Abstract
In mitochondria, the hydrolytic activity of ATP synthase is regulated by an inhibitor protein, IF1. Its binding to ATP synthase depends on pH, and below neutrality, IF1 is dimeric and forms a stable complex with the enzyme. At higher pH values, IF1 forms tetramers and is inactive. In the 2.2 Å structure of the bovine IF1 described here, the four monomers in the asymmetric unit are arranged as a dimer of dimers. Monomers form dimers via an antiparallel α-helical coiled coil in the C-terminal region. Dimers are associated into oligomers and form long fibres in the crystal lattice, via coiled-coil interactions in the N-terminal and inhibitory regions (residues 14–47). Therefore, tetramer formation masks the inhibitory region, preventing IF1 binding to ATP synthase.
Keywords: ATP hydrolysis/bovine inhibitor protein/coiled coil/F1Fo-ATPase/pH-dependent oligomerization
Introduction
The F1Fo-ATP synthase complex plays a central role in energy transformation in most living organisms. In mitochondria, the synthesis of ATP requires an electrochemical proton gradient across the inner membrane to drive protons back into the matrix through the membrane domain of the F1Fo-ATPase, releasing energy that is coupled to ATP synthesis. When a cell is deprived of oxygen, its electrochemical gradient collapses, and the enzyme switches from ATP synthesis to ATP hydrolysis. In mitochondria, this hydrolytic activity is regulated by the natural inhibitor protein, IF1. Under these conditions, glycolysis becomes the only source of cellular ATP. The high rate of glycolysis results in reduction of cytosolic pH (Rouslin, 1983, 1987), which is transmitted to the mitochondrial matrix (Rouslin and Broge, 1989), promoting inhibition of ATP hydrolysis by IF1 in order to preserve ATP. The binding of IF1 to ATP synthase depends on the pH value, and below neutrality its inhibitory capacity increases (Panchenko and Vinogradov, 1985).
Bovine IF1 is a basic protein 84 amino acids in length (Pullman and Monroy, 1963), and homologues have been characterized in mitochondria from rats (Cintron and Pedersen, 1979), Saccharomyces cerevisiae (Hashimoto et al., 1981) and plants (Norling et al., 1990). Its primary sequence is well conserved, particularly over residues 14–47 (bovine numbering), which have been defined as the inhibitory region (the ‘minimal inhibitory sequence’; Van Raaij et al., 1996). Bovine IF1 has been shown to have two oligomeric states, tetramer and dimer, favoured by pH values above and below 6.5, respectively (Cabezón et al., 2000a). Dimerization of IF1 occurs by formation of an antiparallel α-helical coiled coil in its C-terminal region, placing the inhibitory regions at opposite ends of the dimer, allowing the active dimeric IF1 to bind two F1 domains simultaneously (Cabezón et al., 2000b). The formation of residues 44–84 into a symmetrical dimer of antiparallel α-helical coiled coils was observed by proton NMR experiments (Gordon-Smith et al., 2001). The 2.2 Å resolution structure of bovine IF1 containing the mutation H49K (Schnizer et al., 1996) described here shows that the full-length IF1 dimerizes in a similar way.
Results and discussion
Overall protein structure
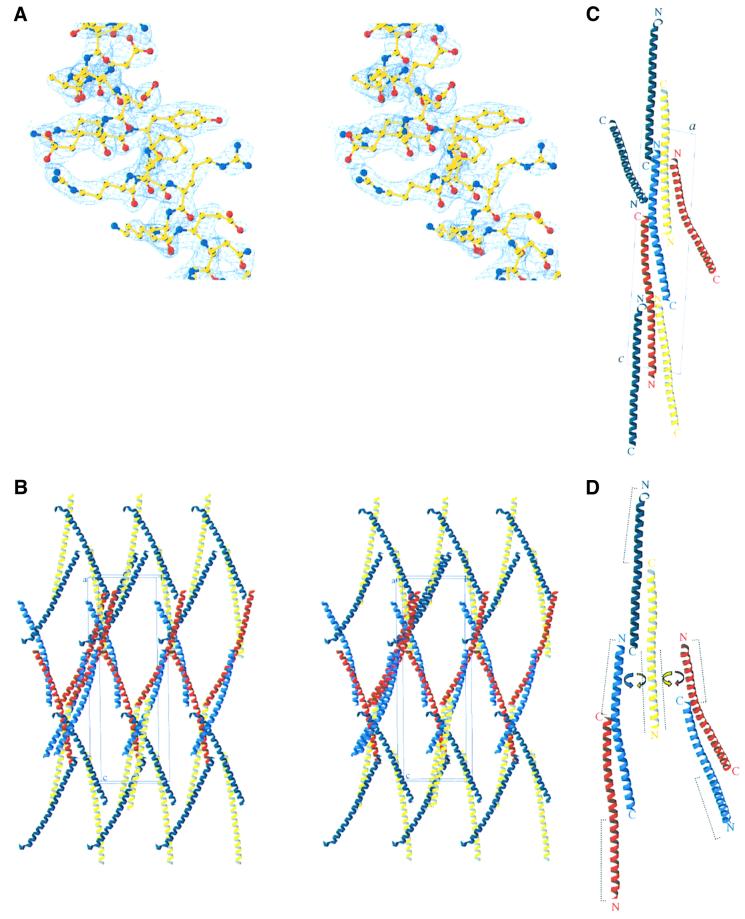
