Abstract
The Escherichia coli RecF, RecO and RecR pro teins have previously been implicated in bacterial recombinational DNA repair at DNA gaps. The RecOR-facilitated binding of RecA protein to single-stranded DNA (ssDNA) that is bound by single-stranded DNA-binding protein (SSB) is much faster if the ssDNA is linear, suggesting that a DNA end (rather than a gap) facilitates binding. In addition, the RecOR complex facilitates RecA protein-mediated D-loop formation at the 5′ ends of linear ssDNAs. RecR protein remains associated with the RecA filament and its continued presence is required to prevent filament disassembly. RecF protein competes with RecO protein for RecR protein association and its addition destabilizes RecAOR filaments. An enhanced function of the RecO and RecR proteins can thus be seen in vitro at the 5′ ends of linear ssDNA that is not as evident in DNA gaps. This function is countered by the RecF/RecO competition for association with the RecR protein.
Keywords: DNA repair/RecA/RecF/RecO/RecR
Introduction
Replication fork progress is halted by encounters with DNA lesions, strand breaks or other barriers. Repair of stalled replication forks requires recombination, using one of many proposed pathways as dictated by the structure of the damaged fork (Kuzminov, 1999; Cox et al., 2000; Marians, 2000; Cox, 2001a,b).
Encounters with base lesions can derail the fork and leave the lesion in a single-strand gap (Cordeiro-Stone et al., 1999). In bacteria, a variety of pathways have been proposed for the repair of lesions in such gaps (West et al., 1981; Whitby et al., 1993; Cox et al., 2000; McGlynn and Lloyd, 2000; Michel, 2000) and the RecF, RecO and RecR proteins are thought to play a significant role. Genetic analyses have led to the inclusion of the RecF, RecO and RecR proteins within the RecF recombination pathway (Horii and Clark, 1973; Kolodner et al., 1985; Mahdi and Lloyd, 1989; Smith, 1989; Clark and Sandler, 1994). In vitro analysis has revealed that RecR protein forms alternative complexes with either RecF or RecO proteins (Umezu and Kolodner, 1994; Shan et al., 1997; Webb et al., 1997). The RecF protein has a weak ATP hydrolysis activity (Webb et al., 1995) leading to dissociation of double-stranded DNA (dsDNA; Webb et al., 1999). The RecO protein binds to single-stranded DNA (ssDNA) and dsDNA, promotes the renaturation of complementary ssDNA molecules and catalyzes assimilation of ssDNA into superhelical dsDNA (Luisi-DeLuca and Kolodner, 1994; Luisi-DeLuca, 1995). No DNA-binding function has been detected in the Escherichia coli RecR protein.
At least part of the function of the RecFOR proteins involves a modulation of RecA protein filament formation. At neutral pH and above, filament nucleation is largely limited to ssDNA. RecA will nucleate within a single-stranded gapped region and polymerize in the 5′ to 3′ direction, encompassing the adjacent duplex DNA (Register and Griffith, 1985; Shaner and Radding, 1987; Shan et al., 1997). Single-stranded DNA-binding protein (SSB) inhibits the nucleation step in RecA filament assembly on ssDNA, but facilitates the subsequent filament extension in vitro. A net end-dependent disassembly of RecA protein filaments has also been observed on dsDNA (Lindsley and Cox, 1990a) and ssDNA (Shan et al., 1997) at neutral and higher pHs. Net dissociation from ssDNA is dependent upon the presence of SSB and ATP (Shan et al., 1997), and proceeds uniquely 5′ to 3′ (Bork et al., 2001).
The exact compositions of the RecOR and RecFR complexes have not been determined. The RecOR and RecFR complexes modulate the assembly and disassembly of RecA filaments on ssDNA in vitro. The RecOR complex promotes the nucleation of RecA filaments on SSB-bound ssDNA and also prevents a net disassembly from linear ssDNA (Umezu and Kolodner, 1994; Shan et al., 1997). The RecOR proteins have no effect on RecA protein disassembly from dsDNA (J.M.Bork, unpublished data). The RecFR complex binds to dsDNA and limits the extension of RecA filaments (Webb et al., 1995, 1997).
The RecOR proteins are part of a large family of functional homologs that modulate the activities of recombinases in every class of organism (Sung, 1997a,b; New et al., 1998; Shinohara and Ogawa, 1998; Bleuit et al., 2001). Our working hypothesis has been that the RecOR and RecFR complexes act together, largely to maintain RecA filaments within DNA gaps. However, our efforts to demonstrate a gap-constrained RecA filament in vitro with all three of these proteins present have been unsuccessful to date. Instead, this report documents an enhanced function of RecOR at the free 5′ ends of linear ssDNA, as well as direct competition between DNA-bound RecF and RecO proteins for association with RecR. We also show that RecR protein, having facilitated the formation of a RecA filament, remains associated with that filament.
Results
Experimental design
This study was initiated to investigate further the effects of the RecF, RecO and RecR proteins on RecA filament assembly and disassembly. The activities of these proteins were studied in pairs, reflecting the RecOR and RecFR protein complexes characterized previously. We also initiated efforts to determine how these alternate pairings functioned together. A combination of an indirect, but real-time, ATPase assay and electron microscopy (EM) was used to assess RecA protein binding to DNA. An immunoaffinity gold-labeling procedure was utilized in some experiments to qualitatively confirm the presence of additional proteins within multi-protein complexes.
RecOR proteins promote RecA filament assembly more efficiently on SSB-coated linear ssDNA than on SSB-coated circular ssDNA
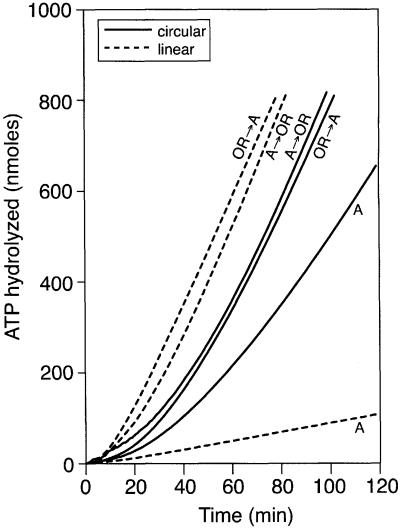
In separate experiments, SSB was bound either to linear or circular ssDNA prior to RecA binding. The effects of the RecOR complex in overcoming the inhibition of RecA binding by SSB were then examined (Figure 1). In the absence of the RecOR proteins, the rate of ATP hydrolysis for RecA on circular ssDNA did not reach its maximum and the rate on linear ssDNA was reduced due to net end-dependent dissociation (Shan et al., 1997). The addition of the RecOR proteins stimulated the binding of RecA and allowed complete binding of RecA protein to both types of ssDNA, as determined by the observed rates of DNA-dependent ATP hydrolysis. However, the lag for RecA to reach its maximal rate of ATP hydrolysis was reduced by at least 10 min when linear rather than circular ssDNA was used.
Fig. 1. The RecOR proteins promote RecA filament assembly more efficiently on SSB-coated linear ssDNA than on SSB-coated circular ssDNA. The ssDNA-dependent ATPase assay was used to monitor RecA binding to ssDNA. The SSB protein (0.3 µM) was pre-bound to either M13mp8.1037(+) circular ssDNA (3 µM) or M13mp8.1037(+)xPstI linear ssDNA (3 µM). After a 10 min pre-incubation at 37°C, ATP (3 mM) and either RecA protein (4 µM) or the RecO (0.03 µM) and RecR (0.15 µM) proteins were added, as indicated. After another 10 min, the RecO and RecR proteins were added to the RecA reactions and vice versa. The addition of RecA protein defines t = 0. The circular ssDNA reactions are indicated with solid lines and the linear ssDNA reactions are indicated with dashed lines.
