Abstract
The bluetongue virus core is a molecular machine that simultaneously and repeatedly transcribes mRNA from 10 segments of viral double-stranded RNA, packaged in a liquid crystalline array. To determine how the logistical problems of transcription within a sealed shell are solved, core crystals were soaked with various ligands and analysed by X-ray crystallography. Mg2+ ions produce a slight expansion of the capsid around the 5-fold axes. Oligonucleotide soaks demonstrate that the 5-fold pore, opened up by this expansion, is the exit site for mRNA, whilst nucleotide soaks pinpoint a separate binding site that appears to be a selective channel for the entry and exit of substrates and by-products. Finally, nucleotides also bind to the outer core layer, providing a substrate sink.
Keywords: bluetongue virus/Reoviridae/transcription/virus structure/X-ray crystallography
Introduction
Bluetongue is an infectious disease of ungulate mammals, caused by bluetongue virus (BTV) and transmitted by a number of species of blood-feeding Culicoides insect vectors (Mellor, 1990). BTV is currently causing disease in southern Europe and the Mediterranean region (in Turkey, Greece, Italy, Tunisia, Algeria and Spain), with vaccination programmes in implementation and many animals being slaughtered in attempts to control the spread of the disease.
BTV is the type species (one of 20 different species) of the genus Orbivirus, within the family Reoviridae (Mertens, 2000). The orbiviruses characteristically have a 10-segmented double-stranded RNA (dsRNA) genome contained within a double-shelled icosahedral protein capsid. The outer shell, made up of viral proteins VP2 and VP5, is lost upon cell entry, releasing transcriptionally active core particles into the cytoplasm. Further disassembly of cores is not thought to be a functional part of the replication process. BTV cores consist of two protein layers. The surface is made up of 780 copies of VP7(T13), which completely encloses the innermost subcore layer of the capsid. The subcore itself is composed of 120 copies of VP3(T2), forming a relatively thin layer of protein that encloses the 10 dsRNA genome segments (Grimes et al., 1998) (Figure 1). The subcore also contains transcriptase complexes (TCs) thought to be situated on the inner surface of the VP3(T2) shell, at the vertices of the icosahedron (Gouet et al., 1999), and by analogy with cypoviruses, each is thought likely to be associated with a particular genome segment (Yazaki and Miura, 1980).
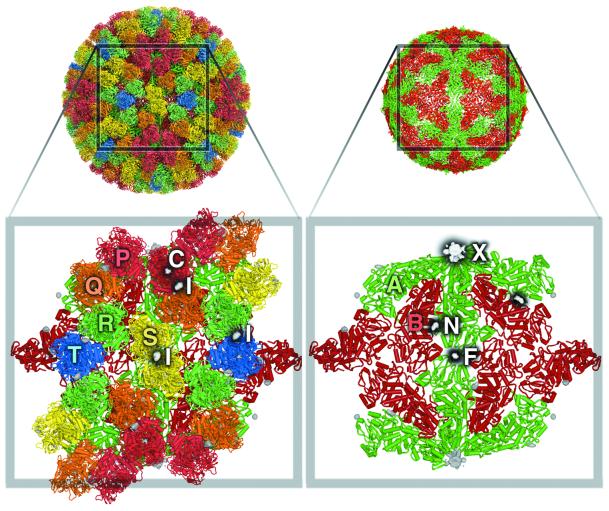
Fig. 1. Binding sites on the core particle of BTV-1. The left panel shows key features in the VP7(T13) layer. The close-up reveals some of the 260 VP7(T13) trimers that form the core surface, with the VP3(T2) layer beneath. The five VP7 trimers that form the icosahedral asymmetric unit are marked P, Q, R, S and T, coloured red, orange, green, yellow and blue, respectively. Trimer T sits on the icosahedral 3-fold axis and thus contributes a monomer to the unique portion. Difference electron density is drawn in white, with black shadows. NTPs bind at the inter-trimer site I, located on the local 2-fold dyad between trimers S–S′, P–Q and R–T. Cations bind along the 3-fold axes of all VP7(T13) trimers (C sites). For clarity, site C is drawn only on the P trimer (it is present on all trimers) and marks the putative Na+ ion binding site. Helices are shown as rods and β-strands as arrows. The right panel shows binding sites on the VP3(T2) scaffold. The icosahedrally unique molecules A and B are coloured in green and red, respectively. The difference electron density for three binding sites is drawn: mRNA leaves the particle at site X, nucleotides are thought to traverse the particle via the pore marked N and density due to a phosphate is marked F.
Upon infection, the intact core particle carries the viral genome and TCs into the host cell, thus preventing the dsRNA from making contact with the cytoplasm and activating host defence mechanisms. In the cytoplasm, the core transcribes and caps viral mRNAs from each of the 10 genome segment templates, which are thought to be packaged as liquid crystalline bundles within its centre (Gouet et al., 1999). The TCs are composed of three minor structural proteins: VP1(Pol), identified as the polymerase (Roy et al., 1988; Urakawa et al., 1989); VP4(Cap), the capping enzyme (Mertens et al., 1992); and VP6(Hel), the helicase (Stauber et al., 1997). During transcription, the polymerase, which requires Mg2+ for activity, takes the negative sense (–ve) RNA template strand and uses nucleo side triphosphate (NTP) substrates to synthesize a positive sense (+ve) daughter strand, producing pyrophosphate (PPi) as by-product. This daughter strand is capped by VP4(Cap), which has the guanylyltransferase, nucleotide phosphohydrolase and two transmethylase activities required for synthesis of Cap 1 structures (Le Blois et al., 1992; Mertens et al., 1992; Martinez-Costas et al., 1998; Ramadevi and Roy, 1998; Ramadevi et al., 1998). It is thought that each dsRNA genome segment is in some way attached to a TC and must move through the polymerase active site during transcription. This imposes topological requirements; not only must the parental dsRNA segment be unwound prior to transcription (to allow the –ve sense template strand to enter the polymerase active site), but since the BTV transcriptase is fully conservative, the resulting parental–daughter strand duplex must also be separated, the daughter strand exported from the core particle and the parental strands of the dsRNA genome segment re-annealed. It is thought that VP6(Hel) is involved in one or both of these activities, using the energy released on ATP hydrolysis to ADP and phosphate (Pi) to separate the strands of dsRNA. The mRNAs produced during transcription are exported from the particle, probably through pores in the VP3(T2) subcore shell at the 5-fold axes (Lawton et al., 1997; Grimes et al., 1998; Gouet et al., 1999).
The core faces several serious logistical problems during transcription: it must provide entry routes and mechanisms to continuously feed the vital NTP and AdoMet substrates to the internal enzymes, maintaining appropriate levels of certain metal ions whilst allowing the reaction by-products to escape and simultaneously propelling the 10 nascent mRNA molecules into the infected cell. At the same time, the dsRNA must be secluded from the cytoplasm of the infected cell. The viral core achieves this seclusion mechanically, by constructing a shell, which appears almost hermetically sealed (Grimes et al., 1998). Here we use X-ray crystallography to address the problem of how a core can exist as a compartment within the infected cell, translocating substrate and product to and from the cytoplasm whilst containing and organizing the gene segments.