The experimental electron density map based on phases from a platinum derivative (Table I) shows extensive regions of α-helical structure (Figure 1A). There are four monomers (A–D) in the crystallographic asymmetric unit, arranged as dimers AB and CD, which in turn associate via extended antiparallel coiled-coil interactions to form oligomeric rope-like structures (Figure 1B–D). This association takes place in the N-terminal part of the protein and involves the minimal inhibitory sequence (residues 14–47). The association between different ‘ropes’ is based mainly on contacts between helices B and D (Table II), leading to a very unusual rhomboid packing (Figure 1B).
Table I. Data collection and phasing statistics.
| Native 1 | Native 2 | K2Pt(CN)6 | |
|---|---|---|---|
| Cell parameters | |||
| a, b, c (Å) | 32.0, 53.5, 156.3 | 32.0, 53.3, 156.9 | 31.9, 53.1, 156.7 |
| α, β, γ (°) | 90, 95.1, 90 | 90, 95.9, 90 | 90, 95.5, 90 |
| Resolution (Å) | 2.53 | 2.23a | 2.6 |
| Number of reflections | 16 887 | 18 758 | 15 387 |
| Completenessa,b (%) | 95.0 (89.1) | 72.3 (10.8)c | 95.6 (89.2) |
| Multiplicityb | 1.9 (1.8) | 2.4 (2) | 1.9 (1.9) |
| <I/σ(I)>b | 11.2 (4.3) | 10.5 (3.7) | 12.1 (5.0) |
| Rmerge (%)b | 5.1 (18.0) | 6.5 (22.7) | 5.3 (27.9) |
| Rderivative/native (%) | – | – | 20.8 (28.3) |
| Phasing power | – | – | 1.2 (1.0) |
| FOMd | – | – | 0.36 (0.17) |
aThe resolution limits were set to 2.6, 2.6 and 2.2 Å in a*, b* and c*, respectively.
bThe values for the highest resolution shell are given in parentheses.
cWithin the anisotropic resolution limits used for the integration, the data set is 96% complete.
dFigure of merit prior to density modification with SOLOMON.
Fig. 1. Crystal structure of bovine IF1-H49K. (A) Stereo view of the 2.2 Å resolution 2Fo – Fc electron density map calculated with CNS (contoured at 1.5 σ) from residues 27 to 41 of the protein. (B) Stereo view of the crystallographic packing. The four IF1 monomers in the asymmetric unit (A–D) are represented in red, sky blue, yellow and dark blue, respectively. The origin of the unit cell and the a, b and c axes are labelled ‘o’, ‘a’, ‘b’ and ‘c’, respectively. (C) View along the crystallographic b-axis. (D) Interactions between the two types of dimers. Dashed lines represent the minimal inhibitory sequence of IF1, which are masked in the tetramers. C- and N-termini are indicated with the letters C and N, respectively.
Table II. Intersubunit interfacesa.
| Interface | Area in interface (Å2) | Interface surface area of each subunitb (%) | Positions involved in the main interactions (%) | Hydrophobicityc (%) |
|---|---|---|---|---|
| A/B | 846 | 14.0 | a, d/a, d (81.3) | 83.2 |
| C/D | 842 | 13.7 | a, d/a, d (83.4) | 84.7 |
| B/C | 820 | 13.3 | b, e/c, g (73.7) | 62.4 |
| A/D_symm1d | 730 | 12.0 | b, e/c, g (72.9) | 63.4 |
| A/C_symm1e | 747 | 12.4 | c, g/b, e (73.2) | 65.0 |
| B/D_symm2f | 447 | 7.3 | c, d, g/a, b, e, fg | 65.0 |
aAll the helices were reduced to the size of the shortest one (residues 23–78).
bRelative to total surface area.
cSurface area involving carbon atoms is considered to be hydrophobic.
dThe crystallographic symmetry operator for a D_symm1 molecule is a translation of one unit cell in x, y and z.
eThe crystallographic symmetry operator for a C_symm1 molecule is 2 – x, y + 1/2, 1 – z.
fThe crystallographic symmetry operator for a D_symm2 molecule is –x, y + 1/2, –z.
gThe positions involved in the main interactions between these two helices do not follow a regular pattern.
