initiator, etc.), then the replication rate per copy in dimer-only cells would be 2r(2x), where x is the dimer concentration. The first ‘2’ reflects that two independent origins can trigger replication and r is now a function of 2x since every dimer consists of two basic replicons. The steady-state 2r(2x̄) = 1 gives:
The ratio between monomer and isogenic dimer copy numbers is thus:
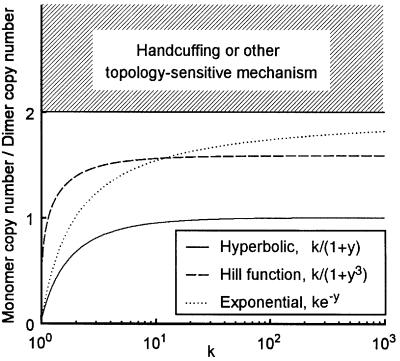
A basic calculus theorem states that if r is a decreasing function, then so is r–1. Negative replication control therefore means that r–1 (1/2)> r–1 (1) because 1/2<1. The copy number ratio R is then limited to 0<R<2. Furthermore, if r is sensitive, so that it works well as a replication control mechanism, then r–1 is insensitive and, vice versa. As a result, for threshold type r, the ratio between r–1 (1) and r–1 (1/2) is close to 1 so that R≈2, while for insensitive r, R can be very low.
In conclusion, insensitive control can give an arbitrarily low ratio between monomer and isogenic dimer copy numbers, but with increasing sensitivity one approaches the theoretical limit of 2-fold higher monomer copy number. To get above this limit, the replication frequency at an origin must depend on a radical cell response to plasmid dimerization or, more probably, on the different topological state of dimers. A mechanism such as handcuffing is certainly consistent with the latter alternative. Some concrete examples of inhibition mechanisms are shown in Figure 7.
Fig. 7. Ratio between average copy numbers in monomer-only and dimer-only cells. The curves correspond to Equation (4) where r is given in the figure inset. Three mechanisms are compared: hyperbolic inhibition, the Hill function with Hill coefficient –3, and exponential inhibition. Parameter k is the maximal replication frequency per copy. All mechanisms become increasingly sensitive with higher k, which in turn affects the monomer–dimer ratio. The shaded area corresponds to cases that violate the assumption that only numbers of regulatory elements matter, and not their presence on monomers versus dimers.
Discussion
The hallmark of plasmids belonging to the iteron family is the presence of iterons in the origin that are essential for initiation of DNA replication. Additionally, these plasmids can have iterons outside of the origin solely for the purpose of limiting copy number. Here we show that the origin iterons can also serve as negative regulators from their natural context. We have considered three parameters that determine how the copy number can be limited when there are no iterons outside of the origin: (i) origin concentration; (ii) host factor limitation; and (iii) plasmid-encoded initiator (RepA) concentration. The assay system described here, which involves comparison of copy numbers of monomeric and dimeric forms of a miniP1 plasmid from two otherwise isogenic cultures, allows us to favor parameter (i) as the main regulator. We find that the dimer copy number is only one-quarter that of monomers (Figure 1). Since the copy number decreased simply by increasing the local origin concentration in a dimer, it is likely that the origin DNA concentration itself rather than some trans-acting host factor normally limits the copy number. Excessive RepA cannot be the reason either, since its concentration was lower in dimer-carrying cells than in monomer-carrying cells (Figure 2). Finally, the possibility that RepA supply was insufficient from the dimer was ruled out since the monomer–dimer copy number difference persisted even when extra RepA were supplied in trans (Figure 2). These results suggest that, as copy number increases, it is primarily the negative regulatory effect of ori DNA that limits a further increase of copy number. We also show that specifically the iteron sequences of ori are responsible for the control (Figure 4), and that handcuffing is a likely mechanism (Figure 6).