It seemed possible that the evident specificity of the RecOR proteins for linear, as opposed to circular, ssDNA reflects the 5′-phosphoryl end group of the linear DNA. However, the times taken to reach maximal ATP hydrolysis of RecA on linear ssDNA and 5′ dephosphorylated linear ssDNA were not significantly different (data not shown).
RecR protein is associated with linear ssDNA coated with SSB
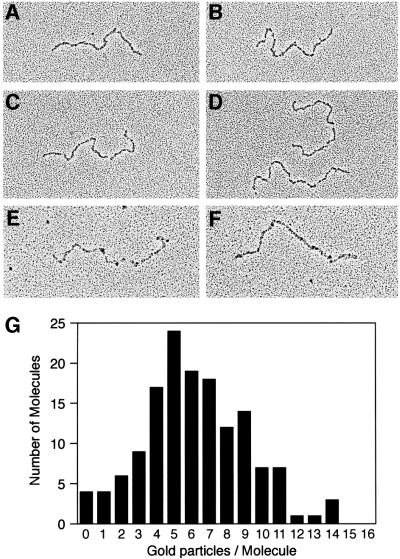
We attempted to determine whether the association of the RecO and/or RecR proteins with linear ssDNA–SSB molecules was transient or continued for significant periods of time, using EM. Linear ssDNA was incubated with SSB for 10 min at 37°C, then the RecO and RecR proteins were added and incubated for an additional 15 min. The appearance of SSB-coated linear ssDNA is similar in the absence (Figure 2A and B) and presence (Figure 2C and D) of the RecOR proteins. Owing to cross-linking, the SSB-bound molecules do not exhibit the typical nucleosome-like structure typically seen when SSB is bound to ssDNA. However, the presence of SSB bound to the linear ssDNA has been confirmed by an immunoaffinity gold-labeling procedure, with antibodies directed against SSB (data not shown). To determine whether the RecO and/or RecR proteins are associated with the SSB-coated linear ssDNA molecules, we used the same immunoaffinity gold-labeling procedure, except with antibodies directed against either the RecO or RecR proteins. Although our RecO polyclonal antibodies recognize purified RecO protein on a western blot, significant binding of gold-labeled antibodies to RecO attached to SSB-coated linear ssDNA molecules on EM grids has not been observed (see Discussion).
Fig. 2. The RecR protein is associated with linear ssDNA coated with SSB. M13mp8.1037(+)xEcoRI (12 µM) was incubated with SSB (1.2 µM) in HEPES buffer and the PEP/PK regeneration system. The samples were cross-linked, dialyzed and spread by the cytochrome C method, as described in Materials and methods. (A–D) Representative molecules are shown in the micrographs for the reaction in the absence (A and B) or presence (C and D) of the RecO (0.12 µM) and RecR (0.6 µM) proteins. (E and F) An immunoaffinity labeling procedure identified RecR protein associated with linear ssDNA bound by SSB in the presence of the RecO and RecR proteins. RecR antibodies were diluted 1000-fold and protein A–gold was diluted 20-fold. (G) The number of gold particles per molecule were counted on a sample similar to those in (E) and (F) except that the RecR antibody was diluted 500-fold. The average number of gold particles per molecule for this labeling was six RecR-labeled gold particles per molecule.
Using antibodies directed against the RecR protein, the presence of RecR was readily detected among the SSB-coated linear ssDNA molecules with protein A–gold (Figure 2E and F). Figure 2G shows a histogram of the number of gold-labeled RecR proteins on each molecule combined from two identical reactions, with a 500-fold antibody dilution and 20-fold protein A–gold dilution. At these antibody and gold concentrations, each molecule has ∼6 RecR-labeled gold particles associated with the SSB-coated linear ssDNA. However, we have not determined the efficiency of the gold labeling procedure and these numbers are likely to represent an underestimate of the RecR protein present.
RecR protein remains associated with a RecA filament after its formation
Stable RecA nucleoprotein filaments formed in the presence of the RecOR proteins were examined by EM. We wanted to determine whether the RecO and/or RecR proteins remained associated with the RecA filament or dissociated during filament formation. Significant binding of gold-labeled antibodies to the RecO protein was not observed (see Discussion). The presence of the RecR protein was detected using RecR antibody (5000-fold dilution) and protein A–gold (20-fold dilution; Figure 3). Gold-labeled RecR proteins associated with the linear ssDNA–RecAOR filament are shown in the micrographs of Figure 3. Judgements were made on the location and abundance of the RecR-directed gold molecules. Dividing the molecule into quarters and counting the number of gold particles in each segment of each molecule, we find that the majority of the RecR-labeled gold particles are located near one end of the linear ssDNA–RecA molecule, although some are clearly present in the filament interior. The end with the majority of the gold particles is oriented to the left in the micrographs and is designated quarter A in the histogram (Figure 3A). Another histogram (Figure 3B) details the distribution for the number of gold particles per molecule. With a 5000-fold antibody dilution, ∼3.5 RecR-labeled gold particles per molecule were recorded. When the antibody dilution is increased to 10 000- or 25 000-fold, the abundance of gold particles per molecule decreases. However, the distribution of gold particles is not altered (data not shown). We were unable to determine whether the favored end was the same in each molecule or varied from one to another.
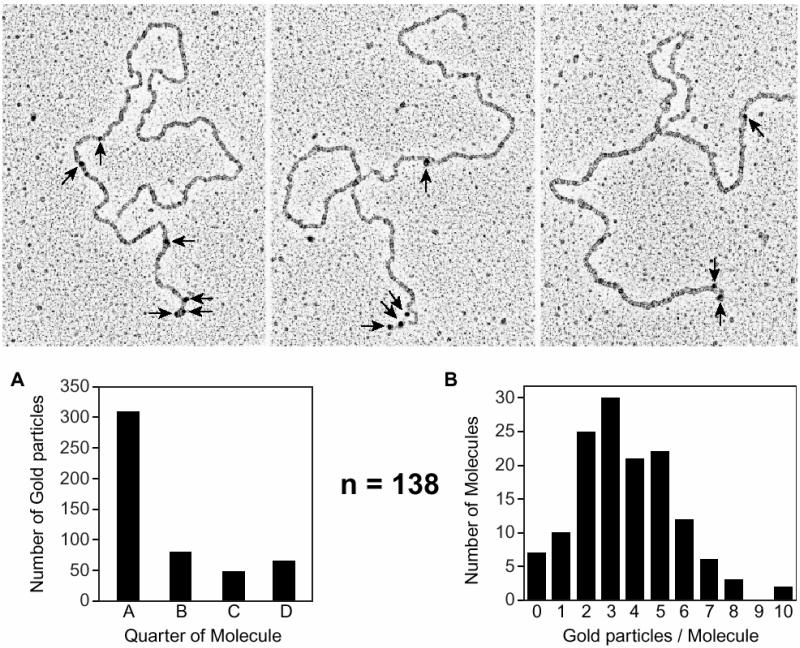
Fig. 3. The RecR protein remains associated with a RecA filament after its formation. The RecA protein (5 µM) was incubated with M13mp8.1037(+) xEcoRI (12 µM) in HEPES buffer with the PEP/PK regeneration system. The SSB protein (1.2 µM), RecO protein (0.2 µM) and RecR protein (0.6 µM) were added to the reaction after an initial 10 min incubation at 37°C. After 15 min, the reaction was stopped with ATPγS and diluted 60-fold before spreading on Alcian-activated grids (described in Materials and methods). An immunoaffinity labeling procedure identified RecR protein that is associated with the RecA filament, as seen in the micrographs, with a 5000-fold dilution of RecR antibody and 20-fold dilution of protein A–gold (arrows). One hundred and thirty-eight molecules were analyzed. (A) Judgements were made as to where the gold-labeled RecR protein was found along the molecule. Molecules were divided into quarters and the number of gold particles in each segment of each molecule was counted. The majority of gold particles were found to be located within one end segment of the molecules, which is oriented to the left on the micrographs and designated quarter A in the histogram. The only gold particles found in quarter D were on molecules that had gold particles associated at both ends. If a molecule did not have any gold particles located in an end segment, then defining the placement of the golds into either quarter B or C was arbitrary. (B) The histogram shows the number of gold particles per molecule, the average being 3.5 RecR-labeled gold particles per molecule.