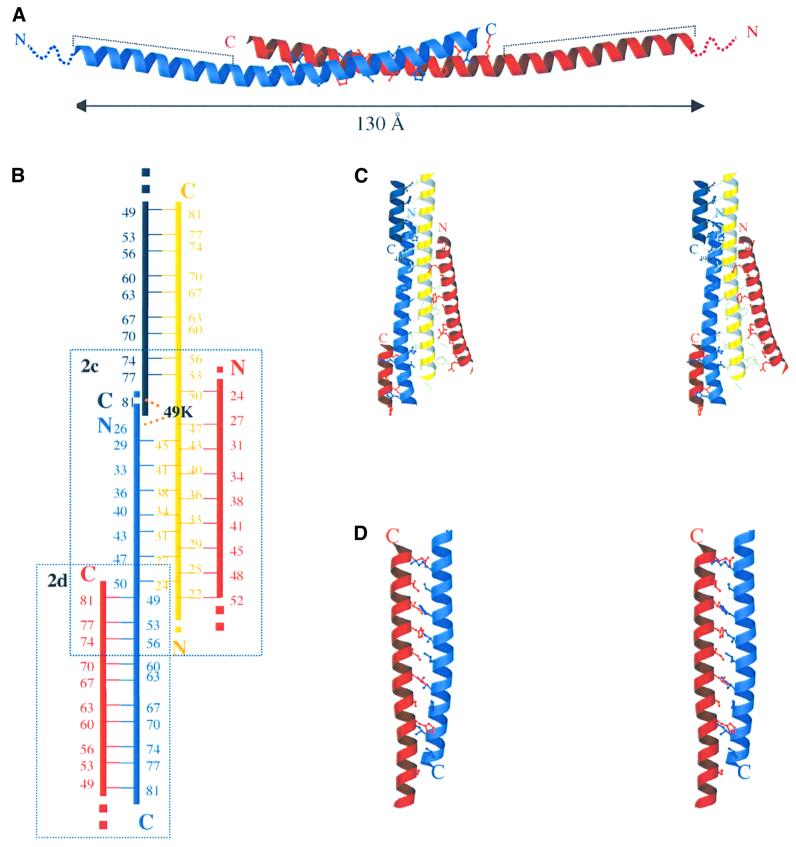
The final model consists of residues 19–83, 20–79, 20–78 and 23–78 in chains A, B, C and D, respectively. The protomer is α-helical along most of its length (Figure 2A). This α-helix participates in three different antiparallel double-stranded coiled-coil units, one in the C-terminal part and two, with different helices, in the N-terminal part of the protein (Figure 2B). In the IF1 dimer, two monomers associate through an extended antiparallel coiled coil in the C-terminal region. The active dimeric IF1 has an end–end distance of ∼130 Å. The arrangement of helices in dimer AB differs clearly in curvature from that in dimer CD [root mean square (r.m.s.) difference in Cα coordinates between the dimers is 4.2 Å]. This difference is localized in the N-terminal region and probably arises from the packing of the helices in the crystal structure. The r.m.s. difference in Cα coordinates between the dimers in the C-terminal part (residues 50–78) is only 0.5 Å.
Fig. 2. Interhelical packing in IF1-H49K structure. (A) Ribbon diagram of the active dimer. Dashed lines represent the minimal inhibitory sequence. The disordered N-terminal residues 1–18 are shown as dotted lines. (B) Schematic representation of the interhelical packing. Residues involved in forming the coiled coils are represented with their sequence number. Helices are coloured as in Figure 1. Dotted lines represent the continuity of the helices. The position of lysine 49 in helix C (yellow) is shown in black, indicating the contacts with helices B (sky blue) and D (dark blue). Dashed boxes show the two areas of the protein represented in more detail in (C) and (D). (C) Stereo view of the interhelical packing in the N-terminus of the protein. (D) Stereo view of the interhelical packing in the C-terminus of the AB dimer.
