Abstract
Retroviral integrase, an essential enzyme for replication of human immunodeficiency virus type-1 (HIV-1) and other retroviruses, contains three structurally distinct domains, an N-terminal domain, the catalytic core and a C-terminal domain. To elucidate their spatial arrangement, we have solved the structure of a fragment of HIV-1 integrase comprising the N-terminal and catalytic core domains. This structure reveals a dimer interface between the N-terminal domains different from that observed for the isolated domain. It also complements the previously determined structure of the C-terminal two domains of HIV-1 integrase; superposition of the conserved catalytic core of the two structures results in a plausible full-length integrase dimer. Furthermore, an integrase tetramer formed by crystal lattice contacts bears structural resemblance to a related bacterial transposase, Tn5, and exhibits positively charged channels suitable for DNA binding.
Keywords: crystal structure/HIV-1/integrase/ transposon Tn5
Introduction
Retroviruses integrate a DNA copy of the viral genome into host DNA as an obligatory step in their replication cycle. DNA integration occurs by a specialized recombination reaction mediated by the viral integrase protein (reviewed in Asante-Appiah and Skalka, 1997; Brown, 1997; Hindmarsh and Leis, 1999; Craigie, 2001). In the first step, 3′-end processing, two nucleotides are cleaved from each 3′ end of the viral DNA to form the DNA substrate for integration. In the next step, DNA strand transfer, the 3′ hydroxyls at the ends of the viral DNA attack a pair of phosphodiester bonds in the target DNA. The sites of attack on the two target DNA strands are separated by five nucleotides in the case of human immunodeficiency virus type-1 (HIV-1) integrase. To complete the integration process, the two unpaired nucleotides at the 5′ ends of the viral DNA are removed, the single strand gaps between viral and target DNA are filled and the 3′ ends of the viral DNA are ligated to the 5′ ends of the target DNA. These latter steps are likely to be accomplished by cellular enzymes. Integrase is sufficient to carry out both 3′ processing and DNA strand transfer in vitro in the presence of a divalent metal ion that can be either Mg2+ or Mn2+. Stereochemical experiments have established that both of these reactions occur by a one-step trans-esterification mechanism (Engelman et al., 1991).
Integrase is composed of three domains based on par tial proteolysis and functional and structural studies (Engelman and Craigie, 1992; Engelman et al., 1993; van Gent et al., 1993). The central core domain contains a triad of acidic residues, the D,D-35-E motif, that is conserved in integrase proteins encoded by retroviruses and retrotransposons, and transposase proteins of many DNA transposons. Mutation of any of these residues abolishes or severely diminishes all catalytic activities of the protein, demonstrating their key role in catalysis (Engelman and Craigie, 1992; Kulkosky et al., 1992; van Gent et al., 1992; Leavitt et al., 1993). By analogy with DNA polymerases, these acidic residues were proposed to coordinate a divalent metal ion (Kulkosky et al., 1992). The role of these acidic residues in binding a divalent metal ion has been demonstrated directly in the structures of the catalytic domain of HIV-1, simian immunodeficiency virus (SIV) and Rous sarcoma virus (RSV) integrases, which have been determined by X-ray crystallography either as a single domain (Dyda et al., 1994; Bujacz et al., 1995, 1996; Goldgur et al., 1998) or together with the C-terminal domain (J.C.H.Chen et al., 2000; Z.G.Chen et al., 2000; Yang et al., 2000). All structures of the catalytic domain are dimeric and structurally homologous.
The solution structures of the isolated N- and C-terminal domains have also been determined by NMR (Eijkelenboom et al., 1995, 1997; Lodi et al., 1995; Cai et al., 1997). The N-terminal domain consists of a bundle of three α-helices, with coordination of zinc by conserved histidine and cysteine residues, the HHCC motif, stabilizing the interaction between the helices. The C-terminal domain has an all β-strand SH3 fold and has been shown to bind DNA non-specifically (Kahn et al., 1991; Vink et al., 1993; Woerner and Marcus-Sekura, 1993; Engelman et al., 1994; Puras-Lutzke et al., 1994). The isolated N- and C-terminal domains are dimeric in solution, but the C-terminal domain of HIV-1 integrase is monomeric when linked to the catalytic core (J.C.H.Chen et al., 2000).
The arrangement of the three structural domains in a full-length integrase remains unknown, even though the structures of each of the domains have been determined as an isolated entity. Direct physical and structural studies of full-length integrase have been impeded by its propensity to form large aggregates under reaction conditions. Two-domain structures comprising the catalytic and C-terminal domains have been determined for HIV-1, SIV and RSV integrase (J.C.H.Chen et al., 2000; Z.G.Chen et al., 2000; Yang et al., 2000). The arrangement of the C-terminal domain relative to the catalytic core, however, differs among these structures, indicating considerable flexibility in the linkage between the catalytic and C-terminal domains. For instance, the C-terminal domain exists either as a dimer and contacts the core domain, as in the case of RSV integrase, or as an independent monomeric domain, as in the case of HIV-1 integrase. These variations make definitive modeling of the complete integrase structure from three separated domains difficult.
To process and integrate the two viral DNA ends into the host genome, two active sites theoretically are required. In the domain structures of HIV-1, SIV and RSV integrase, each catalytic core resembles a hemisphere that dimerizes via the extended flat surface to form a nearly spherical structure. The active sites are located on the opposite faces of the sphere and are separated by >50 Å, an arrangement that is incompatible with the 5 bp spacing between the sites of integration on the two target DNA strands. Rearrangement of this catalytic core dimer is unlikely because the dimer interface is very hydrophobic and conserved in all five independently determined retroviral integrase structures. Studies of the Mu transposase, which is functionally and structurally homologous to HIV-1 integrase (Rice and Mizuuchi, 1995), have revealed that only two of the four active sites present in a Mu transposase tetramer actually participate in the chemical reactions (Aldaz et al., 1996; Williams et al., 1999). It is thus proposed that a tetramer of integrase (dimer-of-dimers) is required for the integration reaction and, within this tetramer, only one of the two active sites in each dimer is actually involved in the chemical reactions. A remaining question is how two integrase core dimers associate to form an active tetramer.
Here we report the structure of the catalytic domain of HIV-1 integrase connected to the N-terminal domain. The results clarify the ambiguity regarding the domain arrangement and provide constraints on possible models of the active integrase tetramer with DNA substrate.
Results and discussion
Structure determination of IN1–212
Residues 1–212 of HIV-1 integrase (IN1–212), comprising the N-terminal and catalytic domains, is a well-behaved dimer in solution at physiological ionic strength (Jenkins et al., 1996). IN1–212 proteins containing a series of point mutations were screened for their ability to crystallize. IN1–212 with the mutations W131D, F139D and F185K readily crystallized out of phosphate buffer. The F185K mutation improves the solubility of integrase (Jenkins et al., 1996). Many mutations of the exposed W131 and F139 were tested because the W131D, W131E and F139D mutations have been shown to facilitate crystallization of fragments of HIV-1 integrase without altering protein structure (Goldgur et al., 1998; J.C.H.Chen et al., 2000). Since IN1–212 only exhibits disintegration activity, the same mutations were introduced in full-length HIV-1 integrase to assess their effects on 3′-end processing and strand transfer activities. The triple mutant was found to be fully active in vitro (data not shown).