The present results are also consistent with the observation that iterons have a stronger effect in cis than in trans. For example, when a single iteron is added to a miniP1 plasmid with origin iterons only, the copy number drops 2-fold. The presence of the same iteron in a multicopy vector (pBR322-derived) reduces the copy number at best by 20% (Pal et al., 1986). These results support handcuffing, but not as directly as the present results. Iterons supplied on a compatible plasmid do not respond to fluctuations in the copy number of the P1 plasmid. The dimerization experiment automatically addresses these problems and makes it possible to draw conclusions about handcuffing without relying on a detailed description of the replication control mechanism.
Although our data are consistent with the handcuffing mechanism, we have not obtained any evidence for intermolecular pairing that the handcuffing mechanism calls for. Some of the obligatory cis reactions involved in site-specific recombination and transcriptional activation can be made to act in trans provided the local concentration of the interacting sites is kept high by catenane formation (Wedel et al., 1990). These results indicate that cis–trans reactions do not have to be qualitatively different.
Understanding of copy number control by the handcuffing mechanism also requires knowledge of how the handcuffing is overcome. In several situations, copy number reduction of miniP1 plasmids by extra iterons has been overcome by increasing initiator concentration (Chattoraj et al., 1984; Pal and Chattoraj, 1988; Park et al., 1998; Mukhopadhyay and Chattoraj, 2000). Thus, initiators have the potential to overcome handcuffing. We suggest that initiators provide the positive force to ensure initiation, and handcuffing is there to prevent overinitiation. The positive and negative forces must balance for plasmid maintenance. The failure to detect control activity from intact origins in vitro (Abeles and Austin, 1991), in contrast to the situation in vivo (Figure 2; Park et al., 1998), perhaps can be rationalized assuming that the in vitro system was optimized towards the initiation mode of the origin DNA. This discrepancy between in vitro and in vivo results remains to be understood.
When ColE1 plasmid monomer and dimer are present in the same cell, as can happen in a wild-type (recombination-proficient) bacterium, the dimers are chosen preferentially for replication (assuming random choice of origins for replication) and this can lead to an ever-increasing fraction of dimers (and higher oligomers) in the plasmid population. As a consequence, the number of partitionable plasmid copies decreases, which can cause plasmid instability in a system where plasmids are distributed randomly to daughter cells. ColE1 plasmids have evolved ingenious mechanisms to prevent drifting of the plasmid population to an ever-increasing size (Hodgman et al., 1998). Here we show that dimer origins initiate replication less frequently than monomer origins. The handcuffing mechanism therefore may help to prevent accumulation of plasmid multimers.
From studies on Bacillus subtilis, it is becoming clear that the chromosomal replication control has similarities with that of plasmids such as miniP1 (Mukhopadhyay and Chattoraj, 2000; Ogura et al., 2001): in both cases, the initiator genes, dnaA and repA, are induced by replication and not by titration, and the binding sites for these proteins, i.e. DnaA boxes and iterons, serve as incompatibility elements. It appears that handcuffing may also apply to B.subtilis replication control.
Materials and methods
Plasmids and strains
Dimers of miniP1 plasmid pSP102 (Pal et al., 1986) were obtained by linearizing the plasmid with HindIII, subsequent ligation and transformation of a recA host, DH5Δlac (Sozhamannan and Chattoraj, 1993). Except for pVM11, all other miniP1 plasmids used in this study are isogenic to pSP102 except for a single mutation in repA (Figure 3) or the additional par locus (pBEF120; Funnell, 1988; Figure 1). pVM11 (Figure 2) was derived from pSP102. The BamHI–PstI fragment carrying the cat gene of pSP102 was replaced with the BamHI–PstI fragment carrying the spec gene (Taylor and Cohen, 1979) of plasmid pSE418 (S.Elledge, unpublished data). From the resultant miniP1 plasmid, pDKC409, ∼1 kb of DNA between PvuII sites was deleted that included the C-terminal half of the repA open reading frame (codons 63–286) and sequences upstream of the spec gene. pVM11 (∼3 kb) is SpecR and replicates only in the presence of a trans source of RepA.