The antiparallel dimer assembly in active IF1
In the IF1 dimer, the antiparallel two-stranded coiled coil in the C-terminal region is stabilized by complementary hydrophobic interactions between the helices, involving residues 49–81 (Figure 2A). A stereo view of these interactions in the AB dimer is shown in Figure 2D. They involve residues at positions a and d of the heptad repeats where only histidine, leucine and isoleucine residues are present. According to the program SOCKET (Walshaw and Woolfson, 2001), the side chains are packed in a type 4 coiled-coil knobs-into-holes structure. In addition, charged side chains and polar residues in positions e and g may help to stabilize the coiled-coil structure by local charge compensation. These interactions involve mainly basic side-chains in the C-terminal region (Lys71-Ser73-Lys78) and acidic side-chains on the opposite face (Glu66-Glu59-Glu52). It seems likely that these opposite charges near the ends of the dimer interface play an important role in favouring the antiparallel orientation. In the dimer interface ∼850 Å2 (14%) of molecular surface area per monomer is buried (Table II).
Superposition of the NMR structure of residues 44–84 with the same residues in the crystal structure of intact IF1 shows that the structures are in reasonable agreement. The r.m.s. difference in Cα coordinates with AB and CD dimers is 2.3 and 1.9 Å, respectively.
The higher oligomeric assembly of inactive IF1
The dimers assemble into higher oligomers by forming antiparallel coiled coils in the N-terminal regions. In ideal coiled coils, the heptad motif a–b–c–d–e–f–g is repeated n times and the a and d residues make up the hydrophobic core of the two helix bundles (Lupas, 1996). In the dimer–dimer interactions in the structure presented here, the buried surface consists of the alternation of three and four residues in the heptad repeat, but compared with other known coiled-coil structures, this assembly has a higher content of polar residues. Furthermore, these residues cannot correspond to positions a and d, since the same helix is involved in two coiled coils in which it interacts with two different helices (Figure 2B and C). This high frequency of favourable electrostatic interactions has not been observed previously in coiled-coil structures. It might underlie the pH dependency of the association/dissociation of dimers (Cabezón et al., 2000a).
In the crystal matrix, association between dimers occurs in distinctly different ways. Helix C from dimer CD interacts with both helices A and B from dimer AB. However, in the CD dimer, helix D only presents coiled-coil interactions with helix A from the AB dimer, since contacts with helix B only involve a few residues and are probably due to the packing in the crystal structure. Depending on which assembly of dimers is considered, the interface buries ∼820, ∼730 or ∼750 Å2, respectively, of molecular surface area (Table II). Figure 1D, showing the assembly of the dimer into higher order aggregates, reflects the different possibilities.
Role of His49 in oligomerization of IF1
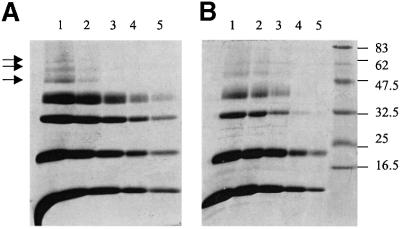
Native IF1 is an active dimer at a pH of ∼6.5 and an inactive tetramer at somewhat higher pH values, whereas the mutant IF1-H49K is active and dimeric at all the pH values investigated (Cabezón et al., 2000a). However, the oligomerization state in solution also depends on the protein concentration (Figure 3). At low protein concentrations and high pH, the mutant IF1-H49K is dimeric, but at higher protein concentrations higher oligomers form in both native and mutant IF1, indicating that observations in the crystal probably reflect the behaviour in solution. The difference between native and mutant proteins is the concentration required for tetramer formation. Therefore, it seems very likely that the mutation H49K affects the oligomerization by shifting the equilibrium, but the way in which the oligomerization is affected is unclear. Lysine 49 in helix C has a different conformation from that in other helices (Figure 2B and C). Apart from interacting with helix D to form one of the active dimers, it also makes contacts with glutamate 26 in helix B and, therefore, with the AB dimer. Another interesting point concerns the curvature of the helices. Although this curvature is localized to the region of residues 47–50 in all four helices, it is more pronounced in helices A and B. Lysine 49 is within this region and might be responsible for promoting this curvature, having an effect on the oligomerization process. At present, however, it is not possible to discern whether the different packing of lysine 49 in helix C and the differences in curvature have any biological relevance or if they are a consequence of crystal packing.
Fig. 3. Covalent cross-linking of IF1 and IF1-H49K with dimethyl suberimidate. Experiments were carried out as described in Materials and methods. Samples treated with the cross-linking reagent were removed after 3 h of incubation and analysed by SDS–PAGE. Lanes 1–5 correspond to protein concentrations of 2, 1, 0.5, 0.2 and 0.1 mg/ml, respectively. IF1 and IF1-H49K are shown in (A) and (B), respectively. The positions of molecular weight markers are shown on the right in kDa. Arrows indicate the formation of higher oligomers.