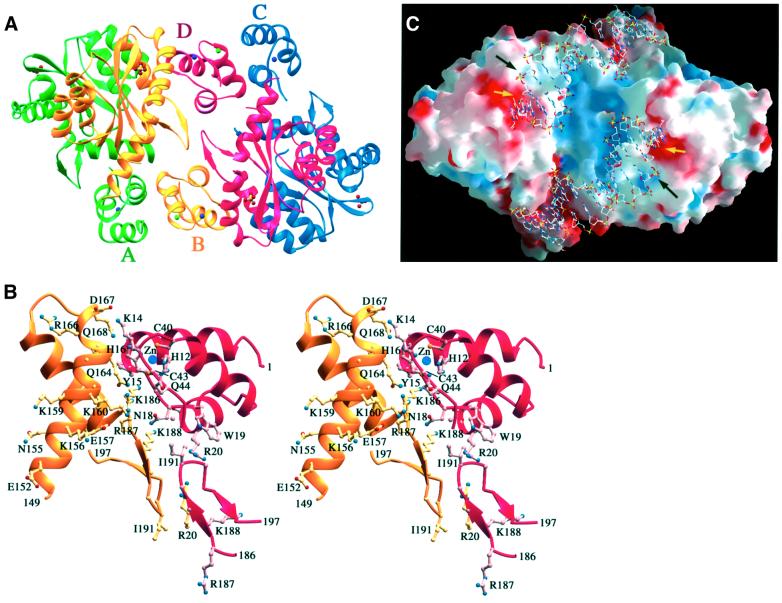
IN1–212 was crystallized in the P43212 space group and diffracted X-rays to 2.4 Å using a synchrotron radiation source. The structure was solved by molecular replacement using the HIV-1 integrase catalytic core dimer as a search model (see Materials and methods). Two dimers of IN1–212, which are related by a non-crystallographic 2-fold axis and designated as AB and CD, are found in each asymmetric unit. The structure has been refined to an R-value of 23.3% and an Rfree of 26.0% with good geometry (see Materials and methods; Table I). The final protein model consists of residues F1–K46, C56–D139 and G149–E212, and a phosphate ion near each active site defined by the D,D-35-E motif.
Table I. Summary of crystallographic data.
| Native 1 (in house) | Native 2 (BNL) | |
|---|---|---|
| Space group | P43212 | P43212 |
| Cell constants (Å) | a = 103.3, c = 281.0 | a = 02.71, c = 280.6 |
| Diffraction dataa (Å) | 30.0–2.90 (3.0–2.90) | 30.0–2.40 (2.44–2.40) |
| Completenessa (%) | 96.1 (94.6) | 90.1 (84.4) |
| Rmergea (%) | 11.9 (37.7) | 8.4 (35.6) |
| Unique reflections | 32 581 | 53 869 |
| Refinement | ||
| Protein atoms | 5298 | |
| Solvent and ligand atoms | 204 | |
| R | 0.233(30–2.40 Å) | |
| Rfreeb | 0.260(2597 reflections) | |
| R.m.s. bond length | 0.006 Å | |
| R.m.s. bond angle | 1.34° | |
| Average B (Å2) | 32.8 |
aData of the highest resolution bin are shown in parentheses.
bRfree is monitored with the reflections excluded from refinement.
Rmerge = ΣhΣi|Ihi – <Ih>|/Σ<Ih>
R = Σ||Fobs| – |Fcalc||/Σ|Fobs|
Structure of the IN1–212 dimer
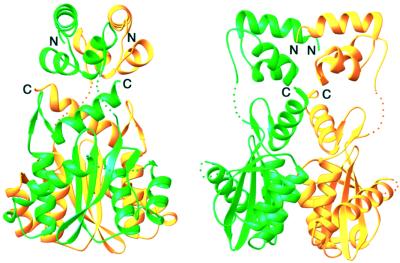
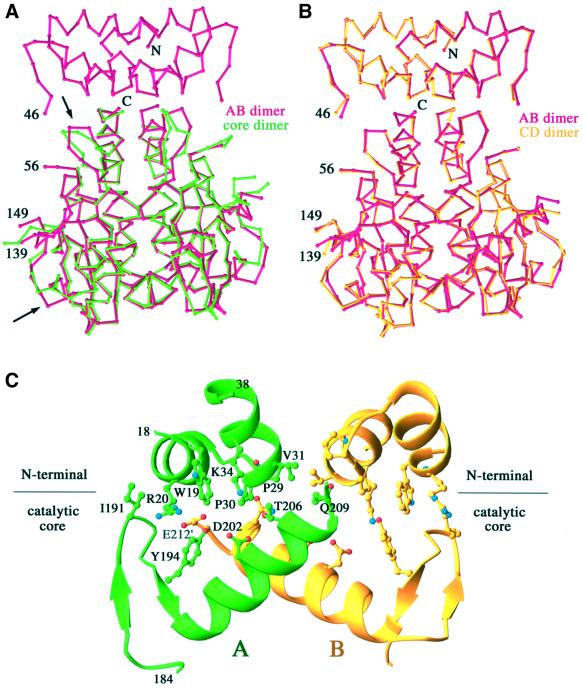
The N-terminal and core domains are packed against each other and share a similar pseudo-dyad axis to form the nearly identical AB or CD dimers (Figure 1). The catalytic core dimer has fundamentally the same structure as previously determined except for the loop comprising residues 188–194, which is at the interface with the N-terminal domain (Figure 2).
Fig. 1. Ribbon diagram of HIV-1 IN1–212. Two orthogonal views of the IN1–212 dimer. The A subunit is colored green, and the B subunit yellow. Disordered loops are indicated by the dotted lines.
Fig. 2. Conservation of the catalytic core domain structure. (A) Superposition of the Cα traces of the AB dimer of IN1–212 with the dimer of IN50–212 (PDB code 1BIS) (B) Superposition of AB and CD dimers of IN1–212 in one asymmetric unit. (C) The interface between the N-terminal and catalytic core domain. Residues 18–38 and 184–209 are shown in ribbon diagrams and the residues involved specifically in domain interactions are shown in ball-and-stick.