RecOR proteins facilitate RecA binding to gapped DNA–SSB molecules
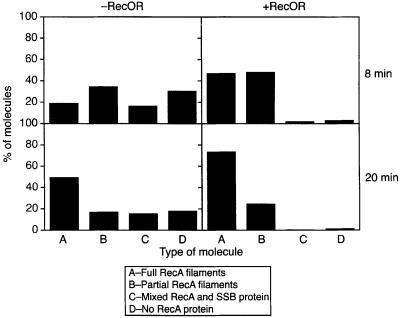
The RecOR proteins enable RecA to bind to circular and linear ssDNA coated with SSB (Umezu et al., 1993; Umezu and Kolodner, 1994; Shan et al., 1997). To put this into the context of recombinational repair, we used a circular gapped DNA (gDNA) substrate with 2089 nucleotides (nt) of ssDNA (GD2089). SSB was pre-bound to the gDNA, then RecA was added either in the presence or absence of the RecOR proteins. EM was used to estimate the percentage of different types of molecules found in the reaction at different times. Histograms shown in Figure 4 depict the number of full RecA filaments (A), partial RecA filaments (B), molecules with both RecA and SSB (C), and molecules without any RecA (D) that were counted after 8 and 20 min incubations. In the absence of the RecOR proteins, RecA filament assembly on the SSB-bound gDNA was inefficient. Even after a 20 min incubation, >25% of the molecules counted still had SSB present and visible. RecA filaments formed much more rapidly in the presence of the RecOR proteins. After an 8 min incubation, only 5% of the molecules had any visible SSB remaining. The vast majority of the molecules were RecA filaments encompassing all or most of the gapped DNA circles. After a 20 min incubation, >70% of the molecules were completely filamented with RecA protein. The lengths of the partial RecA filamented sections were significantly shorter for reactions in the absence of the RecOR proteins than those formed in their presence, as were the lengths at 8 min shorter than at 20 min. As with earlier experiments, the RecOR complex facilitates the nucleation of RecA onto SSB-coated ssDNA, but does not restrict extension of the filaments onto the contiguous duplex DNA outside of the gap.
Fig. 4. The RecOR proteins facilitate RecA binding to gDNA–SSB molecules. Electron microscopy allowed visualization of RecA filaments formed on circular gDNA in the presence or absence of the RecOR proteins in Tris buffer. Reactions were analyzed after 8 and 20 min of incubation. SSB (0.051 µM or one monomer per 10 nt of ssDNA) was pre-incubated with GD2089 (3 µM total nt) in the presence of the PC/CPK regeneration system, as described in Materials and methods. The pre-incubation was 5 min at 37°C, at which point RecA protein (3 µM) and ATP (3 mM) were added. In the absence of the RecOR proteins, samples were taken after 8 and 20 min. Otherwise, RecA was incubated with GD2089–SSB for 5 min, then RecO (0.03 µM) and RecR (0.15 µM) proteins were added. Samples were taken after 8 and 20 min. Reactions were stopped with ATPγS, diluted 10-fold and spread on BSA-activated grids, as described in Materials and methods. The numbers of molecules with full RecA filaments (A), partial RecA filaments (B), mixed RecA and SSB proteins (C) and no RecA protein bound (D) were counted and are shown as percentages of all the molecules counted. For the 8 min samples, n = 299 in the absence of RecOR and n = 249 in the presence of RecOR. For the 20 min samples, n = 209 in the absence of RecOR and n = 133 in the presence of RecOR.
RecF protein destabilizes RecAOR filaments
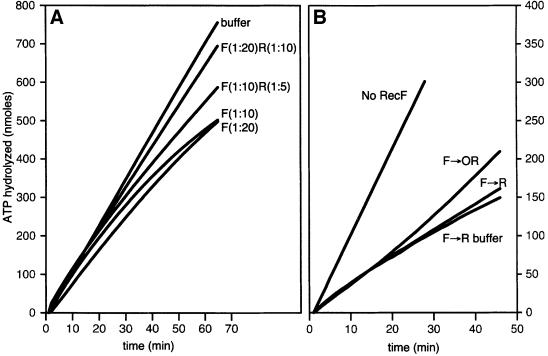
In order to understand how the RecF, RecO and RecR proteins interact, we examined the effects of the RecF protein on the capacity of the RecOR protein to stabilize RecA filaments. Note that the term ‘stable’ means that no net dissociation of RecA filaments is observed, although some exchange of RecA between free and bound forms may be detected (Shan et al., 1997). Stable RecA filaments were formed on linear ssDNA in the presence of the RecOR proteins. Increasing amounts of the RecF protein or RecF storage buffer were added to the reactions at t = 0 (Figure 5A). In the absence of RecF protein, a stable linear ssDNA–RecAOR filament is formed and persisted throughout the experiment, as indicated by a high, steady rate of ATP hydrolysis. The addition of one RecF protein monomer per 80 nt of ssDNA decreased the ATP hydrolysis rate of the linear ssDNA–RecAOR filament by only 10%. Adding RecF at higher concentrations, up to one RecF per 5 nt of ssDNA, resulted in a time-dependent decrease in ATPase rate by >60%, indicative of significant net RecA filament disassembly.
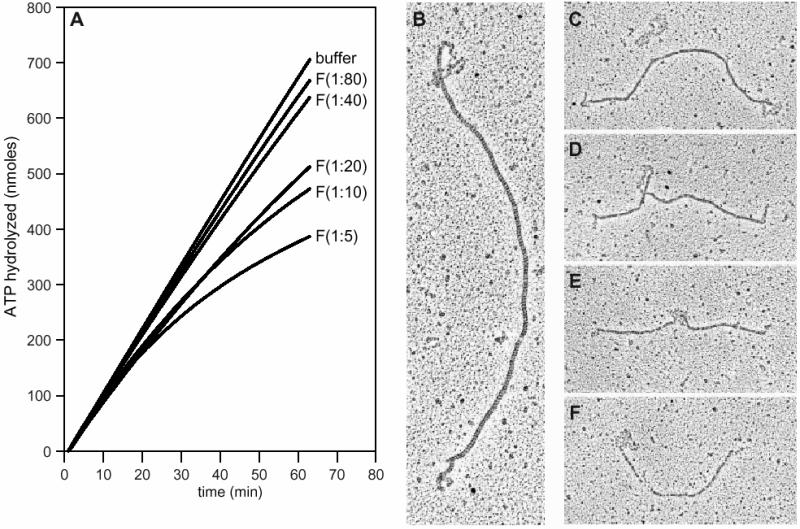
Fig. 5. The RecF protein destabilizes linear ssDNA–RecAOR filaments. (A) The ssDNA-dependent ATPase assay was used to monitor the stability of RecA protein filaments on linear ssDNA. A stable RecA filament (3 µM) was formed on M13mp8.1037(+)xPstI (3 µM) in the presence of SSB (0.3 µM), RecO (0.03 µM) and RecR (0.15 µM) proteins. The reaction was carried out as described in Materials and methods. RecF protein concentrations are expressed as monomer per nucleotide of ssDNA. At t = 0, the stable linear ssDNA–RecAOR filament was challenged by the addition of increasing amounts of RecF protein: no RecF or RecF at 1:80, 1:40, 1:20, 1:10 and 1:5. (B–F) EM allowed visualization of linear ssDNA–RecAOR filaments in the presence of the RecF protein. A stable RecA filament (5 µM) was formed on M13mp8.1037(+)xPstI (12 µM) in the presence of SSB (1.2 µM), RecO (0.12 µM) and RecR (0.6 µM) proteins with the PEP/PK regeneration system for 15 min at 37°C. The RecF protein (1.2 µM) was added at 1:10. After a 20 min incubation, reactions were stopped with ATPγS, diluted 60-fold and spread as described in Materials and methods.