pSP102 dimers with one of the origins partially deleted (Figure 4) were made by partial digestion with HindIII and gel isolation of the full-length (8.5 kb) HindIII fragment. The fragment was partially digested with PvuI, and the 7.5 kb PvuI–HindIII fragment with one of the origins entirely deleted was gel purified. This fragment is ligated to two partially deleted origin fragments. The fragment deleted for the AT-rich half of the origin (called ΔAT) was obtained from pRJM358 (Sozhamannan and Chattoraj, 1993). First the ScaI–SspI region containing the bla-p1 promoter was deleted from pRJM358 to avoid any possible influence of the vector promoter on miniP1 copy number. The deleted DNA subsequently was digested with PvuI and HindIII, and a 1 kb PvuI–HindIII fragment containing pBR322 sequences between PvuI and EcoRI (without the ScaI–SspI region) and the five origin iterons (P1 coordinates 504–606) was ligated to the 7.5 kb PvuI–HindIII fragment. The origin fragment without the iterons (called ΔincC) was obtained from pPP101. The plasmid was constructed by cloning the 216 bp RsaI–NruI fragment of the P1 origin (P1 coordinates 287–503) into the HincII site of pUC19. The P1 origin region was removed from pPP101 by digestion with PvuI and HindIII, and the 1 kb PvuI–HindIII fragment was ligated to the 7.5 kb fragment. To generate plasmid Δ(AT+incC), the ΔincC plasmid was digested with SphI and XbaI that flanked the AT-rich half and the ends ligated after they were made blunt with T4 polymerase.
Plasmid stability and copy number
The dimers of both pSP102 and pBEF120 were slightly unstable compared with monomers without selection. After ∼20 generations of growth in the absence of selection at 37°C, plasmid-carrying cells were 100% for pSP102 monomer, 91 ± 5% for pSP102 dimer, 100% for pBEF120 monomer and 92 ± 8% for pBEF120 dimer. Therefore, copy numbers were measured under selection only (Sozhamannan and Chattoraj, 1993).
Characterization of replication intermediates
The procedure to isolate intracellular DNA and its analysis in two-dimensional gels have been described (Park and Chattoraj, 2001).
Determination of topoisomer distribution by two-dimensional gel electrophoresis
A drug-permeable host (AS19; Wu and Liu, 1992) was transformed separately with monomer and dimer size plasmid DNA, and the transformants, after one round of purification on plates, were used to innoculate cultures for plasmid isolation. In this way, the majority of the plasmid DNA could be obtained in its original size. Cultures of AS19 cells (20 ml) with the desired plasmid(s) were grown to an OD600 of 0.2. A fresh solution of novobiocin was added to a final concentration of 100 µg/ml and the culture was grown for another 30 min and chilled on ice. A total of four OD600 units of cells was used for DNA isolation using a QIAprep Spin Miniprep kit (Qiagen Inc.). DNA was recovered in 50 ml of TE. A 4 µl aliquot was used for gel electrophoresis. The gel was 20 cm × 20 cm × 0.6 cm 1% agarose in 0.5 TPE buffer and was run for ∼600 V-h (typically at 40 V overnight). The gel was soaked in 0.5 TPE + 15 µM chloroquine (Sigma) for 2 h in the dark and run in the second dimension using a fresh solution of 0.5 TPE + 15 µM chloroquine for ∼300 V-h (typically at 70 V). The gel was dried under vacuum without heat for 30 min and for another 30 min at 60°C. The dried gel was placed between two sheets of plastic mesh (FM-100 FlowMesh, Diversified Biotech) for protection during subsequent handling (Lueders and Fewell, 1994). The gel was soaked in 0.25 M HCl for 20 min, rinsed in H2O, soaked in 0.5 M NaOH + 0.15 M NaCl for 20 min, and rinsed in H2O for 5 min (twice) and in 0.5 M Tris pH 8.0 + 0.15 M NaCl for 20 min. Hybridization with radiolabeled probes (pAO or pSP102) was in 6× SSPE, 0.5% SDS, 0.05% sodium pyrophosphate, for 18 h at 55°C (SSPE is 0.15 M NaCl, 10 mM NaH2PO4 pH 7.7). No pre-hybridization was necessary. Washing was in 2× SSC + 0.1% SDS for 15 min at room temperature and in 0.1× SSC + 0.1% SDS for 30 min at 60°C and a further 60 min at room temperature with two changes. The gel was transferred from the plastic screens to a sheet of 3M paper, covered with Saran wrap and exposed to phosphoimaging plates (Fuji) for subsequent analysis in a Fujix BAS2000 image analyzer.