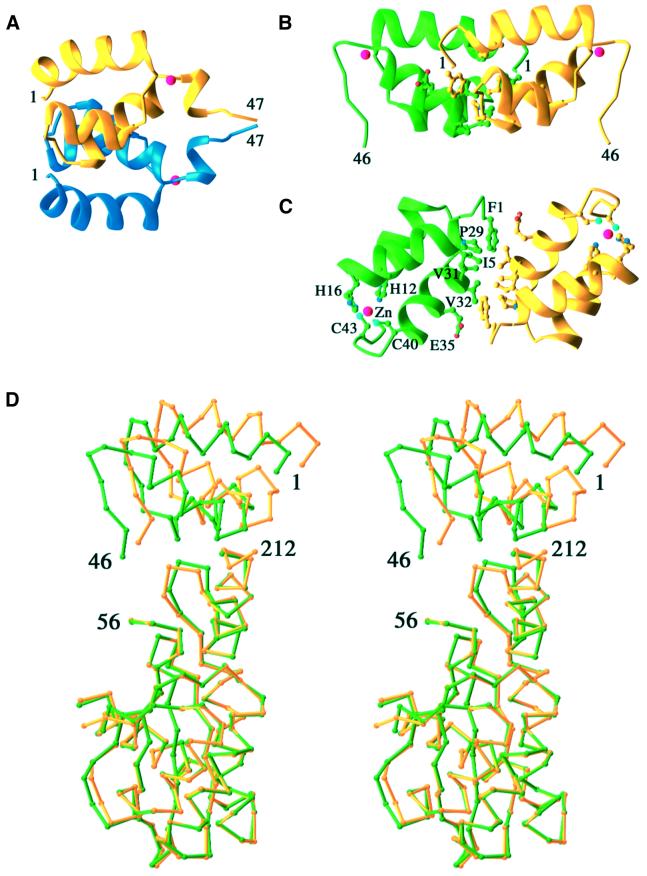
The structure of the N-terminal domain at the monomeric level is very similar to the previously determined NMR structure of the isolated domain, but the dimer interface is entirely different (Figure 3A and B). Whereas it is dominated by interactions between the third helix in the solution structure, the N-terminal domain interface in the IN1–212 crystal structure comprises the N-termini of the first and third helix. The dimer interface between the N-terminal domains in the two-domain structure is smaller and more hydrophobic than in the dimer of the isolated domain (Figure 3C). Part of the dimer interface, including residues F1, L2, P29, V31 and V32, is common to the NMR and crystal structures. Rotation of one subunit by ∼180° centered on the hydrophobic dimeric interface coupled with a slight translation can transform the solution structure of the N-terminal domain alone to the new dimer observed in the structure of IN1–212 (Figure 3A and B). However, because the hydrophobic contacting surfaces are rotated and translated with respect to one another in the two structures, the atomic contacts are quite different.
Fig. 3. Analysis of domain interactions. (A) Ribbons diagram of the N-terminal domain structure as an isolated entity determined by NMR (PDB code 1WJA) and (B) in the context of IN1–212 by X-ray crystallography after superimposing the yellow subunits of the two structures. Residue numbers at the N- and C-termini are labeled. (C) A near orthogonal view from that of (B) looking down the 2-fold axis. Residues involved in the dimerization, which are rather hydrophobic, are shown in ball-and-stick. Zn2+ ions are shown as red spheres. (D) The N-terminal domain is oriented differently relative to the core domain in the A and B subunit of IN1–212 as revealed by superposition of the Cα traces of the core domains.
Residues 47–55, which connect the N-terminal and catalytic domains, are disordered in all four monomers. These nine residues, despite being mobile, are sufficient to serve as a linker to span the distance of 9 Å (A subunit) or 13 Å (B subunit) and to prevent the N-terminal domain from dimerizing as observed with the isolated domain. The linkage of the N-terminal and core domains is determined unambiguously by distance constraints. The alternative connectivity between different subunits, such as A to B or B to A, would require the nine unresolved residues to span 35 Å, and tunneling through the protein, which is clearly impossible. The interface between the N-terminal and the core domains is rather hydrophilic and involves the side chains of R20, K34, Q209, T206 and E212 (Figure 2C); it buries a total of ∼900 Å2 molecular surface, which is comparable with the interface between an antibody and antigen.
The A and B subunits are not structurally identical, as revealed by superposition. The orientation of the N-terminal domain relative to the core differs by 15° (Figure 3D). The interactions between the N-terminal and core domains in the A and B subunits are, therefore, asymmetric (Figure 3D). As the crystallographically independent AB and CD dimers are structurally identical (Figure 2B), the asymmetric arrangement of the N-terminal and core domains is unlikely to be an artifact of crystal lattice packing and is probably functionally relevant.
A phosphate ion binds HIV-1 IN1–212 near the active site
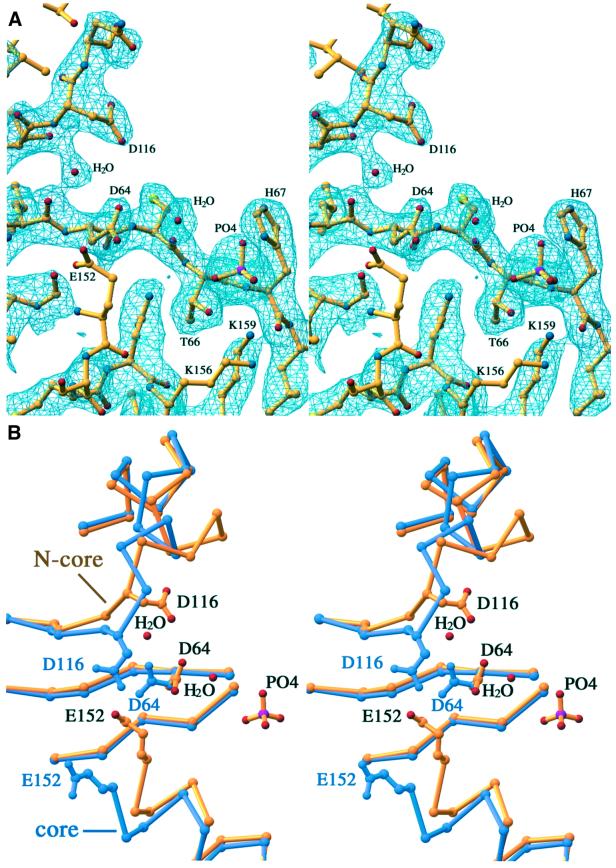
Each of the four copies of HIV-1 IN1–212 contains a phosphate ion ∼7 Å from the active site, which matches the distance between adjacent phosphates in the DNA backbone (Figure 4A). Each phosphate ion is coordinated by the side chains of T66, H67 and K159, the main chain nitrogen of H67 and a water molecule. This water molecule is probably substituted by a divalent metal ion, as indicated by preliminary soaking experiments with Mn2+ (data not shown). Even though the phosphate ions are relatively exposed to solvent, the location of each phosphate relative to the adjacent catalytic carboxylates D64 and D116 remains the same in the AB and CD dimers. The phosphates are refined at full occupancies. Two of them are stabilized by crystal lattice contacts and thus have lower B-values than the other two, which are exposed to solvent.