EM was also used to look at these protein–DNA complexes. In the absence of any RecF protein, RecA formed filaments on linear ssDNA in the presence of the RecOR proteins which uniformly extended across the entire length of the DNA (Shan et al., 1997 and data not shown). When RecF protein was added at one monomer per 10 nt, the RecA filament stability was altered. A wide variety of molecules were found in the reaction, with representative molecules shown in the micrographs (Figure 5B–F). Figure 5C also includes a linear ssDNA molecule coated with SSB, with the characteristic nucleosome-like structure. Some molecules had a tightly packed RecA filament with short SSB-coated single-stranded ends (Figure 5B and C), while others had SSB-coated sections within the interior of the RecA filament (Figure 5D and E). A number of the molecules with shorter RecA filament lengths had very segmented RecA filaments (Figure 5E and F). The disassembly of RecA filaments at interior positions might reflect loss of interior RecOR complexes, combined with the binding of RecFR complexes that block filament extension (Webb et al., 1997).
RecA stability on linear ssDNA was further explored in the presence of the RecF, RecO and RecR proteins using different combinations as well as orders of addition. A stable RecA filament formed in the presence of the RecOR proteins was again indicated by a high, steady rate of ATP hydrolysis. The addition of RecF protein destabilized the filament, as detected by a slower, attenuating rate of ATP hydrolysis. The RecA filament instability can be attributed to the loss of RecR from the RecOR complex, as it associates with RecF protein. The destabilizing effect of the RecF protein was diminished by adding the RecF and RecR proteins together at the time of challenge, (t = 0; Figure 6A). We surmise that the additional RecR protein forms a complex with the RecF and inhibits its interaction with the RecR protein already in the RecA filaments. The destabilization by RecF can also be lessened by the addition of excess RecOR proteins once the attenuation has begun (Figure 6B). After the addition of extra RecOR, the rate of ATP hydrolysis increased, indicating the rebinding of RecA protein, albeit not to the extent observed when RecF protein was not added.
Fig. 6. Competition between RecF and RecO proteins for RecR protein. The ssDNA-dependent ATPase assay was used to monitor the stability of RecA protein filaments on linear ssDNA. A stable RecA filament (3 µM) was formed on M13mp8.1037(+)xPstI (3 µM) in the presence of SSB (0.3 µM), RecO (0.03 µM) and RecR (0.15 µM) proteins. Reactions were carried out as described in Materials and methods. Challenging protein concentrations are expressed as monomer per nucleotide of ssDNA. (A) At t = 0, the linear ssDNA–RecAOR filament was challenged with either RecF protein alone (1:20) or (1:10) or with the RecFR proteins together (1:20; 1:10) or (1:10; 1:5). (B) The linear ssDNA–RecAOR filament was challenged with RecF protein (1:10) or RecF buffer (no RecF). At t = 0, RecR (1:20), RecOR (1:100; 1:20) or RecR buffer (no RecR) were added to the three reactions that were challenged with RecF protein.
RecO and RecR proteins reduce the 3 ′ end bias in RecA protein-mediated DNA pairing reactions
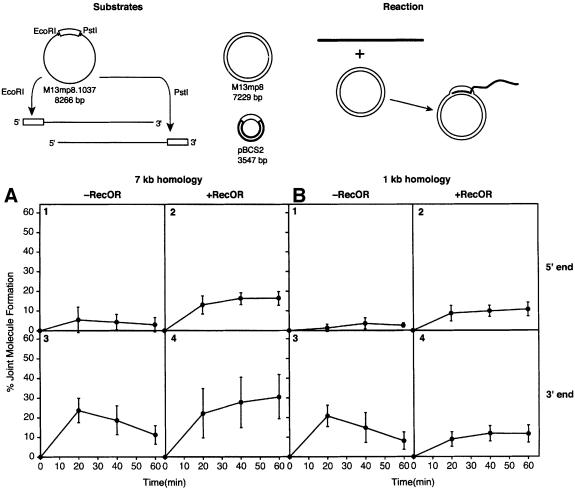
The end-dependent disassembly of RecA filaments was prevented by the use of dATP and/or the RecOR proteins. We carried out a series of experiments to see whether these same reagents would affect RecA-mediated DNA pairing at DNA ends. In vitro joint molecule formation assays were used to determine the efficiency of DNA pairing at the 5′ and 3′ ends of linear ssDNA molecules. Reactions were carried out between a linear ssDNA and a supercoiled duplex DNA. Homology between substrates (1 or 7 kb) was restricted to either the 5′ or 3′ end of the linear ssDNA by employing appropriate restriction endonucleases when linearizing the circular ssDNA. DNA substrates are illustrated in Figure 7.
Fig. 7. Effects of the RecO and RecR proteins on RecA-mediated DNA pairing. The DNA substrates and the reaction are illustrated at the top. M13mp8.1037(+) circular ssDNA was linearized with either EcoRI or PstI to place the 1037 nt insert (open rectangle) at the 5′ or 3′ end, respectively. The supercoiled M13mp8 dsDNA substrate has 7229 bp of homology to the 3′ end of M13mp8.1037(+)xEcoRI linear ssDNA and the 5′ end of M13mp8.1037(+)xPstI linear ssDNA. The supercoiled pBCS2 dsDNA substrate has 1034 bp of homology (open rectangle) to the 5′ end of M13mp8.1037(+)xEcoRI linear ssDNA and the 3′ end of M13mp8.1037(+)xPstI linear ssDNA. (A and B) Four reactions using dsDNA substrates with 7229 bp (M13mp8) and 1034 bp (pBCS2) of homology, respectively, with the linear ssDNA substrate. Within each set of four, reactions 1 and 2 show results with the homology located at the 5′ end of the linear ssDNA, while reactions 3 and 4 show results with the homology located at the 3′ end. In addition, reactions 1 and 3 lack the RecOR proteins, while reactions 2 and 4 contain them. For (A), n = 7, 7, 9 and 8 for reactions 1–4, respectively. For (B), n = 7, 7, 9 and 8 for reactions 1–4, respectively. Reactions were carried out as described in Materials and methods for the agarose gel assay.
Joint molecule formation was monitored with an agarose gel assay. Results in the presence or absence of the RecO and RecR proteins are shown in Figure 7. Whether the 5′ end of the linear ssDNA contained 7 (Figure 7A) or 1 kb (Figure 7B) of homology to the supercoiled DNA substrate, few stable joint molecules were formed in the absence of the RecOR proteins. When homology was limited to the 3′ end, joint molecules were initially formed at high levels, but gradually disappeared over a 60 min time course in the absence of the RecOR proteins (reaction 3).