Results
Diffraction data collected from BTV-1 core crystals that had been soaked with different molecules of interest were used to calculate a number of electron density maps. The molecules included several nucleoside mono-, di- and triphosphates, DNA and RNA oligonucleotides, and various ions. The exquisite sensitivity of maps calculated using differences between these data sets and relevant reference data sets, enhanced by difference attenuation (see Materials and methods) and 30-fold real space averaging, has allowed us to identify binding at several distinct sites around the core particle as well as conformational changes in the core.
Acquiring appropriate reference data sets involved developing a protocol for the stepwise transfer of crystals into two new reference buffers [DMT: 9 mM MgCl2, 6 mM dithiothreitol (DTT), 0.1 M Tris–HCl pH 8.0; and DT: 6 mM DTT, 0.1 M Tris–HCl pH 8.0] without causing them to redissolve or fragment. The crystals in DMT retained enzyme activity, a prerequisite for analysis of biologically relevant complexes (Mertens et al., 1987). However, these conditions are not ideal for maintaining the higher resolution ordering of the crystals, and the diffraction data obtained were weaker and did not extend to as high a resolution as data from crystals maintained under native conditions (Grimes et al., 1998). As with native crystals, we were unable to obtain data from frozen crystals and in general were able to collect only a single small angle diffraction image per crystal position.
A summary of the statistics for the 13 data sets described in this paper is given in Table I. These represent the key data sets, from over 30 collected, which provide the pivotal evidence for the different substrate/product binding sites described. For clarity and conciseness, the other data sets are not presented here; they represent parallel experiments and the results from them simply reinforce the conclusions drawn. From these experiments we have identified a number of different binding sites: some are on the outer surface of the core, some on the inner surface, and some identify entrance and exit pores in the structure. Figure 1 summarizes these results in a composite of the difference maps. Trivial names are assigned to the different sites in these figures and these will be used in the detailed analysis below. Throughout the paper, the notation for a 30-fold real space averaged difference map between a data set from a crystal soaked in molecule A and a reference data set R is |FA| – |FR|.
Table I. Data processing statistics and difference map calculations.
| Data set | No. of crystals | No. of images | Resolution range | Total reflections (in 000s) | Unique reflections (in 000s) | Completeness ofdata (%) | I/σ(I)(%) | Rsyma(%) | MFID versus Nativeb | No. of refs common with DMT (in 000s) | % completeness of common reflections | MFID versus DMT | Binding sitesc | Max peak in σd |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Native | 587 | 1045 | 105–3.5 | 25012 | 3524 | 58 | 2.5 | 23 | – | 2420 | 68 | 27 | – | 15 |
| DMT | 206 | 332 | 104–4.2 | 7401 | 2879 | 81 | 2.2 | 26 | 27 | – | – | – | – | – |
| CTP | 28 | 74 | 102–6.1 | 563 | 426 | 35 | 3.9 | 11 | 17 | 413 | 34 | 21 | XNI | 16 |
| ADP | 87 | 136 | 85–5.2 | 1563 | 871 | 46 | 4.4 | 11 | 19 | 829 | 43 | 14 | XNIC | 30 |
| GDP | 47 | 47 | 81–5.0 | 1292 | 951 | 46 | 3.3 | 19 | 23 | 856 | 41 | 14 | XNIC | 18 |
| AMP | 131 | 193 | 82–4.2 | 6316 | 2876 | 82 | 2.7 | 27 | 26 | 2384 | 67 | 17 | XIC | 14 |
| DNA | 36 | 34 | 69–5.0 | 917 | 739 | 36 | 2.5 | 22 | 25 | 665 | 32 | 16 | X | 22 |
| RNA | 31 | 30 | 74–5.0 | 835 | 671 | 32 | 2.8 | 20 | 25 | 605 | 29 | 16 | X | 23 |
| PPi | 29 | 25 | 80–6.3 | 240 | 211 | 20 | 3.6 | 12 | 28 | 205 | 20 | 16 | XC | 12 |
| Pi | 45 | 106 | 105–4.5 | 1297 | 902 | 31 | 2.5 | 18 | 27 | 137e | 5e | 19e | F | 9 |
| NaCl | 37 | 96 | 85–4.2 | 2397 | 1670 | 47 | 2.3 | 23 | 26 | 1423 | 41 | 17 | – | 8 |
| DT | 13 | 19 | 79–4.2 | 682 | 349 | 10 | 2.4 | 22 | 31 | 307 | 8 | 36 | – | 6 |
aRsym = ∑j∑h(|Ij,h – <Ih>)|/∑j∑h(<Ih>), where h are unique reflection indices, Ij,h are intensities of symmetry-related reflections and <Ih> is the mean intensity.
bMFID = (|FSoak| – |FBuffer|)/|FSoak|. The limits on the high- and low-resolution data in difference map calculations and data statistics were always imposed by the soak data.
cTrivial names for branching sites are given in Figure 1.
dStandard deviation (σ) is taken as the standard deviation of the averaged electron density within the averaging envelope.
eThe buffer used in the Pi soaks was DT rather than DMT, due to the insolubility of magnesium phosphate. Hence, all statistics shown for the difference map calculation were calculated against the DT dataset.
Effects of changing buffer
An 81% complete data set to 4.2 Å resolution was collected from 206 crystals washed in DMT buffer. Examination of an |FDMT| – |FNative| difference map showed the effect of washing the crystals from the original mother liquor into DMT buffer (Figure 2). Virtually all of the observed difference density can be explained by a slight expansion of the core, particularly around the icosahedral 5-fold axis. These observations are in keeping with the higher ‘flexibility’ observed previously around the 5-fold axis (Grimes et al., 1998). Further specific, localized changes will be discussed below. This experiment also demonstrates the importance of using data from the ‘correct’ reference condition in any such difference calculations, to ensure that the differences observed are due solely to ligand binding.
Fig. 2. Conformational changes on changing to DMT buffer. The 6 Å resolution electron density map for the native core is shown in grey. Positive (green) and negative (red) difference densities are shown for the |FDMT| – |FNative| difference electron density map (contoured at levels that are arbitrary but equal in magnitude). The largest changes are localized to the 5-fold axis (marked as a purple line). The close-up shows that the change represents a concerted rigid body radial expansion of the capsid; the helices move up locked together as rigid bodies, resulting in alternating layers of positive and negative difference density.