Implications for the regulation of F1Fo-ATPase
The structure shows that residues 32–44 are involved in tetramer and higher oligomer formation in agreement with earlier work (Cabezón et al., 2000a) and it reveals that the interaction between the dimers involves the minimal inhibitory sequence. Therefore, it seems very likely that, at high pH, the minimal inhibitory sequence (residues 14–47) is masked by tetramer formation as seen in the crystal, preventing the binding of this region to ATP synthase. Recently, a model of the interconversion between the active dimeric and inactive tetrameric states of IF1 was proposed with parallel interactions between dimers (Cabezón et al., 2000a). The structure reveals that the interactions between the dimers are antiparallel, but a similar mechanism of oligomerization applies. A decrease in pH might affect the polar interactions between both dimers, freeing the inhibitory regions to interact with the F1 domain of the ATP synthase. An important feature of the structure is that the inhibitory N-terminal regions are at opposite ends of the dimer, with an end–end distance of at least 130 Å, and therefore the protein can bind two F1 domains simultaneously. Dimerization of bovine F1 by the binding of the inhibitor protein has been shown (Cabezón et al., 2000b) and it is reasonable to assume that dimerization of intact F1Fo-ATPase occurs in a similar way, although this remains to be demonstrated.
The two types of dimers in the crystal (AB and CD) have markedly different curvatures, suggesting that they are flexible in solution. The disorder in the N-terminal regions is probably due to the high content of glycine residues, which allows the protein to be flexible. Such flexibility might be functionally relevant for the interaction with the F1 domain of the ATP synthase. Cross-linking experiments with IF1 and bovine F1-ATPase indicate that IF1 binds in the C-terminal region of the β-subunit (Klein et al., 1980; Jackson and Harris, 1988; Hashimoto et al., 1995), which is a bundle of six α-helices, at the interface with the α-subunit. Sequence alignment of IF1 with residues 368–459 of the β-subunit reveals an identity of 18% (data not shown) and most of this region (residues 395–420), including the DELSEED sequence (residues 398–404) is in contact with the central γ-subunit. Therefore, it is possible that part of IF1 acts as a functional mimetic of this region of the β-subunit, interfering with the contacts between β- and γ-subunits. In support of this idea, synthetic peptides corresponding to this region of the bovine β-subunit have been found to inhibit F1-ATPase (Stout et al., 1993).
The packing of coiled-coil regions of regulatory proteins against helical segments of the protein to be regulated appears to be more general. For example, the structure of arfaptin, a mediator between Rac and Arf signalling pathways, is also a highly elongated antiparallel dimer, each monomer consisting of three α-helices (Tarricone et al., 2001). The regulatory subunit H of the V-ATPase from S.cerevisiae (a distant relation of F-ATPases) is similarly predominantly α-helical (Sagermann et al., 2001). The C-terminal region of the bovine subunit H (residues 397–477), which is a well-conserved region in different species, is 21% identical in sequence to bovine IF1.
Homologues of IF1 are not present in chloroplasts or bacteria. The ATPase activity of the chloroplast enzyme is inhibited by another mechanism involving a redox switch in the γ-subunit (Walker, 1994). In Escherichia coli, it has been proposed that the ε-subunit of F1Fo-ATPase (equivalent to the bovine δ-subunit) has two conformations and that it functions as a ‘clutch’ or ‘ratchet’ to regulate differentially ATP hydrolysis and synthesis (Tsunoda et al., 2001). From the structures of the bovine and yeast mitochondrial δ-subunits, determined in F1 (Gibbons et al., 2000) and F1-c10 (Stock et al., 1999) complexes, respectively, it is unlikely that the protein can rearrange its structure as proposed for the bacterial ε-subunit. There fore, it has been suggested that IF1, rather than acting as an inhibitor of ATP hydrolysis, may fulfil a ‘ratchet’ and ‘clutch’ role in regulating mitochondrial ATP synthesis and hydrolysis in the mitochondrial enzyme. There is currently no evidence to support this idea and, in the absence of detailed knowledge of the interaction of IF1 with F1-ATPase, it is difficult to evaluate its structural basis at present.