Fig. 4. The active site. (A) Stereo view of the phosphate ion bound near the active site composed of three conserved carboxylates, D64, D116 and E152. The D subunit, which is chosen for the active site representation, is completely exposed to solvent and is not affected by lattice contacts even though E152 of this subunit is more mobile compared with E152 of the A, B and C subunits. A 2Fo – Fc electron density map contoured at 1.0σ is superimposed on the final refined structure represented by the stick-and-ball model. Two water molecules are also shown; one is associated with the phosphate and the other stabilizes the main chain configuration at the active site. (B) Comparison of the loop containing D116 in the IN1–212 D subunit, shown in gold, and the IN52–210 structures (PDB code 1EXQ), shown in purple. The phosphate ion is ∼7 Å away from the mid-point between D64 and D116.
The conformation of the loop containing D116, one of the D,D-35-E triad, is quite different from that in the HIV-1 core domain crystallized in the presence of cacodylate (Dyda et al., 1994), but is similar to that in the Mg2+-bound structures of the HIV-1 and RSV integrase core domains (Goldgur et al., 1998; Lubkowski et al., 1998). The binding of phosphate induces a change in the conformation of this loop in comparison with the core domain structure containing the same three mutations (J.C.H.Chen et al., 2000) (Figure 4B). A water molecule, which is hydrogen bonded to the main chain nitrogen atom of D116 and the carbonyl oxygen of L63, keeps the two adjacent β-strands containing D64 and D116 from diverging from each other (Figure 4B). In addition, the helix containing the third carboxylate, E152, is also shifted.
The arrangement of the three domains in the full-length integrase
Unlike the structure of HIV-1 IN1–212, which is on the whole rather compact (Figure 1), the structure of HIV-1 IN52–288 (J.C.H.Chen et al., 2000) is extended and consists of a globular structure of the core dimer and two separated C-terminal domains (Figure 5). Residues 195–222 form a pair of α-helices that cross the 2-fold axis relating the catalytic dimers and extend away from the catalytic core. The C-terminal domains are located on the ends of these helices and make no contact with the catalytic domain or with each other. Interestingly, the spatial arrangement of the catalytic and N-terminal domains in our structure readily accommodates the structure of the C-terminal domain as observed together with the catalytic core. When the dimeric catalytic cores of the HIV-1 IN1–212 and HIV-1 IN52–288 structure are superimposed, the N-terminal domains fit between the diverging C-terminal domains with no steric clash (Figure 5), and the extended α-helices linking the core and C-terminal domain may be stabilized by interactions with the N-terminal domains.
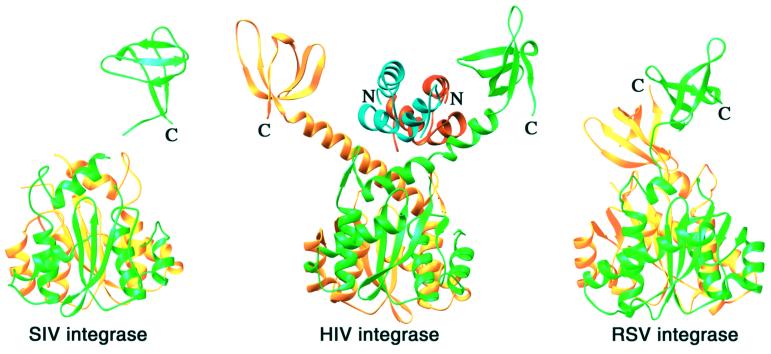
Fig. 5. Location of the C-terminal domain. Ribbons diagram of SIV IN50–293 (PDB code 1C6V), HIV-1 IN52–288 (PDB code 1EX4) and RSV IN49–286 (PDB code 1C0M) after superposition of the catalytic core of each structure. Each dimer is colored green and yellow. The C-terminal domains of the green subunit of SIV, RSV and HIV-1 integrase are located similarly relative to the catalytic core dimer. Superposition of the catalytic cores of HIV-1 IN52–288 and IN1–212 structures positions the N-terminal domain dimer between the C-terminal domains without any steric clash. The modeled N-terminal domain dimer is colored red and blue for the yellow and green subunits, respectively.
The spatial relationship between the core and the C-terminal domain was confusing at first because different arrangements were observed for functionally identical RSV, HIV-1 and SIV integrase. For instance, in the crystal structure of the SIV IN50–293 (Z.G.Chen et al., 2000), only one of the four C-terminal domains accompanying the two catalytic dimers was traceable, and the linker between the catalytic and C-terminal domain (residues 208–215) was disordered. One out of the four possible connections between the core and C-terminal domains was chosen because the two domains are in contact, but the arrangement differs drastically from that of the HIV-1 IN52–288. However, if the C-terminal domain of SIV integrase is allowed to connect to the dimer other than the one previously suggested, its spatial relationship to the catalytic domain is very similar to that of the HIV-1 IN52–288 structure (Figure 5). In the RSV IN49–286 structure (Yang et al., 2000), the two eight-residue linkers connecting the C-terminal and the core domain are in an extended conformation and pair with one another in the dimer (Figure 5). As a result, the C-terminal domains form a dimer with an interface that differs from that of the dimer observed in the case of the isolated HIV-1 integrase C-terminal domain (Eijkelenboom et al., 1995; Lodi et al., 1995). More puzzlingly, the 2-fold axis relating the C-terminal dimer in the RSV structure is inclined by ∼50° towards the 2-fold axis of the core dimer (Figure 5). However, the position of one of the two C-terminal domains relative to the catalytic core in RSV IN49–286 is similar to that of the C-terminal domains in the structure of the HIV-1 IN52–288. The other C-terminal domain in the RSV structure is packed against the surface of the core domain (Figure 5), which may be an artifact resulting from the absence of the N-terminal domain. If the two-domain structure observed for HIV-1 IN1–212 is conserved in the full-length RSV integrase, the N-terminal domains would stabilize each of the two eight-residue linkers connecting the catalytic and the C-terminal domains and keep the two C-terminal domains apart. Clearly, the connection between the core and C-terminal domains is flexible and differs in length, orientation and conformation among integrase homologs. Yet in each of the three C-terminal two-domain structures, at least one C-terminal domain is positioned similarly with respect to the core dimer (Figure 5, shown in green). This arrangement is compatible with the structure of HIV-1 IN1–212, and the domain organization in full-length integrase can be modeled without a steric clash.