Binding at the icosahedral 5-fold axis (site X: exit)
A variety of reagents are seen to bind to a site in the 5-fold pore of the core particle when soaked into BTV-1 core crystals. The electron density difference map for a 10 mM AMP (sodium salt) soak (|FAMP| – |FDMT|) illustrates just how clearly ligand binding can be observed using this technique. Peaks of positive difference density are clearly visible in the pores formed at the 5-fold axes (Figure 3A). The absence of similar peaks in comparable NaCl difference map soaks (|FNa| – |FDMT|) confirms that it is AMP that has bound (map not shown). Note that the peak is far too extensive to be accounted for by a single molecule of AMP (Figure 3B). The pore at the 5-fold axis is surrounded by the apical domains of five VP3(T2)A molecules, each of which contributes three arginines (R317, R413 and R431), asparagine (N411) and threonine (T412) to the wall of the pore. This strongly polar and rather basic environment suggests that the AMP molecules that form this density peak are oriented with the phosphate moiety closest to the wall of the pore. This places the nucleotide bases centrally along the 5-fold axis and suggests that base stacking may be important in stabilizing the observed AMP binding. Additional densities extend from the sides of the main AMP density, in the region of R413, which appears to be due to an ordering of this loop of the protein induced by AMP binding.
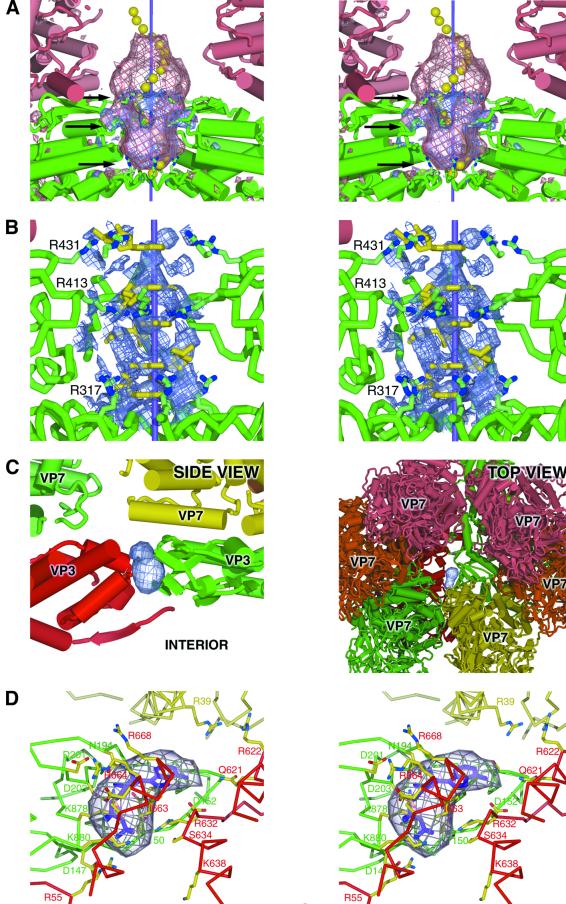
Fig. 3. Nucleotide and oligonucleotide binding in the VP3(T2) layer. (A) The exit site for mRNA (site X). The stereo figure shows positive difference density for both the AMP soak (blue, arbitrary contour level) and the 20 base DNA oligomer soak (pink, arbitrary contour level). Molecule VP3(T2)A is shown in green, VP7(T13)P in pink and the icosahedral 5-fold axis is represented by a purple line. Three arginines, implicated in interacting with the mRNA, are shown (indicated by black arrows). Superimposed is a grossly simplified representation of an ssRNA oligomer, in the A form conformation, with 20 yellow spheres marking the centre of masses of the nucleotides. The ‘ears’ of density in both difference maps (close to the central black arrow) are due to a conformational change in the protein. (B) Stereo diagram of the 5-fold pore region, with five AMP molecules (yellow) modelled into the cyclically density-modified 2|FAMP| – |Fav| map (blue, arbitrary contour level, for clarity drawn only within 5 Å of the AMP molecules). Also shown are the rings of arginines from each of the five copies of VP3(T2)A lining the pore. (C) The pore through the VP3(T2) layer (site N). The left panel shows a side view of a slice through the capsid, showing parts of the VP3(T2) (A, bright green; B, red) and VP7(T13) (R, dull green; S, yellow) layers. Positive difference density from the |FADP| – |FDMT| map is shown in blue. The right panel shows the same difference density viewed from outside the particle, looking through the VP7(T13) layer towards the interior. (D) Stereo diagram of the pore (site N) through the VP3(T2) layer. An ADP molecule (shown in purple) has been fitted into the difference density. The side chains of key residues that interact with the nucleotide and are implicated in facilitating movement across the protein layer are shown, coloured according to the scheme in (C).
The AMP data set was sufficiently complete (82%) to be used alone in cyclic averaging and solvent flattening, resulting in a 2|FO_AMP| – |FAV_AMP| map with an R factor of 27.1% including data extending to 4.2 Å. This map shows the large density peak in the 5-fold pore in considerably greater detail (Figure 3B), allowing five AMP moieties to be modelled into distinct layers within this feature, supporting the base-stacking hypothesis. Almost identical difference densities were also seen on the icosahedral 5-fold axes in the soaks of GMP, NDPs and NTPs as well as NTP analogues. As a consequence of icosahedral averaging, the orientation of each base around the 5-fold axis cannot be uniquely defined. For this reason, no refinement of the modelled nucleotide positions has been attempted.
In addition to nucleoside mono-, di- and triphosphate soaking experiments, data were collected from crystals soaked in 20 base DNA and RNA oligonucleotides. The difference density for these soaks (|FDNA| – |FDMT| and |FRNA| – |FDMT|) (Figure 3A and map not shown) show a significant extension of the 5-fold density, compared with the AMP difference density, demonstrating that the oligomer also binds in the 5-fold pore of the BTV core. No oligonucleotide binding was seen elsewhere on the core particle. A model, based on A form single-stranded (ss) RNA, has been fitted to the density (Figure 3A). The difference density accounts for ∼15 of the 20 bases in the oligomer. It is possible that the oligomer can partially enter the core, where reduced icosahedral ordering would make it much harder to detect; however, the outermost portion of the oligonucleotide is also likely to be free of any constraints imposed by the narrow pore and could therefore be disordered.
Several small volumes of density, lining the edge of the pore at the 5-fold axis, were observed in the 3.5 Å averaged electron density map for the BTV core particle (Grimes et al., 1998). These densities probably represent ordered sulfate ions (present in high concentration in the mother liquor) interacting with the charged and polar side chains, thus mimicking the interaction with the phosphates of the AMP and oligonucleotides. Further evidence for the proposed model of ssRNA at the 5-fold axis comes from an experiment in which crystals were soaked in PPi. The resulting pattern of density at the icosahedral 5-fold axes was different to that observed in AMP soaks. Rather than filling the pore, two smaller volumes of density were detected, one at the top and one at the bottom, both of which match with the rings of arginine side chains lining the pore (Table I, map not shown). PPi is large enough to make contact with two of these 5-fold related side chains, providing a relatively good binding site, but compared with AMP cannot benefit from the base-stacking interactions of nucleotides within the pore itself.