Materials and methods
Expression, purification and crystallization
Expression and purification of recombinant bovine IF1-H49K was carried out as described previously (Cabezón et al., 2000a). The purified protein was fully active as judged by the inhibition assay of F1-ATPase (Lutter et al., 1993). Crystals were grown by equilibrating a IF1-H49K solution, at 6 mg/ml in buffer 10 mM Tris–HCl pH 8.0, against a reservoir containing 0.8 M mono-sodium dihydrogen phosphate, 0.8 M mono-potassium dihydrogen phosphate and 0.1 M HEPES-Na buffer pH 8, at 23°C, in sitting-drop vapour-diffusion trays. The crystallization droplets consisted of 2 µl of protein and 2 µl of reservoir solutions; clusters of crystals appeared within a few days and grew to maximum dimensions of ∼500 × 300 × 200 µm.
Data collection, phasing and refinement
Crystals were cryoprotected with 15% glycerol and 2 M sodium-potassium phosphate, harvested with a cryoloop (Hampton Research, Laguna Niguel, CA), plunged into liquid nitrogen and stored at 100 K. Native and derivative diffraction data were collected at 100 K in-house using CuKα radiation from a Rigaku H3R generator and a MAR 345 image plate. Additional native data to 2.2 Å resolution were collected at 100 K on beamline ID14-1 (λ = 0.934 Å) at the ESRF, Grenoble, France, on a MAR CCD detector. All data were processed using MOSFLM (Leslie, 1992) and the CCP4 program suite (CCP4, 1994). The crystals belong to the monoclinic space group P21 (a = 32.0 Å, b = 53.3 Å, c = 156.9 Å, α = 90°, β = 95.9°, γ = 90°) with four molecules per asymmetric unit (solvent content 64.5%).
Heavy-atom derivative data were collected from crystals soaked in 1 mM K2Pt(CN)6 for ∼18 h before soaking for 10 min in cryo-solution. The positions of the two highest occupancy heavy-atom sites were revealed by an isomorphous difference Patterson. Additional sites were found from difference Fourier maps, and heavy atom parameters were refined initially with MLPHARE (Otwinowski, 1991) and then with SHARP (De La Fortelle and Bricogne, 1997). The SIRAS phased electron density map was subjected to density modification with SOLOMON (Abrahams and Leslie, 1996) and the resulting map allowed tracing of ∼60% of the four independent molecules using the program O (Jones et al., 1991). Refinement using the torsion angle simulated annealing option of the program CNS (Brunger et al., 1998) with the maximum likelihood target, together with manual revisions of the structure in O, allowed a more complete model of the protein to be constructed. The Wilson B-factor for the native data used in refinement was 33.1 Å2. Refinement applying non-crystallographic symmetry restraints reduced the Rfree to 28.0% and the working R to 25.8%. Residues 1–18 and 84 in chain A, 1–19 and 80–84 in chain B, 1–19 and 79–84 in chain C and 1–22 and 79–84 in chain D were disordered and were not visible in the electron density map. There were no features in the electron density map that could be reliably interpreted as solvent molecules. Almost all the main-chain hydrogen bonding groups were involved in intramolecular α-helical hydrogen bonding. The stereochemistry of the final model was verified using the program PROCHECK (Laskowski et al., 1993). The r.m.s. values for bonds and angles were 0.009 Å and 1.16°, respectively. Intersubunit interactions were analysed using the program Areaimol from the CCP4 suite. The data processing and refinement statistics are presented in Table I. Figures 1 and 2A, C and D were generated with BOBSCRIPT (Esnouf, 1997).
Covalent cross-linking of IF1 and IF1H49K
Cross-linking of amino groups in IF1 and IF1-H49K with dimethyl-suberimidate was carried out essentially according to Davies and Stark (1970). Samples were dialysed overnight against 20 mM HEPES pH 8.0, 1 mM EDTA and 0.001% phenylmethylsulfonyl fluoride buffer. The protein concentration was adjusted by dilution with dialysis buffer, and dimethyl-suberimidate (freshly dissolved at 20 mg/ml in the same buffer) was added to a final concentration of 1 mg/ml. The mixture was kept at room temperature for 3 h. Samples (10 µl) were removed, dissolved in sample buffer and analysed by SDS–PAGE (Cabezón et al., 2000a).
Coordinates
The atomic coordinates and structure factors have been deposited in the Protein Data Bank (accession numbers 1gmj and r1gmjsf, respectively).
Acknowledgments
Acknowledgements
We thank the staff of beamline ID14 at European Synchrotron Radiation Facility (ESRF), Grenoble, for help with data collection. E.C. was supported during part of this work by a European Molecular Biology Organization Fellowship and by a TMR Marie Curie Research Training Grant from the European Community.