Acknowledgments
Acknowledgements
The authors are indebted to Hai-Young Wu for advice on experiments of Figure 6, Vandana Madhavan, Peter Papp and Steve Elledge for plasmids pVM11, pPP101 and pSE418, respectively, Subrata Pal and Peter Papp for isolating repA41 and repAc23, Sue Wickner for RepA antibody, and David Summers, Michael Lichten, Richard Fekete and Michael Yarmolinsky for thoughtful comments on the manuscript.
References
- Abeles A.L. and Austin,S.J. (1991) Antiparallel plasmid–plasmid pairing may control P1 plasmid replication. Proc. Natl Acad. Sci. USA, 88, 9011–9015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeles A.L., Snyder,K.M. and Chattoraj,D.K. (1984) P1 plasmid replication: replicon structure. J. Mol. Biol., 173, 307–324. [DOI] [PubMed] [Google Scholar]
- Austin S.J., Mural,R.J., Chattoraj,D.K. and Abeles,A.L. (1985) Trans- and cis-acting elements for the replication of P1 miniplasmids. J. Mol. Biol., 183, 195–202. [DOI] [PubMed] [Google Scholar]
- Blasina A., Kittel,B., Toukdarian,A.E. and Helinski,D.R. (1996) Copy-up mutants of the plasmid RK2 replication initiation protein are defective in coupling RK2 replication origins. Proc. Natl Acad. Sci. USA, 93, 3559–3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendler T., Sawitzke,J., Sergueev,K. and Austin,S. (2000) A case for sliding SeqA tracts at anchored replication forks during Escherichia coli chromosome replication and segregation. EMBO J., 19, 6249–6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewer B.J. and Fangman,W.L. (1993) Initiation at closely spaced replication origins in a yeast chromosome. Science, 262, 1728–1731. [DOI] [PubMed] [Google Scholar]
- Chattoraj D.K. (2000) Control of plasmid DNA replication by iterons: no longer paradoxical. Mol. Microbiol., 37, 467–476. [DOI] [PubMed] [Google Scholar]
- Chattoraj D., Cordes,K. and Abeles,A. (1984) Plasmid P1 replication: negative control by repeated DNA sequences. Proc. Natl Acad. Sci. USA, 81, 6456–6460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chattoraj D.K., Mason,R.J. and Wickner,S.H. (1988) Mini-P1 plasmid replication: the autoregulation–sequestration paradox. Cell, 52, 551–557. [DOI] [PubMed] [Google Scholar]
- Chiang C.S. and Bremer,H. (1988) Stability of pBR322-derived plasmids. Plasmid, 20, 207–220. [DOI] [PubMed] [Google Scholar]
- Durland R.H. and Helinski,D.R. (1990) Replication of the broad-host-range plasmid RK2: direct measurement of intracellular concentrations of the essential TrfA replication proteins and their effect on plasmid copy number. J. Bacteriol., 172, 3849–3858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M., McEachern,M.J. and Helinski,D.R. (1986) Positive and negative roles of an initiator protein at an origin of replication. Proc. Natl Acad. Sci. USA, 83, 9645–9649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman K.L. and Brewer,B.J. (1995) Analysis of replication intermediates by two-dimensional agarose gel electrophoresis. Methods Enzymol., 262, 613–627. [DOI] [PubMed] [Google Scholar]
- Funnell B.E. (1988) Mini-P1 plasmid partitioning: excess ParB protein destabilizes plasmids containing the centromere parS. J. Bacteriol., 170, 954–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helinski D.R., Toukdarian,A.E. and Novick,R.P. (1996) Replication control and other stable maintenance mechanisms of plasmids. Escherichia coli and Salmonella, 2, 2295–2324. [Google Scholar]
- Hodgman T.C., Griffiths,H. and Summers,D.K. (1998) Nucleoprotein architecture and ColE1 dimer resolution: a hypothesis. Mol. Microbiol., 29, 545–558. [DOI] [PubMed] [Google Scholar]