A nucleotide binding pore (site N: entry)
Examination of an averaged difference density map between the DMT data set and the native data set (|FDMT| – |FNative|) revealed a region of negative difference density, located between the VP3(T2)A and B molecules, suggesting that a ligand had been washed out of the core during transfer of crystals to DMT. However, re-examination of the 3.5 Å averaged native map (Grimes et al., 1998) gave no clues as to its identity. This region is accessible both from inside and outside the core, suggesting a pore rather than a binding pocket. The equivalent positive density can be seen in several of the difference maps calculated between data gathered from nucleotide-soaked crystals and the DMT reference, e.g. in |FADP| – |FDMT| (Figure 3C) and |FGDP| – |FDMT| (Table I, map not shown). A slightly larger volume of difference density is seen in the |FCTP| – |FDMT| (Table I, map not shown), suggesting that NTPs can also bind in the pore, although it is possible that sufficient nucleoside diphosphate (NDP) for detectable binding could be generated by NTP hydrolysis, particularly as the BTV core possesses an NTP phosphohydrolase activity. In contrast, no density is seen in this region in the |FAMP| – |FDMT| difference map, demonstrating that the number of phosphates plays a significant role in accessing or binding within this pore. This suggests that loss of ordered sulfate ions could be responsible for the negative difference density seen in the |FDMT| – |FNative| map. However, the |FPi| – |FNative| map also shows the negative difference density, even though phosphate could also be expected to occupy sites accessible to sulfate. An alternative explanation is that the lack of available counter-ions, due to the absence of divalent cations in the native buffer, may trap endogenous NDP in the pore, such that it can be readily detected. NDP molecules would be free to leave the pore when the crystals are moved into a buffer containing Mg2+ (during transfer to DMT), and could be washed out of the crystal during this process.
The NDP/NTP binding pore is located approximately one-third of the distance from the 5-fold axis to the 2-fold axis, along the interface between the VP3(T2)A and B molecules (Figures 1B and 3C). It lies just under one corner of the VP7(T13)S trimer, near to the VP7(T13)R trimer, at a point where there are few close contacts between VP3(T2) and VP7(T13) (Grimes et al., 1998). This gives the pore a wide mouth, clearly accessible from the bulk solvent (Figure 3C). There are a number of arginine residues around the pore mouth; VP3(T2)B R668 is positioned such that it could attract a negative charge into the mouth, with VP3(T2)B R622 and VP7(T13)S R39 also contributing to the general positive charge around the entrance to the pore (Figure 3D). The entrance to the pore is marked by VP3(T2)B R664, with VP3(T2)A D201 available nearby for an ionic interaction. Hydrogen-bonding possibilities are presented by several residues at the entrance, whilst in the middle of the VP3(T2) layer the pore is lined by a series of mainly polar side chains. The pore exit, to the interior of the core, is defined by VP3(T2)A T150 (sequenced as alanine, but the electron density and sequence conservation suggest threonine) and K880, and by a series of charged and polar side chains (see Figure 3D).
External nucleotide binding sites (sites I: inter-trimer)
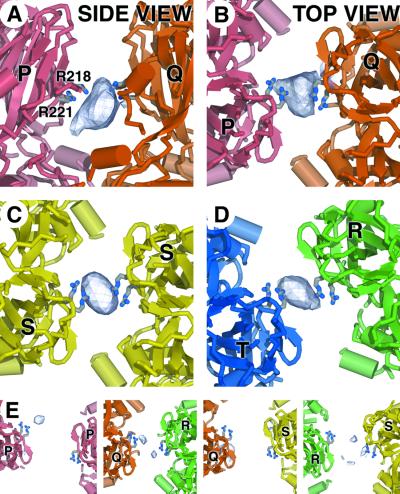
As well as the sites discussed above, several other volumes of positive density were repeatedly detected in |FNTP| – |FDMT| difference maps. These regions occur in the outer layer of the core, on the pseudo 2-fold axis between some (but not all) of the adjacent VP7(T13) trimers. The strength of the difference density appears to be sensitive to both the distance between adjacent trimers and the quality of the inter-trimer 2-fold axis, being greatest between trimers P and Q, situated close to the icosahedral 5-fold axis (Figure 4A and B). Similar, but weaker densities are also observed between trimers T and R, and between adjacent copies of trimer S, related by the icosahedral 2-fold axis of the particle itself (Figure 4C and D). This binding site is closely associated with two arginine residues from a single monomer of VP7(T13) (R218 and R221), which are located just above the neck region of each trimer.
Fig. 4. Nucleotide storage site between VP7(T13) trimers (site I). (A) Side view (tangential to the capsid surface) of the positive density (arbitrary contour level) from the |FCTP| – |FDMT| difference map, showing CTP bound between the upper domains of VP7(T13)P and Q trimers. The side chains of R211 and R218 are drawn. (B) A view orthogonal to that shown in (A), looking towards the centre of the particle along the approximate dyad axis relating trimers P and Q, showing the same electron density map. (C) A view down the icosahedral 2-fold axis of the same CTP difference density map as in (A) and (B). Density between the two VP7(T13)S trimers is shown. (D) A similar view to that shown in (B) and (C) of the CTP density between the VP7(T13)T and R trimers. (E) A panel of four views of the remaining pairs of trimers of VP7(T13), once again looking along the dyad axes, showing the relatively weak level of binding of NTPs. Note the substantial differences in relative separation of the nucleotide ligands.
A binding site for HPO42– and SO42– (site F: anion)
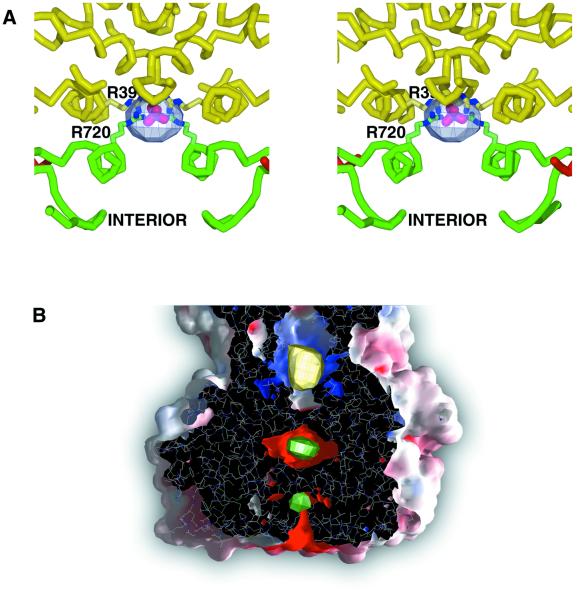
A further site of positive difference density is seen on the icosahedral 2-fold axis in the |FPi| – |FDT| map (Figure 5A; Table I). The site lies between the VP3(T2) layer and 2-fold related S trimers of the outer VP7(T13) layer, and is easily accessible from the interior of the core. Re-examination of the high-resolution native map (Grimes et al., 1998) revealed that this site is occupied in the native crystallization conditions. Residues R720 from VP3(T2)A and R39 from VP7(T13)S, from both of the 2-fold related copies, interact through a portion of density not occupied by protein. The nature of the site and the crystal liquor suggest that this density is due to SO42–. HPO42–, present in the Pi soak, presumably binds in a similar manner. The only contact between 2-fold related VP3(T2)A molecules is across this site, through the bound sulfate or phosphate molecules, suggesting that molecules at this site could play a significant structural role.