References
- Abrahams J.P. and Leslie,A.G.W. (1996) Methods used in the structure determination of bovine mitochondrial F1-ATPase. Acta Crystallogr. D, 52, 30–42. [DOI] [PubMed] [Google Scholar]
- Brunger A.T. et al. (1998) Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D, 54, 905–921. [DOI] [PubMed] [Google Scholar]
- Cabezón E., Butler,P.J.G., Runswick,M.J. and Walker,J.E. (2000a) Modulation of the oligomerization state of bovine F1-ATPase inhibitor protein, IF1, by pH. J. Biol. Chem., 275, 25460–25464. [DOI] [PubMed] [Google Scholar]
- Cabezón E., Arechaga,I., Butler,P.J.G. and Walker,J.E. (2000b) Dimerization of bovine F1-ATPase by binding the inhibitor protein, IF1. J. Biol. Chem., 275, 28353–28355. [DOI] [PubMed] [Google Scholar]
- CCP4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D, 50, 760–763. [DOI] [PubMed] [Google Scholar]
- Cintron N.M. and Pedersen,P.L. (1979) A protein inhibitor of the mitochondrial adenosine triphosphatase complex of rat liver. Purification and characterization. J. Biol. Chem., 254, 3439–3443. [PubMed] [Google Scholar]
- Davies G.E. and Stark,G.R. (1970) Use of dimethyl suberimidate, a cross-linking reagent, in studying the subunit structure of oligomeric proteins. Proc. Natl Acad. Sci. USA, 66, 651–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaFortelle E. and Bricogne,G. (1997) Maximum likelihood heavy-atom parameter refinement for multiple isomorphous replacement and multiwavelength anomalous diffraction. Methods Enzymol., 276, 472–493. [DOI] [PubMed] [Google Scholar]
- Esnouf R.M. (1997) An extensively modified version of MolScript that includes greatly enhanced coloring capabilities. J. Mol. Graph. Model., 15, 132–134. [DOI] [PubMed] [Google Scholar]
- Gibbons C., Montgomery,M.G., Leslie,A.G.W. and Walker,J.E. (2000) The structure of the central stalk in bovine F1-ATPase at 2.4 Å resolution. Nature Struct. Biol., 7, 1055–1061. [DOI] [PubMed] [Google Scholar]
- Gordon-Smith D.J., Carbajo,R.J., Yang,J.C., Videler,H., Runswick,M.J., Walker,J.E. and Neuhaus,D. (2001) Solution structure of a C-terminal coiled-coil domain from bovine IF1: the inhibitor protein of F1-ATPase. J. Mol. Biol., 308, 325–339. [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Negawa,Y. and Tagawa,K. (1981) Binding of intrinsic ATPase inhibitor to mitochondrial ATPase–stoichiometry of binding of nucleotides, inhibitor and enzyme. J. Biochem., 90, 1151–1157. [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Yamamoto,Y., Yoshida,Y. and Tagawa,K. (1995) Cleavage of bovine mitochondrial ATPase inhibitor with endopeptidases and binding of the resulting peptides to the interface between the α- and β-subunits of F1-ATPase. J. Biochem., 117, 641–647. [DOI] [PubMed] [Google Scholar]
- Jackson P.J. and Harris,D.A. (1988) The mitochondrial ATP synthase inhibitor protein binds near the C-terminus of the F1 β-subunit. FEBS Lett., 229, 224–228. [DOI] [PubMed] [Google Scholar]
- Jones T.A., Zou,J.Y., Cowan,S.W. and Kjeldgaard,M. (1991) Improved methods for binding protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A, 47, 110–119. [DOI] [PubMed] [Google Scholar]
- Klein G., Satre,M., Dianoux,A.C. and Vignais,P.V. (1980) Radiolabeling of natural adenosine triphosphatase inhibitor with phenyl [14C]isothiocyanate and study of its interaction with mitochondrial adenosine triphosphatase. Localization of inhibitor binding sites and stoichiometry of binding. Biochemistry, 19, 2919–2925. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., Macarthur,M.W., Moss,D.S. and Thornton,J.M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr., 26, 283–291. [Google Scholar]
- Leslie A.G.W. (1992) Joint CCP4 and EACMB Newsletter Protein Crystallography, 26. Daresbury Laboratory, Warrington, UK.