Including the RecOR proteins in the reaction resulted in a general increase in the yield of joint molecules that were formed in most reactions (reactions 2 and 4). The percentage of joint molecules formed at the 5′ end increased over 3-fold for both 7 and 1 kb of homology when the RecOR proteins were present (Figure 7, reactions 1 and 2). With 7 kb of homology, the percentage of joint molecules formed at the 3′ end was enhanced, especially with longer incubation times (Figure 7A, reaction 4). The reaction at the 5′ end was increased even more, such that the difference in reaction efficiency at the two ends was greatly reduced (Figure 7, reaction 4). With 1 kb of homology, the presence of RecOR rendered the reactions at the two ends equivalent (Figure 7B, reactions 2 and 4). The effects of the RecOR proteins in eliminating 3′ end bias in these reactions was confirmed in separate trials using a filter binding assay (data not shown).
In contrast, the effects of substituting dATP for ATP were more modest (data not shown). With 7 kb of homology, the presence of dATP increased the amount of joint molecules formed at both DNA ends, although a 3′ end bias was still evident. With 1 kb of homology, reactions at the 3′ end were enhanced by dATP, but dATP had little effect on reactions at the 5′ end, with the overall result that the 3′ end bias was actually increased.
Discussion
Four major conclusions can be derived from these data that enhance our understanding of the function of the RecFOR proteins in recombination processes. First, the RecO and RecR proteins facilitate RecA nucleation on any SSB-bound ssDNA. However, RecOR-facilitated loading of RecA protein onto linear ssDNA is significantly faster than the comparable loading onto circular ssDNA, suggesting that the RecOR complex is particularly effective at a ssDNA end. Secondly, RecR protein appears not to act transiently. Instead, it remains associated with the RecA filaments whose assembly it has facilitated. A persistent association is seen with other mediator proteins such as UvsY of bacteriophage T4, and may be a general property of such proteins (Beernink and Morrical, 1999). The majority of the gold-labeled RecR protein is found near an end of the ssDNA. For reasons detailed below, we infer that the concentration is near the 5′ end. Thirdly, the RecF and RecO proteins compete for RecR protein in the functional formation of heteroprotomers. The destabilizing effect of RecF on the observed stability of RecAOR filaments attests to the importance of the continued presence of RecR protein and probably RecO protein in the complexes. Finally, the RecOR proteins largely eliminate the 3′ end bias in RecA protein-promoted DNA pairing reactions.
We were unable to detect an association between RecO and SSB-coated linear ssDNA molecules or RecA filaments using the immunoaffinity gold-labeling procedure on EM grids. A physical interaction between the RecO protein and the SSB protein (and not between RecR and SSB) has previously been demonstrated in vitro (Umezu and Kolodner, 1994). The lack of a RecO signal in the present experiments may reflect RecO dissociation from the SSB-coated linear ssDNA molecule or experimental problems ranging from inadequate antibody sensitivity to masking by other proteins.
The RecOR-facilitated assembly of RecA filaments at the 5′ ends of linear single strands is a theme of these, and previous, results. RecOR promotes the rapid loading of RecA protein, particularly onto linear ssDNAs (implying an interaction with one end), and RecR (at least) remains associated with the filament concentrated near one end. It is reasonable to speculate that the concentration is near the 5′ end, since RecOR also prevents a net 5′ to 3′ end-dependent disassembly of RecA filaments and facilitates RecA-mediated pairing reactions at the 5′ end. RecOR-mediated facilitation of a fast nucleation of RecA protein on linear ssDNA does not depend on the free 5′-phosphoryl group and we do not know how an end bias might be created for these proteins. If RecOR must interact with both DNA and SSB to facilitate RecA protein binding, it is possible that a DNA end might provide accessible sites more readily.
The RecOR and RecFR complexes are distinct, and the RecFOR proteins do not all interact under the conditions we have employed to date. We find that the RecF and RecO proteins appear to compete for the RecR protein. RecA filaments formed with only the RecOR complexes are destabilized by the addition of RecF, suggesting a transfer of RecR protein from RecO to RecF. The RecF protein provides no detectable stimulation of any RecA-mediated function. Instead, the RecF and RecA proteins compete for DNA binding sites (Webb et al., 1997). Therefore, in addition to the RecF protein sequestering RecR from RecOR complexes, leading to RecA filament disassembly from an end, the RecFR proteins may bind to the DNA and prevent RecA filament extension, leading to the segmentation of the RecA filament. All of this may require some rethinking of the role of these proteins in the cell. If the same competition between RecO and RecF proteins for interaction with RecR exists in the cell, then RecF protein may compromise the function of RecOR in the establishment of RecA filaments. It is possible that RecF protein has a switching function that disrupts the function of RecOR when it is no longer needed (releasing RecA filaments that have completed their molecular mission). This could be part of a broader function in which RecF mediates the interface between replication and recombination in the repair of stalled replication forks, facilitating needed transitions between sets of replication and recombination proteins.
The effect of RecOR on RecA protein-promoted DNA pairing is particularly notable and may have implications for model building in recombinational DNA repair. The properties of RecA filament assembly and disassembly lead directly to an easily demonstrable 3′ end bias in DNA pairing reactions. The RecO and RecR proteins eliminate most, if not all, of this end bias. Previously, it has been shown that the end bias is simply a function of RecA protein assembly and disassembly, and that the inherent capacity of RecA protein to pair DNA at DNA ends is not different for the 5′ and 3′ ends (Dutreix et al., 1991; McIlwraith and West, 2001). The RecOR proteins should, in principle, allow the 5′ end of linear ssDNAs to participate actively in DNA pairing simply by stabilizing RecA protein complexes at that end. It is also possible that the RecOR protein complex actively participates in DNA pairing reactions. The retention of RecR protein in RecA filaments suggests that RecOR acts as an integral part of the RecA filament, and previous work has demonstrated that the RecO protein promotes the annealing of complementary ssDNAs and the formation of D-loops (Luisi-DeLuca and Kolodner, 1994; Luisi-DeLuca, 1995). We note that the substitution of dATP for ATP, previously shown to stabilize RecA filaments (Shan et al., 1997), does not enhance 5′ D-loop formation.
The effect of RecOR on RecA protein-mediated strand invasion permits a consideration of some new pathways for recombinational DNA repair. In vivo, the detailed roles of 3′ and 5′ DNA ends have not yet been elucidated. A lack of RecBCD activity or simply the RecD subunit can be compensated for by the action of 5′ to 3′ exonucleases, suggesting that the creation of single strands with 3′ ends is important (Templin et al., 1972; Lovett et al., 1988). The 3′ ends can serve the critical function of replication primers in many situations. However, examination of the effects of the elimination of exonuclease activities with different polarities suggests that 5′ ends also play a role in recombination and recombinational DNA repair (Razavy et al., 1996). Rosenberg and Hastings (1991) have proposed a split-end model for recombination (see also Razavy et al., 1996) that might be applied to double-strand break repair at a damaged fork. RecBCD requires nearly flush ends with <25 bp of ssDNA (Taylor and Smith, 1985). Any broken end with a 5′ extension longer than this, as might be generated by the action of exonuclease III or by the occurrence of a strand break within a single-strand gap, would require a different repair path. A RecAOR filament could provide the alternative to RecBCD needed for the repair of such ends. The restart of replication during replication fork repair must include both the leading and lagging strands, and 5′ end invasions are reasonable starting points for lagging strand synthesis.