Fig. 5. Ion binding sites (sites F and C). (A) Stereo diagram showing phosphate binding on the icosahedral 2-fold axis between the VP3(T2) and VP7(T13) layers (site F). Positive difference density (blue, arbitrary contour level) is shown from the |FPi| – |FDT| map. The side chains of R720 from VP3(T2)A) and R39 from VP7(T13)S are also shown, and a phosphate ion (purple and red) is positioned in the density. (B) A transverse section through a GRASP (Nicholls et al., 1991) solvent-accessible surface coloured by electrostatic potential (blue, positive; red, negative) for a trimer of VP7(T13). Superimposed are the difference density maps for the putative Na+ ion (positive |FPPi| – |FDMT| difference density is shown in yellow) and the two Mg2+ ions (positive |FDMT| – |FNative| difference density is shown in green), which define the C sites.
Putative Na+ binding site on VP7(T13) (site C component)
Another region of positive difference density was detected in the neck region of VP7(T13), in a position internal to the VP7(T13) trimers, lying on the molecular 3-fold axis. This density, shown in the |FPPi| – |FDMT| difference map (Figure 5B), matches that of a structural zinc ion reported in the equivalent VP6(T13) structure from rotavirus (Mathieu et al., 2001), raising the possibility that a cation may also be bound in BTV. The cyclically averaged 2|FO_AMP| – |FAV_AMP| map shows binding in more than one position (map not shown). The side chains of residues R123 and N258 and the backbone carbonyl oxygen of residue P125 derived from each monomer (and therefore acting in triplicate) are candidate coordinating groups, possibly assisted by water molecules. This site is occupied when the crystals are soaked in Na+ nucleotides, but not with Tris+ or Li+ preparations, and may therefore represent a sodium ion binding site. Surprisingly, the density was absent in the |FNaCl| – |FDMT| maps, suggesting that the site itself may be dependent on a conformational change induced by nucleotide binding at the inter-trimer sites (site I). However the change required must be modest since tetrasodium pyrophosphate soaks, which only produce weak inter-trimer difference density, produce strong intra-trimer binding.
Binding sites for Mg2+ (site C components)
Direct analysis of Mg2+ binding should be possible by inspection of |FDMT| – |FDT| difference maps, where the only difference between the two buffers is 9 mM MgCl2. Unfortunately, the low completeness of overlap (8%) coupled with the high mean fractional isomorphous difference (MFID) (36%) results in a map that is too noisy to interpret reliably. However, Mg2+ is also the only additional component of DMT compared with the native buffer (which is distinguished from DT by possessing in addition 25% saturated ammonium sulfate and 25% ethylene glycol). Therefore, positive difference density peaks in the |FDMT| – |FNative| difference map can only arise from either conformational changes or the binding of Mg2+.
In addition to the pattern of difference density in |FDMT| – |FNative| maps due to capsid expansion, two sharp peaks of positive difference density were observed situated approximately on the local 3-fold axis of the VP7(T13) trimers (Figure 5B). The uppermost peak is some 15 Å below the putative Na+ ion site, in a pocket that is lined by three copies of the side chains of residues E108 and E266, along with the hydroxyl group of Y260. The second peak, situated midway between the upper site and the VP3(T2) shell, is in a smaller pocket and is coordinated to the side chains of E104, E261 and N283. The acidic nature of the ligands strongly suggests that Mg2+ is bound in both sites. Binding at these sites is likely to have a structural role, modulating the stability of VP7(T13)–VP3(T2) interactions.
Discussion
The BTV-1 core particle provides a beautifully concise model system for how a biological transcriptional machine solves a number of housekeeping issues, such as compartmentalization of its segmented dsRNA genome from the cellular defence mechanisms, localization and sequestration of substrates to and through a capsid barrier, with subsequent release of mRNA product and by-products. Crystal soaking experiments and the calculation of difference maps incorporating difference attenuation and 30-fold averaging have allowed us to obtain a clear picture of the capsid expansion, subsequent binding of both substrates and products to the BTV core, and have also revealed previously unrecognized binding sites for small ions (see Figure 6).
Fig. 6. Cartoon showing the movement of the NTP raw material into the core particle, and the site of egress of the product mRNA transcript and by-products PPi, Pi and NDPs. NTPs are sequestered at inter-trimer sites I. NTPs are thought to cross the protein barrier via the pore at site N. The product of transcription leaves via the pore at the 5-fold axis of the particle, site X. By-products leave via the pores at site X and site N.
On internalization within the cell cytoplasm, the core encounters an enviroment rich in Mg2+ and other ions. Mimicking this by transfer of BTV-1 core crystals into a buffer containing magnesium at approximately physiological concentration (in which it is known that the core particle is activated) results in an overall expansion of the capsid, particularly around the 5-fold axes. Since in native buffer conditions the cores are not active, this capsid expansion is very likely to have biological relevance. Indeed, since the cores are packed in a crystal lattice, held together by contacts between particles, this expansion may be greater in solution, allowing the ‘looser’ viral shell to become more permeable to nucleotide substrates and ions, as well as facilitating the release of mRNA and by-products.
The pore at the icosahedral 5-fold axis (site X) is the largest opening through the VP3(T2) subcore layer and therefore is the most likely exit site for nascent ssRNA from the transcribing particle (Grimes et al., 1998). Previous electron microscope studies had suggested that mRNA exited close to, but not at, the 5-fold axis of actively transcribing rotavirus particles (Lawton et al., 1997). Addition of nucleotides and oligonucleotides to the magnesium-rich enzyme buffer has allowed us to demonstrate that the pore on the 5-fold icosahedral axis is the site of exit of mRNA in BTV core particles, and rules out the suggestion of egress at a site displaced from the 5-fold axis. We can also pinpoint key amino acids involved in interacting with, and directing, mRNA movement. The pore is formed by the apical domains of VP3(T2) and has a distinctly polar environment. PPi soaks and the previous ‘native’ map show that both phosphate and sulfate groups interact with the set of three arginine residues that line the pore. Given that the size constraints of the pore will prevent the formation of RNA secondary structure, these observations, along with the plugs of positive electron density seen with nucleotide and oligonucleotide soaks, suggest that the major conformational determinants imposed on the RNA are the connectivity of the RNA backbone itself, base stacking and the interaction of the backbone phosphates with the amino acid side chains that line the pore. AMP effectively mimics both the stacking and phosphate interactions, albeit without the constraint of the backbone connectivity. The model derived from the interpretation of the cyclically averaged 2|FAMP| – |Fav| map is, therefore, likely to be a reasonable representation of newly transcribed ssRNA during exit from the core.