- Ingmer H. and Cohen,S.N. (1993) Excess intracellular concentration of the pSC101 RepA protein interferes with both plasmid DNA replication and partitioning. J. Bacteriol., 175, 7834–7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland D.E. Jr, Goldbeter,A. and Stock,J.B. (1982) Amplification and adaptation in regulatory and sensory systems. Science, 217, 220–225. [DOI] [PubMed] [Google Scholar]
- Krause M., Ruckert,B., Lurz,R. and Messer,W. (1997) Complexes at the replication origin of Bacillus subtilis with homologous and heterologous DnaA protein. J. Mol. Biol., 274, 365–380. [DOI] [PubMed] [Google Scholar]
- Lobner-Olesen A. (1999) Distribution of minichromosomes in individual Escherichia coli cells: implications for replication control. EMBO J., 18, 1712–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lueders K.K. and Fewell,J.W. (1994) Hybridization of DNA in dried gels provides increased sensitivity compared with hybridization to blots. Biotechniques, 16, 66–67. [PubMed] [Google Scholar]
- Martin-Parras L., Hernandez,P., Martinez-Robles,M.L. and Schvartzman,J.B. (1992) Initiation of DNA replication in ColE1 plasmids containing multiple potential origins of replication. J. Biol. Chem., 267, 22496–22505. [PubMed] [Google Scholar]
- McEachern M.J., Bott,M.A., Tooker,P.A. and Helinski,D.R. (1989) Negative control of plasmid R6K replication: possible role of intermolecular coupling of replication origins. Proc. Natl Acad. Sci. USA, 86, 7942–7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miron A., Patel,I. and Bastia,D. (1994) Multiple pathways of copy control of γ replicon of R6K: mechanisms both dependent on and independent of cooperativity of interaction of π protein with DNA affect the copy number. Proc. Natl Acad. Sci. USA, 91, 6438–6442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Patel,I. and Bastia,D. (1985) Conformational changes in a replication origin induced by an initiator protein. Cell, 43, 189–197. [DOI] [PubMed] [Google Scholar]
- Mukherjee S., Erickson,H. and Bastia,D. (1988a) Detection of DNA looping due to simultaneous interaction of a DNA-binding protein with two spatially separated binding sites on DNA. Proc. Natl Acad. Sci. USA, 85, 6287–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Erickson,H. and Bastia,D. (1988b) Enhancer–origin interaction in plasmid R6K involves a DNA loop mediated by initiator protein. Cell, 52, 375–383. [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay S. and Chattoraj,D.K. (2000) Replication-induced transcription of an autorepressed gene: the replication initiator gene of plasmid P1. Proc. Natl Acad. Sci. USA, 97, 7142–7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay G., Sozhamannan,S. and Chattoraj,D.K. (1994) Relaxation of replication control in chaperone-independent initiator mutants of plasmid P1. EMBO J., 13, 2089–2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muraiso K., Mukhopadhyay,G. and Chattoraj,D.K. (1990) Location of a P1 plasmid replication inhibitor determinant within the initiator gene. J. Bacteriol., 172, 4441–4447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y., Imai,Y., Ogasawara,N. and Moriya,S. (2001) Autoregulation of the dnaA–dnaN operon and effects of DnaA protein levels on replication initiation in Bacillus subtilis. J. Bacteriol., 183, 3833–3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oram M., Marko,J.F. and Halford,S.E. (1997) Communications between distant sites on supercoiled DNA from non-exponential kinetics for DNA synapsis by resolvase. J. Mol. Biol., 270, 396–412. [DOI] [PubMed] [Google Scholar]