- Lupas A. (1996) Coiled coils: new structures and new functions. Trends Biochem. Sci., 21, 375–382. [PubMed] [Google Scholar]
- Lutter R., Abrahams,J.P., van Raaij,M.J., Todd,R.J., Lundqvist,T., Buchanan,S.K., Leslie,A.G.W. and Walker,J.E. (1993) Crystalliz ation of F1-ATPase from bovine heart mitochondria. J. Mol. Biol., 229, 787–790. [DOI] [PubMed] [Google Scholar]
- Norling B., Tourikas,C., Hamsur,B. and Glaser,E. (1990) Evidence for an endogenous ATPase inhibitor protein in plant mitochondria. Purification and characterization. Eur. J. Biochem., 188, 247–252. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z. (1991) Maximum likelihood refinement of heavy atom parameters. In Wolf,W., Evans,P.R. and Leshe,A.G.W. (eds), Isomorphous Replacement and Anomalous Scattering: Proceedings of the CCP4 Study Weekend, 25–26 January, 1991. SERC Daresbury Laboratory, Warrington, UK, pp. 23–38.
- Panchenko M.V. and Vinogradov,A.D. (1985) Interaction between the mitochondrial ATP synthetase and ATPase inhibitor protein. Active/inactive slow pH-dependent transitions of the inhibitor protein. FEBS Lett., 184, 226–230. [DOI] [PubMed] [Google Scholar]
- Pullman M.E. and Monroy,G.C. (1963) A naturally occurring inhibitor of mitochondrial adenosine triphosphatase. J. Biol. Chem., 238, 3762–3769. [PubMed] [Google Scholar]
- Rouslin W. (1983) Protonic inhibition of the mitochondrial oligomycin-sensitive adenosine 5′-triphosphatase in ischemic and autolyzing cardiac muscle. Possible mechanism for the mitigation of ATP hydrolysis under nonenergizing conditions. J. Biol. Chem., 258, 9657–9661. [PubMed] [Google Scholar]
- Rouslin W. (1987) Factors affecting the reactivation of the oligomycin-sensitive adenosine 5′-triphosphatase and the release of ATPase inhibitor protein during the re-energization of intact mitochondria from ischemic cardiac muscle. J. Biol. Chem., 262, 3472–3476. [PubMed] [Google Scholar]
- Rouslin W. and Broge,C.W. (1989) Regulation of mitochondrial matrix pH and adenosine 5′-triphosphatase activity during ischemia in slow heart-rate hearts. J. Biol. Chem., 264, 15224–15229. [PubMed] [Google Scholar]
- Sagermann M., Stevens,T.H. and Matthews,B.W. (2001) Crystal structure of the regulatory subunit H of the V-type ATPase of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA, 98, 7134–7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnizer R., Van Heeke,G., Amaturo,D. and Schuster,S.M. (1996) Histidine-49 is necessary for the pH-dependent transition between active and inactive states of the bovine F1-ATPase inhibitor protein. Biochim. Biophys. Acta, 1292, 241–248. [DOI] [PubMed] [Google Scholar]
- Stock D., Leslie,A.G.W. and Walker,J.E. (1999) Molecular architecture of the rotary motor in ATP synthase. Science, 286, 1700–1705. [DOI] [PubMed] [Google Scholar]
- Stout J.S., Partridge,B.E., Dibbern,D.A. and Schuster,S.M. (1993) Peptide analogs of the beef heart mitochondrial F1-ATPase inhibitor protein. Biochemistry, 32, 7496–7502. [DOI] [PubMed] [Google Scholar]
- Tarricone C., Xiao,B., Justin,N., Walker,P.A., Rittinger,K., Gamblin,S.J. and Smerdon,S.J. (2001) The structural basis of Arfaptin-mediated cross-talk between Rac and Arf signalling pathways. Nature, 411, 215–219. [DOI] [PubMed] [Google Scholar]
- Tsunoda S.P., Rodgers,A.J.W., Aggeler,R., Wilce,M.C.J., Yoshida,M. and Capaldi,R.A. (2001) Large conformational changes of the ε-subunit in the bacterial F1Fo-ATP synthase provide a ratchet action to regulate this rotary motor enzyme. Proc. Natl Acad. Sci. USA, 98, 6560–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanRaaij M.J., Orriss,G.L., Montgomery,M.G., Runswick,M.J., Fearnley,I.M., Skehel,J.M. and Walker,J.E. (1996) The ATPase inhibitor protein from bovine heart mitochondria: the minimal inhibitory sequence. Biochemistry, 35, 15618–15625. [DOI] [PubMed] [Google Scholar]
- Walker J.E. (1994) The regulation of catalysis in ATP synthase. Curr. Opin. Struct. Biol., 4, 912–918. [DOI] [PubMed] [Google Scholar]
- Walshaw J. and Woolfson,D.N. (2001) SOCKET: A program for identifying and analysing coiled-coil motifs within protein structures. J. Mol. Biol., 307, 1427–1450. [DOI] [PubMed] [Google Scholar]