Relevance of the dimer-of-dimers—the ABCD tetramer
The active form of integrase is likely to be a tetramer, as suggested by previous biochemical and structural studies. In the crystal structure of HIV-1 IN1–212, the AB and CD dimers are related by a non-crystallographic 2-fold axis to form a tetramer (Figure 6A). The Zn2+ coordination region of the N-terminal domain in the B subunit (residues 13–26 and 40–45) interacts extensively with residues 150–196 in the catalytic core of the D subunit (Figure 6B). The B and D subunits also contact one another reciprocally at the β-hairpins consisting of residues 187–196. The AB and CD interface is considerable and buries >1800 Å2 of molecular surface. The contacts, however, are mostly polar, for instance K14, Q18, Q44, K160, Q168, K186 and K188. At physiological salt concentrations, HIV-1 IN1–212 is exclusively dimeric, as judged by gel filtration (Jenkins et al., 1996), equilibrium centrifugation and cross-linking (data not shown), which raises the question of whether this tetramer is functionally relevant.
Fig. 6. The ABCD tetramer of IN1–212. (A) Ribbons diagram of the AB and CD dimers from one asymmetric unit. A, B, C and D subunits are colored green, yellow, blue and red, respectively. Purple and green spheres represent zinc and potassium ions bound to the protein. Balls and sticks represent the ordered phosphates located near the active sites. (B) Details of the upper half of the dimer-of-dimer interface. The N-terminal domain, residues 186–195 of the D subunit and residues 149–197 of the B subunit are shown as ribbon diagrams. Side chains contributing to the interface are shown in ball-and-stick models. The lower half of the interface is essentially a repeat of the upper one. (C) Molecular surface of the ABCD tetramer viewed with a slight rotation from the view shown in (A). Positive and negative electrostatic potentials are shown in blue and red (saturation at ∼15 kT/e). The image was created with GRASP (Nicholls et al., 1991). Viral DNA ends are superimposed on the ABCD tetramer guided by the phosphate, active site and Q148. The black arrow points to the ordered inorganic phosphate in our structure, and the yellow arrow points to the scissile phosphate. The most positively charged region is the central hinge between the AB and CD dimers, which would accommodate target DNA in this model. The negatively charged patch centered around the active site would normally be neutralized by divalent metal ions. Binding of viral DNA would be stabilized by a C-terminal domain serving as a ‘clamp’ (not shown), and possibly by a positively charged patch on the side of the core domain.
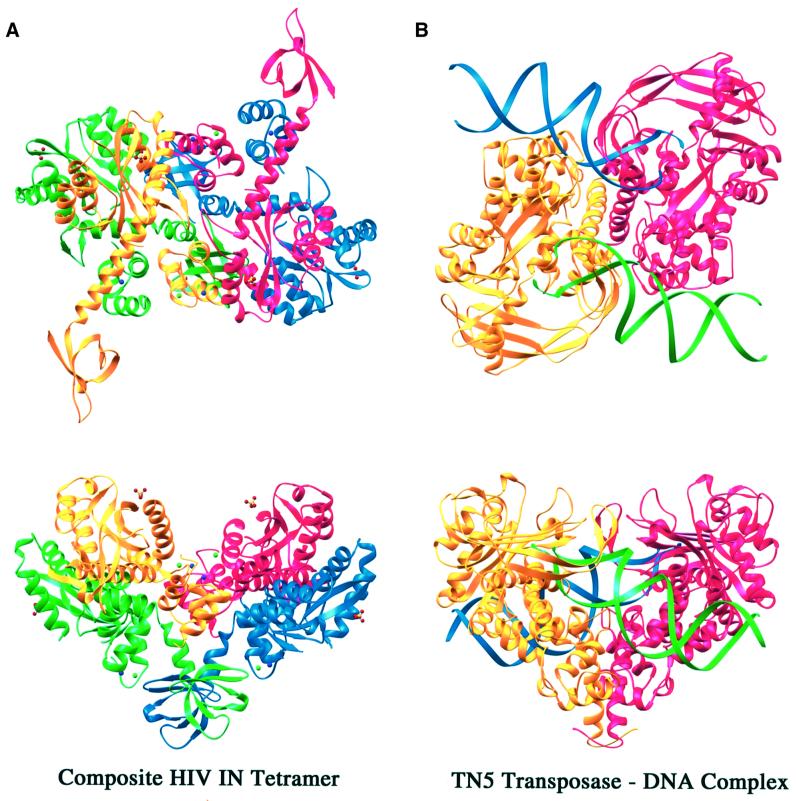
Bacterial Tn5 transposase and HIV-1 integrase share a conserved catalytic domain structure and phosphatidyl transfer mechanism; therefore, it is reassuring to find that the overall structure of the ABCD tetramer is reminiscent of the structure of Tn5 transposase dimer in complex with its DNA substrate (Davies et al., 2000) (Figure 7). Interestingly, the active sites of the B and D subunits in the ABCD tetramer lie on a concave surface with an electrostatic potential that appears to be favorable for interaction with DNA (Figure 6C) and are reminiscent of the two active sites found in Tn5 transposase (Figure 7). The other pair of active sites lie on the opposite convex surface of the tetramer, are >90 Å apart and the subunits containing these active sites probably play a structural supporting role. The B and D active sites of the HIV-1 IN1–212 tetramer are separated by ∼40 Å, which is the same as in Tn5 transposase. This distance is still much longer than the 5 bp spacing of the sites of insertion of the two viral DNA ends into target DNA, but the two active sites conceivably are brought into closer proximity by ‘hinging’ of the dimer interface. However, the active sites are on the same concave surface of the complex, greatly simplifying the conformational changes needed to bring them close enough for the final strand transfer reaction. Alternatively, concerted insertion may occur sequentially, assisted by protein reorientation.
Fig. 7. Comparison between the composite integrase tetramer and Tn5 transposase in complex with DNA substrate. (A) Orthogonal views of a Ribbons diagram of the integrase tetramer with the C-terminal domain modeled in. The integrase full-length tetramer is composed by superposition of the catalytic core domain of the HIV-1 IN52–288 structure (PDB code 1EX4) onto the catalytic domains of the AB and CD dimers of IN1–212. The four subunits are colored the same as in Figure 6A. (B) Orthogonal views of the Tn5 transposase–DNA complex (PDB code 1F3I). The protein dimer is shown in yellow and red, and the two DNA duplexes associated with Tn5 transposase are shown in green and blue ribbons. All figures except those displaying molecular surface were created with Ribbons (Carson, 1987).
The ABCD tetramer, although not observed with HIV-1 IN1–212 in solution, may exist with the full-length protein. The C-terminal two-domain protein (IN50–288) forms both dimers and tetramers in solution (Jenkins et al., 1996), pointing to a crucial role for the C-terminal domain in tetramerization. Interestingly, the full-length HIV-1 integrase dimer resulting from superposition of the N- and C-terminal two-domain structures as shown in Figure 5 can fit into this dimer-of-dimers arrangement without any main-chain steric clashes (Figure 7). Moreover, the C-terminal domains of the A and C subunits extend towards each other on the convex surface, forming a coiled-coil between the helical stalks, and potentially stabilize the tetramer (Figure 7). The resulting heart-shaped tetrameric structure of integrase closely resembles the structure of Tn5 transposase (Figure 7). The other pair of C-terminal domains, from the B and D subunits, project from the substrate-binding surface defined by the active sites and extend the positively charged groove at the dimer–dimer interface (Figures 6C and 7), and therefore potentially participate in binding viral DNA ends. Since the ABCD tetramer is formed without the C-terminal domain, and has only been observed in the crystals, we speculate that an active integrase tetramer may be similar in overall configuration, but there are likely to be adjustments of the tetramer interface.