- Pal S.K. and Chattoraj,D.K. (1988) P1 plasmid replication: initiator sequestration is inadequate to explain control by initiator-binding sites. J. Bacteriol., 170, 3554–3560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S.K., Mason,R.J. and Chattoraj,D.K. (1986) P1 plasmid replication. Role of initiator titration in copy number control. J. Mol. Biol., 192, 275–285. [DOI] [PubMed] [Google Scholar]
- Park K. and Chattoraj,D.K. (2001) DnaA boxes in the P1 plasmid origin: the effect of their position on the directionality of replication and plasmid copy number. J. Mol. Biol., 310, 69–81. [DOI] [PubMed] [Google Scholar]
- Park K., Mukhopadhyay,S. and Chattoraj,D.K. (1998) Requirements for and regulation of origin opening of plasmid P1. J. Biol. Chem., 273, 24906–24911. [DOI] [PubMed] [Google Scholar]
- Paulsson J. and Ehrenberg,M. (2001) Noise in a minimal regulatory network: plasmid copy number control. Q. Rev. Biophys., 34, 1–59. [DOI] [PubMed] [Google Scholar]
- Paulsson J., Nordström,K. and Ehrenberg,M. (1998) Requirements for rapid plasmid ColE1 copy number adjustments—a mathematical model of inhibition modes and RNA turnover rates. Plasmid, 39, 215–234. [DOI] [PubMed] [Google Scholar]
- Skarstad K., Lueder,G., Lurz,R., Speck,C. and Messer,W. (2000) The Escherichia coli SeqA protein binds specifically and co-operatively to two sites in hemimethylated and fully methylated oriC. Mol. Microbiol., 36, 1319–1326. [DOI] [PubMed] [Google Scholar]
- Sozhamannan S. and Chattoraj,D.K. (1993) Heat shock proteins DnaJ, DnaK and GrpE stimulate P1 plasmid replication by promoting initiator binding to the origin. J. Bacteriol., 175, 3546–3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers D. (1998) Timing, self-control and a sense of direction are the secrets of multicopy plasmid stability. Mol. Microbiol., 29, 1137–1145. [DOI] [PubMed] [Google Scholar]
- Summers D.K. (1996) The Biology of Plasmids. Blackwell Science, Oxford, UK.
- Swack J.A., Pal,S.K., Mason,R.J., Abeles,A.L. and Chattoraj,D.K. (1987) P1 plasmid replication: measurement of initiator protein concentration in vivo. J. Bacteriol., 169, 3737–3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor D.P. and Cohen,S.N. (1979) Structural and functional analysis of cloned DNA segments containing the replication and incompatibility regions of a miniplasmid derived from a copy number mutant of NR1. J. Bacteriol., 137, 92–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsui H., Fujiyama,A., Murotsu,T. and Matsubara,K. (1983) Role of nine repeating sequences of the mini-F genome for expression of F-specific incompatibility phenotype and copy number control. J. Bacteriol., 155, 337–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga H., Matsunaga,F. and Wada,C. (1999) Regulation of DNA replication by iterons: an interaction between the ori2 and incC regions mediated by RepE-bound iterons inhibits DNA replication of mini-F plasmid in Escherichia coli. EMBO J., 18, 3856–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viguera E., Hernandez,P., Krimer,D.B., Boistov,A.S., Lurz,R., Alonso,J.C. and Schvartzman,J.B. (1996) The ColE1 unidirectional origin acts as a polar replication fork pausing site. J. Biol. Chem., 271, 22414–22421. [DOI] [PubMed] [Google Scholar]
- Wedel A., Weiss,D.S., Popham,D., Droge,P. and Kustu,S. (1990) A bacterial enhancer functions to tether a transcriptional activator near a promoter. Science, 248, 486–490. [DOI] [PubMed] [Google Scholar]
- Wu H.Y. and Liu,L.F. (1992) Topological approaches to studies of protein-mediated looping of DNA in vivo. Methods Enzymol., 212, 346–351. [DOI] [PubMed] [Google Scholar]
- Zechiedrich E.L., Khodursky,A.B. and Cozzarelli,N.R. (1997) Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes Dev., 11, 2580–2592. [DOI] [PMC free article] [PubMed] [Google Scholar]