Materials and methods
Enzymes and biochemicals
The E.coli RecA protein was purified by a procedure developed for the RecA K72R mutant protein (Shan et al., 1996). Escherichia coli RecO (Shan et al., 1997), RecR (Webb et al., 1995), RecF (Webb et al., 1995) and SSB (Lohman et al., 1986; Shan et al., 1997) proteins were purified as described. The concentration of each protein was determined by absorbance at 280 nm using their respective extinction coefficients: ε280 = 2.23 × 104 M–1 cm–1 for RecA (Craig and Roberts, 1981), ε280 = 5.6 × 103 M–1 cm–1 for RecR (Shan et al., 1997), ε280 = 3.87 × 104 M–1 cm–1 for RecF (Webb et al., 1999) and ε280 = 2.83 × 104 M–1 cm–1 for SSB (Lohman and Overman, 1985). The concentration of the RecO protein was determined by the method of Bradford (1976) using bovine serum albumin (BSA) as a standard.
RecO and RecR polyclonal antibodies were produced in rabbit using purified proteins as antigens (Animal Care Unit, University of Wisconsin-Madison). Polyclonal antibodies directed against SSB (rabbit) were a generous gift from Dr Timothy Lohman (Washington University). Restriction endonucleases (unless noted otherwise), T4 ligase and T4 DNA polymerase were purchased from New England BioLabs. The PstI restriction endonuclease was purchased from Promega. SeaPlaque low melting temperature agarose was from FMC BioLabs. Bacterial alkaline phosphatase (BAP) was purchased from Gibco BRL. Dithiothreitol (DTT) was from Research Organics. Inorganic salts were from Fisher. All other biochemical reagents were purchased from Sigma.
DNA
The supercoiled circular duplex DNA and circular ssDNA from bacteriophage M13mp8 and M13mp8.1037 were prepared as described previously (Neuendorf and Cox, 1986). Bacteriophage M13mp8.1037 DNA (8266 bp) is M13mp8 (7229 bp) with a 1037 bp insert (EcoRV fragment from E.coli galT gene) at the SmaI site (Lindsley and Cox, 1990b). The concentrations of dsDNA and ssDNA stock solutions were measured by absorbance at 260 nm, using 50 and 36 µg ml–1 A260–1, respectively, as conversion factors. DNA concentrations are expressed in terms of total nucleotides. Linear ssDNA substrates were generated by digestion of M13mp8.1037(+) circular ssDNA (8266 bp) with either EcoRI, as described previously (Shan et al., 1997), or PstI and an oligo with the sequence 5′-TTGGCTGCAGGTCGACGGAT-3′. Complementary oligos are incubated with the circular ssDNA at 70°C for 15 min, then cooled slowly to room temperature to allow annealing. The restriction enzyme used for linearization was incorporated into the nomenclature for the linear ssDNA as either M13mp8.1037(+)xEcoRI or M13mp8.1037(+)xPstI. BAP was used to remove the 5′-phosphoryl end group of M13mp8.1037(+)xPstI at 37°C for 30 min. The dephosphorylated DNA was extracted, ethanol precipitated and resuspended in TE. In some cases, the linear ssDNAs were purified and 3′ end-labeled with [α-32P]ddATP using terminal transferase. Unincorporated nucleotides were removed using Sephadex G-50 spun columns. Labeled linear ssDNAs were ethanol precipitated, resuspended in TE and used within 7 days of their preparation.
The plasmid pBCS2 (3547 bp) is a pBR322 derivative constructed by replacing the PvuII–EcoRI fragment of pBR322 with a 1037 bp fragment (EcoRV fragment from the E.coli galT gene) using SalI linkers (Schutte and Cox, 1988). The pBCS2 plasmid purification was adapted from a published procedure (Garger et al., 1983) to start with 200 ml of cell culture. [3H]pBCS2 was generated for the filter binding assays by adding 2 mCi of [methyl-3H]thymidine (Amersham) and chloramphenicol (3.4 mg) during amplification of the plasmid.
Circular DNA molecules with precisely defined gaps were prepared by a method developed by Mara Robu in this laboratory (unpublished results). M13mp8(+) circular ssDNA, sequence-specific oligos, T4 ligase and T4 DNA polymerase were used to generate a gDNA with 2089 bases of ssDNA (GD2089). In brief, two oligos were annealed to a ssDNA circle. Only one of the oligos included a 3′-hydroxyl end suitable to act as a primer and T4 DNA polymerase was used to fill in one of the gaps separating the oligos. Residual protein was removed. The gDNA was resuspended in TE, concentrated using a Microcon-50 Ultrafilter (Amicon), purified by separation in 1.0% SeaPlaque low melting agarose and extraction from the gel with a QIAquick gel extraction kit (Qiagen). The conversion factor used to calculate the concentration of gDNA was based on the fractions of ssDNA and dsDNA in the molecule.
ATPase assay
The DNA-dependent ATP hydrolysis activity of RecA protein was measured by a coupled spectrophotometric enzyme assay (Morrical et al., 1986; Lindsley and Cox, 1990a). Absorbance measurements were taken with a Varian Cary 100 dual beam spectrophotometer equipped with a temperature controller and 12 position cell chamber. The cell path length and band pass were 0.5 cm and 2 nm, respectively. The regeneration of ATP from ADP and phosphoenolpyruvate with the oxidation of NADH can be followed by the decrease in absorbance. Although the absorbance maximum for NADH occurs at 340 nm, absorbances were measured at 380 nm to remain within the linear range of the spectrophotometer for the duration of the experiment. Concentrations of NADH were calculated using an extinction coefficient of ε380 = 1.21 mM–1 cm–1 at 380 nm.
Rates of ssDNA-dependent ATP hydrolysis were measured at 37°C in a reaction mixture (400 µl) containing 25 mM Tris–acetate pH 7.5 (80% cation), 10 mM Mg(OAc)2, 5% (w/v) glycerol, 3 mM ATP and 1 mM DTT. An ATP regeneration system (10 mM phosphoenolpyruvate, 10 U/ml pyruvate kinase, 3 mM potassium glutamate) and a coupling system (3 mM NADH, 4.5 U/ml lactic dehydrogenase) were also included. The final pH after addition of all reaction components was pH 7.6. Concentrations of DNA and proteins, as well as orders of addition, are reported in the text and figure legends. When present together, the RecO and RecR proteins were always added to the reaction as a mixture.
Agarose gel assay for joint molecule formation
Standard reaction mixtures (60 µl) were incubated at 37°C in a buffer containing 25 mM Tris–acetate (80% cation), 10 mM magnesium acetate, 3 mM potassium glutamate, 1 mM DTT and 5% (w/v) glycerol. The reaction also contained an ATP regeneration system consisting of 20 U/ml pyruvate kinase and 10 mM phosphoenolpyruvate. Linear ssDNA (10 µM) was pre-incubated with RecA protein (3.33 µM) for 10 min before the addition of SSB (1.0 µM), ATP (3 mM) and, when noted, RecO protein (0.133 µM) and RecR protein (0.5 µM). Incubation was continued for 10 min and supercoiled DNA (20 µM) was added to initiate pairing. Aliquots (12 µl) of the reactions were removed at each time point indicated and the reactions were stopped by the addition of 0.5 vol. of gel loading buffer [25 mM EDTA, 25% (w/v) glycerol, 5% SDS, 0.025% Bromophenol Blue]. Samples were electrophoresed on 0.8% agarose gel. After electrophoresis, gels were dried and DNA bands visualized using a PhosphorImager 425e (Molecular Dynamics).