Complementation and cross-linking strategies have elucidated some aspects of the organization of the active complex of integrase with DNA substrates. Complemen tation experiments with integrase proteins that lack the N-terminal domain, or have an inactivated catalytic domain, demonstrate that the integrase subunit containing an intact active site functions in trans with the subunit containing the N-terminal domain (Engelman et al., 1993; van Gent et al., 1993; Ellison et al., 1995). The C-terminal domain has been reported to function in cis (Engelman et al., 1993) and in trans (Gao et al., 2001). Cross-linking experiments also indicate that that both viral and target DNA substrates interact primarily in trans with the subunit containing the functional active site (Heuer and Brown, 1998). The physical interpretation of these results on cis versus trans interactions is complicated because the active multimer of integrase is likely to be at least a tetramer. For example, does trans complementation of the catalytic and N-terminal domains occur between two monomers within a dimer unit, or between two dimers that form a tetramer? A further complication is that the in vitro assay for integrase activity is a ‘half-site’ reaction that integrates a single viral DNA end into a single strand of target DNA. Proteins that complement for the half-site reaction may not be competent for the complete reaction. Thus, although the ABCD tetramer is consistent with most of the above data, the tests are not particularly rigorous.
Models for interaction of HIV-1 integrase with DNA
The non-specific nature of the binding of both viral and target DNA to HIV-1 integrase presents a major challenge to direct determination of the structure of a complex. However, the crystal structure of IN1–212, together with other biochemical and structural data, places constraints on viable models. The presence of an ordered phosphate ∼7 Å from the active site residues, which equals the distance between adjacent phosphates along a DNA backbone, in all four monomers in the asymmetric unit identifies a potential binding site of a backbone phosphate near the viral DNA ends. Indeed, this phosphate contacts the side chain of K159, a residue that has been identified by photo-cross-linking as being in close proximity to the A of the CA dinucleotide adjacent to the scissile phosphate (Jenkins et al., 1997). In the structure of Tn5 transposase in complex with DNA substrate (Davies et al., 2000), the phosphate 5′ to the scissile phosphate is located at the equivalent position in the structure and is in contact with K333, a counterpart of K159 of HIV-1 integrase based on structural and sequence alignment. We therefore propose that the ordered phosphate in our structure mimics the phosphate immediately 5′ to the scissile phosphodiester bond. The orientation of the viral DNA at the active site is also constrained by the location of the active site residues and the likely interaction of the two terminal nucleotides at the 5′ ends of the viral DNA with residues in the vicinity of Q148. Incubation of HIV-1 integrase with viral DNA substrate in the presence of a divalent metal ion results in the formation of a stable complex that is resistant to challenge with competitor DNA (Ellison and Brown, 1994). Removal of the two 5′ nucleotides from the viral DNA substrate (Ellison and Brown, 1994) or mutation of Q148 to leucine (Gerton et al., 1998) abrogates stable complex formation, suggesting that the two entities normally interact. Given these constraints, the orientation of the viral DNA in the active site region can be modeled (Figure 6C). The terminal nucleotides are expected to be unstacked, as judged by the hyper-reactivity of modified DNA substrates that favor unstacking of the terminal bases (Scottoline et al., 1997).
Although we are reasonably confident of the orientation of the viral DNA with respect to the active site residues, the path that the viral DNA follows on the surface of integrase away from the active site is less apparent. In Figure 6C, it is depicted as a straight helix on the surface of the ABCD tetramer. The linkers connecting the catalytic and the C-terminal domains of the B and D subunits extend from the concave surface as depicted in Figure 7 such that the C-terminal domains may anchor the viral DNA. Such an arrangement agrees well with biochemical evidence that the C-terminal domain binds a subterminal region of the viral DNA ∼5–10 bp internal to the site of cleavage (Heuer and Brown, 1997; Esposito and Craigie, 1998). This arrangement of binding the viral DNA ends leaves a basic groove between the active sites that may accommodate target DNA, as has been proposed for Tn5 (Davies et al., 2000). However, significant adjustments to the structure would be required to bind both viral and target DNA. Distortion of the target DNA in the vicinity of each active site is also required to fit the target DNA into this groove, consistent with the finding that bent DNA is preferred as a target for integration (Pryciak and Varmus, 1992; Muller and Varmus, 1994; Bor et al., 1995).
Several models have been proposed for the interaction of DNA with a multimer of integrase (e.g. Heuer and Brown, 1998; J.C.H.Chen et al., 2000; Z.G.Chen et al., 2000; Gao et al. 2001). A common feature of these models is that each viral DNA end interacts with an active site and C-terminal domain contributed by the same integrase dimer. In contrast, in the model shown in Figure 6C, the active site and C-terminal domain are donated by different dimers. Although further work is required to test these and other models, the growing body of structural and biochemical information increasingly constrains viable models for the complex of integrase with DNA substrates.
Concluding remarks
Although structures of each individual domain of HIV-1 integrase have been available for some time, their spatial organization and interaction with DNA substrate have remained enigmatic. The new structure of the catalytic domain with the N-terminal domain, together with previous results, places constraints on possible models of the active complex of integrase with its DNA substrates. In particular, the nearly identical organization of two IN1–212 dimers in different crystal packing environments lends credence to the biological relevance of this dimer. Furthermore, superposition of this dimer with the previously determined HIV-1 IN52–288 structure results in a plausible model for the structure of a dimer of intact HIV-1 integrase. A higher order multimer is probably required for the full integration reaction, and a dimer-of-dimers observed in the crystal lattice packing of IN1–212 is a candidate for an active tetrameric unit of integrase. Finally, an ordered phosphate near each active site probably mimics the phosphate 5′ to the scissile phosphate and provides an initial glimpse of the interaction of the active site with DNA. The presence of bound DNA has been shown to influence strongly the binding of inhibitors, and future structures of the integrase active site with bound nucleotide replacing the phosphate should greatly facilitate understanding of inhibitor binding.
Materials and methods
Expression constructs
Construction of the plasmid for expression of the N-terminal doamin together with the catalytic domain of HIV-1 IN1–212 with the F185K mutation in pET15B has been described previously (Jenkins et al., 1996). The additional mutations W131D and F139D were introduced using a QuikChange kit (Stratagene). All constructs were confirmed by DNA sequencing.