BTV core particles can catalyse the addition of Cap 1 structures to exogenous ssRNA (Mertens et al., 1992). This suggests that either the 5′ terminus of the ssRNA can enter the core or that VP4(Cap) is present on the external surface of the core, yet remains an integral part of the core structure. Since there is no evidence of extra protein density on the core surface and the oligonucleotide difference maps show that the ends of the oligomers come close to penetrating into the internal cavity of the core, we suggest that the active site of VP4(Cap) is located beneath and relatively close to the 5-fold pore (however, given the icosahedral averaging imposed upon the |FDNA| – |FDMT| density, we cannot identify the polarity of the bound oligonucleotide). The model for the egress of the ssRNA is based on the topological constraint of the linear phosphate–sugar backbone, stabilized base-stacking interactions, and coordination by the set of aliphatic but positively charged arginine side chains, which provide favourable but non-sequence-specific interactions. This explains how the core particle is able to ensure that transcription does not stall due to the build-up of significant tertiary structure at the 5′ end of the mRNA, which would physically prevent its exit from the core.
In members of the Reoviridae, transcription of ssRNA occurs simultaneously from each genome segment (Gillies et al., 1971; Yazaki and Miura, 1980; Lawton et al., 1997). Since the 12 pores at the 5-fold axes are therefore likely to become blocked by emerging ssRNA strands, particularly in those viruses that contain 12 genome segments (Mertens, 2000), another route is presumably necessary for the entry of substrates (NTPs and S-adenosyl-methionine) and the release of reaction by-products (Pi, PPi, S-adenosyl-homocysteine and ADP). The pore between the A and B molecules of VP3(T2) (site N), identified in NDP and NTP soaks, serves this role. As a result of ionic interactions between the arginines/lysines and aspartates, this pore carries little overall charge. However, the amphipathic nature of arginine and lysine side chains is such that they might easily re-orient to present either a hydrophobic or charged face to the pore. This would give the pore the ability to accept both the negatively charged phosphate and the hydrophobic base of a nucleotide, through small changes in side chain conformation as the nucleotide passes. VP3(T2)B I663 presents a small hydrophobic patch approximately halfway through, and there are various hydrogen-bonding possibilities (Figure 3D). The negatively charged VP3(T2)A E157 may lower the binding affinity of the pore for negatively charged nucleotides, thereby increasing the rate at which they can pass through. This description of a pore that allows the internalization of nucleotide provides, for the first time, a molecular explanation of how a sealed compartment such as the BTV core might selectively import substrate molecules required for mRNA synthesis.
In addition to the sites of exit of mRNA at the 5-fold pore and possible entry/exit pore for substrates and products, a variety of nucleotide soaks have revealed a large number of further binding sites in the VP7(T13) layer (the I sites). These occur between monomers of VP7(T13) that are related by local 2-fold axes between adjacent trimers, and are defined by four arginine residues, two from each monomer. Not all local 2-folds have equivalent occupancies, since binding appears to be sensitive to the distance between trimers and deviations from exact 2-fold symmetry. Since nucleotides bind across a 2-fold axis, the symmetry of the binding site is not matched by the symmetry of the nucleotide. Once again, the particular physiochemical nature of flexible arginine side chains appears to be important. The delocalized positively charged nitrogens at the end of arginine side chains would allow the negative charge of the nucleoside phosphates to be balanced, whilst the hydrophobic nature of the aliphatic carbon side chain would facilitate binding of the nucleoside base. The binding of NTPs also appears to cause slight conformational changes in the capsid structure, possibly associated with particle/polymerase activation, and could be a factor in stabilizing the more open active comformation of the core. The presence of relatively large numbers of nucleotides on the particle surface may act as a substrate sink, increasing the local concentration and supply of nucleotides around the core particle, facilitating polymerase activity. This increase in substrate concentration by sequestration of NTP to the core surface allows the core particle to compete with other cellular activities for limited nucleotide resources.
These changes in capsid structure are also correlated with ion binding sites (the C sites). In the expanded capsid, two cation sites are observed at the base of the VP7(T13) trimers, which appear to be occupied by Mg2+ ions, and are likely to have a role in stabilizing the expanded state of the core structure. A further ion site has been observed in a position that matches that of a Zn2+ ion in the corresponding protein in rotavirus VP6, suggesting the binding of a cation in BTV. This may be a Na+ ion, which binds on a further subtle capsid expansion following nucleotide sequestration. A general anionic binding site has also been observed between the VP3(T2) and VP7(T13) layers on the icosahedral 2-fold axis (the F site), and plays an important role in bridging the interaction between VP3(T2)As. This site can be occupied by either phosphate or sulfate ions. If bound phosphate molecules can exit this site towards the outside of the particle, it may also act as a pore, specifically allowing product phosphate to leave the interior of the core.
In summary, the BTV core particle is a model system for how a subcellular organelle orchestrates its structural and functional components and biochemical activities. These studies have shed light on the logistics of the supply of raw materials and the removal of products and by-products from the transcription factory, which is a dsRNA virus core. Mg2+ ions are not only essential for polymerization, but cause structural changes on binding that result in an expansion of the capsid structure. Further results have enabled the delineation of nucleotide and oligonucleotide binding sites, which allow us to put together a picture of how core particles sequester substrates, the modes of entry and exit of substrates and products, and the structural role of ions, such that efficient and repetitive transcription can occur. The substrate pore, which permits the selective transport of nucleotides into the core centre, enables the virus to replenish the raw materials required by the transcription complex for synthesis of capped mRNA. In addition, the results presented here suggest that a series of other subtle interactions probably favour the transition to the expanded state; a state in which the selective transfer of substrates and products into and out of the core compartment would be facilitated.
Materials and methods
Crystal preparation
Crystals of BTV-1 cores were grown under ‘native’ conditions (Burroughs et al., 1995) using the sitting drop method. For crystal mounting, crystals were washed into ‘native’ mother liquor composed of 25% saturated ammonium sulfate and 25% ethylene glycol in 0.1 M Tris–HCl pH 8.0. The crystals were transferred into the appropriate reference buffer (usually DMT, see Results) by serial addition to and subtraction from the drop volume, in several steps over a number of hours. The final transfer into the ligand solutions (described below) was then made as a single step and the crystals were left to equilibrate for at least 1 h before mounting. The crystals were mounted in quartz capillaries and sealed prior to removal from the secure laboratories at the Institute for Animal Health, Pirbright (in compliance with DEFRA disease security regulations).
Soaking experiments
A variety of possible ligands were examined, as shown in Table I. CTP [selected as the least likely NTP to be hydrolysed in the crystal (Stauber et al., 1997; Ramadevi and Roy, 1998)] and the NTP analogues AMPPNP and GMPPNP (data not shown) were all soaked into crystals individually. Binding of ssRNA and ssDNA was examined using soaks of 20 base RNA and DNA oligomers representing the first 20 bases of BTV-1 ssRNA from genome segment 7 (sequence GUUAAAAAUCUAUAGAGAUG) and its DNA equivalent [DDBJ/EMBL/GenBank accession No. X53740 (Wade-Evans, 1990)]. AMP, ADP, GDP, PPi and Pi (Ramadevi and Roy, 1998) were all soaked into crystals. NaCl and LiCl soaks (data not shown) were also performed because sodium and lithium salts of the nucleotides were used. Most of the ligands were soaked into crystals at a concentration of 10 mM in DMT, with the exception of the RNA oligomer (250 µM) and the DNA oligomer (180 µM). DT (see Results) was used as the reference buffer for the Pi soaks since Mg3(PO4)2 precipitate formed in DMT.