Joint molecule formation was quantitated (ImageQuant software) as the fraction of total label in each lane present as joint molecules. The background at t = 0 was determined and subtracted from all other data points. This background correction in no case changed the final value by >15%. The number of assays (n) averaged to obtain the data for each reaction is noted in the figure legends. A few failed assays in which no joint molecule signal was obtained (<10% of the total assays) were omitted in tabulating the data.
Preparation of denatured BSA-coated carbon film for electron microscopy
A solution containing BSA (0.1%) and 0.5% sarkosyl was heated in a boiling water bath for 10 min and then stored at 4°C. Carbon-coated electron microscopy grids were then floated (carbon side down) on the denatured BSA solution for 5 min. Finally, the grids were picked up with clean forceps and immersed in three consecutive beakers containing 100 ml of water for 10 s each, touched to filter paper to absorb most of the liquid from the grid, dried under a heat lamp and stored in a desiccator.
Electron microscopy
Reaction mixtures contained 25 mM Tris–acetate pH 7.5 (80% cation) or 25 mM HEPES pH 7.5, 10 mM Mg(OAc)2, 5% (w/v) glycerol and 3 mM ATP. An ATP regeneration system of 10 mM phosphoenolpyruvate, 20 U/ml pyruvate kinase and 3 mM potassium glutamate (PEP/PK) or 12 mM phosphocreatine and 10 U/ml creatine phosphokinase (PC/CPK) was included. Concentrations of DNA and proteins, as well as orders of addition, are included in the text and figure legends. Reactions were stopped by adding adenosine 5′-o-(3-thio)triphosphate (ATPγS) to 1 mM and incubating at 37°C for 5 min. Reactions were spread by a modified adsorption procedure onto carbon films ‘activated’ by either Alcian Blue (Webb et al., 1995) or denatured BSA (as described above), with the following changes. The samples were diluted into 200 mM ammonium acetate, 10 mM HEPES pH 8.0 and 10% (w/v) glycerol, then adsorbed to carbon films on an EM grid for 3 min. The grids were washed by touching to a drop of 50 mM ammonium acetate containing 10 mM HEPES pH 8.0 and 10% glycerol, and then floating on a drop of the same solution for 30 s. Glycerol [5% (w/v)] was included in the 5.0% uranyl acetate staining solution and subsequent water washes. The grids were dried under a heat lamp for 10 min before circularly shadowing with platinum. This protocol is designed so that complete reaction mixtures can be visualized; however, it has the disadvantage of higher than normal background because of unreacted protein.
Immunoaffinity gold labeling
The identification of proteins associated with DNA molecules was carried out using an immunoaffinity labeling procedure. Reaction mixtures were cross-linked to ensure stabilization of the proteins on DNA. Cross-linking was accomplished by the addition of 0.5% glutaraldehyde with consecutive incubations at room temperature and then on ice for 20 min. Cross-linked samples were dialyzed against 20 mM NaCl, 5 mM EDTA and 10 mM HEPES pH 7.5 at 4°C for 1 h, and diluted 60-fold before adsorption onto the EM grid. Some reactions were spread by a modification of the cytochrome C method (Inman and Schnös, 1970), while others were deposited and dried on Alcian-activated carbon films. The grids were then incubated with anti-RecR antibodies and protein A–gold sequentially as described (Webb et al., 1997). Antibodies and protein A–gold were diluted as needed (empirically) in phosphate-buffered saline with 0.1% BSA. Although this procedure is useful for demonstrating the presence of a protein, the identification is qualitative.
Acknowledgments
Acknowledgements
We are grateful to Maria Schnös and David Inman for technical assistance. This work was supported by grants GM52725 (to M.M.C.) and GM14711 (to R.B.I.) from the National Institutes of Health.
References
- Beernink H.T.H. and Morrical,S.W. (1999) RMPs: recombination/replication mediator proteins. Trends Biochem. Sci., 24, 385–389. [DOI] [PubMed] [Google Scholar]
- Bleuit J.S., Xu,H., Ma,Y.J., Wang,T.S., Liu,J. and Morrical,S.W. (2001) Mediator proteins orchestrate enzyme–ssDNA assembly during T4 recombination-dependent DNA replication and repair. Proc. Natl Acad. Sci. USA, 98, 8298–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bork J.M., Cox,M.M. and Inman,R.B. (2001) RecA protein filaments disassemble in the 5′ to 3′ direction on single-stranded DNA. J. Biol. Chem., 276, in press. [DOI] [PubMed] [Google Scholar]
- Bradford M.M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Clark A.J. and Sandler,S.J. (1994) Homologous genetic recombination: the pieces begin to fall into place. Crit. Rev. Microbiol., 20, 125–142. [DOI] [PubMed] [Google Scholar]
- Cordeiro-Stone M., Makhov,A.M., Zaritskaya,L.S. and Griffith,J.D. (1999) Analysis of DNA replication forks encountering a pyrimidine dimer in the template to the leading strand. J. Mol. Biol., 289, 1207–1218. [DOI] [PubMed] [Google Scholar]
- Cox M.M. (2001a) Historical overview: searching for replication help in all of the rec places. Proc. Natl Acad. Sci. USA, 98, 8173–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox M.M. (2001b) Recombinational DNA repair of damaged replication forks in Escherichia coli: questions. Annu. Rev. Genet., 35, 53–82. [DOI] [PubMed] [Google Scholar]
- Cox M.M., Goodman,M.F., Kreuzer,K.N., Sherratt,D.J., Sandler,S.J. and Marians,K.J. (2000) The importance of repairing stalled replication forks. Nature, 404, 37–41. [DOI] [PubMed] [Google Scholar]
- Craig N.L. and Roberts,J.W. (1981) Function of nucleoside triphosphate and polynucleotide in Escherichia coli recA protein-directed cleavage of phage λ repressor. J. Biol. Chem., 256, 8039–8044. [PubMed] [Google Scholar]
- Dutreix M., Rao,B.J. and Radding,C.M. (1991) The effects on strand exchange of 5′ versus 3′ ends of single-stranded DNA in RecA nucleoprotein filaments. J. Mol. Biol., 219, 645–654. [DOI] [PubMed] [Google Scholar]
- Garger S.J., Griffith,O.M. and Grill,L.K. (1983) Rapid purification of plasmid DNA by a single centrifugation in a two-step cesium chloride–ethidium bromide gradient. Biochem. Biophys. Res. Commun., 117, 835–842. [DOI] [PubMed] [Google Scholar]
- Horii Z. and Clark,A.J. (1973) Genetic analysis of the RecF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J. Mol. Biol., 80, 327–344. [DOI] [PubMed] [Google Scholar]
- Inman R.B. and Schnös,M. (1970) Partial denaturation of thymine- and 5-bromouracil-containing λ DNA in alkali. J. Mol. Biol., 49, 93–98. [DOI] [PubMed] [Google Scholar]
- Kolodner R., Fishel,R.A. and Howard,M. (1985) Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J. Bacteriol., 163, 1060–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzminov A. (1999) Recombinational repair of DNA damage in Escherichia coli and bacteriophage λ. Microbiol. Mol. Biol. Rev., 63, 751–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley J.E. and Cox,M.M. (1990a) Assembly and disassembly of RecA protein filaments occurs at opposite filament ends: relationship to DNA strand exchange. J. Biol. Chem., 265, 9043–9054. [PubMed] [Google Scholar]