Protein expression and purification
HIV-1 IN1–212 with mutations F185K/W131D/F139D was expressed and purified as described previously (Jenkins et al., 1996). The histidine tag was removed by addition of thrombin (Sigma) to a concentration of 10 NIH U/mg of protein and incubation at 26°C for 30 min, followed by addition of a further 10 NIH U/mg and incubation at 26°C for another 30 min. Thrombin was removed by adsorption to benzamidine–Sepharose 6B (Pharmacia). The protein was then dialyzed against 25 mM HEPES pH 8.0, 50 mM NaCl, 10% (w/v) glycerol, 1 mM EDTA, 5 mM dithiothreitol (DTT), loaded onto a MonoQ HR 10/10 column (Pharmacia) and eluted with a linear gradient of 0.05–0.4 M NaCl containing 25 mM HEPES pH 8.0, 10% (w/v) glycerol, 1 mM EDTA, 5 mM DTT. Peak fractions were pooled and dialyzed overnight against 20 mM HEPES pH 7.5, 0.5 M NaCl, 100 µM ZnCl2, 5% (w/v) glycerol, 10 mM DTT. The purified protein was filtered (0.22 µm filter; Millipore) and then concentrated using a Centriprep YM-10 (Millipore). The final protein concentration ranged from 6 to 10 mg/ml.
Crystallization and data collection
Crystals were grown by the vapor diffusion method in hanging or sitting drops at 4°C, which resulted in crystals of better quality than at room temperature. Sitting drops were set up by mixing 6 µl of protein at 7 mg/ml in 0.5 M NaCl, 20 mM HEPES pH 7.2, 50 µM ZnCl2, 5 mM DTT with 6 µl of well solution containing 0.7 M NaH2PO4, 1.0 M K2HPO4 and 0.1 M acetate pH 4.6. Tetragonal crystals reached a final size of 0.3 × 0.3 × 0.6 mm in several days.
Crystals were transferred to cryo-protectant buffer containing 0.8 M NaH2PO4/1.2 M K2HPO4 pH 7.0, 0.2 M NaCl, 20% glycerol, then flash frozen in liquid propane. The initial 2.8 Å native data set was collected at 100 K using an R-axis IPII detector mounted on a Rigaku RU 200 generator. Subsequently, a 2.4 Å resolution data set was collected on beamline X9B at Brookhaven National Laboratory (BNL). The crystals belong to space group P41212 or P43212, with cell dimensions of a = b = 102.71 Å and c = 280.56 Å. Diffraction data were processed using HKL (Otwinowski and Minor, 1997) (Table I).
Structure determination
The structure of HIV-1 IN1–212 was solved by molecular replacement (Brünger et al., 1998) using the 2.8 Å in-house data. The catalytic core dimer (IN50–212) was used successfully as a search model, while the N-terminal domain IN1–50 HIV-1 integrase produced no solution either as the dimer found in solution or as a monomer. Two core dimers were found in each asymmetric unit by rotation and translation search. The translation search indicated that P43212 was the correct space group. After solvent flipping and density modification (CCP4, 1994), the N-terminal domains were located individually in the electron density map with the phases from the core domains and structure factors from the 2.4 Å synchrotron data. There were only two IN1–212 dimers in each asymmetric unit and the solvent content was 69.6%. It became clear that the N-terminal domain in IN1–212 has a different dimer interface from that of the NMR structure of the isolated domain. This must have caused the failure of molecular replacement using the dimer as the search model. The model was fitted manually with the program O and refined using CNS (Brünger et al., 1998). The final model, which includes four integrase core domains, four integrase N-terminal domains, four phosphate ions, four K+ ions and 200 water molecules, was refined to R and Rfree of 0.23/0.26 with all the data from 20 to 2.4 Å.
Acknowledgments
Acknowledgements
We thank David Davies and Fred Dyda for helpful advice, and D.Leahy for comments on the manuscript. This work was supported in part by the NIH Intramural Aids Targeted Antiviral Program. Coordinates reported in this paper have been deposited in the Protein Data Bank with the ID code 1K6Y.
References
- Aldaz H., Schuster,E. and Baker,T.A. (1996) The interwoven architecture of the Mu transposase couples DNA synapsis to catalysis. Cell, 85, 257–269. [DOI] [PubMed] [Google Scholar]
- Asante-Appiah E. and Skalka,A.M. (1997) Molecular mechanisms in retrovirus DNA integration. Antiviral Res., 36, 139–156. [DOI] [PubMed] [Google Scholar]
- Bor Y.C., Bushman,F.D. and Orgel,L.E. (1995) In-vitro integration of human-immunodeficiency-virus type-1 cDNA into targets containing protein-induced bends. Proc. Natl Acad. Sci. USA, 92, 10334–10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.O. (1997) Integration. In Coffin,J.M., Hughes,S.H. and Varmus,H.E. (eds), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 161–203.
- Brünger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Bujacz G., Jaskolski,M., Alexandratos,J., Wlodawer,A., Merkel,G., Katz,R.A. and Skalka,A.M. (1995) High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol., 253, 333–346. [DOI] [PubMed] [Google Scholar]
- Bujacz G., Jaskolski,M., Alexandratos,J., Wlodawer,A., Merkel,G., Katz,R.A. and Skalka,A.M. (1996) The catalytic domain of avian sarcoma virus integrase: conformation of the active-site residues in the presence of divalent cations. Structure, 4, 89–96. [DOI] [PubMed] [Google Scholar]
- Cai M., Zheng,R., Caffrey,M., Craigie,R., Clore,G.M. and Gronenborn,A.M. (1997) Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nature Struct. Biol., 4, 567–577. [DOI] [PubMed] [Google Scholar]
- Carson M. (1987) Ribbon models of macromolecules. J. Mol. Graphics, 5, 103–106. [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Chen J.C.H., Krucinski,J., Miercke,L.J.W., Finer-Moore,J.S., Tang,A.H., Leavitt,A.D. and Stroud,R.M. (2000) Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. Proc. Natl Acad. Sci. USA, 97, 8233–8238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z.G. et al. (2000) X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50–293)—an initial glance of the viral DNA binding platform. J. Mol. Biol., 296, 521–533. [DOI] [PubMed] [Google Scholar]
- Craigie R. (2001) HIV integrase, a brief overview from chemistry to therapeutics. J. Biol. Chem., 276, 23213–23216. [DOI] [PubMed] [Google Scholar]
- Davies D.R., Goryshin,I.Y., Reznikoff,W.S. and Rayment,I. (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science, 289, 77–85. [DOI] [PubMed] [Google Scholar]
- Dyda F., Hickman,A.B., Jenkins,T.M., Engelman,A., Craigie,R. and Davies,D.R. (1994) Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science, 266, 1981–1986. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom A.P., Lutzke,R.A., Boelens,R., Plasterk,R.H., Kaptein,R. and Hard,K. (1995) The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nature Struct. Biol., 2, 807–810. [DOI] [PubMed] [Google Scholar]
- Eijkelenboom A.P., van den Ent,F.M., Vos,A., Doreleijers,J.F., Hard,K., Tullius,T.D., Plasterk,R.H., Kaptein,R. and Boelens,R. (1997) The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: a three-helix bundle stabilized by zinc. Curr. Biol., 7, 739–746. [DOI] [PubMed] [Google Scholar]