Data collection and processing
X-ray diffraction data were collected on stations ID2 and ID14 EH1, EH3 and EH4 at the European Synchrotron Radiation Facility (ESRF), France, using a MAResearch 345 image plate, MAResearch CCD detectors (135 and 165 mm diameters) and an ADSC Quantum4 2×2 CCD detector, respectively, with X-ray wavelengths in the range 0.92–1.00 Å. The oscillation range varied between 0.3 and 0.4°. Exposure times varied between 3 and 30 s. The crystal-to-detector distance was generally set to give a resolution of ∼5 Å at the edge of the detector, although some data were collected to 4.2 Å Bragg spacing. Crystals were mounted and transferred to the ESRF within 24 h to minimize any problems of crystal instability. The temperature at the crystal during the diffraction experiment was ∼8°C.
Data were indexed and scaled using the package HKL (Otwinowski and Minor, 1997). The resolution of each image was dynamically determined using the program TRIM-DENZO (D.I.Stuart, unpublished). After an initial round of scaling, partially recorded reflections more than 70% measured were scaled up using the program POST (D.I.Stuart, unpublished). A final round of scaling gave a set of merged intensities. Implausibly strong reflections were removed (program TIDY; D.I.Stuart, unpublished) and structure factor amplitudes calculated (program TRUNCATE; CCP4, 1994). The unit cells were all set to the native cell (a = 795.6 Å, b = 821.8 Å, c = 753.3 Å, α = β = γ = 90°) (Grimes et al., 1998); observed values differed by less than half the minimum Bragg spacing of the data. Data processing statistics are summarized in Table I.
Map calculation
The ligand soaks were evaluated by calculation of an averaged difference map. In each case, the map was calculated as the difference between the observed structure factor amplitudes of the ligand soak (|FSoak|) and those of the buffer alone (|FBuffer|).
A set of structure factor amplitudes was calculated from the atomic model of the core particle (|Fcalc|) (Grimes et al., 1998) using the package CNS (Brunger et al., 1998). The |FBuffer| were scaled to these |Fcalc| by application of a scale factor and B factor to give a sharpened dataset (program SCALEIT; CCP4, 1994). The |FSoak| were then scaled in the same way to the sharpened |FBuffer|.
Final scaling of |FSoak| to |FBuffer| used SHELLSCALE (D.I.Stuart, unpublished), which also performed difference attenuation to reduce noise levels. Pairs of reflections with a large combined standard error relative to some fraction of their mean amplitude were both significantly adjusted toward their mean value, whereas pairs of reflections with a small combined standard error were altered only slightly. The error due to the incorrect assumption of phase relationships inherent in difference Fourier calculations was allowed for by the incorporation of a basal error term σbasal, calculated as a fraction of the mean |F| (0.25 gave satisfactory results). Thus, for a pair of observed reflections |F1|,σ1 and |F2|,σ2:
|F|mean = (|F1| + |F2|)/2 and Δ|F| = |F1| – |F2|
σobserved = √(σ12 + σ22) and σbasal = |F|mean × 0.25
σtotal = √(σbasal2 + σobserved2)
weight = (σbasal/σtotal)2
Δ|F|attenuated = Δ|F| × weight
|F1|attenuated = |F|mean + (Δ|F|attenuated/2)
|F2|attenuated = |F|mean – (Δ|F|attenuated/2)
Finally, Rayment weights (Rayment, 1983) were applied to account for the variation in the reliability of the phase estimates.
An |FSoak| – |FBuffer| map was calculated using no resolution limits (actual resolution ranges and completeness are given in Table I). Phases were taken from the most recent round of cyclic averaging of the native dataset (J.M.Diprose, unpublished results). The difference map was then 30-fold averaged in the program GAP (D.I.Stuart, J.M.Grimes, J.M.Diprose, unpublished) (the NCS operators used are essentially those given in the Protein Data Bank, entry code 2BTV). A sphere enclosing the entire core particle was used as the averaging envelope. Maps were examined using O (Jones et al., 1991). The height of the interpreted peaks in each of the averaged difference maps is given in standard deviations (σ) in Table I; these are usually well above 10 σ. For comparison, the highest uninterpreted feature in the difference maps is usually between 6 and 8 σ.
In addition to the difference maps, both the native conditions and the AMP soak gave data of sufficient quality for cyclic real space refinement. A set of structure factor amplitudes (|Fcalc|) complete to 3.5 Å was calculated from the atomic model of the core (Grimes et al., 1998) in CNS (Brunger et al., 1998). |Fobs| were scaled to these by application of a scale factor and a negative isotropic B factor (program ISOSCALE; D.I.Stuart, J.M.Grimes, J.M.Diprose, unpublished) to give a set of sharpened |Fobs|. A final scaling in shells was performed (mainly to take out solvent effects) and Rayment weighting applied (program SHELLSCALE; D.I.Stuart, unpublished). Phases were derived from the atomic model. In order to minimize the known failings of the atomic model, the low-resolution limit was set to 25 Å and a 2|Fobs| – |Fcalc| map calculated. This map was then averaged in the program GAP (D.I.Stuart, J.M.Grimes, J.M.Diprose, unpublished). The solvent region was flattened to its average value, the protein region 30-fold averaged, and the disordered interior averaged and smoothed using a Gaussian function (Gouet et al., 1999). The resultant map was back-transformed (program MAPTOSF; R.Bryan, unpublished) to give a new set of structure factors, Fav. A starting 2|Fobs| – |Fav| map was calculated using phases from the inverted density-modified map and again using only reflections at resolutions higher than 25 Å. After 10 cycles of averaging, |Fav| were included in place of unobserved |Fobs| to estimate missing data. After 15 cycles, low-resolution phase extension was initiated and reflections beyond the low-resolution limit of the observed data (105 Å) were modelled from Fcalc. Finally, unit weights were imposed. The averaging R factor (R = (||Fobs| – |Fav||/|Fobs|) and correlation coefficient (CC = ((<|Fobs|> – |Fobs|)(<|Fav|> – |Fav|))/[((<|Fobs|> – |Fobs|)2 – (<|Fav|> – |Fav|)2)]2) for the native data improved from R = 21.5%, CC = 86.0% to R = 18.8%, CC = 89.0%. For the AMP data, these statistics improved from R = 29.6%, CC = 72.6% to R = 27.1%, CC = 77.1%. The quality of the data appears to be the limiting factor. The final averaged map was examined in O (Jones et al., 1991).
Figures were drawn using Bobscript (Esnouf, 1999) and rendered using Raster-3D (Merritt and Bacon, 1997) unless stated otherwise.