- Lindsley J.E. and Cox,M.M. (1990b) On RecA protein-mediated homologous alignment of 2 DNA molecules—3 strands versus 4 strands. J. Biol. Chem., 265, 10164–10171. [PubMed] [Google Scholar]
- Lohman T.M. and Overman,L.B. (1985) Two binding modes in Escherichia coli single strand binding protein–single stranded DNA complexes. Modulation by NaCl concentration. J. Biol. Chem., 260, 3594–3603. [PubMed] [Google Scholar]
- Lohman T.M., Green,J.M. and Beyer,R.S. (1986) Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under λ PL control. Biochemistry, 25, 21–25. [DOI] [PubMed] [Google Scholar]
- Lovett S.T., Luisi,D.C. and Kolodner,R.D. (1988) The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics, 120, 37–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi-DeLuca C. (1995) Homologous pairing of single-stranded DNA and superhelical double-stranded DNA catalyzed by RecO protein from Escherichia coli. J. Bacteriol., 177, 566–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luisi-DeLuca C. and Kolodner,R. (1994) Purification and characterization of the Escherichia coli RecO protein. Renaturation of complementary single-stranded DNA molecules catalyzed by the RecO protein. J. Mol. Biol., 236, 124–138. [DOI] [PubMed] [Google Scholar]
- Mahdi A.A. and Lloyd,R.G. (1989) Identification of the recR locus of Escherichia coli K-12 and analysis of its role in recombination and DNA repair. Mol. Gen. Genet., 216, 503–510. [DOI] [PubMed] [Google Scholar]
- Marians K.J. (2000) PriA-directed replication fork restart in Escherichia coli. Trends Biochem. Sci., 25, 185–189. [DOI] [PubMed] [Google Scholar]
- McGlynn P. and Lloyd,R.G. (2000) Modulation of RNA polymerase by (p)ppGpp reveals a RecG-dependent mechanism for replication fork progression. Cell, 101, 35–45. [DOI] [PubMed] [Google Scholar]
- McIlwraith M.J. and West,S.C. (2001) The efficiency of strand invasion by Escherichia coli RecA is dependent upon the length and polarity of ssDNA tails. J. Mol. Biol., 305, 23–31. [DOI] [PubMed] [Google Scholar]
- Michel B. (2000) Replication fork arrest and DNA recombination. Trends Biochem. Sci., 25, 173–178. [DOI] [PubMed] [Google Scholar]
- Morrical S.W., Lee,J. and Cox,M.M. (1986) Continuous association of Escherichia coli single-stranded DNA binding protein with stable complexes of RecA protein and single-stranded DNA. Biochemistry, 25, 1482–1494. [DOI] [PubMed] [Google Scholar]
- Neuendorf S.K. and Cox,M.M. (1986) Exchange of RecA protein between adjacent RecA protein–single-stranded DNA complexes. J. Biol. Chem., 261, 8276–8282. [PubMed] [Google Scholar]
- New J.H., Sugiyama,T., Zaitseva,E. and Kowalczykowski,S.C. (1998) Rad52 protein stimulates DNA strand exchange by Rad51 and replication protein A. Nature, 391, 407–410. [DOI] [PubMed] [Google Scholar]
- Razavy H., Szigety,S.K. and Rosenberg,S.M. (1996) Evidence for both 3′ and 5′ single-strand DNA ends in intermediates in Chi-stimulated recombination in vivo. Genetics, 142, 333–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Register J.C. III, and Griffith,J. (1985) The direction of RecA protein assembly onto single strand DNA is the same as the direction of strand assimilation during strand exchange. J. Biol. Chem., 260, 12308–12312. [PubMed] [Google Scholar]
- Rosenberg S.M. and Hastings,P.J. (1991) The split-end model for homologous recombination at double-strand breaks and at Chi. Biochimie, 73, 385–397. [DOI] [PubMed] [Google Scholar]
- Schutte B.C. and Cox,M.M. (1988) Homology-dependent underwinding of duplex DNA in RecA protein generated paranemic complexes. Biochemistry, 27, 7886–7894. [DOI] [PubMed] [Google Scholar]
- Shan Q., Cox,M.M. and Inman,R.B. (1996) DNA strand exchange promoted by RecA K72R. Two reaction phases with different Mg2+ requirements. J. Biol. Chem., 271, 5712–5724. [DOI] [PubMed] [Google Scholar]
- Shan Q., Bork,J.M., Webb,B.L., Inman,R.B. and Cox,M.M. (1997) RecA protein filaments: end-dependent dissociation from ssDNA and stabilization by RecO and RecR proteins. J. Mol. Biol., 265, 519–540. [DOI] [PubMed] [Google Scholar]
- Shaner S.L. and Radding,C.M. (1987) Translocation of Escherichia coli RecA protein from a single-stranded tail to contiguous duplex DNA. J. Biol. Chem., 262, 9211–9219. [PubMed] [Google Scholar]
- Shinohara A. and Ogawa,T. (1998) Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature, 391, 404–407. [DOI] [PubMed] [Google Scholar]
- Smith G.R. (1989) Homologous recombination in E.coli: multiple pathways for multiple reasons. Cell, 58, 807–809. [DOI] [PubMed] [Google Scholar]
- Sung P. (1997a) Function of yeast Rad52 protein as a mediator between replication protein A and the Rad51 recombinase. J. Biol. Chem., 272, 28194–28197. [DOI] [PubMed] [Google Scholar]
- Sung P. (1997b) Yeast Rad55 and Rad57 proteins form a heterodimer that functions with replication protein A to promote DNA strand exchange by Rad51 recombinase. Genes Dev., 11, 1111–1121. [DOI] [PubMed] [Google Scholar]
- Taylor A.F. and Smith,G.R. (1985) Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J. Mol. Biol., 185, 431–443. [DOI] [PubMed] [Google Scholar]
- Templin A., Kushner,S.R. and Clark,A.J. (1972) Genetic analysis of mutations indirectly suppressing recB and recC mutations. Genetics, 72, 105–115. [PMC free article] [PubMed] [Google Scholar]
- Umezu K. and Kolodner,R.D. (1994) Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem., 269, 30005–30013. [PubMed] [Google Scholar]
- Umezu K., Chi,N.W. and Kolodner,R.D. (1993) Biochemical interaction of the Escherichia coli RecF, RecO and RecR proteins with RecA protein and single-stranded DNA binding protein. Proc. Natl Acad. Sci. USA, 90, 3875–3879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B.L., Cox,M.M. and Inman,R.B. (1995) An interaction between the Escherichia coli RecF and RecR proteins dependent on ATP and double-stranded DNA. J. Biol. Chem., 270, 31397–31404. [DOI] [PubMed] [Google Scholar]
- Webb B.L., Cox,M.M. and Inman,R.B. (1997) Recombinational DNA repair—the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell, 91, 347–356. [DOI] [PubMed] [Google Scholar]
- Webb B.L., Cox,M.M. and Inman,R.B. (1999) ATP hydrolysis and DNA binding by the Escherichia coli RecF protein. J. Biol. Chem., 274, 15367–15374. [DOI] [PubMed] [Google Scholar]
- West S.C., Cassuto,E. and Howard-Flanders,P. (1981) Mechanism of E.coli RecA protein directed strand exchanges in post-replication repair of DNA. Nature, 294, 659–662. [DOI] [PubMed] [Google Scholar]
- Whitby M.C., Ryder,L. and Lloyd,R.G. (1993) Reverse branch migration of Holliday junctions by RecG protein: a new mechanism for resolution of intermediates in recombination and DNA repair. Cell, 75, 341–350. [DOI] [PubMed] [Google Scholar]