- Ellison V. and Brown,P.O. (1994) A stable complex between integrase and viral DNA ends mediates human immunodeficiency virus integration in vitro. Proc. Natl Acad. Sci. USA, 91, 7316–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison V., Gerton,J., Vincent,K.A. and Brown,P.O. (1995) An essential interaction between distinct domains of HIV-1 integrase mediates assembly of the active multimer. J. Biol. Chem., 270, 3320–3326. [DOI] [PubMed] [Google Scholar]
- Engelman A. and Craigie,R. (1992) Identification of conserved amino acid residues critical for human immunodeficiency virus type 1 integrase function in vitro. J. Virol., 66, 6361–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A., Mizuuchi,K. and Craigie,R. (1991) HIV-1 DNA integration: mechanism of viral DNA cleavage and DNA strand transfer. Cell, 67, 1211–1221. [DOI] [PubMed] [Google Scholar]
- Engelman A., Bushman,F.D. and Craigie,R. (1993) Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J., 12, 3269–3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelman A., Hickman,A.B. and Craigie,R. (1994) The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol., 68, 5911–5917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esposito D. and Craigie,R. (1998) Sequence specificity of viral end DNA binding by HIV-1 integrase reveals critical regions for protein–DNA interaction. EMBO J., 17, 5832–5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao K., Butler,S.L. and Bushman,F. (2001) Human immunodeficiency virus type 1 integrase: arrangement of protein domains in active cDNA complexes. EMBO J., 20, 3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerton J.L., Ohgi,S., Olsen,M., Derisi,J. and Brown,P.O. (1998) Effects of mutations in residues near the active site of human immunodeficiency virus type 1 integrase on specific enzyme–substrate interactions. J. Virol., 72, 5046–5055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgur Y., Dyda,F., Hickman,A.B., Jenkins,T.M., Craigie,R. and Davies,D.R. (1998) Three new structures of the core domain of HIV-1 integrase: an active site that binds magnesium. Proc. Natl Acad. Sci. USA, 95, 9150–9154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer T.S. and Brown,P.O. (1997) Mapping features of HIV-1 integrase near selected sites on viral and target DNA molecules in an active enzyme–DNA complex by photo-cross-linking. Biochemistry, 36, 10655–10665. [DOI] [PubMed] [Google Scholar]
- Heuer T.S. and Brown,P.O. (1998) Photo-cross-linking studies suggest a model for the architecture of an active human immunodeficiency virus type 1 integrase–DNA complex. Biochemistry, 37, 6667–6678. [DOI] [PubMed] [Google Scholar]
- Hindmarsh P. and Leis,J. (1999) Retroviral DNA integration. Microbiol. Mol. Biol. Rev., 63, 836–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins T.M., Engelman,A., Ghirlando,R. and Craigie,R. (1996) A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem., 271, 7712–7718. [DOI] [PubMed] [Google Scholar]
- Jenkins T.M., Esposito,D., Engelman,A. and Craigie,R. (1997) Critical contacts between HIV-1 integrase and viral DNA identified by structure-based analysis and photo-crosslinking. EMBO J., 16, 6849–6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn E., Mack,J.P.G., Katz,R.A., Kulkosky,J. and Skalka,A.M. (1991) Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res., 19, 851–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkosky J., Jones,K.S., Katz,R.A., Mack,J.P. and Skalka,A.M. (1992) Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol., 12, 2331–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leavitt A.D., Shiue,L. and Varmus,H.E. (1993) Site-directed mutagenesis of HIV-1 integrase demonstrates differential effects on integrase functions in vitro. J. Biol. Chem., 268, 2113–2119. [PubMed] [Google Scholar]
- Lodi P.J., Ernst,J.A., Kuszewski,J., Hickman,A.B., Engelman,A., Craigie,R., Clore,G.M. and Gronenborn,A.M. (1995) Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry, 34, 9826–9833. [DOI] [PubMed] [Google Scholar]
- Lubkowski J. et al. (1998) Structure of the catalytic domain of avian sarcoma virus integrase with a bound HIV-1 integrase-targeted inhibitor. Proc. Natl Acad. Sci. USA, 95, 4831–4836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller H.P. and Varmus,H.E. (1994) DNA bending creates favored sites for retroviral integration: an explanation for preferred insertion sites in nucleosomes. EMBO J., 13, 4704–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association—insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol., 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pryciak P.M. and Varmus,H.E. (1992) Nucleosomes, DNA-binding proteins and DNA sequence modulate retroviral integration target site selection. Cell, 69, 769–780. [DOI] [PubMed] [Google Scholar]
- Puras-Lutzke R.A., Vink,C. and Plasterk,R.H. (1994) Characterization of the minimal DNA-binding domain of the HIV integrase protein. Nucleic Acids Res., 22, 4125–4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P. and Mizuuchi,K. (1995) Structure of the bacteriophage Mu transposase core: a common structural motif for DNA transposition and retroviral integration. Cell, 82, 209–220. [DOI] [PubMed] [Google Scholar]
- Scottoline B.P., Chow,S., Ellison,V. and Brown,P.O. (1997) Disruption of the terminal base pairs of retroviral DNA during integration. Genes Dev., 11, 371–382. [DOI] [PubMed] [Google Scholar]
- van Gent D.C., Groeneger,A.A. and Plasterk,R.H. (1992) Mutational analysis of the integrase protein of human immunodeficiency virus type 2. Proc. Natl Acad. Sci. USA, 89, 9598–9602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D.C., Vink,C., Groeneger,A.A. and Plasterk,R.H. (1993) Complementation between HIV integrase proteins mutated in different domains. EMBO J., 12, 3261–3267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink C., Oude Groeneger,A.M. and Plasterk,R.H. (1993) Identification of the catalytic and DNA-binding region of the human immunodeficiency virus type I integrase protein. Nucleic Acids Res., 21, 1419–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams T.L., Jackson,E.L., Carritte,A. and Baker,T.A. (1999) Organization and dynamics of the Mu transpososome: recombination by communication between two active sites. Genes Dev., 13, 2725–2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woerner A.M. and Marcus-Sekura,C.J. (1993) Characterization of a DNA binding domain in the C-terminus of HIV-1 integrase by deletion mutagenesis. Nucleic Acids Res., 21, 3507–3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.N., Mueser,T.C., Bushman,F.D. and Hyde,C.C. (2000) Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J. Mol. Biol., 296, 535–548. [DOI] [PubMed] [Google Scholar]