Acknowledgments
Acknowledgements
We thank the ESRF and the EMBL outstation, Grenoble, for their assistance in data collection, in particular the staff of ID14; the disease security officers S.Williams, A.Meyer and D.Goodridge; R.Esnouf and J.Dong for computing; and R.Bryan for software. This work has been supported by the Biotechnology and Biological Sciences Research Council, Medical Research Council and European Community. J.M.G. was funded by the Royal Society and D.I.S. by the Medical Research Council. P.P.C.M. and J.N.B. are supported by the Department for Environment, Food and Rural Affairs.
References
- Brunger A.T. et al. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Burroughs J.N., Grimes,J.M., Mertens,P.P.C. and Stuart,D.I. (1995) Crystallization and preliminary X-ray analysis of the core particle of bluetongue virus. Virology, 210, 217–220. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1999) Further additions to Molscript version 1.4, including reading and contouring of electron density maps. Acta Crystallogr. D, 55, 938–940. [DOI] [PubMed] [Google Scholar]
- Gillies S., Bullivant,S. and Bellamy,A.R. (1971) Viral RNA poly merases: electron microscopy of reovirus reaction cores. Science, 174, 694–696. [DOI] [PubMed] [Google Scholar]
- Gouet P., Diprose,J.M., Grimes,J.M., Malby,R., Burroughs,J.N., Zientara,S., Stuart,D.I. and Mertens,P.P.C. (1999) The highly ordered double-stranded RNA genome of bluetongue virus revealed by crystallography. Cell, 97, 481–490. [DOI] [PubMed] [Google Scholar]
- Grimes J.M., Burroughs,J.N., Gouet,P., Diprose,J.M., Malby,R., Zientara,S., Mertens,P.P.C. and Stuart,D.I. (1998) The atomic structure of the bluetongue virus core. Nature, 395, 470–478. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,Y.J., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for building protein models in electron density and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Lawton J.A., Estes,M.K. and Prasad,B.V.V. (1997) Three-dimensional visualisation of mRNA release from actively transcribing rotavirus particles. Nature Struct. Biol., 4, 118–121. [DOI] [PubMed] [Google Scholar]
- Le Blois H., Mertens,P.P.C., French,T., Burroughs,J.N. and Roy,P. (1992) The expressed VP4 protein of bluetongue virus is the guanylyl transferase. Virology, 189, 757–761. [DOI] [PubMed] [Google Scholar]
- Martinez-Costas J., Sutton,G., Ramadevi,N. and Roy,P. (1998) Guanylyltransferase and RNA 5′-triphosphatase activities of the purified expressed VP4 protein of bluetongue virus. J. Mol. Biol., 280, 859–866. [DOI] [PubMed] [Google Scholar]
- Mathieu M., Petitpas,I., Navaza,J., Lepault,J., Kohli,E., Pothier,P., Prasad,B.V., Cohen,J. and Rey,F.A. (2001) Atomic structure of the major capsid protein of rotavirus: implications for the architecture of the virion. EMBO J., 20, 1485–1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor P.S. (1990) The replication of bluetongue virus in Culicoides vectors. Curr. Top. Microbiol. Immunol.,162, 143–161. [DOI] [PubMed]
- Merritt E.A. and Bacon,D.J. (1997) Raster3D: photorealistic molecular graphics. In Carter,J.W.,Jr and Sweet,R.M. (eds), Macromolecular Crystallography. Vol. 277. Academic Press, San Diego, CA, pp. 505–524. [DOI] [PubMed]
- Mertens P.P.C. (2000) Orbiviruses and coltiviruses—general features. In Webster,R.G. and Granoff,A. (eds), Encyclopedia of Virology. Academic Press, London, UK.
- Mertens P.P.C., Burroughs,J.N. and Anderson,J. (1987) Purification and properties of virus particles, infectious subviral particles and cores of bluetongue virus serotypes 1 and 4. Virology, 157, 375–386. [DOI] [PubMed] [Google Scholar]
- Mertens P.P.C., Burroughs,J.N., Wade-Evans,A.M., Le Blois,H., Oldfield,S., Basak,A., Loudon,P. and Roy,P. (1992) Analysis of guanyltransferase and transmethylase activities associated with bluetongue virus cores and recombinant baculovirus-expressed core-like particles. In Walton,T.E. and Osburn,B.I. (eds), Bluetongue, African Horse Sickness and Related Orbiviruses: Proceedings of the Second International Symposium. CRC Press, Boca Raton, FL, pp. 404–415.
- Nicholls A., Sharp,K.A. and Honig,B. (1991) Protein folding and association: insights from the interfacial and thermodynamic properties of hydrocarbons. Proteins, 11, 281–296. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. and Minor,W. (1997) Processing of X-ray diffraction data collected in oscillation mode. In Carter,J.W.,Jr and Sweet,R.M. (eds), Methods in Enzymology. Vol. 276. Academic Press, San Diego, CA, pp. 307–326. [DOI] [PubMed]
- Ramadevi N. and Roy,P. (1998) Bluetongue virus core protein VP4 has nucleoside triphosphate phosphohydrolase activity. J. Gen. Virol., 79, 2475–2480. [DOI] [PubMed] [Google Scholar]
- Ramadevi N., Burroughs,N.J., Mertens,P.P.C., Jones,I.M. and Roy,P. (1998) Capping and methylation of mRNA by purified recombinant VP4 protein of bluetongue virus. Proc. Natl Acad. Sci. USA, 95, 13537–13542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayment I. (1983) Molecular replacement method at low resolution: optimum strategy and intrinsic limitations as determined by calculations on icosahedral virus models. Acta Crystallogr. A, 39, 102–116. [Google Scholar]
- Roy P., Fukusho,A., Ritter,G.D. and Lyon,D. (1988) Evidence for genetic relationship between RNA and DNA viruses from the sequence homology of a putative polymerase gene of bluetongue virus with that of vaccinia virus: conservation of RNA polymerase genes from diverse species. Nucleic Acids Res., 16, 11759–11767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber N., Martinez-Costas,J., Sutton,G., Monastyrskaya,K. and Roy,P. (1997) Bluetongue virus VP6 protein binds ATP and exhibits an RNA-dependent ATPase function and a helicase activity that catalyze the unwinding of double-stranded RNA substrates. J. Virol., 71, 7220–7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urakawa T., Ritter,D.G. and Roy,P. (1989) Expression of largest RNA segment and synthesis of VP1 protein of bluetongue virus in insect cells by recombinant baculovirus: association of VP1 protein with RNA polymerase activity. Nucleic Acids Res., 17, 7395–7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade-Evans A.M. (1990) The complete nucleotide sequence of genome segment 7 of bluetongue virus, serotype 1 from South Africa. Nucleic Acids Res., 18, 4919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki K. and Miura,K. (1980) Relation of the structure of cytoplasmic polyhedrosis virus and the synthesis of its messenger RNA. Virology, 105, 467–479. [DOI] [PubMed] [Google Scholar]